Abstract
Osteoporosis is a common complex disorder with reduced bone mineral density (BMD) and increased susceptibility to fracture. Peak BMD is one of the primary determinants of osteoporotic fracture risk, and is under substantial genetic control. Extracellular matrix, a major component of bone, influences BMD by regulating mineral deposition and maintaining cellular activity. It contains several SIBLING family proteins, null mutations of which cause mineralization defects in humans. In this study, we tested 59 single-nucleotide polymorphisms (SNPs) located in the 5 SIBLING family genes (DSPP, DMP1, IBSP, MEPE and SPP1) for association with normal variation in peak BMD in healthy men and women. We measured femoral neck (FN) and lumbar spine (LS) areal BMD by dual energy x-ray absorptiometry (DXA) in 1,692 premenopausal European-American women, 512 premenopausal African-American women and 715 European-American men. SNPs were tested for association with FN and LS BMD in the 3 subsamples. In the European-American women, we observed association (p≤0.005) with LS-BMD for SNPs in DSPP, IBSP and MEPE, and for FN-BMD with SNPs in DMP1 and IBSP. Allele specific regulation of gene expression (ASE) is an important mechanism in which an allele giving rise to modest influence in transcript abundance might result in a predisposition to disease. To identify whether there was ASE of SIBLING family genes at these SNPs, we examined 52 human bone samples obtained from the femoral neck during surgical hip replacement (27 female, 25 male; 44 European-American and 8 African-American). We observed unidirectional ASE for the IBSP gene, with lower expression of the G allele compared to the A allele for SNP rs17013181. Our data suggest that SNPs within the SIBLING genes may contribute to normal variation of peak BMD. Further studies are necessary to identify the functional variants and to determine the mechanisms underlying the differences in ASE and how these differences relate to the pathophysiology of osteoporosis.
Keywords: Bone mineral density, Osteoporosis, Genetic association, SIBLING genes, Allele specific expression
1 Introduction
Osteoporosis is a common multifactorial disorder with reduced bone mineral density (BMD) and increased susceptibility to fracture. Peak BMD is one of the primary skeletal determinants of osteoporotic fracture risk. Twin and family studies have shown that as much as 80% of the normal variation of peak BMD is influenced by genetic factors [1,2]. Genome-wide association studies (GWAS) identified several single-nucleotide polymorphisms (SNPs) that influence BMD variation but the overall variation of BMD explained by these polymorphisms is small, indicating that there are many more genes and genetic variants that regulate this complex trait which remain to be discovered [3,4]. Identifying the specific genes and causal allelic variants underlying peak BMD will help reveal the pathogenesis of osteoporosis and ultimately improve diagnosis, prevention, and treatment strategies of this complex disease.
Extracellular matrix is a major component of bone and is a highly dynamic structure [5]. It provides a well-organized framework for bone mineralization and orchestrates many cellular processes required for maintaining BMD. ECM contains several SIBLING (Small Integrin-Binding LIgand N-linked Glycoprotein) family proteins including dentin sialophosphoprotein (DSPP), dentin matrix protein 1 (DMP1), integrin-binding sialoprotein (IBSP), matrix extracellular phosphoglycoprotein (MEPE), and secreted phosphoprotein 1(SPP1) or osteopontin (OPN) [5]. SIBLING proteins are encoded by 5 identically oriented tandem genes clustered within an approximately 375 kb region on human chromosome 4q21-25. Previously, genome-wide association studies identified associations of DMP1, IBSP and MEPE with BMD and/or fracture risk [3,4,6–12]. However, most of these studies were performed in elderly (≥65 years) individuals of Caucasian ancestry. Moreover, it is unknown which of these SIBLING gene/s underlie the association with peak BMD. Previously, we identified genetic variants in IBSP contributing to normal variation of femoral neck BMD in premenopausal women [10]. In this study, we expanded our analysis to evaluate whether SNPs in the 5 SIBLING genes (DSPP, IBSP, DMP1, SPP1and MEPE) were associated with normal variation of peak BMD in a sample of healthy young (≤45 years) European-American women, African-American women and European-American men.
Genetic variation or polymorphism in populations can determine phenotypic variations in complex traits and diseases by regulating the level of gene expression. One approach to detecting cis-acting effects on gene expression is to compare the relative expression of the two alleles in the same heterozygous individual, known as allele specific expression (ASE). ASE is quite common (~30%) in humans [13] and the imbalance of this expression has been shown to contribute to the variation of heritable traits such as breast cancer, colorectal tumor and familial ovarian cancer [14–16]. Thus, identification of allele specific gene expression difference due to functionally important regulatory variation is important for understanding the divergence of phenotypes among individuals, including differences in disease susceptibility. Also, detection of alleles that modify disease risk has important implications for how genetic variation in human may affect response to different treatments. The power of this approach is that both alleles within the heterozygous individuals are exposed to the same environmental, genetic and hormonal influences, which permits testing for even small expression differences.
