Abstract
Osteoclasts are bone-resorbing multinucleated cells differentiated from monocyte/macrophage lineage precursors. A novel osteoclast precursor cell line, 4B12 was established from Mac-1+c-Fms+RANK+ cells from calvaria of 14-day-old mouse embryos using immunofluorescence and cell-sorting methods. Like M-CSF-dependent bone marrow macrophages (M-BMMs), M-CSF is required for 4B12 cells to differentiate into TRAP-positive multinucleated cells [TRAP(+) MNCs] in the presence of RANKL. Bone-resorbing osteoclasts differentiated from 4B12 cells on dentine slices possess both a clear zone and ruffled borders and express osteoclast-specific genes. Bone-resorbing activity, but not TRAP, was enhanced in the presence of IL-1α. The number of TRAP(+) MNCs and the number of pits formed from 4B12 cells on dentine slices was four-fold higher than that from M-BMMs. 4B12 cells were identified as macrophages with Mac-1 and F4/80, yet lost these markers upon differentiation into osteoclasts as determined by confocal laser scanning microscopy. The 4B12 cells do not have the potential to differentiate into dendritic cells indicating commitment to the osteoclast lineage. 4B12 cells are readily transfectable with siRNA transfection before and after differentiation. These data show that 4B12 cells faithfully replicate the properties of primary cells and are a useful and powerful model for analyzing the molecular and cellular regulatory mechanisms of osteoclastogenesis and osteoclast function.
Keywords: osteoclast precursor, cell line, macrophage, differentiation, transfection
Introduction
Osteoclasts are multinucleated giant cells with the capacity to resorb mineralized tissues. It is believed that circulating monocyte/macrophage lineage cells differentiate into mature osteoclasts at bone surfaces through several steps including chemotaxis and attachment of the cells to bone, differentiation into multinucleated cells, the formation of both a sealing zone and ruffled borders, followed by active resorption [1]. Two cytokines have been identified as essential factors for the differentiation of osteoclast precursor cells into mature osteoclasts. One is macrophage colony-stimulating factor (M-CSF) produced by osteoblast/stromal cells [2]. It is also known that the M-CSF functions as an important factor for the proliferation of osteoclast progenitor cells and the survival of mature osteoclasts [3]. The effects of M-CSF are mediated by c-Fms (M-CSF receptor) [4]. c-Fms is expressed on primitive multipotent hematopoietic cells, monocytes, and osteoclasts [5]. M-CSF/c-Fms signaling in osteoclast precursors is indispensable for osteoclastogenesis [6]. The other is the receptor activator of NF-κB (RANK) ligand (RANKL) expressed on the osteoblast/stromal cell membrane [7]. RANKL is an anchored cell membrane factor that binds to RANK, a RANKL receptor [8]. RANK is mainly expressed in osteoclast progenitor cells, osteoclasts, dendritic cells (DCs), and T-lymphocytes [8,9]. RANKL/RANK signaling in osteoclast precursors is indispensable for osteoclastogenesis [10]. The discovery of these signaling pathways enables us to investigate the regulation of osteoclast differentiation and function.
Although M-CSF-dependent bone marrow macrophages (M-BMMs) have been commonly used to study the mechanism of osteoclast differentiation, they are not a homogeneous population. The development of a maintainable cell line with characteristics of osteoclast precursors would allow further studies of the molecular and cellular biology of osteoclasts. Several cell lines representing osteoclast progenitors have been established from in vivo or in vitro immortalized cells [11–14], while previously established macrophage cell lines such as human leukemic cell line (FLG 29.1), murine macrophage cell line (BDM-1, RAW 264.7), have also been used to study osteoclastogenesis [9,15,16]. However, an osteoclast precursor cell line recapitulating the features of primary osteoclast differentiation and function has not been established. In the present study, we established and characterized a novel osteoclast precursor cell line, 4B12, from 14-day-old (E14) mouse embryo calvarial cells. Previously we had devised an assay system utilizing devitalized bone slices for the study of osteoclast formation [17]. We hypothesized that it would be possible to isolate osteoclast precursors from calvaria-derived cells of E14 mouse embryos used in this system. Using this approach, a Mac-1 and F4/80 positive osteoclast progenitor cell line was created, 4B12, which gives rise to Mac-1 and F4/80 negative bone-resorbing osteoclasts expressing osteoclast-specific genes and possessing both a clear zone and ruffed borders. In addition, 4B12 cells were transfectable with siRNA. 4B12 cells will be a useful and powerful model for studying the cellular and molecular regulatory mechanisms of osteoclast differentiation and function.
Materials and Methods
Reagents and antibodies
M-CSF and sRANKL were purchased from R&D Systems (Minneapolis, MN). Mouse SCF (mSCF), mouse GM-CSF (mGM-CSF), and mouse IL-4 (mIL-4) were purchased from Peprotech (Rocky Hill, NJ). Human IL-1 α (hIL-1α) was kindly supplied by Otsuka Pharmaceutical Co. Ltd. (Tokushima, Japan). Monoclonal antibodies: R-phycoerythrin (R-PE)-conjugated rat anti-mouse CD11b (Mac-1 α chain, M1/70.15), CD45R (RA3-6B2), CD117 (c-Kit, 2B8), F4/80 (CI:A3-1), and CD34 (MEC14.7) were from Caltag (Burlingame, CA). Fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse MOMA-2 was from Beckman-Coulter (Palo Alto, CA). FITC-conjugated anti-mouse CD11c (HL3), R-PE-conjugated anti-mouse CD14 (rmC5-3), and anti-mouse CD16/CD32 (Fc Block, 2.4G2) were from BD Biosciences PharMingen (San Diego, CA). Polyclonal antibodies: Anti-Fms (CSF-1 R) was from Upstate Biotechnology (Lake Placid, NY). Secondary antibodies: Alexa Fluor 647-conjugated anti-rabbit IgG was from Molecular Probes (Eugene, OR). FITC-conjugated anti-rat IgG was from Beckman-Coulter. Escherichia coli 0111:B4 lipopolysaccharide (LPS) was purchased from InvivoGen (San Diego, CA).
Preparation of recombinant mouse soluble RANKL fusion protein (rmsRANKL)
The rmsRANKL fusion protein expression vector was prepared by fusing the extracellular domain of RANKL (Lys158–Asp316) [7] to the C-terminal end of His using the pBAD-TOPO Expression System (Invitrogen, Carlsbad, CA). The rmsRANKL-His tag fusion protein was purified by using a high-performance liquid chromatography C8 reverse-phase column, μBONDASPHERE (Waters, Milford, MA). LPS contamination of the purified rmsRANKL was measured by use of a colorimetric endotoxin determinant reagent (Seikagaku Kogyou, Tokyo, Japan), and the endotoxin concentration was only 0.001 pg/mg protein. The rmsRANKL was biotinylated by using a FluoReporter Mini-biotin-XX protein labeling kit (Molecular Probes).
