Summary
Recently we prepared sulfated, low molecular weight lignins (LMWLs) to mimic the biological activities of heparin and heparan sulfate. Chemo-enzymatically prepared sulfated LMWLs represent a library of diverse non-sugar, aromatic molecules with structures radically different from the heparins, and have been found to potently inhibit thrombin and factor Xa. To assess their effect on the fibrinolytic system, we studied the interaction of LMWLs with human plasmin. Enzyme inhibition studies indicate that the three sulfated LMWLs studied inhibit plasmin with IC50 values in the range of 0.24 and 1.3 μM, which are marginally affected in the presence of antithrombin. Similarly, plasmin degradation of polymeric fibrin is also inhibited by sulfated LMWLs. Michaelis – Menten kinetic studies indicates that maximal velocity of hydrolysis of chromogenic substrates decreases nearly 70% in the presence of LMWLs, while the effect on Michaelis constant is dependent on the nature of the substrate. Competitive binding studies indicate that the sulfated LMWLs compete with full-length heparin. Comparison with thrombin–heparin crystal structure identifies an anionic region on plasmin as a plausible sulfated LMWL binding site. Overall, the chemo-enzymatic origin coupled with coagulation and fibrinolysis inhibition properties of sulfated LMWLs present novel opportunities for designing new pharmaceutical agents that regulate complex pathologies in which both systems are known to play important roles such as disseminated intravascular coagulation.
Keywords: Plasmin, lignins, heparin mimetics, enzyme inhibition, allostery, anticoagulant, anti-fibrinolytic agent
Introduction
Heparin and heparan sulfate are members of an interesting family of sulfated, linear mucopolysaccharides that play important roles in a number of physiological and pathological processes including coagulation, immune response, angiogenesis, inflammation, and viral invasion (1-3). Both molecules are composed of uronic acid and glucosamine residues that are variably sulfated and are biosynthesized with massive polydispersity and structural heterogeneity (Fig. 1A) (3-5). The pharmaceutical version of heparin, called unfractionated heparin (UFH1), has been in clinical use as an anticoagulant for the past 8 decades. The past decade has seen the introduction of several low molecular weight (LMW) variants of UFH, which are gaining greater acceptance for a number of indications. However, both UFH and LMW heparins are known to be associated with several adverse effects, primarily hemorrhagic risk and variable patient response.
Figure 1. Comparison of structures of heparins (A) and sulfated LMWLs (B).
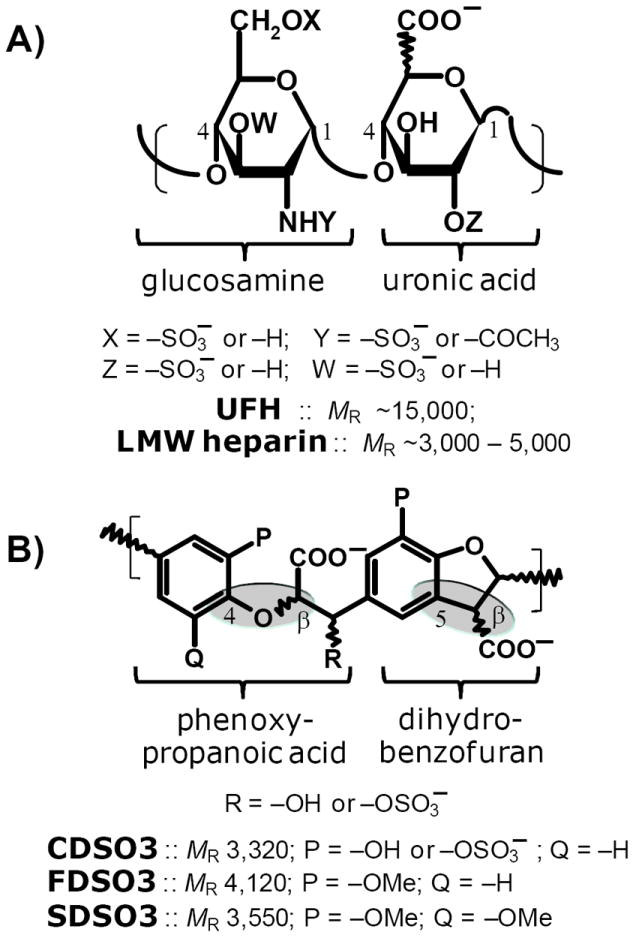
A) UFH or LMW heparin is a polysaccharide composed of alternating glucosamine and uronic acid (either iduronic or glucuronic) residues that are linked to each other in 1→ 4 manner. The saccharides residues are variably sulfated resulting in phenomenal structural heterogeneity. The average molecular weight of UFH is ~15,000, while it is 3,000–5,000 for LMW heparins. Heparins contain an average of ~2.6 sulfates per disaccharide unit. B) CDSO3, FDSO3 and SDSO3 are chemo-enzymatically prepared oligomers, which can also be thought of as a complex polymer composed of dihydrobenzofuran and phenoxy propanoic acid units connected through β-O-4 or β-5 linkages (shown as shaded ovals). The average molecular weight of sulfated LMWLs is in the range of 3,000–4,000. On average these molecules contain one sulfate group every 2 – 3 monomer units.
We reasoned that scaffolds radically different from the sulfated polysaccharides may lead to new anticoagulants with reduced side effect profile in comparison to heparins. These scaffolds were proposed to be significantly less polyanionic and considerably more hydrophobic than UFH and LMW heparins. A less anionic and more hydrophobic scaffold can be expected to recognize the target protein with greater non-ionic binding energy, which may reduce non-specific recognition of numerous proteins and cells typically found in the case of heparins. Yet, engineering such polymeric scaffolds is difficult considering that hydrophobic macromolecules of the size of UFH or LMW heparins that enable introduction of limited number of critical sulfate groups are not readily available.