To identify whether there was preferential allelic expression in SIBLING genes, we examined human bone samples obtained from the femoral neck collected during surgical hip replacement. To our knowledge, this is the first study of ASE involving human bone samples to identify whether any imbalance of allelic expression of SIBLING genes might influence peak BMD.
2 Subjects and Methods
2.1 Subjects
Families consisting of healthy siblings were recruited from the State of Indiana to identify genes contributing to BMD [17,18]. Recruitment focused on families with two or more healthy sisters or brothers. Sisters were required to be premenopausal and within 10 years of each other in age. Parents of the siblings were recruited for genotyping only and did not undergo any phenotypic assessment. A detailed medical history of the siblings was obtained through administration of health and lifestyle questionnaires. Studies were performed at the Clinical Research Center of Indiana University School of Medicine. Informed written consent was obtained from all subjects prior to their participation in the study. The study was approved by the institutional review board of Indiana University.
Characteristics of study cohorts are summarized in Table 1. The sample of sisters and brothers ranged in age from 27 to 40 and 22 to 45 years, respectively. Women who had irregular menses or a history of pregnancy or lactation within three months prior to enrollment were excluded; women taking oral contraceptives were not excluded. Additional exclusion criteria included a history of chronic disease, use of medications known to affect bone mass or metabolism, an inability to have BMD measured because of obesity, and abnormal blood biochemistry (hypo- or hypercalcemia, abnormal thyroid function, serum alkaline phosphatase and serum creatinine level).
Table 1.
Sample Characteristics
| European-American Women | African- American Women |
European- American Men |
|
|---|---|---|---|
| Number of Sibling subjects | 1692 | 512 | 715 |
| Number of Families | 766 | 236 | 336 |
| Number of Genotyped Parents | 634 | 69 | 233 |
| Age (yr) | 33.1 ± 7.2 | 33.0 ± 6.6 | 33.7 ± 10.9 |
| Height (cm) | 165.4 ± 6.0 | 164.4 ± 6.2 | 178.1 ± 6.9 |
| Weight (kg) | 69.8 ± 16.4 | 81.9 ± 19.8 | 87.1 ± 16.9 |
| Spine aBMD (g/cm2) | 1.27 ± 0.14 | 1.34 ± 0.14 | 1.27 ± 0.16 |
| Femoral Neck aBMD (g/cm2) | 1.02 ± 0.13 | 1.11 ± 0.15 | 1.09 ± 0.17 |
Mean ± SD
2.2 BMD, Height and Weight Measurements
Areal BMD (g/cm2) at the lumbar spine (vertebrae L2–L4) and hip (femoral neck) were measured by dual energy x-ray absorptiometry (DXA), using DPX-L and Prodigy instruments (GE Lunar Corp., Madison, WI). These instruments were cross-calibrated weekly using a custom-made step-wedge phantom. There was no detectable systematic difference between these machines over the course of the study. Siblings BMD were measured on the same instrument, usually at the same visit. The coefficient of variation for duplicate measurements after repositioning the subject was 0.5% for lumbar spine and 1.0% for femoral neck. Height and weight were measured using a Harpenden stadiometer and a Scale-Tronix weighing scale, respectively.
2.3 Association Analysis
SNPs located within the 375-kb SIBLING region were chosen for genotyping using the SNP Tagger software [19] (Supplementary Table 1), and were designed to tag at r2 >0.9 all variants with a minor allele frequency of at least 25% in the CEPH Utah population. Genotyping was performed using iPLEX™ assays on the MassARRAY® platform (Sequenom Inc., San Diego, CA). The average missing rate for the genotyping assays was 1.43%. Using one randomly selected individual in each family, each SNP was tested for significant (p<0.001) deviation from Hardy-Weinberg equilibrium. In addition, parental genotypes were used to identify Mendelian inconsistencies, using the program PedCheck [20]. Inconsistent genotypes were individually reviewed and removed before data analysis.
In this sample of men and women, only age and weight were significant covariates of BMD. Together they accounted for 18.4% of lumbar spine and 13.9% of femoral neck BMD variation in the European-American women. Corresponding values were 17.4% and 17.5% in the European-American men, and 10.4% and 15.0% in the African-American women, for spine and femoral neck BMD, respectively. Height and years of oral contraceptive use were also considered as potential covariates, but were not significant in regression models (p>0.10) when age and weight were included. Therefore, regression residuals, representing age- and weight-adjusted BMD, were computed separately in each of these subgroups and used in all subsequent analyses.