Isolation and cloning of osteoclast precursors from Mac-1+c-Fms+RANK+ cell population from calvaria of E14 mouse embryos
All animal experiments were performed in accordance with the Guidelines of the Animal Center of the Meikai University School of Dentistry. DDY mouse embryos at the age of 14 days (Sankyo, Tokyo, Japan) were dissected. Their calvariae were digested at room temperature for 30 min in 10 ml of phosphate-buffered saline (PBS) (pH 7.2) containing 0.1% bacterial collagenase, 0.05% trypsin, and 4 mM EDTA. The digested calvarial cells were plated at a cell density of 1×107 cells/10 ml in 90-mm culture dishes, and cultured for 7 days in α-Eagle’s minimum essential medium (α-MEM; Sigma, St. Louis, MO) containing 10% fetal bovine serum (FBS; Sigma). Upper-layer cells containing osteoclast progenitors separated by centrifugation on 1.70 Percoll density gradients were cultured for 7 days. These cells were then used for isolation of Mac-1+c-Fms+RANK+ cells. Mac-1+c-Fms+RANK+ cells were sorted by using an EPICS ALTRA flow cytometer (Beckman-Coulter). The purity of the Mac-1+c-Fms+RANK+ cells was 96%. Cellular viability was determined to be greater than 90% by trypan blue exclusion. The Mac-1+c-Fms+RANK+ cell population was maintained for 1 month in α-MEM supplemented with 10% FBS, 30% calvaria-derived stromal cell conditioned media (CSCM) from, mM-CSF (1.5 ng/ml), and rmsRANKL (1.5 ng/ml). After testing for TRAP-positive (TRAP(+)) cell-forming activity, the cells were plated at a density of 1/well in 96-well culture plates by limiting dilution. We obtained 50 clonal cells from 1000 wells. Nineteen of 50 cloned cells showed both TRAP(+) cell-forming and pit-forming abilities in the presence of mM-CSF (10 ng/ml) and rmsRANKL (100 ng/ml). The cell line with the highest levels of these abilities was 4B12. The 4B12 cells were maintained in α-MEM supplemented with 10% FBS and 30% CSCM. The concentration of M-CSF in the CSCM was 9 ng/ml. The 4B12 cells were used for the following studies.
Proliferation assay
The CyQUANT Cell Proliferation Assay Kit (Molecular Probes) was used according to the manufacturer’s instructions. The observed fluorescence was converted to a cell number using a cell number standard curve generated from the same 4B12 cells. Results are expressed as the mean ± standard error (SE) of triplicate cultures.
Culture of macrophage cell line and osteoclast precursor cell line
RAW 264.7 cells were kindly provided by Dr. Toshio Kukita (Kyushu University) MOCP-5 cells obtained from Dr. Yi-ping Li have been described previously [12]. These cell lines were maintained in α-MEM containing 10% FBS.
M-BMMs culture
Bone marrow cells were obtained by flushing femuri from 6-week-old DDY male mice. For the formation of M-BMMs, stroma free bone marrow cells were cultured in the presence of M-CSF (10 ng/ml) for 3 days. M-BMMs were suspended in α-MEM containing 10% FBS.
Detection of TRAP(+) cells and TRAP-solution assay
Cells were fixed with 10% formalin-ethanol after cultivation with samples, and stained for TRAP as previously described [17]. TRAP(+) cells were counted under a light microscope. Enzyme activity in a ten-fold dilution of the culture medium was measured by the TRAP-solution assay as described previously [18]. These results were expressed as the mean ± SE of quadruplicate cultures.
RNA preparation and semiquantitative RT-PCR analyses
Total RNA was extracted using the QIAquick PCR purification kit (Qiagen, Valencia, CA). PCR was performed using PCR Master Mix (Promega, Madison, WI) for 17~32 cycles on single-strand complementary DNA prepared from total RNA (10 ng) using a Superscript III preamplification system (Invitrogen). Primers used are described in Table 1. The following conditions were used for PCR: denaturation at 94°C for 30 sec, annealing at 60°C for 1 min, polymerization at 72°C for 1 min, and elongation at 72°C for 15 min. For semiquantitative estimation, the signals of each cDNA were normalized using the values of the corresponding products from the GAPDH amplification, and the expression of these factors was compared at the logarithmic phase of the PCR reaction.
Table 1.
Nucleotide sequence of primers used for reverse transcription- polymerase chain reaction
| Gene | Nucleotide sequence (5′–3′) | Fragment size (bp) | Accession No. |
|---|---|---|---|
| TRAP | GTCTCTGGGGGACAATTTCTACT GTTTGTACGTGGAATTTTGAAGC |
241 | M99054 |
| MMP-9 | TTGCCCCTACTGGAAGGTATTAT GAGAATCTCTGAGCAATCCTTGA |
172 | NM013599 |
| CAII | TCAGGGAGCCCATTACTGTC TCCAAATCACCCAGCCTAAC |
234 | BC055291 |
| CTK | TAACAGCAAGGTGGATGAAATCT CTGTAGGATCGAGAGGGAGGTAT |
195 | NM007802 |
| Integrin alpha V | ACAATGTAAGCCCAGTTGTGTCT TTTGTAAGGCCACTGGAGATTTA |
236 | NM008402 |
| Integrin beta 3 | CTGCTCATCTGGAAGCTACTCAT CACACACACACAAATTGTCCTCT |
233 | AF026509 |
| OPN | AGTTTCCAGGTTTCTGATGAACA CTCTTTGGAATGCTCAAGTCTGT |
219 | AF515708 |
| CR-1a | TGCGGCGGGATCCTATAA AGCCAGCAGTTGTCGTTGTA |
238 | AF056329 |
| c-fos | CCAGTCAAGAGCATCAGCAA TAAGTAGTGCAGCCCGGAGT |
248 | BC029814 |
| PU.1 | GAGAAGCTGATGGCTTGGAG TTGTGCTTGGACGAGAACTG |
175 | M32370 |
TRAP, tartrate-resistant acid phosphatase; MMP, matrix metalloproteinase; CAII, carbonic anhydrase II; CTK, cathepsin K; OPN, osteopontin; CR, calcitonin receptor
Pit formation assay
Cells on dentine slices in 24-well culture plates were cultured with or without test samples. After TRAP staining, the pits on the dentine slices were observed with an incident light microscopy with metallurgic objectives (Olympus, Tokyo, Japan) as described previously [19]. The number and total area of the pits were measured. The results were expressed as the mean ± SE of six cultures.
Transmission electron microscopy (TEM)
4B12 cells on dentine slices were fixed, decalcified, and postfixed as previously described [17]. After dehydration in ethanol, the cells were embedded in Araldite 502. Ultrathin sections were stained with lead citrate and uranyl acetate and examined under a JSM-6360LV microscope (Japan Electronics, Tokyo, Japan) at 15 kV.
Flow cytometry
Cells were blocked with anti-mouse CD16/CD32 for 15 min before the addition of the appropriate monoclonal or polyclonal antibodies. Cell surface fluorescence was determined using antibodies conjugated directly with FITC and PE. For unconjugated primary antibodies, Alexa Fluor 647- or FITC-conjugated secondary antibody was used. A minimum of 5 ×104 cells were analyzed using an EPICS ALTRA flow cytometer. The percentage of positive cells was determined by setting the lower limit over the nonspecific fluorescence with a suitable control.
