Abstract
A Gram-negative, rod-shaped bacterium, designated H63T, was isolated from aortic valve tissue of a patient with native valve endocarditis. 16S rRNA gene sequencing revealed that H63T belongs to the genus Legionella, with its closest neighbours being the type strains of Legionella brunensis (98.8 % similarity), L. londiniensis (97.0 %), L. jordanis (96.8 %), L. erythra (96.2 %), L. dresdenensis (96.0 %) and L. rubrilucens, L. feeleii, L. pneumophila and L. birminghamensis (95.7 %). DNA–DNA hybridization studies yielded values of <70 % relatedness between strain H63T and its nearest neighbours in terms of 16S rRNA gene sequence similarity, indicating that the strain represents a novel species. Phylogenetic analysis of the 16S rRNA, macrophage infectivity potentiator (mip) and RNase P (rnpB) genes confirmed that H63T represents a distinct species, with L. brunensis being its closest sister taxon. Fatty acid composition and biochemical traits, such as the inability to ferment glucose and reduce nitrate, supported the affiliation of H63T to the genus Legionella. H63T was distinguishable from its neighbours based on it being positive for hippurate hydrolysis. H63T was further differentiated by its inability to grow on BCYE agar at 17 °C, its poor growth on low-iron medium and the absence of sliding motility. Also, H63T did not react with antisera generated from type strains of Legionella species. H63T replicated within macrophages. It also grew in mouse lungs, inducing histopathological evidence of pneumonia and dissemination to the spleen. Together, these results confirm that H63T represents a novel, pathogenic Legionella species, for which the name Legionella cardiaca sp. nov. is proposed. The type strain is H63T ( = ATCC BAA-2315T = DSM 25049T = JCM 17854T).
Legionellae are Gram-negative bacteria that are ubiquitous in freshwater environments as well as man-made water systems (Diederen, 2008; Fields et al., 2002). Many legionellae have been associated with human disease, and the most common mode of transmission is through inhalation of aerosolized water droplets containing the bacteria (König et al., 2005; Muder & Yu, 2002; Whiley & Bentham, 2011). Inside the lung, legionellae utilize alveolar macrophages and epithelial cells for intracellular replication, resulting in a severe pneumonia referred to as Legionnaires’ disease (Fields et al., 2002). In rare cases, Legionella species can be isolated from extrapulmonary sites such as the heart (Lowry & Tompkins, 1993). At the time of writing, the genus Legionella comprised 54 species with validly published names and one genomospecies (Benson et al., 1996; Edelstein et al., 2012; Euzéby, 1997; Yang et al., 2012).
We previously described a rare case of native valve endocarditis due to a novel Legionella strain (Pearce et al., 2011). The strain, designated H63T, was isolated from resected aortic valve tissue of a patient requiring aortic valve replacement for treatment of congestive heart failure related to infective native valve endocarditis. Sequencing of the 16S rRNA gene and preliminary blast analysis suggested that the strain represented either a novel clinical strain of Legionella brunensis or a novel Legionella species (Pearce et al., 2011). L. brunensis was first isolated from cooling tower water in Czechoslovakia and has been isolated once from a case of Legionnaires’ disease in Europe (Ricketts et al., 2007; Wilkinson et al., 1988). Based on DNA–DNA hybridization values of less than 70 % and sequence analysis of multiple gene targets, we now report that H63T represents a novel Legionella species. Phenotypic profiling revealed a number of differences between H63T and its phylogenetically nearest neighbours. Furthermore, H63T replicated in a human macrophage cell line and in the murine lung, indicating that H63T represents a virulent strain.
To establish the placement of the novel strain in the genus Legionella, the almost-complete 16S rRNA gene (1423 bp) was amplified from purified genomic DNA of H63T and sequenced using primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′), 806R (5′-GGACTACCAGGGTATCTAAT-3′), 515F (5′-TGCCAGCAGCCGCGGTAA-3′) and rP1 (5′-GGTTACCTTGTTACGACTT-3′) (Relman et al., 1992; Weisburg et al., 1991). The EzTaxon service was used to determine the nearest neighbours in the genus Legionella on the basis of 16S rRNA gene sequence similarity as recommended for the calculation of pairwise percentage similarity values (Chun et al., 2007; Tindall et al., 2010). H63T shared highest similarity with the type strains of L. brunensis (98.80 %), followed by Legionella londiniensis (97.03 %), L. jordanis (96.76 %), L. erythra (96.20 %), L. dresdenensis (95.99 %), L. rubrilucens (95.73 %), L. feeleii (95.72 %), L. pneumophila (95.71 %) and L. birminghamensis (95.71 %) (Table 1). To determine whether H63T represents a novel Legionella species, DNA–DNA hybridization was performed at 30 °C using the microplate technique with photobiotin-labelled DNA as described previously (Ezaki et al., 1989, 1990) and modified (Willems et al., 2001), with the exception that the DNA was sheared prior to biotin labelling as opposed to after. Strain H63T was only 17 and 20 % related to its nearest neighbour, L. brunensis ATCC 43878T, in reciprocal relationships, well below the 70 % cut-off for species delineation (Table 1). Furthermore, H63T exhibited less than 20 % relatedness to all eight of the remaining type strains tested (Table 1). The difference between reciprocal hybridizations was within 20 % and the standard deviation among replicates was ≤7 %, both of which are acceptable deviations for the microplate technique (Ezaki et al., 1990; Kuroki et al., 2007; Willems et al., 2001).
Table 1. Genetic analysis of strain H63T compared with its nearest neighbours based on 16S rRNA gene sequences.
| Strain | Source | Reference | 16S rRNA gene sequence similarity (%) to strain H63T | DNA–DNA hybridization (%)* with H63T DNA as: | DNA G+C content (mol%) | |
| Probe | Covalent DNA | |||||
| L. cardiaca sp. nov. H63T | Clinical | Pearce et al. (2011) | (100) | (100) | (100) | 41.8 |
| L. brunensis ATCC 43878T | Environmental | Wilkinson et al. (1988) | 98.8 | 17.3±5.8 | 20.3±7.2 | 40.5 |
| L. londiniensis ATCC 49505T | Environmental | Dennis et al. (1993) | 97.0 | 5.7±2.4 | 2.4±1.1 | 43.0 |
| L. jordanis ATCC 33623T | Environmental | Cherry et al. (1982) | 96.8 | 15.7±1.8 | 4.5±3.0 | 45.0 |
| L. erythra ATCC 35303T | Environmental | Brenner et al. (1985) | 96.2 | 4.3±0.6 | 7.5±6.6 | 51.0 |
| L. dresdenensis NCTC 13409T | Environmental | Lück et al. (2010) | 96.0 | 5.0±0.4 | 8.7±2.6 | 42.5 |
| L. rubrilucens ATCC 35304T | Environmental | Brenner et al. (1985) | 95.7 | 6.2±2.6 | 9.3±4.7 | 52.0 |
| L. feeleii ATCC 35072T | Environmental | Herwaldt et al. (1984) | 95.7 | 3.7±2.2 | 6.4±2.8 | 46.0 |
| L. pneumophila ATCC 33152T | Clinical | Brenner et al. (1979) | 95.7 | 5.3±2.3 | 15.1±5.7 | 39.0 |
| L. birminghamensis ATCC 43702T | Clinical | Wilkinson et al. (1987) | 95.7 | 6.3±2.0 | 4.5±2.6 | 42.9 |
Means±sd of at least triplicate hybridization experiments are shown.