Association testing was performed using a linear mixed model framework [21]. SNP genotype was modeled as a fixed effect (taking on three levels corresponding to the observed genotypes), and family (sibship) as a random effect in the mixed model. Correlation between subjects in the same family was modeled by assuming an error covariance matrix exhibiting compound symmetry. The model was fitted using the MIXED procedure in the SAS statistical software (version 9.1, SAS Institute, Cary, NC). The mixed model association test employs data from all siblings with both genotype and phenotype. The proportion of BMD variation explained by each SNP was estimated by the R2 measure for the mixed model framework [21]. This measure is analogous to the common R2 measure in traditional ANOVA. The significant threshold for association was defined as a p-value of 0.05, correcting for independent comparisons within each of the 3 regions (Supplementary Table 1). This corresponds to 4–6 observed SNP groupings per region (r2>0.9 for grouped SNPs, r2<0.2 between groups). Linkage disequilibrium was calculated from the study sample. These results suggest a Bonferroni-corrected significance threshold of 0.01 for each region and a conservative threshold of 0.005 across all regions.
2.4 Allele-specific Expression (ASE)
A total of 52 de-identified (information available for only age, race, sex and diagnosis) human femoral neck bone samples were obtained during surgical hip replacement due to osteoarthritis (n=42), degenerative joint disease (n=9) and osteoporotic fracture (n=1). The samples included 44 European-American (24 male and 20 female) and 8 African-American (3 male and 5 female) individuals. The bones were snap frozen in liquid nitrogen and stored at −80°C. The bones were crushed and homogenized in liquid nitrogen using a mortar and pestle. Genomic DNA (gDNA) was extracted from these bone samples using Gentra Puregene Tissue kit (Qiagen, Valencia, CA). Total RNA was isolated from these bone tissues using Trizol (Life Technologies, Grand Island, NY) followed by DNA-free (Life Technologies, Grand Island, NY) treatment to minimize gDNA contamination. RNA was further purified using RNeasy Mini columns (Qiagen, Valencia, CA) and tested for gDNA contamination by PCR. Single stranded complementary DNA (cDNA) was synthesized from total RNA with High Capacity cDNA Kit (Life Technologies, Grand Island, NY) using random hexamers. Duplicate sets (4 replicates for each set) of cDNA samples were prepared with or without reverse-transcriptase (RT). cDNA from ten nanograms of RNA and same amount of gDNA were used for ASE assays using the MassArray system (Sequenom, Inc., San Diego, CA).
ASE can only be measured for SNPs within the RNA, and only for heterozygous individuals. We chose 9 SNPs located in the exons of SIBLING genes (3 for IBSP, 2 each for DSPP and SPP1, 1 for DMP1 and MEPE) using the dbSNP database (http://www.ncbi.nih.gov/SNP/; Build 137) based on the following criteria: 1) SNP present in the transcribed region, 2) MAF >0.05, and 3) PCR-primer design that avoided introns and did not include adjacent SNPs that could confound results (Table 2 and Supplementary Table 2). A multiplex iPLEX assay was designed using SpectroDesigner (Sequenom, Inc. San Diego, CA) and analyzed by MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight) mass spectrometry. Genotyping and allelotyping were performed by the SpectroTyper 4.0 (Sequenom, Inc. San Diego, CA). For each sample, we measured 4 replicates for gDNA and a total of 8 replicates for cDNA (4 each from two independent reverse transcription reactions). The mean allelic ratio for the cDNAs was normalized by the mean allelic ratio for the heterozygous gDNAs to account for differences in the mass spectrometric properties of the extended oligonucleotides (which are present in exactly equal amounts in the DNA). Significance was determined by a t-test comparing the allelic ratios of the 8 duplicated cDNA samples from an individual to the ratios for all heterozygous genomic DNAs.
Table 2.
SNPs significantly associated with unidirectional allele specific expression in human bone samples
| Gene | SNPa | Sample ID |
Gender | Age | Race | T-test | Allele 1b | Allele 2 | Relative Expression of Allele 1 |
Relative Expression of Allele 2 |
Ratio of expression Allele 2/Allele 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IBSP | rs17013181 | ASE-50 | Male | 51 | European-American | 2.61E-03 | A | G | 0.52 | 0.48 | −1.10 |
| ASE-39 | Male | 55 | European-American | 2.49E-03 | A | G | 0.52 | 0.48 | −1.10 | ||
| ASE-9 | Male | 62 | European-American | 2.38E-03 | A | G | 0.52 | 0.48 | −1.10 | ||
| ASE-25 | Male | 63 | European-American | 1.02E-02 | A | G | 0.52 | 0.48 | −1.10 | ||