Immunofluorescence and confocal laser scanning microscopy
Cells on dentine slices were washed twice in PBS and then fixed with 10% formalin-ethanol. After blocking using 5% normal goat serum in PBS, cells were incubated with R-PE-conjugated anti-mouse CD11b, F4/80, or CD14 antibodies, anti-mouse c-Fms followed by Alexa Fluor 647-conjugated anti-rabbit IgG, and anti-mouse CD16/CD32 followed by FITC-conjugated anti-rat IgG. For visualization of the F-actin, cells were permeabilized in 0.1% Triton X-100 in PBS and then incubated with FITC phalloidin or rhodamine phalloidin. Samples were viewed with a LSM 510 confocal laser scanning microscope (Carl Zeiss, Jena, Germany).
Short-interfering RNA (siRNA) transfection and quantitative real-time RT-PCR (qRT-PCR)
4B12 cells were transfected with Cy3-labeled negative control #1 siRNA (Ambion, Austin, TX) using TransIT-siQUEST Transfection Reagent (Mirus Bio, Madison, WI) according to the manufacturer’s protocol. After 24 h, tranfection efficiency was estimated by counting the Cy3-positive cells in randomly under a LSM 510 confocal microscope. 4B12 cells were transfected with GFP siRNA (siGFP) (Invitrogen) and TRAF6 siRNA (siTRAF6) (Invitrogen) using TransIT-siQUEST Transfection Reagent. After 72 h, total cellular RNA was extracted by using the RNeasy protect mini kit (Qiagen, Valencia, CA), and treated with deoxyribonuclease I to eliminate genomic DNA. One microgram of total RNA was transcribed into cDNA in a total volume of 20 μl using random primers and MultiScribe reverse transcriptase (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. qRT-PCR was performed with the ABI 5700 real-time PCR system, using the FastStart Universal Probe Master ROX (Roche Applied Science, Indianapolis, IN). The oligonucleotide sequences of primers and identification number for the corresponding Roche mouse universal probe library are listed in Table 2. 18S RNA was used as an endogenous control to normalize quantification of TRAF6, NFATc1 and TRAP. Relative amounts of cDNA were calculated by the relative quantification (ΔΔCt) study method [20].
Table 2.
Primers and probes used for qPCR analysis
| Gene | Primers(5′–3′) | Roche probe No. | Accession No. |
|---|---|---|---|
| TRAF6 | TTGCACATTCAGTGTTTTTGG TGCAAGTGTCGTGCCAAG |
No. 6 | NM009424 |
| NFATc1 | TGCAAGTGTCGTGCCAAG TCCAAATCACCCAGCCTAAC |
No. 50 | NM198429 |
| TRAP | TCTGACCACCTGTGCTTCCT GGAGTGGGAGCCATATGATTT |
No. 3 | NM007388 |
TRAF6, Tnf receptor-associated factor 6; NFATc1, nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1, transcript variant 1; TRAP, tartrate-resistant acid phosphatase, transcript variant 3
Statistics
The significance of differences between groups was determined by use of Student’s t-test. Values of p<0.05 were considered to indicate statistical significance.
Results
4B12 cells established from the Mac-1+c-Fms+RANK+ cell population derived from calvaria of E14 mouse embryos depend on M-CSF for both proliferation and osteoclast differentiation
Yamada et al. [21] reported that the frequency of osteoclast precursors was increased proportional to an increase in Mac-1 and c-Fms double-positive cells. Thus, we tried to establish osteoclast precursor cell lines from Mac-1+c-Fms+RANK+ cell population, and could obtain a novel osteoclast cell line, 4B12. (see Materials and Methods). In this study, we show that 4B12 cells are a useful model for osteoclast precursors.
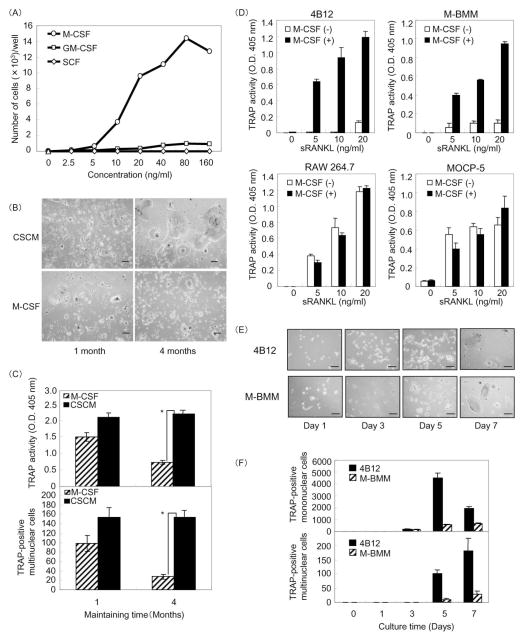
We initially examined the proliferating ability of 4B12 cells in the presence of myeloid growth factors such as M-CSF, GM-CSF, and SCF. 4B12 cells proliferated in response to M-CSF or GM-CSF, but not in response to SCF (Figure 1A). The growth of 4B12 cells was more dependent on M-CSF than on GM-CSF, and cell growth reached a plateau at 80 ng/ml mM-CSF (Figure 1A). Next we examined whether M-CSF treatment could sustain and maintain 4B12 cells as osteoclast precursors. The cells were maintained in α-MEM containing M-CSF (10 ng/ml) or 30% CSCM for 1 or 4 months, and were used for the analysis of TRAP(+) cell-forming activity. TRAP(+) multinucleated (≥ 3 nuclei) cell-forming activity of 4B12 cells maintained in α-MEM containing M-CSF alone was lower than that of the cells maintained in α-MEM containing the CSCM (Figure 1B,C). The concentration of M-CSF in the CSCM was 9 ng/ml. These results suggest that the proliferation of 4B12 cells is dependent on M-CSF, and that the CSCM can sustain 4B12 cells as osteoclast precursors. It is well known that M-CSF is an indispensable factor for osteoclast differentiation. M-BMMs, which are used as primary osteoclast precursors, differentiate into osteoclasts in the presence of M-CSF and RANKL, whereas RAW264.7 or MOCP-5 cells form osteoclasts in the presence of RANKL alone [9,22]. Furthermore we examined the response of 4B12 cells to M-CSF on RANKL-induced TRAP(+) cell formation. M-BMMs, 4B12, RAW264.7 and MOCP-5 cells were treated with sRANKL in the presence or absence of M-CSF. In the presence of M-CSF, all of them formed TRAP(+) cells in response to sRANKL stimulation. In the absence of M-CSF, RAW264.7 and MOCP-5 cells formed TRAP(+) cells by sRANKL stimulation, however 4B12 cells similar to M-BMMs did not (Figure 1D). These results suggest that 4B12 cells are dependent on M-CSF for not only proliferation but also osteoclast differentiation. Next, we compared time courses for TRAP(+) cell formation in 4B12 cells and M-BMMs. TRAP(+) mononuclear and multinuclear cell formation from both M-BMMs and 4B12 cells was detected on days 3 and 5, respectively. On day 7, the number of TRAP(+) mononuclear and multinuclear cells formed from 4B12 cells was three- and four-fold higher, respectively, than that formed from M-BMMs (Figure 1E,F). These results suggest that both 4B12 cells and M-BMMs differentiate into TRAP(+) MNCs at the same time point, although 4B12 cells are more efficient than M-BMMs at TRAP(+) MNC formation.