In one approach to design new anticoagulants with the dual hydrophobic and anionic nature, we recently prepared sulfated low molecular weight variants of lignins (LMWLs, Fig. 1B) as functional mimetics of LMW heparins (6). These designed molecules were prepared in a simple, two-step chemo-enzymatic process involving enzymatic coupling of 4-hydroxycinnamic acids followed by the chemical sulfation of the resulting DHPs. Sulfated LMWLs are composed of oligomeric chains of varying lengths and contain different inter-monomeric linkages, such as β-O-4 and β-5 (Fig. 1B). In this respect, sulfated LMWLs are similar to LMW heparins. Yet, sulfated LMWLs are radically unlike LMW heparins with respect to the nature of their backbone, which is essentially completely aromatic. In fact in terms of structure, sulfated LMWLs are unlike any other class of anticoagulants investigated to-date, including the heparins, the coumarins, the hirudins, the peptidomimetics and the small molecule direct inhibitors. Functionally, the sulfated LMWLs display plasma and blood anticoagulation similar to that of LMW heparins (6,7). Yet mechanistically, the sulfated LMWLs were found to exhibit a novel mechanism of anticoagulation involving an exosite II-mediated allosteric inhibition of thrombin (8). This represents the first example of an exclusive exosite II-dependent inactivation of thrombin’s catalytic function.
To assess whether sulfated LMWLs mimic other interactions of heparin with proteins, we studied the recognition of human plasmin, a key enzyme of the fibrinolysis system. In contrast to heparin, which is known to not inhibit plasmin under physiological conditions, sulfated LMWLs inhibit human plasmin with high potency. The study reveals that plasmin inhibition is not enhanced in the presence of antithrombin, as reported for the case with heparin. Yet, sulfated LMWLs appear to bind in the same region of plasmin as heparin and induce allosteric disruption of the enzyme’s catalytic function. This work supports the concept that sulfated LMWL-like aromatic scaffolds may be exploited to design molecules that simultaneously inactivate plasmin and thrombin for use in pathologies such as disseminated intravascular coagulation, cancer, and others, in which the two enzymes play important roles.
Experimental Procedures
Proteins, Cells and Chemicals
Sulfated LMWLs, CDSO3, FDSO3 and SDSO3 (Fig. 1B), were prepared in two steps from 4-hydroxycinnamic acid monomers, caffeic acid, ferulic acid and sinapic acid as described earlier (6). Human antithrombin (AT) and human plasmin were purchased from Haematologic Technologies (Essex Junction, VT). Chromogenic substrates Spectrozyme PL and Spectrozyme TH were purchased from American Diagnostica (Greenwich, CT). UFH (MR ~15000) was obtained from Sigma (St. Louis, MO). All other chemicals were analytical reagent grade from either Sigma (St. Louis, MO) or Fisher (Pittsburgh, PA) and used without further purification.
Direct and Indirect Inhibition of Plasmin
Both direct and indirect inhibition of human plasmin by sulfated LMWLs was determined through a chromogenic substrate hydrolysis assay (8,9). A 10 μL sample of the sulfated LMWL at concentrations ranging from 0.035 to 10,000 μg/mL was diluted with 930 μL of 50 mM Tris-HCl buffer, pH 7.4, containing 150 mM NaCl at room temperature in a 20K polyethylene glycol-coated polystyrene cuvette, followed by addition of 10 μL of the plasmin solution. After 60 s incubation at room temperature, 50 μL Spectrozyme PL was added. The final concentrations of plasmin and Spectrozyme PL in these experiments were 20 nM and 50 μM, respectively. The residual enzyme activity was determined from the initial rate of increase in absorbance at 405 nm. Relative residual proteinase activity at each concentration was calculated using the activity measured under otherwise identical conditions, except for the absence of the sulfated LMWL. Indirect inhibition of plasmin by sulfated LMWLs was performed at a fixed 20 nM concentration of AT. Except for the presence of AT, the indirect inhibition assay was performed in an otherwise identical manner to the direct inhibition assay. Logistic equation I was used to fit the dose-dependence of residual proteinase activity to obtain the IC50.
| Eq. I |
In this equation Y is the ratio of residual proteinase activity in the presence of inhibitor to its absence (fractional residual activity), YM and YO are the maximum and minimum possible values of the fractional residual proteinase activity, IC50 is the concentration of the inhibitor that results in 50% inhibition of enzyme activity, and HS is the Hill Slope. HS does not represent co-operativity because sulfated LMWLs are highly complex species that may possess multiple binding modes and geometries. Sigmaplot 8.0 (SPSS, Inc. Chicago, IL) was used to perform non-linear curve fitting in which YM, YO, IC50 and HS were allowed to float.
Inhibition of Fibrinolytic Activity of Plasmin by Sulfated LMWLs
A suspension of fibrin was prepared as described in the literature (10). Briefly, a fibrin clot was prepared from thrombin cleavage of fibrinogen in 0.3 M NaCl containing 1 mM EDTA at 37 °C. The clot was washed with water, dissolved in 150 ml of 8M urea, dialyzed against water to remove urea and the fibrin suspension finally heated at 70 °C for 5 minutes. Following cooling, 4 ml of 15% glucose containing 5% gelatin was added to 14 ml of the fibrin suspension and the mixture sonicated for 30 minutes at 42 KHz.
A 10 μl aliquot of plasmin (2 nM) was incubated with 10 μl FDSO3 solution (to give 1.3 – 6.4 μM final concentration) and 680 μl of 20 mM Tris HCl buffer, pH 7.4, containing 100 mM NaCl, 2.5 mM CaCl2 and 0.1% PEG8000 at 37 °C in a 20000 polyethylene glycol-coated polystyrene cuvette. To this solution, 300 μl fibrin suspension as prepared above was mixed and the transmittance at 600 nm monitored. The increase in transmittance as a function of time was fitted by an exponential equation II to derive the observed rate constant of reaction (kOBS) and maximal change in the transmittance (Tmax). In this equation, T0 corresponds to the transmittance value at time 0.