| ASE-30 | Male | 64 | European-American | 1.82E-07 | A | G | 0.55 | 0.45 | −1.21 | ||
| ASE-31 | Male | 68 | European-American | 1.56E-09 | A | G | 0.52 | 0.48 | −1.10 | ||
| ASE-41 | Male | 74 | European-American | 1.56E-08 | A | G | 0.54 | 0.46 | −1.19 | ||
| ASE-36 | Male | 77 | European-American | 3.30E-06 | A | G | 0.54 | 0.46 | −1.19 | ||
| ASE-22 | Female | 40 | European-American | 6.80E-05 | A | G | 0.53 | 0.47 | −1.15 | ||
| ASE-38 | Female | 56 | European-American | 5.76E-08 | A | G | 0.56 | 0.44 | −1.28 | ||
| ASE-17 | Female | 57 | European-American | 6.74E-06 | A | G | 0.53 | 0.47 | −1.15 | ||
| ASE-32 | Female | 59 | European-American | 2.04E-10 | A | G | 0.56 | 0.44 | −1.26 | ||
| ASE-2 | Female | 69 | European-American | 1.83E-05 | A | G | 0.55 | 0.45 | −1.20 | ||
| ASE-24 | Female | 69 | European-American | 2.01E-03 | A | G | 0.52 | 0.48 | −1.08 | ||
| ASE-8 | Female | 83 | European-American | 5.03E-08 | A | G | 0.55 | 0.45 | −1.21 | ||
| ASE-40 | Female | 77 | African-American | 2.39E-10 | A | G | 0.55 | 0.45 | −1.20 | ||
| ASE-43 | Female | 77 | African-American | 4.62E-04 | A | G | 0.53 | 0.47 | −1.15 | ||
| ASE-52 | Female | 80 | African-American | 1.50E-02 | A | G | 0.52 | 0.48 | −1.07 | ||
| SPP1 | rs6812524 | ASE-40 | Female | 77 | African-American | 7.60E-07 | G | A | 0.48 | 0.52 | 1.07 |
| ASE-43 | Female | 77 | African-American | 9.50E-05 | G | A | 0.49 | 0.51 | 1.06 | ||
| ASE-52 | Female | 80 | African-American | 1.2E-09 | G | A | 0.47 | 0.53 | 1.13 |
Chromosome positon, MAF and Function of SNPs rs1701381 (88,732,763; 0.265; Missense) and rs6812524 (88,902,725; 0.063; Synonymous)
Bold represents reference allele
MAF: Minor allele frequency base d on dbSNP database (http://www.ncbi.nih.gov/SNP/; Build 137)
3 Results
Sixty SNPs were genotyped across the SIBLING genes. One SNP, rs2045836, deviated from Hardy-Weinberg equilibrium (p<0.001) in our sample and was not analyzed. For the regions genotyped, linkage disequilibrium (LD) patterns (both D’ and r2) in the samples from European-American and African-American samples were similar to respective HapMap data (CEPH and YRI); however, several SNPs were less informative (MAF<0.25; Supplementary Table 1) in our cohorts than in the CEPH data used for the high-coverage tag SNP selection (r2>0.9).
3.1 SNPs associated with lumbar spine and femoral neck BMD in European-American women, African-American women and European-American men
Eight of the 59 analyzed SNPs showed an association (p≤0.005) with BMD in the sample of European-American women (n=1692). Four of these SNPs were associated with lumbar spine BMD, including one missense SNP (rs3750025) in DSPP, one missense SNP (rs1054627) in IBSP and two SNPs (rs3903347 in intron and rs1477603 at 3’ near end) in MEPE (Figure 1 and Supplementary Table 1). In addition, four SNPs were associated with femoral neck BMD, including one SNP (rs2615492) located near the 5’ end of DMP1, one missense SNP (rs1054627) in IBSP and two SNPs (rs13136331 and rs2616268) located in the intergenic region between DMP1and IBSP genes (Figure 2 and Supplementary Table 1). In the sample of European-American men (n=715), association (p≤0.005) was found with lumbar spine BMD for one SNP (rs13136331) in the intergenic region between DMP1 and IBSP genes and one SNP (rs6532013) for femoral neck BMD in the intergenic region between DSPP and DMP1genes (Figures 1 and 2, Supplementary Table 1).
Fig. 1.
SNPs associated with lumbar spine areal BMD in European-American women and European-American men. Four SNPs were associated (p≤0.005) with lumbar spine BMD in European-American women, including one missense SNP (rs3750025) in DSPP (A), one missense SNP (rs1054627) in IBSP (B) and two SNPs (rs3903347 in intron and rs1477603 at 3’ near end) in MEPE (C). In the sample of European-American men, association (p≤0.005) was found with lumbar spine BMD for one SNP (rs13136331) in the intergenic region between DMP1 and IBSP genes (B). Position and orientation of the genes are indicated by the blue arrows.
Fig. 2.
SNPs associated with femoral neck areal BMD in European-American women and European-American men. Four SNPs were associated (p≤0.005) with femoral neck BMD in European-American women, including one SNP (rs2615492) located near the 5’ end of DMP1 (A), one missense SNP (rs1054627) in IBSP (B) and two SNPs (rs13136331 and rs2616268) located in the intergenic region between DMP1and IBSP genes (B). In the sample of European-American men, association (p≤0.005) was found for one SNP (rs6532013) for femoral neck BMD in the intergenic region between DSPP and DMP1genes (A). Position and orientation of the genes are indicated by the blue arrows.