Figure 1. Proliferation and maintenance of 4B12 cells.
(A) 4B12 cells were cultured in the presence of various concentrations of mM-CSF, mGM-CSF or mSCF in 96-well culture plates. After 7 days, the cells were counted by CyQuant cell proliferation assay. (B) 4B12 cells were maintained in α-MEM containing 10% FBS in the presence of M-CSF (10 ng/ml) or 30% calvaria-derived stromal cell conditioned media (CSCM) for 1 to 4 months. The cells of each group were plated at a cell density of 500 cells/200 μl in 96-well culture plates, and cultured in the presence of mM-CSF (10 ng/ml) and sRANKL (10 ng/ml) for 7 days. Bar = 100μm. (C) TRAP(+) MNCs were counted and TRAP activity in the culture medium was measured. *P<0.01 versus 4B12 cells precultured in the presence of M-CSF. (D) 4B12 cells (500/well), M-BMMs (500/well), RAW264.7 cells (500/well) and MOCP-5 cells (500/well) were plated in 96-well culture plates, and cultured with or without sRANKL (10 ng/ml) in the presence or absence of mM-CSF (10 ng/ml) for 6 days. TRAP activity in the culture medium was measured. Similar results were obtained in two independent experiments. (E) 4B12 cells (500/well) and M-BMMs (500/well) were cultured in the presence of mM-CSF (10 ng/ml) plus sRANKL (10 ng/ml) in 96-well culture plates for the indicated times. Cultured cells were fixed and stained for TRAP. Bar = 100 μm. (F) TRAP(+) mononuclear and multinuclear cells were counted. Similar results were obtained in two independent experiments.
4B12 cells are capable of differentiating into bone-resorbing osteoclasts with both a clear zone and ruffled borders on dentine slices
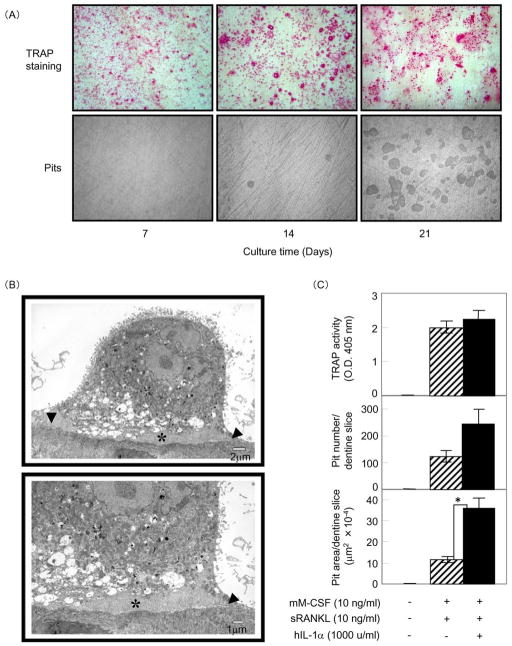
Next, we examined whether 4B12 cells could differentiate into bone-resorbing osteoclasts on dentine slices. It has been reported that IL-1 enhances bone resorption activity [23]. Therefore, 4B12 cells were cultured with M-CSF plus sRANKL plus IL-1α on dentine slices for 7, 14, and 21 days. As shown in Figure 2A, TRAP(+) MNCs were observed on day 7, and typical resorption pits were observed on day 14 using reflected light microscopy. The resorption pits increased in number by day 21. To investigate the efficiency at which 4B12 cells formed mature osteoclasts, we measured both the number of TRAP(+) mononuclear and multinuclear cells and the number of pits (Table 3). The ratio of TRAP(+) MNCs to TRAP(+) mononuclear cells was 0.46% on day 7, 3.93% on day 14, and 4.09% on day 21, suggesting that the differentiation of 4B12 cells from TRAP(+) mononuclear cells into TRAP(+) MNCs reached a plateau by day 14. However, the number of pits increased from 40 on day 14 to 310 on day 21, suggesting that TRAP(+) MNCs efficiently differentiated into bone-resorbing cells and the bone resorption activity of TRAP(+) MNCs enhanced between days 14 and 21. To determine whether the bone-resorbing cells formed from 4B12 cells on the dentine slices had the characteristic features of mature osteoclasts, we used TEM to examine the cells for the existence of both a clear zone and ruffled borders. As expected, the bone-resorbing cells possessed both features (Figure 2B), the same as those of osteoclasts found adjacent to bone in Howship’s lacunae in vivo [24]. This observation demonstrates that 4B12 cells have the potential to differentiate into bone-resorbing osteoclasts with features of native mature osteoclasts on dentine slices. We examined the effect of IL-1α on the bone-resorbing activity of osteoclasts formed from 4B12 cells. There was no difference in TRAP activity between cells cultured in the presence of sRANKL alone and those receiving sRANKL plus hIL-1α stimulation. However 4B12 cells stimulated by sRANKL plus hIL-1α in the presence of M-CSF formed about twice the number of pits and three times the pit area compared to cells stimulated by sRANKL alone in the presence of M-CSF (Figure 2C), suggesting that the bone-resorbing activity of osteoclasts formed from 4B12 cells is directly enhanced by IL-1α stimulation.
Figure 2. Osteoclast differentiation of 4B12 cells on dentine slices.
(A) 4B12 cells (5 × 103) were cultured in the presence of mM-CSF (10 ng/ml) plus sRANKL (10 ng/ml) plus hIL-1α (1000 u/ml) on dentine slices in 24-well culture plates. The culture medium was exchanged for new culture medium every 7 days. After 7, 14, and 21 days of culture, 4B12 cells on dentine slices were stained for TRAP. After removing the 4B12 cells on dentine slices, resorption pits on the dentine surface were observed by reflective light microscopy. (B) Ultrastructural features of bone-resorbing cells formed from 4B12 cells in the presence of mM-CSF (10 ng/ml) plus sRANKL (10 ng/ml) plus hIL-1α (1000 u/ml). The cells exhibit a clear zone (arrowhead) and ruffled borders (asterisk). (C) 4B12 cells (5 × 103) were cultured with or without hIL-1α (1000 u/ml) in the presence of mM-CSF (10 ng/ml) and sRANKL (10 ng/ml) on dentine slices for 21 days. TRAP activity and the number and total area of the resorption pits formed on the dentine slices were measured. *P<0.05 versus the culture without hIL-1α. Similar results were obtained in two independent experiments.
Table 3.
Quantification of TRAP(+) mononuclear and multinuclear cells, and resorption pits formed on the dentine slices shown in Figure 2(A).
| Culture time (days) | No. of TRAP(+)mononuclear cells | No. of TRAP(+)multinuclear | Ratio of multinuclear to mononuclear cells (%) | No. of resorption pits |
|---|---|---|---|---|
| 7 | 37667 ± 444 | 172 ± 111 | 0.46 ± 0.29 | 0 ± 0 |
| 14 | 29536 ± 2850 | 1160 ± 135 | 3.93 ± 0.46 | 40 ± 8.67 |
| 21 | 29294 ± 2440 | 1198 ± 155 | 4.09 ± 0.53 | 310 ± 56.2 |
The results are expressed as the mean ± SE of six cultures. Similar results were obtained in two independent experiments.