To define the relationship between H63T and other Legionella species further, a 584 bp portion of the mip gene and a 327 bp portion of the rnpB gene of H63T were sequenced as described previously (Kuroki et al., 2007; Lück et al., 2010; Ratcliff et al., 1998; Rubin et al., 2005; Yang et al., 2012). The European Working Group for Legionella Infections (EWGLI) Legionella mip gene sequence database was used to determine the similarity based on mip, and NCBI blast was used to determine similarity based on rnpB (Altschul et al., 1990; Fry et al., 2007). Similar to the 16S rRNA gene sequence analysis, the mip gene sequence of strain H63T was most similar to that of L. brunensis ATCC 43878T (85.49 %), followed by Legionella hackeliae ATCC 35250T (85.11 %), L. jamestowniensis ATCC 35298T (85.11 %), L. feeleii ATCC 35072T (83.95 %) and L. lansingensis ATCC 49751T (83.56 %). Based on analysis of rnpB sequences, strain H63T was again most similar to the type strain of L. brunensis (91.2 %), followed by the type strains of L. lansingensis (89.4 %), L. jamestowniensis (89.7 %), L. hackeliae (88.3 %) and L. feeleii (88.3 %). For phylogenetic analyses, the 16S rRNA, mip and rnpB sequences of type strains of Legionella species and the nearest other relative within the Legionellaceae, Coxiella burnetii, were obtained from GenBank (Benson et al., 2008). Trimmed sequences were aligned using the clustal w program (Larkin et al., 2007). Phylogenetic trees were inferred by the neighbour-joining method using topali version 2 and edited using TreeView version 1.6.6 (Milne et al., 2009; Page, 1996). Phylogenetic analysis based on the consensus alignment of 16S rRNA, mip and rnpB gene sequences indicated that strain H63T is most closely related to L. brunensis, followed by the group of L. hackeliae and L. jamestowniensis (Fig. 1). The strength of the association was confirmed by bootstrap values ≥80 based on 100 replicates. For completeness, DNA–DNA hybridizations were performed comparing H63T with both L. hackeliae ATCC 35250T and L. jamestowniensis ATCC 35298T, because they were closely related according to the consensus tree. According to hybridization analysis, L. hackeliae ATCC 35250T was 8.0 % (±0.9) related to H63T when H63T DNA served as the probe and 31.7 % (±0.4) similar when H63T represented the covalent DNA. L. jamestowniensis ATCC 35298T was 10.1 % (±1.2) similar to H63T when H63T DNA was the probe and 7.7 % (±3.3) similar when H63T DNA was the covalent DNA. The topologies of the individual gene trees support the consensus assignment of H63T and L. brunensis as sister taxa (Figs S1–S3, available in IJSEM Online).
Fig. 1.
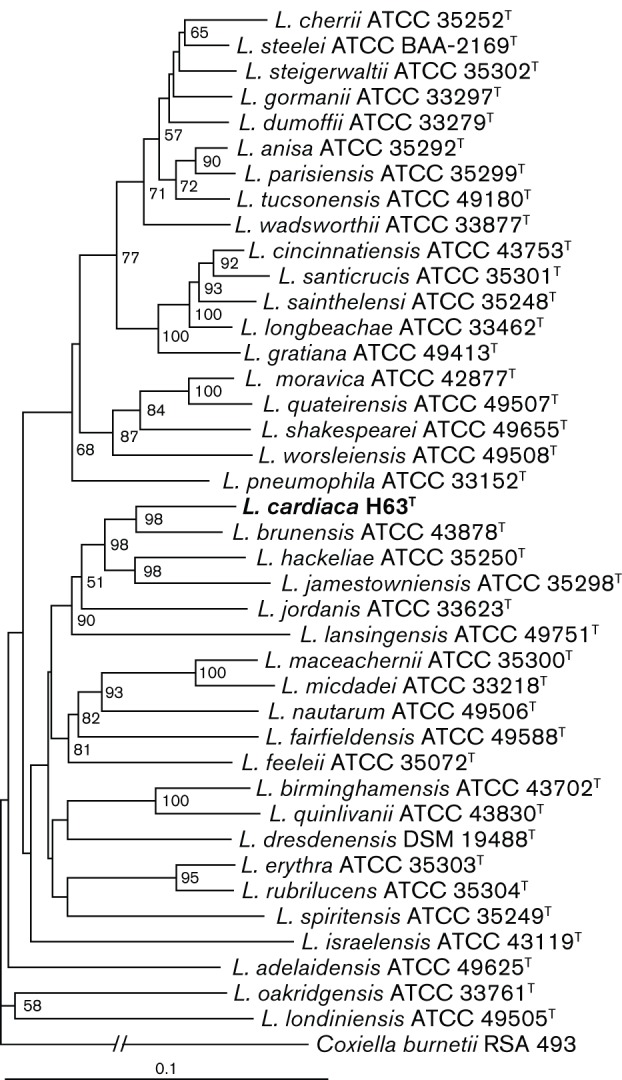
Neighbour-joining tree showing relationships between strain H63T and all previously sequenced type strains of Legionella species based on the consensus sequence of the 16S rRNA, mip and rnpB loci. Bootstrap values greater than 50 (from 100 replicates) are shown. Coxiella burnetii RSA 493 was used as an outgroup. GenBank accession numbers of the individual sequences used to reconstruct the tree are provided in Figs S1–S3. Bar, 0.1 substitutions per nucleotide site.
To complete our genetic analysis, the DNA G+C content of H63T was determined through HPLC analysis performed by the Identification Service of the DSMZ. The DNA G+C content of H63T was 41.8 mol%, within the range of values reported for its neighbours (39.0–52.0 mol%; Table 1).