| Eq. II |
Plasmin Binding Studies
Plasmin interaction studies were performed at 25 °C and in 50 mM Tris-HCl buffer, pH 7.4. Fluorescence experiments were performed using a QM4 fluorometer (Photon Technology International, Birmingham, NJ). Equilibrium dissociation constants (KD) for the sulfated LMWLs – plasmin complex were determined by titrating the LMWL into a solution of plasmin in the absence of UFH and monitoring the decrease in the fluorescence at 340 nm (λEX = 280 nm). Sulfated LMWLs also absorb at 280 nm and induce small inner filter effects. Inner filter effects were calculated from A280 values of sulfated LMWLs and the geometry of fluorometer cell using the equation developed by Parker and Barnes (11,12). These effects were found to be approximately 10 – 20% for FDSO3 and were subtracted from the fluorescence of the complex to obtain the change in fluorescence due to plasmin binding (ΔFOBS). The slit widths on the excitation and emission side were 2 and 2 mm, respectively. The decrease in fluorescence signal due to the formation of the complex was fit to the quadratic equilibrium binding equation III to obtain the KD,App of interaction. In this equation, ΔF represents the change in fluorescence due to the formation of the complex following each addition of the ligand ([LMWL]O) from the initial fluorescence FO and ΔFMAX represents the maximal change in fluorescence observed on saturation of plasmin ([PL]O). A binding stoichiometry of 1:1 was assumed for the LMWL – plasmin interaction.
| Eq. III |
Michaelis-Menten Kinetics of Spectrozyme PL and Spectrozyme TH Hydrolysis by Plasmin in the Presence of CDSO3
The initial rate of Spectrozyme PL hydrolysis by 8 nM thrombin was monitored from the linear increase in absorbance at 405 nm corresponding to less than 10% consumption of the substrate. The initial rate was measured as a function of various concentrations of the substrate (10 to 600 μM) in the presence of fixed concentration of CDSO3 (0–60 nM) in 20 mM Tris-HCl buffer, pH 7.4, at 25 °C. The data was fitted by the Michaelis-Menten equation IV to determine KM,app and VMAX. Hydrolysis of Spectrozyme TH by plasmin in the presence of CDSO3 was also followed in an identical manner. In this experiment, Spectrozyme TH was screened in the range of 25 μM to 1 mM concentration range, while plasmin was present at 16 nM in the final solution.
| Eq. IV |
Competitive Binding of Sulfated LMWLs to Plasmin in the Presence of Heparin
The apparent equilibrium dissociation constant (KD,App) of the sulfated LMWL – plasmin complex was measured in the presence of fixed concentrations of UFH spectrofluorometrically at pH 7.4, 25°C. The decrease in intrinsic tryptophan fluorescence was analyzed in the same manner as above using equation II to obtain the KD,App..
Comparison of Crystal Structures of Plasmin and Thrombin
Structures of plasmin and thrombin for analysis were prepared from the crystal structures of plasmin – streptokinase complex (PDB entry 1BML (13)), thrombin – heparin octasaccharide complex (1XMN (14)) and thrombin – haemadin complex (1E0F (15)). Both heparin octasaccharide and haemadin are exosite II ligands. Sybyl 8.1 was used for visualization, generation of molecular surfaces, and electrostatic potential maps. Other than adding hydrogen atoms in Sybyl, no other structural optimization such as minimization was performed. The side chain of Lys236 residue in 1XMN is not defined (14), however it is defined in the thrombin – haemadin co-complex (15), which served the purpose of identifying the corresponding residue in plasmin. The plasmin present in the plasmin – streptokinase complex contains S741A mutation to eliminate its enzymatic activity (13). This change was not expected to significantly affect the comparison of plasmin and thrombin with regard to heparin binding. Connolly surfaces were generated using a probe radius of 1.4 Å in the standard Molcad algorithm available in Sybyl 8.1. The polypeptide chains of plasmin and thrombin were aligned using the sequence alignment algorithm of Sybyl.
Results and Discussion
Sulfated LMWLs Inhibit Plasmin in the Absence of Antithrombin
The discovery of the interaction of sulfated LMWLs with electropositive domains of trypsin-like enzymes to induce inhibition (6,8) suggested that human plasmin may also be a likely target of these interesting macromolecules. To assess this possibility, the enzymatic activity of plasmin in the presence of three sulfated LMWLs, namely CDSO3, FDSO3 and SDSO3 (Fig. 1B), was studied under pseudo-first order conditions. The enzyme activity was followed by spectrophotometric determination of the initial rate of hydrolysis of chromogenic substrate, Spectrozyme PL. As the concentration of the sulfated LMWL was increased, the residual plasmin activity progressively decreased (Fig. 2). Under similar conditions, UFH and LMW heparin displayed no inhibition of human plasmin even at concentrations as high as 26 μM (not shown). The decrease in enzyme activity by sulfated LMWLs could be fitted using the logistic dose–response equation I to derive the IC50 value of inhibition (Table 1).
Figure 2. Direct inhibition of plasmin by sulfated LMWLs.
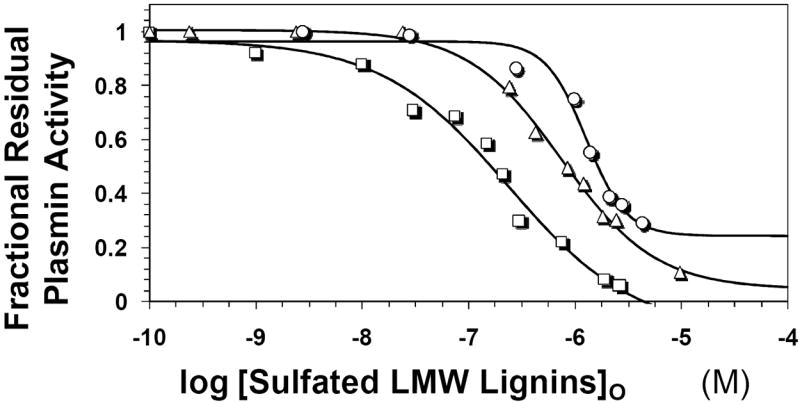
The inhibition of human plasmin by CDSO3 (squares), FDSO3 (triangles) and SDSO3 (circles) was determined spectrophotometrically through a chromogenic substrate hydrolysis assay at pH 7.4 and 25 °C. Solid lines represent sigmoidal fits to the data to obtain IC50, HS, YM, and YO values (equation I), as described in the Experimental Procedures.
Table 1.