Bone-resorbing osteoclasts formed from 4B12 cells express osteoclast marker genes
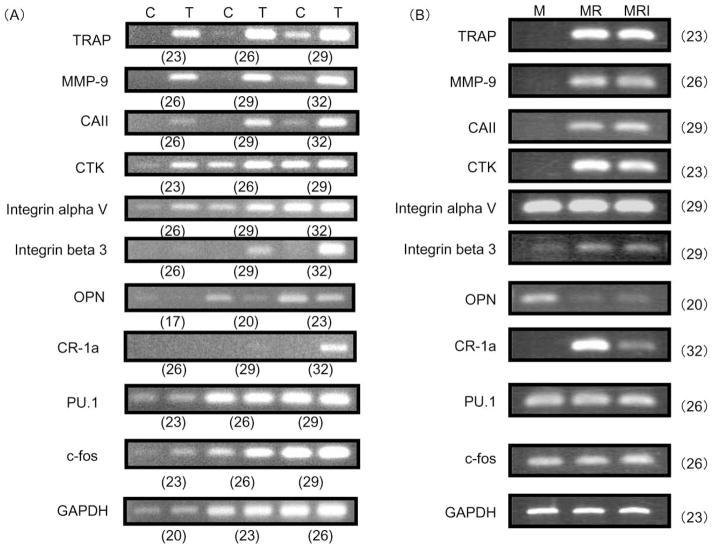
To determine whether bone-resorbing osteoclasts formed from 4B12 cells express osteoclast-specific genes, total RNA was prepared from 4B12 cells treated with M-CSF plus sRANKL plus IL-1α for 21 days on dentine slices, and analyzed by semiquantitative RT-PCR (Figure 3A). Interestingly, non-stimulated 4B12 cells weakly expressed osteoclast marker mRNAs of TRAP, matrix metalloproteinase-9 (MMP-9), and carbonic anhydrase II (CAII), and strongly expressed cathepsin K (CTK) and osteopontin (OPN). In the nontreated 4B12 cells, calcitonin receptor 1a (CR-1a) and integrin beta 3 were not detected even after 32 cycles. 4B12 cells stimulated with factors expressed all of the osteoclast marker mRNAs, TRAP, MMP-9, CAII, CTK, integrin alpha V, integrin beta 3, CR-1a, and OPN. The mRNA expression level of OPN in 4B12 cells was slightly decreased by stimulation with M-CSF plus sRANKL plus IL-1α. We also examined the mRNA levels of transcriptional factors such as PU.1 and c-fos. mRNA of PU.1 was present in nontreated 4B12 cells, and the expression did not change during osteoclast differentiation, whereas the expression of c-fos mRNA was present in nontreated 4B12 cells, and slightly enhanced.
Figure 3. Expression of osteoclast-specific genes in 4B12 cells by semiquantitative RT-PCR analyses.
(A) 4B12 cells (2.5 × 104) were cultured in the presence of mM-CSF alone (C) or in the presence of mM-CSF (10 ng/ml) plus sRANKL (10 ng/ml) plus hIL-1α (1000 u/ml) (T) on dentine slices in 24-well culture plates for 21 days. Total RNA was extracted from the cells and processed by RT-PCR. The PCR reactions were stopped at the indicated number of cycles: 23, 26, and 29 for TRAP, CTK, PU.1, and c-fos; 26, 29, and 32 for CR-1a, MMP-9 CAII, integrin alphaV, and integrin beta 3; 17, 20, and 23 for OPN; and 20, 23, and 26 for GAPDH. The primers described in Table 1 were used for mouse genes of TRAP, MMP-9, CAII, CTK, integrin alpha V, integrin beta 3, OPN, CR-1a, PU.1, and c-fos. Numbers in parentheses indicate the number of PCR cycles. Data are representative of three similar experiments.
(B) 4B12 cells (2.5 × 104) were cultured in the presence of mM-CSF (10 ng/ml) alone (M), mM-CSF (10 ng/ml) plus sRANKL (10 ng/ml) (MR), and mM-CSF (10 ng/ml) plus sRANKL (10 ng/ml) plus hIL-1α (1000 u/ml) (MRI) on dentine slices in 24-well culture plates for 21 days. Total RNA was extracted from the cells and processed by RT-PCR. The PCR reactions were stopped at the indicated number of cycles: 20 for OPN; 23 for TRAP, CTK and GAPDH; 26 for MMP-9, PU.1 and c-fos; and 29 for CAII, integrin alphaV, and integrin beta 3; and 32 for CR-1a. Data are representative of three similar experiments.
The bone-resorbing activity of osteoclasts formed from 4B12 cells was directly enhanced by IL-1α stimulation. Next the effect of IL-1α in the presence of M-CSF and RANKL on osteoclast-specific gene expression was examined. We did not observe any difference in osteoclast-specific gene expression in cells treated with or without IL-1α, except for the expression of CR-1a gene. Interestingly, IL-1α inhibited the expression of CR-1a gene in the osteoclasts formed from 4B12 cells (Figure 3B).
4B12 cells have high TRAP(+) multinuclear cell- and pit-forming capacity
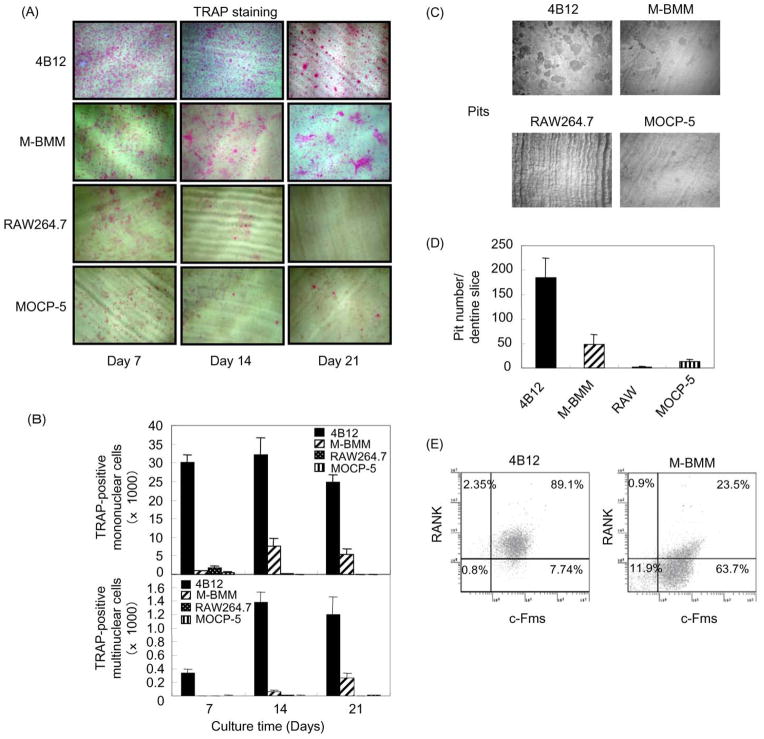
Next, the number of TRAP(+) mononuclear and multinuclear cells and pits formed from M-BMMs, 4B12, RAW264.7 and MOCP-5 cells on dentine slices was compared. The number of TRAP(+) mononuclear and multinuclear cells formed from 4B12 cells was higher than those formed from M-BMMs, RAW264.7 and MOCP-5 cells (Figure 4A,B). The number of pits formed from 4B12 cells was also higher than that formed from M-BMMs, RAW264.7 and MOCP-5 cells (Figure 4C,D). The number of TRAP(+) multinuclear cells and pits formed from 4B12 cells on day 21 was approximately fourfold higher than those formed from M-BMMs (Figure 4B,D). We hypothesized that the difference in the efficiency of osteoclast formation between 4B12 cells and M-BMMs is due to a higher percentage of c-Fms+RANK+ cells in the 4B12 cells. So next the percentage of c-Fms+RANK+ cells in 4B12 cells and M-BMMs was compared. The percentage of c-Fms+RANK+ cells in 4B12 cells was approximately four-fold higher than that in M-BMMs (Figure 2E). These results suggest that 4B12 cells are more efficient in forming osteoclasts than M-BMMs, RAW264.7 and MOCP-5 cells most likely due to the high ratio of c-Fms+RANK+ cells.