3.2 Allelic specific expression (ASE)
To identify whether there was any preferential allelic expression of the SNPs in the SIBLING genes we selected 9 SNPs based on the criteria mentioned in the Methods. Due to the requirement that SNPs be present in the transcribed region and limitations on PCR-primer design we could not use the same SNPs associated with BMD for the ASE study. However, we selected alternative ASE SNPs in the close proximity to our associated SNPs. The number of informative heterozygotes for each SNP varied widely (Table 2 and Supplementary Table 2). One SNP rs3750025 in DSPP had only 1 heterozygote and was excluded from the analysis. The ASE is measured from 8 replicates of cDNA, 4 from each of two different reverse transcription reactions, normalized to multiple replicates of different genomic DNAs, and the significance is determined by a t-test. For the alleles reported as significant, the SEM was in the range of 0.003 to 0.007 and the coefficient of variation (CV) was between 0.01% to 0.06% among samples, indicating high precision and reproducibility of ASE method employed in this study.
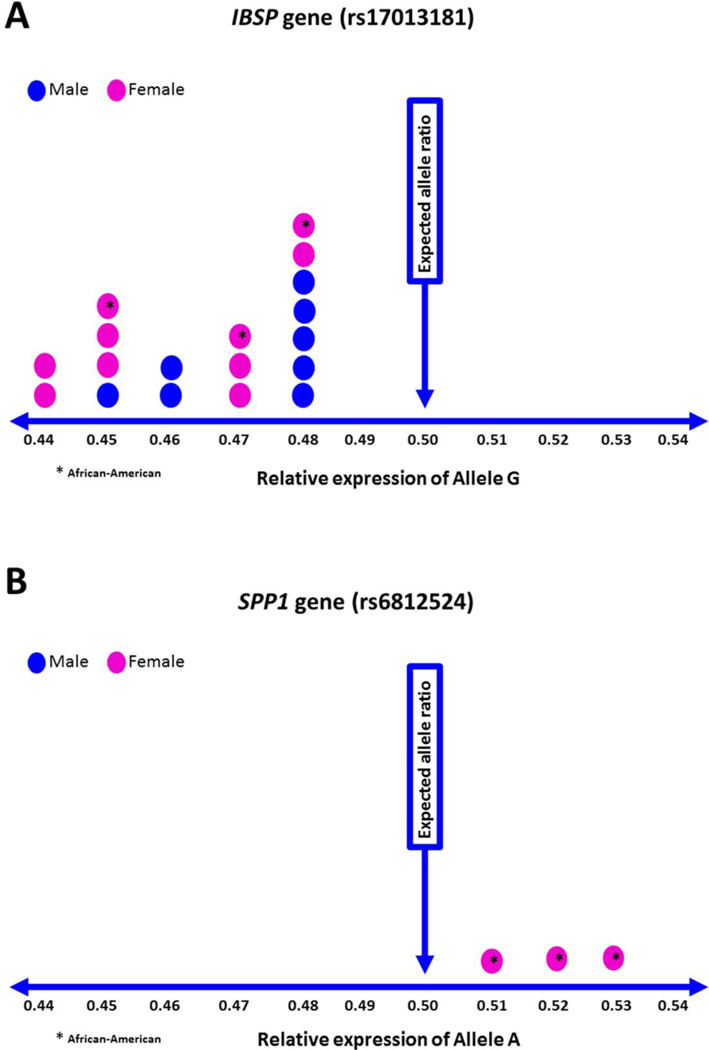
We detected significant unidirectional ASE (all heterozygous samples showing the same allele either lower or higher) for 2 SNPs, one in each IBSP and SPP1. For SNP rs17013181 in IBSP, the minor allele G was expressed at lower levels than the ancestral allele A in all 18 heterozygote individuals (Figure 3A and Table 2). The magnitude of imbalance (ratio of expression of minor and reference alleles; negative 1.08–1.28) was similar in male and female as well as in European-Americans and African-Americans. For SNP rs6812524 in SPP1 we detected higher expression of the minor allele A compared to the reference allele G in all 3 heterozygote African-American females (Figure 3B and Table 2). The magnitude of imbalance was lower (ratio of expression of minor and reference alleles; positive 1.06–1.13) for the SNP in SPP1 compared to the SNP for IBSP gene.
Fig. 3.
Allelic imbalance of mRNA expression in human bone tissue. Significant unidirectional ASE (all heterozygous samples showing the same allele either lower or higher) was observed for 2 SNPs, one in each IBSP and SPP1. For SNP rs17013181 in IBSP, the minor allele G was expressed at lower levels than the ancestral allele A in all 18 heterozygote individuals (A). For SNP rs6812524 in SPP1 higher expression of the minor allele A was observed compared to the reference allele G in all 3 heterozygote African-American females (B).