Using the slide agglutination test as described previously (Thacker et al., 1985), antigen from H63T did not react with antisera generated previously against the type strains of Legionella species, including sera raised against all of the nearest neighbours in the 16S rRNA gene tree as well as the type strains of L. jamestowniensis and L. hackeliae (Table S1).
Initially, we determined the phenotype of strain H63T by examining a set of 13 physiological traits that are standards for Legionella (Table 2) (Hookey et al., 1996). Like most members of the genus Legionella (Dennis et al., 1993; Edelstein et al., 2012; Hookey et al., 1996; Yang et al., 2012), including its nearest neighbours, H63T grew well at 37 °C on buffered charcoal yeast extract (BCYE) agar or in buffered yeast extract (BYE) broth and required supplementary cysteine for growth. Colonies of strain H63T on BCYE agar did not autofluoresce under UV light, distinguishing the strain from its neighbours L. erythra, L. dresdenensis, L. rubrilucens and L. birminghamensis (Table 2). The strain, like many other legionellae (Hookey et al., 1996) but unlike L. dresdenensis and L. birminghamensis, secreted a brown pigment upon entering stationary phase (Table 2) (Chatfield & Cianciotto, 2007). Tests for glucose fermentation, nitrate reduction, urease, catalase, gelatinase and oxidase were performed as described previously (Orrison et al., 1983; Weaver & Feeley, 1979) using stationary-phase bacteria obtained from BCYE agar. β-Lactamase and hippurate hydrolysis activities were assessed by disc assays (Becton Dickinson) as described previously (Kuroki et al., 2007). As expected of a member of the genus Legionella (Dennis et al., 1993; Weaver & Feeley, 1979; Yang et al., 2012), H63T was negative for glucose fermentation, nitrate reduction and urease activity (Table 2). However, the strain was positive for catalase, gelatinase, β-lactamase and hippurate hydrolysis (Table 2). The strongly positive hippurate hydrolysis test distinguished H63T from L. brunensis, L. londiniensis, L. jordanis, L. erythra, L. dresdenensis, L. rubrilucens and L. birminghamensis, and the presence of both gelatinase and β-lactamase differentiated H63T from L. feeleii. The fact that H63T was positive for hippurate hydrolysis and weakly positive for oxidase distinguishes it from L. hackeliae and L. jamestowniensis, the two other species that showed high similarity to H63T based on mip and rnpB sequences (Brenner et al., 1985).
Table 2. Differential characteristics of strain H63T compared with its nearest neighbours based on 16S rRNA gene sequences.
Strains: 1, L. cardiaca sp. nov. H63T (data from this study); 2, L. brunensis ATCC 43878T (unless indicated, data from Wilkinson et al., 1988); 3, L. londiniensis ATCC 49505T (Dennis et al., 1993); 4, L. jordanis ATCC 33623T (Cherry et al., 1982); 5, L. erythra ATCC 35303T (Brenner et al., 1985), 6, L. dresdenensis DSM 19488T (Lück et al., 2010); 7, L. rubrilucens ATCC 35304T (Brenner et al., 1985); 8, L. feeleii ATCC 35072T (Brenner et al., 1985); 9, L. pneumophila ATCC 33152T (Brenner et al., 1979); 10, L. birminghamensis ATCC 43702T (Wilkinson et al., 1987). Reactions are scored as follows unless indicated: +, positive; +w, weakly positive; −, negative; ±, variable; nd, no data available. All strains grow on BCYE at 37 °C but do not grow under these conditions without cysteine, and all strains grow in BYE at 37 °C. All strains are positive for catalase and are negative for glucose fermentation, nitrate reduction and urease activity.
| Characteristics | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Pigment production when grown in BYE/BCYE at 37 °C | + | + | + | + | + | −* | + | +w | + | − |
| Autofluorescence when grown on BCYE at 37 °C | − | − | − | − | + | +w | + | − | − | + |
| Gelatinase | + | + | + | + | + | + | + | − | + | + |
| β-Lactamase | + | + | + | + | + | nd | + | − | + | + |
| Hippurate hydrolysis | + | − | −/+w | − | − | − | − | +w | + | − |
| Oxidase | +w | − | − | + | + | − | − | − | + | ± |
| Protease*† | + | + | +w | + | ++ | − | ++ | − | ++ | ++ |
| Phosphatase*† | + | ++ | − | +w | +w | +w | +w | − | ++ | − |
| Lipase*† | − | +w | +w | − | + | − | + | +w | ++ | +w |
| Swimming motility | + | −* | − | + | + | +* | + | + | + | + |
| Sliding motility* | − | − | − | − | − | − | − | + | + | − |
| Growth on BCYE at 17 °C*‡ | − | + | − | + | +++ | + | + | +++ | +++ | ++ |
| Growth on low-iron BCYE at 37 °C*‡ | + | ++ | + | + | +++ | +++ | +++ | +++ | +++ | +++ |
| Growth in deferrated CDM at 37 °C§ | + | ++ | +/− | +* | + | +* | +++ | +++ | +++ | ++ |
| Siderophore secretion§ | − | ++ | +/− | +* | +/− | −* | +++ | ++ | +++ | ++ |
| Salt sensitivity|| | s | s | r | r | s | s* | s | r | s | r |
Data from this study.
++, Strongly positive; +, positive; +w, weakly positive, slightly above background; −, negative.
Scored as follows in comparison with growth of spot dilutions on BCYE at 37 °C: +++, equal or 1 log less growth; ++, 2–4 logs less growth; +, 5–6 logs less growth; −, no growth.
Scored as follows: +++, high CDM growth/chrome azurol S (CAS) reactivity; ++, moderate CDM growth/CAS reactivity;+, slight CDM growth/CAS reactivity; +/−, CDM growth/CAS reactivity varied between experiments for unknown reasons (Starkenburg et al., 2004).
r, Resistant (similar level of growth on BCYE with or without NaCl); s, sensitive (reduced efficiency of plating on BCYE in the presence of 100 mM NaCl at 37 °C) (O’Connell et al., 1996).