Figure 4. Comparison between 4B12 cells, M-BMMs, RAW264.7 and MOCP-5 cells in osteoclastogenesis.
(A) M-BMMs, 4B12, RAW264.7 and MOCP-5 cells were plated at a cell density of 5 × 103 on dentine slices in 24-well culture plates, and cultured in the presence of mM-CSF (10 ng/ml) plus sRANKL (10 ng/ml). The culture medium was exchanged for new culture medium every 7 days. After 7, 14, and 21 days of culture, cells on dentine slices were stained for TRAP. (B) TRAP(+) mononuclear and multinuclear cells were counted. (C) Resorption pits on the dentine surface on day 21 were observed by reflective light microscopy. (D) The number of the resorption pits formed on the dentine slices was measured. Similar results were obtained in two independent experiments. (E) Percentage of the c-Fms+RANK+ cells in 4B12 cells and M-BMMs was analysed by flow cytometry.
4B12 cells are already committed to a specific subset of macrophages
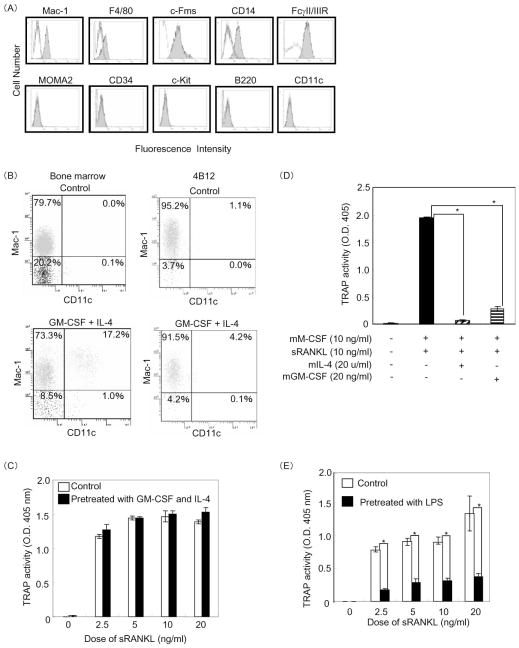
It has been reported that, in addition to CD34-positive hematopoietic stem cells, macrophages expressing Mac-1, F4/80, and c-Fms act as sources of osteoclast precursors [9,25]. To characterize the differentiation stage of the 4B12 cells established from a Mac-1+c-Fms+RANK+ cell population, we initially examined their surface markers by flow cytometry. 4B12 cells expressed macrophage markers such as Mac-1, CD14, and FcγII/IIIR, weakly expressed F4/80, and were negative for MOMA-2, CD34, c-kit, CD45R, and CD11c (Figure 5A). These results suggest that 4B12 cells belong to a macrophage lineage.
Figure 5. Differentiation stages of 4B12 cells along the osteoclast lineage and inhibition by GM-CSF, IL-4 and LPS.
(A) 4B12 cells were stained with various antibodies for the cell surface (solid lines) as described in Materials and Methods. Dotted lines represent the isotype control. (B) Bone marrow or 4B12 cells precultured in the presence of mGM-CSF (10 ng/ml) and mIL-4 (10 u/ml) for 7 days were used for analysis of Mac-1 and CD11c expression by flow cytometry. (C) In addition, 4B12 cells pretreated with mGM-CSF (10 ng/ml) and mIL-4 (10 u/ml) for 7 days were plated at a cell density of 500/well in 96-well culture plates, washed with α-MEM three times, and cultured in the presence of mM-CSF (10 ng/ml) and various doses of sRANKL for 7 days. TRAP activity in the culture medium was measured. (D) The 4B12 cells were plated at a cell density of 500/well in 96-well culture plates and cultured in the presence of mM-CSF (10 ng/ml) and sRANKL (10 ng/ml) with or without mIL-4 (20 u/ml) or mGM-CSF (20 ng/ml) for 7 days. TRAP activity in the culture medium was measured. *P<0.01 versus 4B12 cells cultured in the presence of mM-CSF and sRANKL. (E) 4B12 cells pretreated with LPS (10 ng/ml) in the presence of mM-CSF (10 ng/ml) for 5 days were plated at a cell density of 500/well in 96-well culture plates, washed with α-MEM three times, and cultured in the presence of mM-CSF (10 ng/ml) and various doses of sRANKL for 7 days. TRAP activity in the culture medium was measured. *P<0.05 versus the control. Similar results were obtained in two independent experiments.
It has been shown that osteoclasts and dendritic cells (DCs) share a common origin from the same precursor cell and are separated by a lineage bifurcation process [26]. Therefore, we examined whether 4B12 cells have the potential to differentiate into DCs. It has been reported that the combination of GM-CSF/IL-4 increases the number and function of DCs [27]. Thus, 4B12 cells were exposed to GM-CSF and IL-4 for 7 days. Bone marrow cells treated with or without GM-CSF and IL-4 were used as a control. We analyzed the expression of CD11c and Mac-1 as markers of DCs and macrophages, respectively. Although the percentage of CD11c-positive cells in the bone marrow cells increased from 0.1% to 17.2%, almost no changes were observed in 4B12 cells (Figure 5B). In addition, there was no difference in TRAP(+) cell formation activity between nontreated 4B12 cells and 4B12 cells pretreated with both GM-CSF and IL-4 (Figure 5C). It has been demonstrated that GM-CSF or IL-4 directly inhibits the development of osteoclasts from bone marrow precursors in the presence of both M-CSF and RANKL [28,29]. Thus, we confirmed the response of 4B12 cells to GM-CSF or IL-4 on TRAP(+) cell formation. Such formation was obviously inhibited by GM-CSF or IL-4 treatment (Figure 5D). These results suggest that 4B12 cells have the potential to be inhibited in response to GM-CSF or IL-4 during osteoclastogenesis. However, 4B12 cells in response to GM-CSF and IL-4 do not have the potential to differentiate into DCs.