Inhibition parameters for sulfated LMWLs inhibiting human plasmin in the presence and absence of antithrombin.a
| Parameter | CDSO3 | FDSO3 | SDSO3 | UFH | LMW heparin | |
|---|---|---|---|---|---|---|
| Without Antithrombin | IC50 (nM) | 242 ± 28b | 760 ± 24 | 1285 ± 60 | >26,000c | >40,000c |
| HS | 0.73 ± 0.19 | 0.98 ± 0.10 | 2.30 ± 0.89 | n/ad | n/a | |
| YM | 0.97 ± 0.04 | 1.01 ± 0.01 | 1.96 ± 0.03 | n/a | n/a | |
| YO | -0.15 ± 0.16 | 0.05 ± 0.04 | 0.24 ± 0.09 | n/a | n/a | |
| With Antithrombin | IC50 (nM) | 262 ± 12 | 1040 ± 36 | 1151 ± 66 | -e | >222,000c |
| HS | 1.11 ± 0.20 | 1.60 ± 0.30 | 1.86 ± 0.24 | - | - | |
| YM | 0.99 ± 0.03 | 1.01 ± 0.02 | 0.98 ± 0.03 | - | - | |
| YO | 0.01 ± 0.07 | 0.17 ± 0.05 | 0.03 ± 0.14 | - | - |
The IC50, HS, YM, YO values were obtained following non-linear regression analysis of direct and indirect inhibition assays. The inhibition assays were performed in 50 mM Tris-HCl buffer, pH 7.4, at 25 °C through spectrophotometric measurement of residual plasmin activity following incubation of the enzyme and the inhibitors (see ‘Experimental Procedures’).
Errors represent ± 2 S. E.
Estimated values based on the highest concentration of the inhibitor used in the experiment.
Not applicable.
Not performed.
CDSO3 and FDSO3 displayed IC50 values of 242 and 760 nM with almost complete inhibition of plasmin (Y0 ~0). SDSO3 displayed 1.7 to 5.3-fold reduced potency in comparison the other two and also did not fully inhibit the enzyme (Y0 = 0.24). These results indicate that the sulfated LMWLs studied here are potent direct inhibitors of human plasmin.
These results are similar in principle to direct inhibition of thrombin by LMWLs, wherein the order of potency was found to be CDSO3 ≥ FDSO3 > SDSO3 (6,8). With respect to thrombin, however, the sulfated LMWLs are nearly 13-fold more potency (IC50 = 18 nM for CDSO3 and 94 nM for SDSO3).
Equation I also provides values of Hill slopes (HS) of the inhibition curves. HS values ranged from 0.7 to 2.3 for direction inhibition of plasmin by sulfated LMWLs and correspond to the steepness of the inhibition profile. Although the common interpretation of HS is the level of co-operativity of the system, in the present case this is unlikely because of the highly complex nature of ligands. Each sulfated LMWL studied herein is a complex mixture of structural species, which may possess multiple modes of binding with multiple geometries in same binding site. These multiple sequences may also exhibit competitive phenomenon resulting in further complexity. Thus, a priori a multivalent molecular analysis of Hill-type is not advisable. Despite this complexity, the analysis of direct plasmin inhibition profiles by sulfated LMWLs shows that HS values are generally closer to 1.0, except for a slightly higher value for SDSO3 (Table 2). It is interesting to note that the HS values for plasmin inhibition are similar to that found for thrombin inhibition and possibly suggest similarity of the two systems.
Table 2.
Hydrolysis of Spectrozyme PL and Spectrozyme TH by plasmin in the presence of CDSO3.a
| [CDSO3]O | Spectrozyme PL | Spectrozyme TH | ||
|---|---|---|---|---|
| KM | VMAX | KM | VMAX | |
|
| ||||
| (nM) | (μM) | (mAbsU/min) | (μM) | (mAbsU/min) |
| 0 | 24.3 ± 2.3b | 70.7 ± 1.6 | 239 ± 27c | 210.0 ± 8.5 |
| 30 | 41.1 ± 4.6 | 46.9 ± 1.1 | 210 ± 16 | 153.7 ± 4.0 |
| 60 | 46.5 ± 8.4 | 28.9 ± 1.4 | 192 ± 38 | 101.4 ± 6.7 |
| 90 | 48.6 ± 12.7 | 24.7 ± 1.7 | 149 ± 30 | 60.9 ± 3.7 |
KM and VMAX of Spectrozyme PL and Spectrozyme TH hydrolysis by plasmin were measured as described in ‘Experimental Procedures’.
Error represents ±1 S.E.
Although the three sulfated LMWLs appear to be rather similar in terms of their polydispersity, heterogeneity and level of sulfation (6), subtle structural differences exist between them. Whereas CDSO3 and FDSO3 are primarily composed of β-O-4 and β-5 inter-monomeric linkages (Fig. 1B), the 3,5-dimethoxy substitution present in SDSO3 does not allow the formation of β-5-linked chains. This suggests that β-O-4 linked chains possess plasmin (and thrombin) inhibition property. Yet, higher inhibition potency, as shown by CDSO3 and FDSO3, is likely to arise from the β-5 chains.
Presence of Antithrombin Does Not Affect Plasmin Inhibition By Sulfated LMWLs
AT, a serpin and an important regulator of coagulation, is known to inhibit several enzymes, including plasmin (16-20). To test whether the presence of the serpin enhances the efficacy of the three sulfated LMWLs to inhibit human plasmin, the direct inhibition protocol was modified to include a fixed concentration of AT (200 nM). A sigmoidal decrease in plasmin activity with sulfated LMWL concentrations on a semi-log plot was observed (not shown) in a manner similar to direct inhibition experiments, which yielded IC50 values of 262, 1040, and 1151 nM for CDSO3, FDSO3 and SDSO3, respectively (Table 1). This indicates that the presence of antithrombin marginally affects the IC50 of the three sulfated LMWLs. This result is in contrast to the thrombin system, wherein the AT-mediated pathway is known to be a competing side reaction for CDSO3 and FDSO3, but not for SDSO3 (19).
The presence of AT also did not affect the non-inhibitory nature of LMW heparin (Table 1). In fact, even ~10-fold higher LMW heparin concentrations in comparison to those used in the direct inhibition assays resulted in no increase in the level of inhibition of plasmin (not shown). Literature suggests that heparin accelerates antithrombin inhibition of plasmin some 20 – 100-fold (17,18), however, a majority of these studies appear to have been performed at low salts concentrations ([NaCl] < 50 mM), while our studies were performed at physiological salt concentrations ([NaCl] = 150 mM), which may explain the discrepancy. In fact, previous studies on heparin binding to plasmin estimate an affinity of approximately 100 μM under low salt conditions (19), which further support current results.