Although we were able to detect phenotypic differences between H63T and its nearest neighbours using long-established methods, it can be difficult to distinguish Legionella species based on the biochemical tests that are typically done, because various species give similar reactions in many of the tests. For example, L. lansingensis cannot be distinguished from Legionella micdadei and Legionella maceachernii based on standard biochemical profiling (Hookey et al., 1996; Thacker et al., 1992). For this reason, we examined 10 additional characteristics that we have recently found to be expressed variably within the genus Legionella (Söderberg et al., 2008; Starkenburg et al., 2004; Stewart et al., 2009). To that end, cell-free supernatants from late-exponential BYE broth cultures were analysed for protease, acid phosphatase and lipase activities as measured by azocasein, p-nitrophenyl phosphate and p-nitrophenyl palmitate hydrolysis, respectively (Aragon et al., 2000, 2001; Thorpe & Miller, 1981). Strain H63T was positive for both protease and phosphatase activities but lacked lipase activity, a finding that distinguished it from all nine of its nearest neighbours (Table 2). That H63T had these activities in BYE culture supernatants suggests that the strain has a functional type-II protein secretion system, as has been documented extensively in L. pneumophila (Cianciotto, 2009; Pearce & Cianciotto, 2009). Interestingly, in L. pneumophila, a functional type-II secretion system has also been linked to sliding on low-agar media (Stewart et al., 2009) and growth at low temperature (Söderberg et al., 2008). H63T exhibited swimming motility by wet-mount microscopy of 3-day-old BCYE agar-grown cultures, but did not show sliding motility (surface translocation) and its associated surfactant when grown on 0.5 % agar BCYE plates incubated at 30 °C for 14 days (Stewart et al., 2009). These data indicated further differences between H63T and L. brunensis, L. londiniensis, L. feeleii and L. pneumophila (Table 2). Strain H63T was unable to grow at 17 °C on BCYE agar, differentiating it from eight of its nine nearest neighbours; L. londiniensis was the only other species in the panel that did not grow under this low-temperature condition (Table 2). That H63T did not exhibit sliding motility nor grow at 17 °C on BCYE agar would suggest that it lacks those type-II-dependent factors associated with sliding and low-temperature growth. Unlike L. brunensis, L. erythra, L. dresdenensis, L. rubrilucens, L. feeleii, L. pneumophila and L. birminghamensis, H63T grew very poorly at 37 °C on BCYE agar depleted for iron by the addition of 14 µM deferoxamine mesylate (Table 2) (Chatfield et al., 2011). This result suggested that the strain has a higher-than-average iron requirement and/or a reduced ability to scavenge iron. In support of this hypothesis, H63T, unlike most of its nearest neighbours, showed poor growth in deferrated chemically defined medium (CDM) at 37 °C, and cell-free supernatants obtained 15 h post-inoculation showed no evidence of siderophore activity as measured by the chrome azurol S assay (Table 2) (Chatfield et al., 2011; Liles et al., 2000; Starkenburg et al., 2004). Interestingly, strain H63T secreted a yellow pigment upon culturing in CDM, and its supernatants displayed a green fluorescence under UV light (not shown). We also observed that H63T was more sensitive to the presence of 100 mM NaCl on BCYE agar at 37 °C than were some of the other species (Table 2), as described previously (O’Connell et al., 1996). In L. pneumophila, salt-sensitivity is correlated with a type-IV secretion system known as Dot/Icm (Sadosky et al., 1993; Vogel et al., 1996). In summary, the results from these additional chemotaxonomic assays provide strong evidence for H63T being phenotypically distinct from its phylogenetically nearest neighbours.
The fatty acid composition of H63T was determined after 72 h of incubation at 35 °C on BCYE agar using the Microbial Identification System (MIDI Inc.) and MIDI operating software version 6.0 (Diogo et al., 1999) as described previously (Pearce et al., 2011). Typical of the genus Legionella, the profile of H63T consisted primarily of branched-chain fatty acids and a few hydroxyl fatty acids (Lambert & Moss, 1989; Pearce et al., 2011). The three most abundant fatty acids were anteiso-C15 : 0 (29 %), C16 : 1ω6c and/or C16 : 1ω7c (22 %) and C16 : 0 (21 %); H63T contained only small amounts of 14-carbon and cyclic 17-carbon fatty acids (Table 3).
Table 3. Fatty acid profiles of strain H63T and its nearest neighbours based on 16S rRNA gene sequence similarity.
Strains: 1, L. cardiaca sp. nov. H63T (data from this study); 2, L. brunensis ATCC 43878T (Wilkinson et al., 1988); 3, L. londiniensis ATCC 49505T(Dennis et al., 1993); 4, L. jordanis ATCC 33623T (Cherry et al., 1982); 5, L. erythra ATCC 35303T (Lambert & Moss, 1989); 6, L. dresdenensis DSM 19488T (Lück et al., 2010); 7, L. rubrilucens ATCC 35304T (Lambert & Moss, 1989); 8, L. feeleii ATCC 35072T (Moss et al., 1983); 9, L. pneumophila ATCC 33152T (Brenner et al., 1979; Cherry et al., 1982); 10, L. birminghamensis ATCC 43702T (Wilkinson et al., 1987). Values are percentages of total fatty acids. tr, Trace amount (<1 %); −, not detected/not reported.
| Fatty acid | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| C14 : 0 | 2 | 1 | − | tr | − | − | − | 1 | tr | − |
| iso-C14 : 0 | tr | 1 | tr | tr | tr | − | 2 | 4 | 8 | − |
| anteiso-C14 : 0 | 2 | − | − | − | − | − | − | − | − | − |
| C15 : 0 | − | 1 | 1 | tr | − | − | − | 3 | tr | 3 |
| iso-C15 : 0 | 3 | 2 | tr | − | 1 | − | tr | − | − | − |
| anteiso-C15 : 0 | 29 | 39 | 5 | 42 | 8 | 16 | 12 | 18 | 14 | 30 |
| C15 : 0 2-OH | tr | − | − | − | − | − | − | − | − | − |
| C15 : 1 | − | tr | − | 2 | − | − | − | 3 | tr | − |
| C15 : 1ω6c | tr | − | 2 | − | − | − | − | − | − | − |
| C16 : 0 | 21 | 12 | 15 | 4 | 26 | − | 15 | 21 | 10 | 13 |
| iso-C16 : 0 | 5 | 5 | 11 | 17 | 4 | 25 | 23 | 17 | 32 | 14 |
| C16 : 1 | − | 9 | 19 | 7 | − | − | − | 18 | 13 | 11 |
| iso-C16 : 1 | − | − | tr | 2 | − | − | − | 2 | 2 | − |
| C16 : 1ω7c/C16 : 1ω6c | 22 | − | − | − | 38 | 24 | 27 | − | − | − |
| C17 : 0 | tr | tr | 8 | 2 | − | − | − | 1 | tr | − |
| iso-C17 : 0 | 3 | 1 | 0 | − | 1 | − | tr | − | − | − |
| anteiso-C17 : 0 | 10 | 24 | 23 | 18 | 10 | 9 | 9 | 6 | 11 | 16 |
| C17 : 0 cyclo | 1 | 1 | 0 | 2 | − | 0 | − | − | 3 | 0 |
| C18 : 0 | 1 | 1 | 8 | tr | 4 | − | 2 | 2 | 2 | − |
| C20 : 0 | 2 | 2 | − | 2 | − | − | − | 3 | 2 | − |