We detected significant (p<0.05) bidirectional allele specific expression for 6 other SNPs in 5 SIBLING genes (Supplementary Table 2). The direction of effect for these SNPs was not the same for all subjects, indicating that the cis-acting element causing ASE was not the measured SNP. Significant ASE was observed for SNP rs1054629 in IBSP in eight heterozygous individuals, 4 who showed higher and 4 lower expression of the ancestral allele (Supplementary Table 2). Two individuals had higher and one individual had lower expression of the ancestral allele A for SNP rs17013182 in IBSP. For DMP1, 9 individuals had higher and 5 individuals had lower expression of the ancestral allele G of rs2615497. For SNP rs1126616 in SPP1, 9 individuals had higher and 5 individuals had lower expression of the ancestral allele C. For DSPP, 4 individuals had higher and one individual had lower expression of the minor allele of rs2736982. In addition, 4 individuals had higher expression of the minor allele A for SNP rs17013285 in MEPE whereas 2 individuals had lower expression of the same allele.
4 Discussion
Our results showed that several SNPs in the SIBLING region were associated with both femoral neck and lumbar spine BMD in European-American women. We replicated some SNPs associated with BMD from previous findings [8,11] and identified two novel SNPs associated with femoral neck and spine BMD in this study. We also found evidence of unidirectional allelic-specific expression and imbalance of IBSP and SPP1 genes and detected bidirectional ASE for most of the SIBLING family genes in human bone tissue.
In the European-American women, we observed significant association of SNP rs1054627 located in IBSP with both femoral neck and spinal BMD. This SNP was significantly associated in a genome-wide association study with femoral neck BMD in Icelanders and a replication samples of European descent [8]. In our previous study, the same SNP was associated with femoral neck BMD in premenopausal European- and African-American women [10]. Furthermore, association with SNP rs1054627 was replicated in premenopausal women with either extreme high or low hip BMD [11]. This non-synonymous SNP (G195E) located in exon 8 results in a change from a non-hydrophilic to a hydrophilic residue, which could alter the protein structure and/or function. In addition, we detected 2 novel SNPs including one SNP (rs3750025) in DSPP associated with lumber spine BMD in European-American women and one SNP (rs6532013) in the intergenic region between DSPP and DMP1 genes associated with femoral neck BMD in European-American men.
Regulation of gene expression is fundamental to genetic control of human phenotypic variation and disease susceptibility [13]. Studies demonstrated the allele specific regulation of gene expression is an important mechanism for controlling phenotypic variation associated with complex traits and diseases [13–16]. Even an allele giving rise to modest influence in transcript abundance might result in a predisposition to severe disease [15]. As both alleles experience the same cellular environment and hormonal and other influences, the expression difference is most likely responsible for phenotypic variation. The data from this study also suggest that allele specific gene expression is common in SIBLING family genes and their expression difference might contribute to the variation of peak BMD in humans. SNPs may cause differential allelic expression by several mechanisms including influence on transcription binding sites or mRNA stability, differential mRNA processing, and/or miRNA binding efficiency. These mechanisms may result in differences in mRNA concentrations, which may in turn result in different protein expression and, thereby, influence BMD.
Allele-specific gene expression detects cis-acting effects from nearby variants, because both alleles share the same environment. Although the measured SNP could itself be the regulatory variation, the most common mechanism for allelic imbalance is due to cis-acting regulatory variations in strong linkage disequilibrium (LD) with the assayed SNP [13,24–26]. Genetic variation of these nearby regulatory sequences thus could give higher expression of the same mRNA in one person and lower expression in others. The direction of the allelic imbalance will depend on the relationship between the regulatory variants and the measured alleles; individuals having the allele at the regulatory variant that increases expression will show higher expression of whichever measured (reported) allele is on the same strand. If the allele expression imbalance for a given SNP is unidirectional showing the over-expression of the same allele in all individuals, the cis-acting variant is likely to be in high LD with the regulatory variants or could itself be the regulatory SNP.
Of the 9 SNPs for 5 SIBLING genes selected in this study, we detected bidirectional (both higher and lower than expected allele ratio of 50:50) allele specific expression for 6 SNPs in IBSP, DMP1, DSPP, MEPE and SPP1. This bidirectional ASE was observed not only in subjects from a different ethnic background but also between individuals from the same ethnic group. This might be due to the relationship between the measured and functional alleles. The allele measured by the ASE assay is not necessarily the allele(s) that causes the difference in expression; it is in cis with the functional allele(s). Therefore, in subjects showing higher ASE expression, both measured and higher expressing functional alleles may be on the same strand and in subjects showing lower ASE expression, the measured allele is on the same strand with the lower expressing functional allele. In addition, we observed evidence of unidirectional allele specific expression imbalance for SNP rs17013181 in IBSP and for SNP rs6812524 in SPP1 (Figure 3 and Table 2). We observed lower expression of G allele (minor allele) compared to A allele (reference allele) in all 18 heterozygote individuals (8 male European-American, 7 female European-American and 3 female African-American) for rs17013181. Although SNP rs6812524 in SPP1 was only heterozygous in our African-American samples, we also detected ASE (lower expression of G or minor allele and higher expression of A or reference allele) for this SNP in all 3 samples. The evidence of unidirectional allele specific expression imbalance for SNP rs17013181 in IBSP and for SNP rs6812524 in SPP1 suggest that cis-acting functional polymorphism or variation co-segregating with these SNPs exist within the same gene affecting differential mRNA expression in bone. Further studies are necessary to identify these cis-acting elements i.e. the functional variants and to determine the mechanisms underlying the differences in allele specific expression and how these differences relate to the pathophysiology of osteoporosis – for example by changing levels of the proteins. Importantly, significant association of the missense SNP (rs1054627) on exon 8 in IBSP gene as well as clear evidence of ASE in human bone tissue raises the possibility of important role of its allele specific regulation of BMD in humans.