Sulfated LMWLs Inhibit Plasmin Hydrolysis of Fibrin
The observation that sulfated LMWLs inhibit plasmin hydrolysis of chromogenic substrates does not directly imply that these novel molecules will also inhibit plasmin hydrolysis of polymeric fibrin. To test whether sulfated LMWLs are inhibitors of plasmin hydrolysis of fibrin, a transmittance assay was used. Polymeric fibrin was first prepared from fibrinogen following literature procedures (10) and dispersed in 20 mM Tris-HCl buffer, pH 7.4, at 37 °C. The dispersed polymeric fibrin occludes light, thereby reducing the transmittance of the sample. Plasmin degradation of fibrin increases the transmittance of the sample, which provides a convenient measure of the rate of this enzymatic hydrolysis.
Figure 3 shows change in transmittance at 600 nm of fibrin samples hydrolyzed by human plasmin in the presence of 0 to 6.4 μM FDSO3. A characteristic increase in transmittance was observed, as would be expected for an enzyme – substrate reaction, which could be fitted by an exponential equation II to derive the observed rate constant of the reaction (kOBS) and the maximal change in transmittance (Tmax). The kOBS decreases from 2.8±0.2 ms-1 in the absence of FDSO3 to 0.93±0.16 ms-1 in the presence of 6.4 μM FDSO3. Similarly, the Tmax value reduces from 8.1±0.2 to 5.3±0.3 % as the FDSO3 concentration increases from 0 to 6.4 μM. These results suggest that sulfated LMWLs are effective inhibitors of plasmin hydrolysis of polymeric fibrin.
Figure 3. Inhibition of fibrinolysis by a sulfated LMWL.
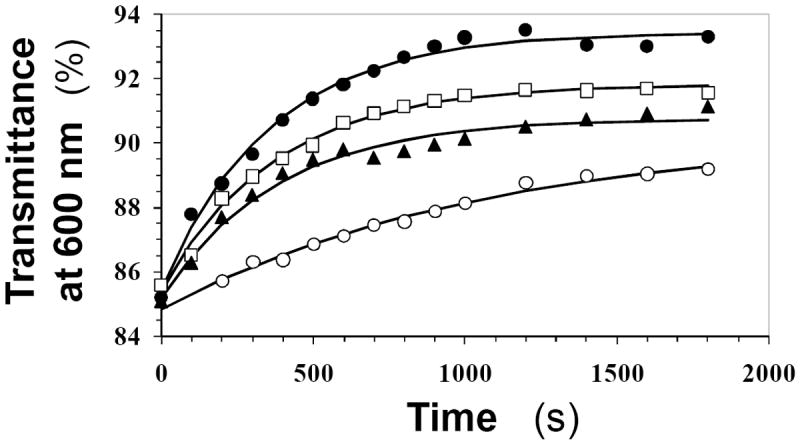
Human plasmin hydrolysis of fibrin was followed in a spectrophotometric assay in the presence of varying concentrations of FDSO3 (●=1.3 μM, □=3.9 μM, ▲=4.5 μM, ○=6.4 μM) at pH 7.4 and 37 °C. The increase in transmittance at 600 nm due to plasmin cleavage of fibrin was monitored as a function of time and fitted by an exponential equation to derive the observed rate constant of inhibition (kOBS) and maximal change in transmittance (Amax). Solid lines represent the exponential fits to the data.
CDSO3 Inhibits Human Plasmin by Disrupting its Catalytic Apparatus
To understand the molecular basis for sulfated LMWLs inhibiting plasmin, we studied the Michaelis-Menten kinetics of Spectrozyme PL hydrolysis at pH 7.4 and 25 °C in the presence of CDSO3, a representative sulfated LMWL. Plots of the initial rates versus Spectrozyme PL concentration were hyperbolic, as expected (Fig. 4A), from which the Michaelis constant (KM) and maximal velocity of the reaction (VMAX) were derived (Table 2). The results show that as the concentration of CDSO3 increased from 0 to 90 nM, the KM value increased nearly 2-fold. This suggests that the presence of CDSO3 disfavors the binding of the chromogenic substrate to the active site of plasmin. In contrast, the VMAX value decreased steadily from a high of 70.7 mAbsU/min in the absence of CDSO3 to a low of 24.7 mAbsU/min at 90 nM CDSO3 (Table 2). Thus, the presence of CDSO3 brings about significant structural changes in the active site of plasmin, which decrease its efficiency of conversion of the Michaelis complex into products.
Figure 4. Michaelis-Menten kinetics of Spectrozyme PL (A) and Spectrozyme (TH) hydrolysis by human plasmin in the presence of CDSO3.
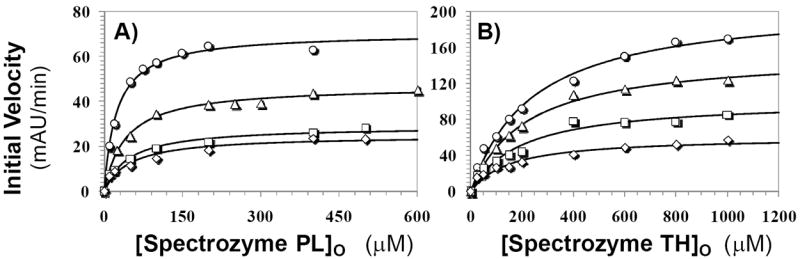
The initial rate of hydrolysis at various substrate concentrations was measured spectrophotometrically in pH 7.4 buffer as described in ‘Experimental Procedures’. The concentrations of CDSO3 chosen for study include 0 (○), 30 (△), 60 (□), and 90 nM (◊). Solid lines represent non-linear regressional fits to the data by the Michaelis-Menten equation III.