L. pneumophila and some of the other Legionella species tested are well known for their capacity to grow in macrophages; indeed, intracellular infection of lung macrophages is critical for the pathogenesis of legionellosis (Brieland et al., 1994; Rossier et al., 2004). Therefore, we assayed the ability of H63T to replicate within human U937 cell (ATCC CRL-1593.2) macrophages as described previously (Rossier et al., 2004). During the first 48 h of infection, H63T replicated by approximately two logs, indicating that H63T is quite adept at intracellular replication (Fig. S4a). In comparison, L. pneumophila ATCC BAA-74 displayed approximately 4 logs of growth during 48 h of infection (Fig. S4a). H63T was next examined for its ability to grow and survive within the lungs of 6- to 8-week-old A/J mice (Jackson Laboratory, Bar Harbor, ME, USA) following intratracheal inoculation with 106 c.f.u. stationary-phase BCYE agar-grown bacteria (Rossier et al., 2004). The numbers of H63T bacteria increased approximately 5-fold by 48 h post-inoculation, indicating that the strain can grow in the mammalian lung (Fig. S4b). This level of growth was comparable to what we and others have observed for L. pneumophila inoculated into A/J mice (Brieland et al., 1994; Rossier et al., 2004). After 48 h post-inoculation, the numbers of H63T bacteria decreased steadily (Fig. S4b), presumably due to bacterial clearance from the lung by the innate immune response. Compatible with the acute infection observed, histopathological examination of haematoxylin/eosin-stained lung sections taken from infected mice at 72 h post-inoculation displayed irregular interstitial inflammation with moderate mononuclear infiltrate (not shown), indicative of pneumonia, as has been observed previously in A/J mice infected with L. pneumophila for 72 h (Brieland et al., 1994). At 24 h post-inoculation, 4.5 logs of H63T were present in the spleen, and the bacterial burden remained at that level for an additional 24 h (Fig. S4c). After 48 h, the numbers of bacteria in the spleen decreased and, by the fifth day, H63T was detectable in the spleen of only one of the five animals. In summary, H63T replicates significantly within both human macrophages and the murine lung, indicating that the strain is pathogenic, compatible with its isolation from a human patient experiencing severe illness.
Although H63T was isolated from a patient and no environmental source was identified in the case history, it is undoubtedly true that the novel species exists naturally in freshwaters, as is the case for most other legionellae. However, it is difficult to predict if and when an environmental isolation of the species would occur, based on what has been seen for other Legionella species. Indeed, most Legionella species were monotypic in their initial description (Brenner et al., 1985; Morris et al., 1980; Thacker et al., 1988, 1989). In the case of Legionella tucsonensis, since the publication of its clinical isolation in the 1980s, additional strains have yet to be described, despite years of environmental sampling and clinical surveillance (Thacker et al., 1989).
Of the 55 previously characterized Legionella species, 26 have been isolated from clinical cases and, in 25 of those instances, they were believed to be the likely causative agent of disease (Berger et al., 2006; Diederen, 2008; Edelstein et al., 2012; Gobin et al., 2009; König et al., 2005; Marrie et al., 2001; Tang & Krishnan, 1993; Yang et al., 2012). Eleven additional species have been linked to disease through serology (Berger et al., 2006; Fang et al., 1989; Lieberman et al., 2002; Marrie et al., 2001; McNally et al., 2000). We therefore conclude that the novel species represents the fifty-sixth Legionella species, and the thirty-seventh to be linked to disease. Although the lung is the typical primary site for Legionella infections, extrapulmonary manifestations do occur, and legionellae have been found in various niches within the body including the spleen, lymph node, blood, kidney, liver, skin, bone, sinus and heart (Edelstein et al., 1979; Evans & Winn, 1981; McClelland et al., 2004; Monforte et al., 1989; Schlanger et al., 1984; Waldor et al., 1993; Watts et al., 1980; Weisenburger et al., 1980). In the heart, Legionella infection can take the form of myocarditis, pericarditis or endocarditis. There have been 18 documented cases of Legionella endocarditis, including the recent isolation of H63T (Leggieri et al., 2012; Pearce et al., 2011). Four Legionella species, L. pneumophila, L. micdadei, L. dumoffii and L. longbeachae, have been implicated in prosthetic valve endocarditis; however, only L. pneumophila has been isolated from a native heart (Leggieri et al., 2012; Samuel et al., 2011; Tompkins et al., 1988). Therefore, the novel species represents the first non-pneumophila species to be isolated from native valve endocarditis.
Description of Legionella cardiaca sp. nov.
Legionella cardiaca (car.di.a′ca. L. fem. adj. cardiaca of or pertaining to the heart, in reference to the isolation of the type strain from aortic valve tissue).
Gram-negative rod. Grows on BCYE agar and requires l-cysteine. Negative in tests for glucose fermentation, nitrate reduction, urease and autofluorescence. Positive in tests for swimming motility, catalase, gelatinase, β-lactamase, hippurate hydrolysis and pigmentation in BYE broth. The fatty acid profile consists primarily of branch-chained fatty acids.
The type strain is H63T ( = ATCC BAA-2315T = DSM 25049T = JCM 17854T), isolated from human aortic valve tissue and the causative agent of endocarditis. The DNA G+C content of the type strain is 41.8 mol%.
Note added in proof
A paper describing two novel species of Legionella, Legionella tunisiensis sp. nov. and Legionella massiliensis sp. nov. by Campocasso et al. (2012), which was accepted for publication shortly after this paper, is published on pp. 3003–3006 of this issue.
Acknowledgements
We acknowledge members of the Cianciotto lab for helpful comments and advice, in particular Kessler McCoy-Simandle and Brendan Mulhern for their technical assistance in murine infections and siderophore assays. We are grateful to Natalia Kosak for her advice and assistance with serology. We thank Christian Lück for kindly providing the L. dresdenensis strain and anti-L. dresdenensis sera. We thank Daniel Garrison, Department of Classics, Northwestern University, Evanston, IL, for his expert assistance in Latin etymology. This work was funded in part by NIH grants AI034937, AI043987 and AI076693 awarded to N. P. C.
Footnotes
Four supplementary figures and a supplementary table are available with the online version of this paper.