IBSP is a major non-collagenous bone matrix protein involved in calcium and hydroxyapatite binding and plays an important role in cell-matrix interactions through its tripeptide RGD (Arg-Gly-Asp) motifs. IBSP is highly expressed in osteoblasts, osteoclasts and hypertrophic chondrocytes. The expression of IBSP is upregulated in osteoporotic bone [22]. Ibsp knockout mice show high trabecular bone density with impaired bone resorption and mineralization [23]. Future functional studies are necessary to determine how SNPs in IBSP gene and their allele specific expression difference might influence bone density.
Our study has several strengths. We included men and women of young age as well as different ethnic groups for our analysis. In addition, measuring the allele specific expression within the same heterozygous individual eliminates the influence of trans-acting elements as well as environmental, physiological and experimental effects as both alleles are influenced by the same cellular environment and hormonal and other influences. This approach also improves sensitivity of the allele-specific imbalance measurements. However, we have several limitations. We tested SNPs with a MAF of >0.25 for association and MAF of >0.05 for ASE analysis and thus cannot exclude the possible contributions of less common (i.e. MAF <0.01) variants. Also, our phenotype is based on DXA measurements of BMD, which do not provide a compartmental measurements of cortical and trabecular BMD. Furthermore, we obtained association in sample of younger individuals whereas ASE was tested in samples from older donors with mostly osteoarthritic anomalies.
In summary, we tested SNPs for association of SIBLING genes with variation in BMD. We found association for lumbar spine and femoral neck BMD for several SNPs in the SIBLING genes in the European-American men and women. In addition, we detected bidirectional ASE for most of the SIBLING family genes in human bone suggesting that allele specific expression difference due to genetic variation in the SIBLING genes is common. Furthermore, we observed clear evidence of unidirectional allele specific expression imbalance as well as association for SNP in IBSP for femoral neck and spine BMD suggesting that expression difference in this gene might contribute to the variation of peak BMD in allele specific manner in humans. Further studies are warranted to determine the regulatory variations for the differential allele specific expression in this gene and the mechanism how they influence the phenotypic variation of bone mineral density.
Supplementary Material
Highlights.
We tested SNPs in the SIBLING family genes for association with normal variation in peak BMD in humans
In the European-American women, we observed association of SNPS in the DMP1, DSPP, IBSP and MEPE with LS-BMD and FN-BMD
Allele specific gene expression (ASE) is important for controlling phenotypic variation associated with complex traits and diseases
We examined preferential ASE for SNPs in the SIBLING family genes in human bone samples
We observed an allelic imbalance in mRNA expression for SNP rs17013181 in IBSP
Acknowledgements
We thank the siblings and their parents who participated in this study, as well as the study coordinators, without whom this work would not have been possible. This work was supported by National Institutes of Health grants P01 AG18397, R01 AG041517 and M01 RR00750. SNP genotyping by MALDI-TOF mass spectrometry used the facilities of the Center for Medical Genomics at Indiana University School of Medicine, which is supported in part by a grant from the Indiana Genomics Initiative (INGEN). INGEN is supported in part by the Lilly Endowment, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: All authors state that they have no conflicts of interest.
Authors’ roles
Study design: IA, LRP, SI, and MJE. Study conduct: IA, LRP, and MA. Data analysis: IA, LRP, DLK, DL, XX, TF, and HJE. Data interpretation: IA, DLK, MP, XX, TF, HJE, and MJE. Drafting manuscript: IA and LRP. Revising manuscript content: IA, LRP, DLK, TF, XX, HJE, and MJE. Approval of final version of manuscript: IA, LRP, SI, MK, DLK, DL, MP, XX, TF, HJE, and MJE.