To confirm that the structural changes induced in plasmin by CDSO3 binding are generic in nature, and not specific to Spectrozyme PL alone, the kinetics of hydrolysis of Spectrozyme TH was studied. Spectrozyme TH is a thrombin substrate, but retains some affinity for plasmin. Presence of CDSO3 decreased the KM and VMAX nearly 1.6- and 3.4-fold (Fig. 4B, Table 2). Thus, in contrast to Spectrozyme PL, the interaction of Spectrozyme TH is more favored in the presence of CDSO3, while the catalytic apparatus is made dysfunctional.
FDSO3 Competes With Heparin for Binding to Plasmin
To determine the site of sulfated LMWL binding to plasmin, we measured the affinity of FDSO3 – plasmin complex in the presence of UFH. Recently the interaction of sulfated LMWLs with AT was studied in detail using fluorescence spectroscopy (19). Binding of sulfated LMWLs to the serpin resulted in nearly 100% decrease in intrinsic tryptophan fluorescence, which could be fitted by a quadratic binding equation III to obtain the equilibrium dissociation constant KD.
Utilizing an identical protocol, plasmin was first titrated against FDSO3 at pH 7.4 and 25 °C in the absence of any competitor. A characteristic decrease in plasmin fluorescence at 340 nm (λEX = 280 nm) was observed, which reached a plateau at approximately 600 nM FDSO3 (Fig. 5). It is possible that this decrease originates from inner filter effect of FDSO3 absorbing at the excitation wavelength (19). However, even at low levels of FDSO3, wherein inner filter effects are non-existent, a characteristic decrease can be noted. Subtraction of inner filter effects due to background absorption, followed by non-linear regression analysis results in a KD of 35 nM (Table 3). Addition of 29 nM UFH in the pH 7.4 buffer containing no added NaCl resulted in a small right shift of the fluorescence profile (Fig. 5), which resulted in an apparent KD of 117 nM, a 3.4–fold increase. Likewise, increasing the concentration of UFH to 296 nM further weakened the affinity of FDSO3 for plasmin to 781 nM. These results suggest that FDSO3 competes with heparin for binding to human plasmin.
Figure 5. Interaction of FDSO3 with human plasmin in pH 7.4 buffer at 25 °C in the presence and absence of heparin.
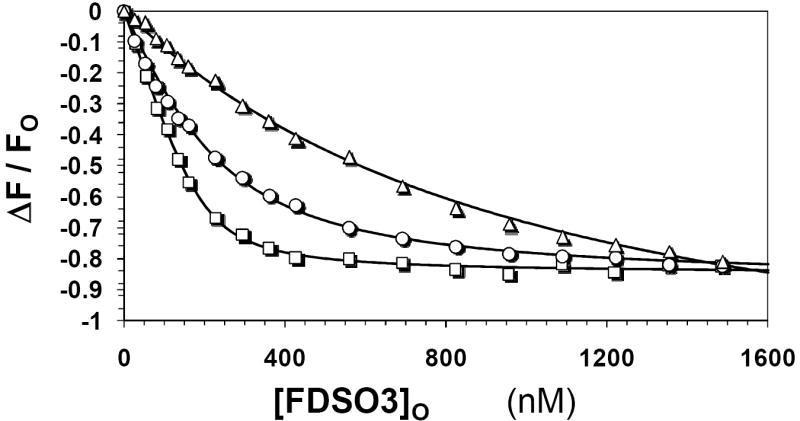
The decrease in intrinsic fluorescence of plasmin (λEX = 280 nm, λEM = 340 nm) that accompanies binding of FDSO3 was used to determine the KD of FDSO3–plasmin complex. UFH was present in the titrations at 0 (□), 29 (○) and 296 nM (△) concentrations. Solid lines represent non-linear fits to the data using quadratic equation II. See Experimental Procedures for details.
Table 3.
Affinity parameters for FDSO3 binding to human plasmin in the presence and absence of unfractionated heparin.a
| KD (nM) | ΔFMAX | |
|---|---|---|
| At 0 mM NaCl and 0 nM UFH | 34.6 ± 6.3b | -0.86 ± 0.02 |
| At 0 mM NaCl and 29 nM UFH | 117 ± 11 | -0.88 ± 0.04 |
| At 0 mM NaCl and 296 nM UFH | 781 ± 108 | -1.29 ± 0.16 |
The KD and ΔFMAX values were obtained following non-linear regression analysis of spectrofluorometric determination of FDSO3 binding to plasmin in 50 mM Tris-HCl buffer, pH 7.4, at 25 °C. (see ‘Experimental Procedures’).
Errors represent ± 1 S. E.
To assess whether the competition between heparin and FDSO3 is ideal, the KUFH of UFH – plasmin interaction in 50 mM Tris-HCl, pH 7.4 at 25 °C was calculated from the Dixon-Webb relationship (equation V) for the two conditions tested.
| Eq. V |
where KUFH refers to the equilibrium dissociation constant of UFH – plasmin complex. The KUFH values from 29 and 296 nM UFH experiments were calculated to be 12.1 and 13.7 nM. These results suggest that a Dixon-Webb-related proportional increase in apparent affinity of FDSO3 for plasmin is observed with the increase in the concentration the competitor. Thus, the competition between heparin and FDSO3 for human plasmin appears to be essentially ideal.
Comparison with Structure of Thrombin–Heparin Octasaccharide Complex Identifies an Exosite-II like Domain in Plasmin as a Plausible Binding Site for Sulfated LMWL
The similarity of the phenomenon of sulfated LMWLs binding to plasmin and thrombin suggested the possibility that the two enzymes may contain a homologous site that recognizes these novel molecules. The observation that sulfated LMWLs compete with heparin suggested that the binding site on plasmin is likely to be a domain homologous to exosite II of thrombin.