It is well known that macrophages are activated by LPS stimulation [30]. We examined whether 4B12 cells activated by LPS could differentiate into TRAP(+) cells. The rate of TRAP(+) cell formation from 4B12 cells was reduced by pretreatment with LPS in the presence of M-CSF for 5 days (Figure 5E). Taken together, these findings demonstrated that 4B12 cells are already committed to a specific subset of macrophages, and that 4B12 cells lose the potential for osteoclastogenesis in the presence of GM-CSF or IL-4, or by pretreatment with LPS.
4B12 cells lose macrophage markers such as Mac-1 and F4/80 during osteoclast maturation
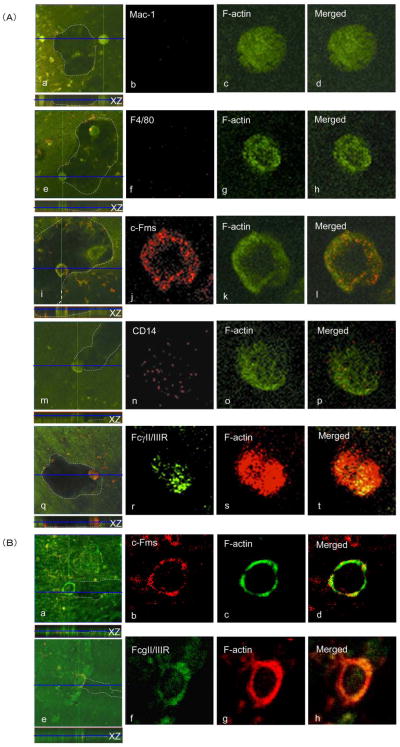
It has been reported that the macrophage phenotype of osteoclast progenitor cells is lost during osteoclastogenesis [31]. Therefore, we examined the expression of Mac-1, F4/80, c-Fms, CD14, and FcγII/IIIR during osteoclast differentiation from 4B12 cells. Bone-resorbing osteoclasts formed from 4B12 cells on dentine slices in the presence of M-CSF plus sRANKL plus IL-1α were observed by confocal laser scanning microscopy. As shown in Figure 6A, both c-Fms, and FcγII/IIIR were detected on the bone-resorbing osteoclasts, whereas CD14 was barely detected, and neither Mac-1 nor F4/80 could be detected. The macrophage markers Mac-1 and F4/80 expressed on 4B12 cells disappeared during osteoclast maturation. Furthermore, both c-Fms and FcγII/IIIR expressed on the bone-resorbing osteoclasts were localized to F-actin, whereas CD14 was not. Co-localization of c-Fms or FcγII/IIIR with F-actin was also observed on bone-resorbing osteoclasts formed from M-BMMs (Figure 6B).
Figure 6. Expression of macrophage markers on 4B12 cells and M-BMMs during osteoclast maturation.
(A) Bone-resorbing osteoclasts formed from 4B12 cells in the presence of mM-CSF (10 ng/ml) plus sRANKL (10 ng/ml) plus hIL-1α (1000 u/ml) on dentine slices were stained with antibodies against Mac-1 (red), F4/80 (red), c-Fms (red), CD14 (red), and FcγII/IIIR (green) as described in Materials and Methods. In the case of staining with antibodies against Mac-1, F4/80, c-Fms, and CD14, FITC-phalloidin was used to detect actin distribution (green). In the case of staining with antibody against FcγII/IIIP, rhodamine-phalloidin was used to detect actin distribution (red). The dentine surface position in the x-z perspective is indicated by the blue line. Dotted lines in a, e, i, m, and q are outlines of pits formed by functional mature osteoclasts differentiated from 4B12 cells, determined by background fluorescence at very high gains. Panel d, h, l, p, and t show merged figures of b+c, f+g, j+k, n+o, and r+s, respectively. Both colocalization of c-Fms (red) with F-actin (green) and colocalization of FcγII/IIIR (green) with F-actin (red) are revealed by yellow/orange color. Data are representative of three similar experiments. (B) Bone-resorbing osteoclasts formed from M-BMMs in the presence of mM-CSF (10 ng/ml) plus sRANKL (10 ng/ml) plus hIL-1α (1000 u/ml) on dentine slices were stained with antibodies against c-Fms (red) and FcγII/IIIR (green). In the case of staining with antibodies against c-Fms, FITC-phalloidin was used to detect actin distribution (green). In the case of staining with antibody against FcγII/IIIP, rhodamine-phalloidin was used to detect actin distribution (red). The dentine surface position in the x-z perspective is indicated by the blue line. Dotted lines in a and e are outlines of pits formed by functional mature osteoclasts differentiated from M-BMMs. Panel d and h show merged figures of b+c and f+g respectively. Both colocalization of c-Fms (red) with F-actin (green) and that of FcγII/IIIR (green) with F-actin (red) are revealed by yellow/orange color. Data are representative of three similar experiments.
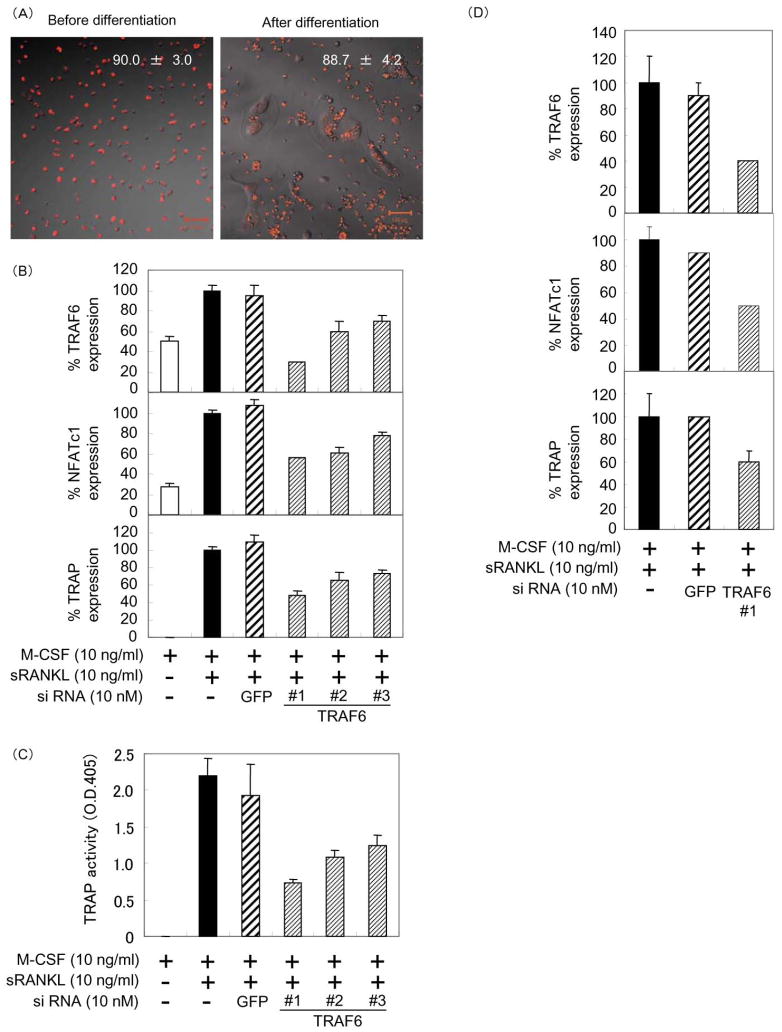
4B12 cells before and after osteoclast differentiation can be used for siRNA transfection experiments
Gene knockdown using siRNA is a powerful tool for studying the biological effects of decreased levels of mRNA and subsequent protein levels. To determine the transfection efficiency, we transfected Cy3-labeled siRNA in the 4B12 cells. The transfection efficiency before and after differentiation was estimated to be approximately 90% (Figure 7A). The RANK signal regulating osteoclast development and function is dependent on TNF receptor-associated factor 6 (TRAF6) [32,33]. Therefore, we examined the effects of TRAF6 knockdown in 4B12 cells on osteoclast formation. The knockdown efficiency of siTRAF6 #1 in the 4B12 cells before osteoclast differentiation was estimated to be approximately 70% (Figure 7B). The expression of NFATc1 and TRAP genes in siTRAF6 transfected 4B12 cells was inhibited relative to the knockdown efficiency of TRAF6 by siTRAF6 #1 to #3 (Figure 7B). TRAP activity on day 5 was also inhibited in proportion to the knockdown efficiency of TRAP gene expression on day 3 (Figure 7C). The knockdown efficiency of TRAF6 in the 4B12 cells after osteoclast differentiation was estimated to be approximately 60%. The expression of NFATc1 and TRAP genes was also inhibited by siTRAF6 #1 transfection (Figure 7D). These results suggest that 4B12 cells are very useful for siRNA knockdown experiments in all stages of differentiation from TRAP-negative mononuclear cells to TRAP(+) MNCs.