References
- 1.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23(3):303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 2.Peacock M, Koller DL, Fishburn T, Krishnan S, Lai D, Hui S, et al. Sex-specific and non-sex-specific quantitative trait loci contribute to normal variation in bone mineral density in men. J Clin Endocrinol Metab. 2005;90(5):3060–3066. doi: 10.1210/jc.2004-2143. [DOI] [PubMed] [Google Scholar]
- 3.Hsu YH, Kiel DP. Clinical review: Genome-wide association studies of skeletal phenotypes: what we have learned and where we are headed. J Clin Endocrinol Metab. 2012;97(10):E1958–E1977. doi: 10.1210/jc.2012-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat Rev Genet. 2012;13(8):576–588. doi: 10.1038/nrg3228. [DOI] [PubMed] [Google Scholar]
- 5.Fisher LW, Fedarko NS. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res. 2003;44(Suppl 1):33–40. [PubMed] [Google Scholar]
- 6.Zmuda JM, Yerges-Armstrong LM, Moffett SP, Klei L, Kammerer CM, Roeder K, et al. Osteoporotic Fractures in Men (MrOS) Study Group. Genetic analysis of vertebral trabecular bone density and cross-sectional area in older men. Osteoporos Int. 2011;22(4):1079–1090. doi: 10.1007/s00198-010-1296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41(1):15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 8.Yerges LM, Klei L, Cauley JA, Roeder K, Kammerer CM, Moffett SP, et al. MrOS Research Group. High-density association study of 383 candidate genes for volumetric BMD at the femoral neck and lumbar spine among older men. J Bone Miner Res. 2009;24(12):2039–2049. doi: 10.1359/JBMR.090524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koller DL, Ichikawa S, Lai D, Padgett LR, Doheny KF, Pugh E, et al. Genome-wide association study of bone mineral density in premenopausal European-American women and replication in African-American women. J Clin Endocrinol Metab. 2010;95(4):1802–1809. doi: 10.1210/jc.2009-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan EL, Danoy P, Kemp JP, Leo PJ, McCloskey E, Nicholson GC, et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011 Apr;7(4):e1001372. doi: 10.1371/journal.pgen.1001372. Epub 2011 Apr 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Styrkarsdottir U, Halldorsson BV, Gudbjartsson DF, Tang NL, Koh JM, Xiao SM, et al. European bone mineral density loci are also associated with BMD in East-Asian populations. PLoS One. 2010 Oct 7;5(10):e13217. doi: 10.1371/journal.pone.0013217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297(5584):1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Weaver J, Bove BA, Vanderveer LA, Weil SC, Miron A, et al. Allelic imbalance in BRCA1 and BRCA2 gene expression is associated with an increased breast cancer risk. Hum Mol Genet. 2008;17(9):1336–1348. doi: 10.1093/hmg/ddn022. [DOI] [PubMed] [Google Scholar]
- 15.Yan H, Dobbie Z, Gruber SB, Markowitz S, Romans K, Giardiello FM, et al. Small changes in expression affect predisposition to tumorigenesis. Nat Genet. 2002;30(1):25–26. doi: 10.1038/ng799. [DOI] [PubMed] [Google Scholar]
- 16.Shen J, Medico L, Zhao H. Allelic imbalance in BRCA1 and BRCA2 gene expression and familial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2011;20(1):50–56. doi: 10.1158/1055-9965.EPI-10-0720. [DOI] [PubMed] [Google Scholar]
- 17.Econs MJ, Koller DL, Hui SL, Fishburn T, Conneally PM, Johnston CC, Jr, et al. Confirmation of linkage to chromosome 1q for peak vertebral bone mineral density in premenopausal white women. Am J Hum Genet. 2004;74:223–228. doi: 10.1086/381401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peacock M, Koller DL, Hui S, Johnston CC, Foroud T, Econs MJ. Peak bone mineral density at the hip is linked to chromosomes 14q and 15q. Osteoporos Int. 2004;15:489–496. doi: 10.1007/s00198-003-1560-7. [DOI] [PubMed] [Google Scholar]
- 19.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 20.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R. Measuring explained variation in linear mixed effects models. Stat Med. 2003;22(22):3527–3541. doi: 10.1002/sim.1572. [DOI] [PubMed] [Google Scholar]
- 22.Trost Z, Trebse R, Prezelj J, Komadina R, Logar DB, Marc J. A microarray based identification of osteoporosis-related genes in primary culture of human osteoblasts. Bone. 2010;46(1):72–80. doi: 10.1016/j.bone.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Malaval L, Wade-Guéye NM, Boudiffa M, Fei J, Zirngibl R, Chen F, et al. Bone sialoprotein plays a functional role in bone formation and osteoclastogenesis. J Exp Med. 2008;205(5):1145–1153. doi: 10.1084/jem.20071294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bray NJ, Buckland PR, Owen MJ, O’Donovan MC. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet. 2003;113(2):149–153. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- 25.Knight JC. Allele-specific gene expression uncovered. Trends Genet. 2004;20(3):113–116. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Buckland PR. Allele-specific gene expression differences in humans. Hum Mol Genet. 2004;13(suppl 2):R255–R260. doi: 10.1093/hmg/ddh227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.