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- Aragon V., Kurtz S., Flieger A., Neumeister B., Cianciotto N. P. (2000). Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect Immun 68, 1855–1863 10.1128/IAI.68.4.1855-1863.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon V., Kurtz S., Cianciotto N. P. (2001). Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect Immun 69, 177–185 10.1128/IAI.69.1.177-185.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson R. F., Thacker W. L., Daneshvar M. I., Brenner D. J. (1996). Legionella waltersii sp. nov. and an unnamed Legionella genomospecies isolated from water in Australia. Int J Syst Bacteriol 46, 631–634 10.1099/00207713-46-3-631 [DOI] [PubMed] [Google Scholar]
- Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Wheeler D. L. (2008). GenBank. Nucleic Acids Res 36 (Database issue), D25–D30 10.1093/nar/gkm929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger P., Papazian L., Drancourt M., La Scola B., Auffray J. P., Raoult D. (2006). Ameba-associated microorganisms and diagnosis of nosocomial pneumonia. Emerg Infect Dis 12, 248–255 10.3201/eid1202.050434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. J., Steigerwalt A. G., McDade J. E. (1979). Classification of the Legionnaires’ disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann Intern Med 90, 656–658 [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Steigerwalt A. G., Gorman G. W., Wilkinson H. W., Bibb W. F., Hackel M., Tyndall R. L., Campbell J., Feeley J. C. & other authors (1985). Ten new species of Legionella. Int J Syst Bacteriol 35, 50–59 10.1099/00207713-35-1-50 [DOI] [Google Scholar]
- Brieland J., Freeman P., Kunkel R., Chrisp C., Hurley M., Fantone J., Engleberg C. (1994). Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. A murine model of human Legionnaires’ disease. Am J Pathol 145, 1537–1546 [PMC free article] [PubMed] [Google Scholar]
- Campocasso A., Boughalmi M., Fournous G., Raoult D., La Scola B. (2012). Legionella tunisiensis sp. nov. and Legionella massiliensis sp. nov., isolated from environmental water samples. Int J Syst Evol Microbiol 62, 3003–3006 10.1128/IAI.00489-07 [DOI] [PubMed] [Google Scholar]
- Chatfield C. H., Cianciotto N. P. (2007). The secreted pyomelanin pigment of Legionella pneumophila confers ferric reductase activity. Infect Immun 75, 4062–4070 10.1128/IAI.00489-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield C. H., Mulhern B. J., Burnside D. M., Cianciotto N. P. (2011). Legionella pneumophila LbtU acts as a novel, TonB-independent receptor for the legiobactin siderophore. J Bacteriol 193, 1563–1575 10.1128/JB.01111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry W. B., Gorman G. W., Orrison L. H., Moss C. W., Steigerwalt A. G., Wilkinson H. W., Johnson S. E., McKinney R. M., Brenner D. J. (1982). Legionella jordanis: a new species of Legionella isolated from water and sewage. J Clin Microbiol 15, 290–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J., Lee J. H., Jung Y., Kim M., Kim S., Kim B. K., Lim Y. W. (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57, 2259–2261 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- Cianciotto N. P. (2009). Many substrates and functions of type II secretion: lessons learned from Legionella pneumophila. Future Microbiol 4, 797–805 10.2217/fmb.09.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P. J., Brenner D. J., Thacker W. L., Wait R., Vesey G., Steigerwalt A. G., Benson R. F. (1993). Five new Legionella species isolated from water. Int J Syst Bacteriol 43, 329–337 10.1099/00207713-43-2-329 [DOI] [PubMed] [Google Scholar]
- Diederen B. M. (2008). Legionella spp. and Legionnaires’ disease. J Infect 56, 1–12 10.1016/j.jinf.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Diogo A., Veríssimo A., Nobre M. F., da Costa M. S. (1999). Usefulness of fatty acid composition for differentiation of Legionella species. J Clin Microbiol 37, 2248–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein P. H., Meyer R. D., Finegold S. M. (1979). Isolation of Legionella pneumophila from blood. Lancet 313, 750–751 10.1016/S0140-6736(79)91207-8 [DOI] [PubMed] [Google Scholar]
- Edelstein P. H., Edelstein M. A., Shephard L. J., Ward K. W., Ratcliff R. M. (2012). Legionella steelei sp. nov., isolated from human respiratory specimens in California, USA, and South Australia. Int J Syst Evol Microbiol 62, 1766–1771 [DOI] [PubMed] [Google Scholar]
- Euzéby J. P. (1997). List of bacterial names with standing in nomenclature: a folder available on the Internet. Int J Syst Bacteriol 47, 590–592 10.1099/00207713-47-2-590 [DOI] [PubMed] [Google Scholar]
- Evans C. P., Winn W. C., Jr (1981). Extrathoracic localization of Legionella pneumophila in Legionnaires’ pneumonia. Am J Clin Pathol 76, 813–815 [DOI] [PubMed] [Google Scholar]
- Ezaki T., Hashimoto Y., Yabuuchi E. (1989). Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39, 224–229 10.1099/00207713-39-3-224 [DOI] [Google Scholar]
- Ezaki T., Hashimoto Y., Yamamoto H., Lucida M. L., Liu S. L., Kusunoki S., Asano K., Yabuuchi E. (1990). Evaluation of the microplate hybridization method for rapid identification of Legionella species. Eur J Clin Microbiol Infect Dis 9, 213–217 10.1007/BF01963841 [DOI] [PubMed] [Google Scholar]
- Fang G. D., Yu V. L., Vickers R. M. (1989). Disease due to the Legionellaceae (other than Legionella pneumophila). Historical, microbiological, clinical, and epidemiological review. Medicine (Baltimore) 68, 116–132 10.1097/00005792-198903000-00005 [DOI] [PubMed] [Google Scholar]
- Fields B. S., Benson R. F., Besser R. E. (2002). Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev 15, 506–526 10.1128/CMR.15.3.506-526.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry N. K., Afshar B., Bellamy W., Underwood A. P., Ratcliff R. M., Harrison T. G., European Working Group for Legionella Infections (2007). Identification of Legionella spp. by 19 European reference laboratories: results of the European Working Group for Legionella Infections External Quality Assessment Scheme using DNA sequencing of the macrophage infectivity potentiator gene and dedicated online tools. Clin Microbiol Infect 13, 1119–1124 10.1111/j.1469-0691.2007.01808.x [DOI] [PubMed] [Google Scholar]