To assess the above possibility, the structures of human plasmin (13) and human thrombin (14,15) were superimposed. The exosite II-like domain in human plasmin was identified as the region corresponding to the octasaccharide binding site in thrombin. This domain retains the “S” shaped exosite II channel of thrombin (Fig. 6), which is lined by electropositive and hydrophobic residues. The basic residues include Lys645, Arg637 and Arg779, which correspond in three-dimensional space to Arg101, Lys87, and Arg233 of thrombin. In addition, Arg776 of plasmin may equate to a protonated His230 of thrombin. The hydrophobic residues include Val704, Leu725, Phe780, Val787, and Trp783 in plasmin corresponding to Ile162, Phe181, Leu234, Val241, and Trp237 of thrombin, respectively. Significant residue level differences do exist. For example, there are no counterparts in plasmin to thrombin’s Arg93, Arg126, Lys236, and Lys240. Likewise, selected aromatic side chains (Phe232, Phe245) are also not present in plasmin. The reduced electropositive and hydrophobic character of the exosite-II like domain in plasmin, as compared to that in thrombin, may be the reason for the reduced inhibition potency of sulfated LMWLs for plasmin. Overall, the three-dimensional structural and topological similarity with exosite II of thrombin suggest the possibility that sulfated LMWLs interact with the corresponding domain in plasmin.
Figure 6. Comparison of crystal structures of human plasmin and thrombin.
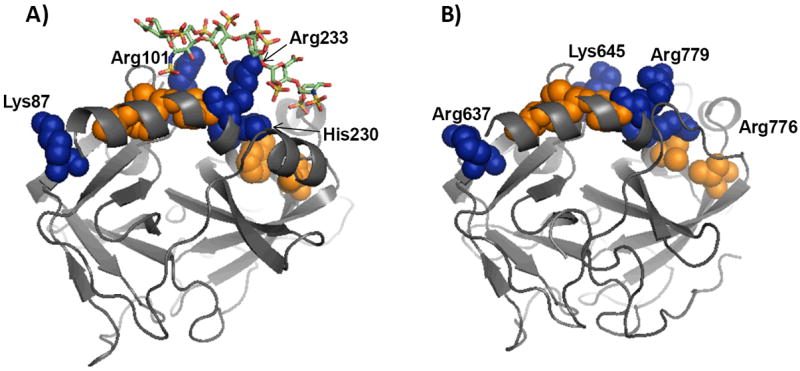
Shown is a comparison of plasmin and thrombin structures from ‘1BML’ (plasmin – streptokinase complex) and 1XMN (thrombin – heparin octasaccharide complex) files. The structures were aligned and the orientation of individual residues in the two structures compared to assess similarities and differences. Several basic (shown in blue van der Waals surfaces) and hydrophobic (shown in orange van der Waals surfaces) residues in exosite II of human thrombin were found to correlate well with comparable residues present in human plasmin. Heparin oligosaccharide present in the 1XMN structure is shown in the stick representation. See text for details.
Significance
The observation that sulfated LMWLs are able to inhibit human plasmin, in addition to thrombin and factor Xa reported previously (6,8), is exciting. Molecules that inhibit both these enzymes in an allosteric manner are rare, primarily because the structures of plasmin and thrombin, although based on the trypsin scaffold, are sufficiently different with respect to their exosites. In fact, UFH, the prototypical exosite modulator, is able to induce plasmin inhibition at sub-physiologic ionic strengths (17,18), but its interaction with exosite II of thrombin is not inhibitory. Thus, the ability of sulfated LMWLs to inhibit both enzymes under physiological conditions is novel and expected to be very useful.
Clinically, several disordered conditions are known in which both thrombin and plasmin are active. In disemminated intravascular coagulation (DIC), the normally tightly regulated processes of coagulation and fibrinolysis escape endogenous control. The result is widespread clotting due to thrombin action followed paradoxically by bleeding due to plasmin action (21,22). Thrombin and plasmin also play important roles in angiogenesis and cancer invasion (23-25). Thus, simultaneously inhibiting both enzymes can be expected to useful in thrombosis as well as thrombosis-dependent cancer.
From the viewpoint of sulfated LMWLs, these are expected to be function better than multiple agents targeting multiple disorders because single-drug poly-pharmacology represents an increase in efficiency, while offering a relative decrease in possible side and/or toxic effects. Additionally, in a prophylactic setting, presence of the multi-effect drug is expected to protect better against follow up attacks. For example, the presence of sulfated LMWLs may automatically protect against bleeding in DIC when used to protect against excessive coagulation.
Yet, challenges exist in engineering sulfated LMWL-based pharmaceutical agents for treating these complex pathologies. For example, CDSO3 is a heterogeneous mixture. Whether a select group of molecules in this mixture is the cause of both plasmin and thrombin inhibition remains to be determined. However, recent studies with homogeneous, chemically synthesized small molecules designed in our laboratory show that the property of inhibiting coagulation enzymes is resident in the common β-5 scaffold present in these sulfated LMWLs (26). Thus, sulfated LMWLs provide the first platform for engineering pharmaceutical agents with allosteric and dual (plasmin and thrombin) inhibition property.
The allosteric mechanism of inhibition of the two targeted enzymes is also of considerable interest. Allosteric modulation enzyme function is especially useful because it affords better regulation. In the case of sulfated LMWL induced, exosite II-mediated inhibition of thrombin, regulation (or reversal of inhibition) can be theoretically achieved with sucrose octasulfate, a heparin mimetic and an exosite II ligand. Whether this ligand can reverse sulfated LMWL inhibition of human plasmin remains to be established. Yet, the possibility of such reversal is likely to offer better outcomes.
Overall, the chemo-enzymatic origin of sulfated LMWLs coupled with dual plasmin and thrombin inhibition properties present novel opportunities for designing new pharmaceutical agents that regulate complex pathologies in which both systems are known to play important roles.
What is known about this topic?
Molecules that possess dual – plasmin and thrombin – inhibition properties are expected to be useful in treating complex pathologies, but are not commonly available
What does this paper add?
This work discovers potent human plasmin inhibition properties of non-sugar heparin mimetics, sulfated low molecular weight lignins
This work identifies the mechanism of plasmin inhibition and pinpoints a plausible domain on plasmin that might serve as the sulfated low molecular weight lignin binding site
Acknowledgments
We thank Dr. Philip Mosier of Virginia Commonwealth University for assisting with structure comparison work. This work was supported by the grants HL090586 and HL099420 from the National Institutes of Health, grant EIA 0640053N from the American Heart Association National Center, grant 6-46064 from the A. D. Williams Foundation and a grant from the Mizutani Foundation for Glycoscience, Japan.