- Gobin I., Newton P. R., Hartland E. L., Newton H. J. (2009). Infections caused by nonpneumophila species of Legionella. Rev Med Microbiol 20, 1–11 10.1097/MRM.0b013e32832e82da [DOI] [Google Scholar]
- Herwaldt L. A., Gorman G. W., McGrath T., Toma S., Brake B., Hightower A. W., Jones J., Reingold A. L., Boxer P. A. & other authors (1984). A new Legionella species, Legionella feeleii species nova, causes Pontiac fever in an automobile plant. Ann Intern Med 100, 333–338 [DOI] [PubMed] [Google Scholar]
- Hookey J. V., Saunders N. A., Fry N. K., Birtles R. J., Harrison T. G. (1996). Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like amoebal pathogens. Int J Syst Bacteriol 46, 526–531 10.1099/00207713-46-2-526 [DOI] [Google Scholar]
- König C., Hebestreit H., Valenza G., Abele-Horn M., Speer C. P. (2005). Legionella waltersii – a novel cause of pneumonia? Acta Paediatr 94, 1505–1507 10.1080/080352505100 [DOI] [PubMed] [Google Scholar]
- Kuroki H., Miyamoto H., Fukuda K., Iihara H., Kawamura Y., Ogawa M., Wang Y., Ezaki T., Taniguchi H. (2007). Legionella impletisoli sp. nov. and Legionella yabuuchiae sp. nov., isolated from soils contaminated with industrial wastes in Japan. Syst Appl Microbiol 30, 273–279 10.1016/j.syapm.2006.11.005 [DOI] [PubMed] [Google Scholar]
- Lambert M. A., Moss C. W. (1989). Cellular fatty acid compositions and isoprenoid quinone contents of 23 Legionella species. J Clin Microbiol 27, 465–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A. & other authors (2007). clustal w and clustal_x version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Leggieri N., Gouriet F., Thuny F., Habib G., Raoult D., Casalta J.-P. (2012). Legionella longbeachae and endocarditis. Emerg Infect Dis 18, 95–97 10.3201/eid1801.110579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman D., Lieberman D., Shmarkov O., Gelfer Y., Ben-Yaakov M., Lazarovich Z., Boldur I. (2002). Serological evidence of Legionella species infection in acute exacerbation of COPD. Eur Respir J 19, 392–397 10.1183/09031936.02.00256702 [DOI] [PubMed] [Google Scholar]
- Liles M. R., Scheel T. A., Cianciotto N. P. (2000). Discovery of a nonclassical siderophore, legiobactin, produced by strains of Legionella pneumophila. J Bacteriol 182, 749–757 10.1128/JB.182.3.749-757.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry P. W., Tompkins L. S. (1993). Nosocomial legionellosis: a review of pulmonary and extrapulmonary syndromes. Am J Infect Control 21, 21–27 10.1016/0196-6553(93)90203-G [DOI] [PubMed] [Google Scholar]
- Lück P. C., Jacobs E., Röske I., Schröter-Bobsin U., Dumke R., Gronow S. (2010). Legionella dresdenensis sp. nov., isolated from river water. Int J Syst Evol Microbiol 60, 2557–2562 10.1099/ijs.0.017863-0 [DOI] [PubMed] [Google Scholar]
- Marrie T. J., Raoult D., La Scola B., Birtles R. J., de Carolis E., Canadian Community-Acquired Pneumonia Study Group (2001). Legionella-like and other amoebal pathogens as agents of community-acquired pneumonia. Emerg Infect Dis 7, 1026–1029 10.3201/eid0706.010619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. R., Vaszar L. T., Kagawa F. T. (2004). Pneumonia and osteomyelitis due to Legionella longbeachae in a woman with systemic lupus erythematosus. Clin Infect Dis 38, e102–e106 10.1086/386322 [DOI] [PubMed] [Google Scholar]
- McNally C., Hackman B., Fields B. S., Plouffe J. F. (2000). Potential importance of Legionella species as etiologies in community acquired pneumonia (CAP). Diagn Microbiol Infect Dis 38, 79–82 10.1016/S0732-8893(00)00181-4 [DOI] [PubMed] [Google Scholar]
- Milne I., Lindner D., Bayer M., Husmeier D., McGuire G., Marshall D. F., Wright F. (2009). TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25, 126–127 10.1093/bioinformatics/btn575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monforte R., Marco F., Estruch R., Campo E. (1989). Multiple organ involvement by Legionella pneumophila in a fatal case of Legionnaires’ disease. J Infect Dis 159, 809 10.1093/infdis/159.4.809 [DOI] [PubMed] [Google Scholar]
- Morris G. K., Steigerwalt A., Feeley J. C., Wong E. S., Martin W. T., Patton C. M., Brenner D. J. (1980). Legionella gormanii sp. nov. J Clin Microbiol 12, 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss C. W., Bibb W. F., Karr D. E., Guerrant G. O., Lambert M. A. (1983). Cellular fatty acid composition and ubiquinone content of Legionella feeleii sp. nov. J Clin Microbiol 18, 917–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muder R. R., Yu V. L. (2002). Infection due to Legionella species other than L. pneumophila. Clin Infect Dis 35, 990–998 10.1086/342884 [DOI] [PubMed] [Google Scholar]
- O’Connell W. A., Dhand L., Cianciotto N. P. (1996). Infection of macrophage-like cells by Legionella species that have not been associated with disease. Infect Immun 64, 4381–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrison L. H., Cherry W. B., Tyndall R. L., Fliermans C. B., Gough S. B., Lambert M. A., McDougal L. K., Bibb W. F., Brenner D. J. (1983). Legionella oakridgensis: unusual new species isolated from cooling tower water. Appl Environ Microbiol 45, 536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. D. (1996). TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12, 357–358 [DOI] [PubMed] [Google Scholar]
- Pearce M. M., Cianciotto N. P. (2009). Legionella pneumophila secretes an endoglucanase that belongs to the family-5 of glycosyl hydrolases and is dependent upon type II secretion. FEMS Microbiol Lett 300, 256–264 10.1111/j.1574-6968.2009.01801.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M. M., Theodoropoulos N., Noskin G. A., Flaherty J. P., Stemper M. E., Aspeslet T., Cianciotto N. P., Reed K. D. (2011). Native valve endocarditis due to a novel strain of Legionella. J Clin Microbiol 49, 3340–3342 10.1128/JCM.01066-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff R. M., Lanser J. A., Manning P. A., Heuzenroeder M. W. (1998). Sequence-based classification scheme for the genus Legionella targeting the mip gene. J Clin Microbiol 36, 1560–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman D. A., Schmidt T. M., MacDermott R. P., Falkow S. (1992). Identification of the uncultured bacillus of Whipple’s disease. N Engl J Med 327, 293–301 10.1056/NEJM199207303270501 [DOI] [PubMed] [Google Scholar]
- Ricketts K. D., Joseph C. A., European Working Group for Legionella Infections (2007). Legionnaires disease in Europe: 2005–2006. Euro Surveill 12, E7–E818076862 [Google Scholar]
- Rossier O., Starkenburg S. R., Cianciotto N. P. (2004). Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires’ disease pneumonia. Infect Immun 72, 310–321 10.1128/IAI.72.1.310-321.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C. J., Thollesson M., Kirsebom L. A., Herrmann B. (2005). Phylogenetic relationships and species differentiation of 39 Legionella species by sequence determination of the RNase P RNA gene rnpB. Int J Syst Evol Microbiol 55, 2039–2049 10.1099/ijs.0.63656-0 [DOI] [PubMed] [Google Scholar]
- Sadosky A. B., Wiater L. A., Shuman H. A. (1993). Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61, 5361–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel V., Bajwa A. A., Cury J. D. (2011). First case of Legionella pneumophila native valve endocarditis. Int J Infect Dis 15, e576–e577 10.1016/j.ijid.2011.04.007 [DOI] [PubMed] [Google Scholar]
- Schlanger G., Lutwick L. I., Kurzman M., Hoch B., Chandler F. W. (1984). Sinusitis caused by Legionella pneumophila in a patient with the acquired immune deficiency syndrome. Am J Med 77, 957–960 10.1016/0002-9343(84)90551-5 [DOI] [PubMed] [Google Scholar]
- Söderberg M. A., Dao J., Starkenburg S. R., Cianciotto N. P. (2008). Importance of type II secretion for survival of Legionella pneumophila in tap water and in amoebae at low temperatures. Appl Environ Microbiol 74, 5583–5588 10.1128/AEM.00067-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkenburg S. R., Casey J. M., Cianciotto N. P. (2004). Siderophore activity among members of the Legionella genus. Curr Microbiol 49, 203–207 10.1007/s00284-004-4342-3 [DOI] [PubMed] [Google Scholar]
- Stewart C. R., Rossier O., Cianciotto N. P. (2009). Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J Bacteriol 191, 1537–1546 10.1128/JB.01531-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P., Krishnan C. (1993). Legionellosis in Ontario, Canada: laboratory aspects. In Legionella: Current Status and Emerging Perspectives, pp. 16–17 Edited by Barbaree J. M., Breiman R. F., Dufour A. P. Washington, DC: American Society for Microbiology [Google Scholar]
- Thacker W. L., Plikaytis B. B., Wilkinson H. W. (1985). Identification of 22 Legionella species and 33 serogroups with the slide agglutination test. J Clin Microbiol 21, 779–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker W. L., Benson R. F., Staneck J. L., Vincent S. R., Mayberry W. R., Brenner D. J., Wilkinson H. W. (1988). Legionella cincinnatiensis sp. nov. isolated from a patient with pneumonia. J Clin Microbiol 26, 418–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker W. L., Benson R. F., Schifman R. B., Pugh E., Steigerwalt A. G., Mayberry W. R., Brenner D. J., Wilkinson H. W. (1989). Legionella tucsonensis sp. nov. isolated from a renal transplant recipient. J Clin Microbiol 27, 1831–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker W. L., Dyke J. W., Benson R. F., Havlichek D. H., Jr, Robinson-Dunn B., Stiefel H., Schneider W., Moss C. W., Mayberry W. R., Brenner D. J. (1992). Legionella lansingensis sp. nov. isolated from a patient with pneumonia and underlying chronic lymphocytic leukemia. J Clin Microbiol 30, 2398–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe T. C., Miller R. D. (1981). Extracellular enzymes of Legionella pneumophila. Infect Immun 33, 632–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall B. J., Rosselló-Móra R., Busse H.-J., Ludwig W., Kämpfer P. (2010). Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60, 249–266 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- Tompkins L. S., Roessler B. J., Redd S. C., Markowitz L. E., Cohen M. L. (1988). Legionella prosthetic-valve endocarditis. N Engl J Med 318, 530–535 10.1056/NEJM198803033180902 [DOI] [PubMed] [Google Scholar]
- Vogel J. P., Roy C., Isberg R. R. (1996). Use of salt to isolate Legionella pneumophila mutants unable to replicate in macrophages. Ann N Y Acad Sci 797, 271–272 10.1111/j.1749-6632.1996.tb52975.x [DOI] [PubMed] [Google Scholar]
- Waldor M. K., Wilson B., Swartz M. (1993). Cellulitis caused by Legionella pneumophila. Clin Infect Dis 16, 51–53 10.1093/clinids/16.1.51 [DOI] [PubMed] [Google Scholar]
- Watts J. C., Hicklin M. D., Thomason B. M., Callaway C. S., Levine A. J. (1980). Fatal pneumonia caused by Legionella pneumophila, serogroup 3: demonstration of the bacilli in extrathoracic organs. Ann Intern Med 92, 186–188 [DOI] [PubMed] [Google Scholar]
- Weaver R. E., Feeley J. C. (1979). Cultural and biochemical characterization of the Legionnaires’ disease bacterium. In Legionnaires’: the Disease, the Bacterium and Methodology, pp. 20–25 Edited by Jones G. L., Hebert G. A. Atlanta, GA: Centers for Disease Control [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173, 697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenburger D. D., Rappaport H., Ahluwalia M. S., Melvani R., Renner E. D. (1980). Legionnaires’ disease. Am J Med 69, 476–482 10.1016/0002-9343(80)90023-6 [DOI] [PubMed] [Google Scholar]
- Whiley H., Bentham R. (2011). Legionella longbeachae and legionellosis. Emerg Infect Dis 17, 579–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson H. W., Thacker W. L., Benson R. F., Polt S. S., Brookings E., Mayberry W. R., Brenner D. J., Gilley R. G., Kirklin J. K. (1987). Legionella birminghamensis sp. nov. isolated from a cardiac transplant recipient. J Clin Microbiol 25, 2120–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson H. W., Drasar V., Thacker W. L., Benson R. F., Schindler J., Potuznikova B., Mayberry W. R., Brenner D. J. (1988). Legionella moravica sp. nov. and Legionella brunensis sp. nov. isolated from cooling-tower water. Ann Inst Pasteur Microbiol 139, 393–402 10.1016/0769-2609(88)90102-0 [DOI] [PubMed] [Google Scholar]
- Willems A., Doignon-Bourcier F., Goris J., Coopman R., de Lajudie P., De Vos P., Gillis M. (2001). DNA–DNA hybridization study of Bradyrhizobium strains. Int J Syst Evol Microbiol 51, 1315–1322 [DOI] [PubMed] [Google Scholar]
- Yang G., Benson R. F., Ratcliff R. M., Brown E. W., Steigerwalt A. G., Thacker W. L., Daneshvar M. I., Morey R. E., Saito A., Fields B. S. (2012). Legionella nagasakiensis sp. nov., isolated from water samples and from a patient with pneumonia. Int J Syst Evol Microbiol 62, 284–288 10.1099/ijs.0.027193-0 [DOI] [PubMed] [Google Scholar]