Footnotes
Abbreviations used: AT, antithrombin; CDSO3, sulfated dehydropolymer of caffeic acid; FDSO3, sulfated dehydropolymer of ferulic acid; IC50, concentration of inhibitor that results in 50% inhibition; LMW, low-molecular-weight; LMWL, low-molecular weight lignin; PEG, polyethylene glycol; SDSO3, sulfated dehydropolymer of sinapic acid; UFH, unfractionated heparin
References
- 1.Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interactions with proteins. Chem Biol Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 2.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 3.Coombe DR, Kett WC. Heparan sulfate-protein interactions: therapeutic potential through structure-function insights. Cell Mol Life Sci. 2005;62:410–424. doi: 10.1007/s00018-004-4293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 5.Rabenstein DL. Heparin and heparan sulfate: structure and function. Nat Prod Rep. 2002;19:312–331. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 6.Monien BH, Henry BL, Raghuraman A, et al. Novel chemo-enzymatic oligomers of cinnamic acids as direct and indirect inhibitors of coagulation proteinases. Bioorg Med Chem. 2006;14:7988–7998. doi: 10.1016/j.bmc.2006.07.066. [DOI] [PubMed] [Google Scholar]
- 7.Henry BL, Thakkar JN, Martin EJ, et al. Characterization of the plasma and blood anticoagulation potential of structurally and mechanistically novel oligomers of 4-hydroxycinnamic acids. Blood Coag Fibrinol. 2009;20:27–34. doi: 10.1097/MBC.0b013e328304e077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henry BL, Monien BH, Bock PE, et al. A novel allosteric pathway of thrombin inhibition: Exosite II mediated potent inhibition of thrombin by chemo-enzymatic, sulfated dehydropolymers of 4-hydroxycinnamic acids. J Biol Chem. 2007;282:31891–31899. doi: 10.1074/jbc.M704257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vörös G, Kolev K, Csomor K, et al. Inhibition of plasmin activity by sulfated polyvinylalcoholacrylate copolymers. Thromb Res. 2000;100:353–361. doi: 10.1016/s0049-3848(00)00329-7. [DOI] [PubMed] [Google Scholar]
- 10.Kanai S, Okamoto H, Tamaura Y, Yamazaki S, Inada Y. Fibrin suspension as a substrate fop plasmin: determination and kinetics. Thromb Haemost. 1979;42:1153–1158. [PubMed] [Google Scholar]
- 11.Lutz H-P, Luisi PL. Correction for inner filter effects in fluorescence spectroscopy. Helv Chim Acta. 1983;66:1929–1935. [Google Scholar]
- 12.Parker CA, Barnes WJ. Some experiments with spectrofluorimeters and filter fluorimeters. Analyst. 1957;82:606–618. [Google Scholar]
- 13.Wang X, Lin X, Loy JA, et al. Crystal structure of the catalytic domain of human plasmin complexed with streptokinase. Science. 1998;281:1662–1665. doi: 10.1126/science.281.5383.1662. [DOI] [PubMed] [Google Scholar]
- 14.Carter WJ, Cama E, Huntington JA. Crystal structure of thrombin bound to heparin. J Biol Chem. 2005;280:2745–2749. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- 15.Richardson JL, Kroger B, Hoeffken W, et al. Crystal structure of the human α-thrombin – haemadin complex: an exosite II binding inhibitor. EMBO J. 2000;19:5650–5660. doi: 10.1093/emboj/19.21.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson ST, Swanson R, Raub-Segall E, et al. Accelerating ability of synthetic oligosaccharides on antithrombin inhibition of proteinases of the clotting and fibrinolytic systems. Comparison with heparin and low-molecular-weight heparin. Thromb Haemost. 2004;92:929–939. doi: 10.1160/TH04-06-0384. [DOI] [PubMed] [Google Scholar]
- 17.Jordan RE, Oosta GM, Gardner WT, et al. The kinetics of hemostatic enzyme-antithrombin interactions in the presence of low molecular weight heparin. J Biol Chem. 1980;255:10081–10090. [PubMed] [Google Scholar]
- 18.Machovich R, Bauer PI, Arányi P, et al. Kinetic analysis of the heparin-enhanced plasmin--antithrombin III reaction. Apparent catalytic role of heparin. Biochem J. 1981;199:521–526. doi: 10.1042/bj1990521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry BL, Connell J, Liang A, et al. Interaction of antithrombin with sulfated, low molecular weight lignins: Opportunities for potent, selective modulation of antithrombin function. J Biol Chem. 2009;284:20897–20908. doi: 10.1074/jbc.M109.013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan RE, Oosta GM, Gardner WT, et al. The binding of low molecular weight heparin to hemostatic enzymes. J Biol Chem. 1980;255:10073–80. [PubMed] [Google Scholar]
- 21.Horan JT, Francis CW. Fibrin degradation products, fibrin monomer and soluble fibrin in disseminated intravascular coagulation. Semin Thromb Hemost. 2001;27:657–666. doi: 10.1055/s-2001-18870. [DOI] [PubMed] [Google Scholar]
- 22.Levi M. Disseminated intravascular coagulation. Crit Care Med. 2007;35:2191–2195. doi: 10.1097/01.ccm.0000281468.94108.4b. [DOI] [PubMed] [Google Scholar]
- 23.Lijnen HR. Plasmin and matrix metalloproteinases in vascular remodeling. Thromb Haemost. 2001;86:324–333. [PubMed] [Google Scholar]
- 24.Tarui T, Majumdar M, Miles LA, et al. Plasmin-induced migration of endothelial cells: a potential target for the anti-angiogenic action of angiostatin. J Biol Chem. 2002;277:33564–33570. doi: 10.1074/jbc.M205514200. [DOI] [PubMed] [Google Scholar]
- 25.Snyder KM, Kessler CM. The pivotal role of thrombin in cancer biology and tumorigenesis. Semin Thromb Hemost. 2008;34:734–741. doi: 10.1055/s-0029-1145255. [DOI] [PubMed] [Google Scholar]
- 26.Verghese J, Liang A, Sidhu PP, et al. First steps in the direction of synthetic, allosteric, direct inhibitors of thrombin and factor Xa. Bioorg Med Chem Lett. 2009;19:4026–4029. doi: 10.1016/j.bmcl.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]