Figure 7. Transfection of siRNA into 4B12 cells.
(A) 4B12 cells were plated on 96-well plates at a cell density of 1 × 103 per well, and cultured in α-MEM supplemented with 10% FBS and 30% CSCM. After 24 h, the cells were transfected with Cy3-labeled negative control #1 siRNA (20 nM) using TransIT-siQUEST transfection reagent (0.09 μl/well) according to the manufacturer’s protocol. TRAP(+) MNCs, which were formed from 4B12 cells in the presence of M-CSF (10 ng/ml) and sRANKL (10 ng/ml) for 7 days, were transfected with Cy3-labeled negative control #1 siRNA in the presence of M-CSF (10 ng/ml) and sRANKL (10 ng/ml). Transfection efficiency was estimated by counting the Cy3-positive cells at 48 h after transfection under a LSM 510 confocal microscope. The results are expressed as the mean ± standard deviation (SD) of quadruplicate cultures. (B) 4B12 cells were plated at a cell density of 5 × 103 in each well of 96-well culture plates and cultured in α-MEM supplemented with 10% FBS and M-CSF (10 ng/ml) for 24 h, and transfected with siGFP (10 nM) and siTRAF6 (10 nM) using TransIT-siQUEST transfection reagent (0.09 μl/well). After 72 h, TRAF6, NFATc1 and TRAP expression were measured by qRT-PCR as described in Materials and Methods. Relative amounts of cDNA were calculated by the relative quantification (ΔΔCt) study method. The results are expressed as the mean ± SD of triplicate cultures. Similar results were obtained in two independent experiments. (C) After 5 days, TRAP activity was measured. Similar results were obtained in two independent experiments. (D) 4B12 cells were plated at a cell density of 5 × 103 in each well of 24-well culture plates and cultured in the presence of M-CSF (10 ng/ml) plus sRANKL (10 ng/ml) plus hIL-1α (1000 u/ml) for 14 days, and transfected with siGFP (10 nM) and siTRAF6 (10 nM) using TransIT-siQUEST transfection reagent (0.09 μl/well). The trasfected cells were cultured in the presence of M-CSF plus sRANKL plus hIL-1α. After 72 h, the expression of TRAF6, NFATc1, and TRAP genes were measured. Similar results were obtained in two independent experiments.
Discussion
In this study, we established a new osteoclast precursor cell line, 4B12, with high potential to differentiate into authentic bone-resorbing osteoclasts in the presence of M-CSF and RANKL. 4B12 is the first osteoclast precursor cell line requiring both M-CSF/c-Fms and RANKL/RANK signals to form TRAP(+) MNCs. Terminal differentiation of 4B12 cells into fully functional bone-resorbing mature osteoclasts occurs during days 14 to 21 of culture. Distinct from macrophage polykaryons, the TRAP(+) MNCs formed from 4B12 cells possess both a clear zone and ruffled borders, identical to native osteoclasts. Even though 4B12 cells progress towards osteoclast differentiation at the same pace as M-BMMs, their frequency of osteoclast differentiation was higher most likely due to the fact that 4B12 cells are homogeneous, while M-BMMs are a heterogeneous population.
Although M-CSF is an indispensable factor for proliferation of osteoclast precursors, M-CSF alone could not maintain the 4B12 cells as osteoclast precursors with a high differentiation potential. Additional unknown soluble factors produced by the calvaria-derived supporting stromal cells in combination with M-CSF were required to maintain 4B12 cells as osteoclast precursors for months in culture. We assume that the unknown factors are normally produced by these supporting stromal cells in vivo. It will be important to identify this unknown factor in future studies.
In this study, we also observed that the bone-resorbing activity of 4B12 cells was enhanced by the addition of IL-1α without an increase in the number of TRAP(+) MNCs, however, an increase in expression of osteoclast-specific genes involved in bone resorption was not observed. The observed increase in pit formation in response to IL-1α was consistent with previous reports [34]. Nakamura et al. [35] showed that IL-1 regulates cytoskeletal organization in osteoclasts via TNF receptor-associated factor 6/c-src complex. The enhancement of bone-resorbing activity by 4B12 cells treated with IL-1α may result from optimal cytoskeletal re-organization.
The point at which osteoclasts branch off from other lineages is ambiguous. Studies have attempted to address the precise stage of differentiation at which osteoclast precursors diverge from other hematopoietic lineages. Several studies suggest that the major population of osteoclasts in bone marrow is derived from c-kit-positive cells [36,37]. However, osteoclast production does not require a signal from the c-kit receptor [36]. On the other hand, Udagawa et al. [38] showed that osteoclasts could be induced from mature monocytes or macrophages. In the present study, we showed that 4B12 cells were positive for macrophage markers F4/80, Mac-1, and c-Fms, but not c-kit. Results from our studies suggest that macrophages are the final branching point for differentiation into osteoclasts, since 4B12 cells do not have the potential to differentiate into dendritic cells, and LPS-activated 4B12 cells lose their potential for osteoclastogenesis. It will be interesting to clarify the transcriptional mechanisms during osteoclast maturation using 4B12 cells.
It has been reported that the osteoclast precursor cell lines MOCP-5 and BDM-1 type-2 subclone cells established from bone marrow macrophages are negative for F4/80, but positive MOMA-2 [12,16] which is expressed on both bone marrow monocyte/macrophage precursors and mature subsets of macrophages in lymphoid organs [39]. In contrast, 4B12 cells are positive for F4/80 but negative for MOMA-2. MOMA-2-positive early monocyte precursors may differentiate into a subset of F4/80 positive macrophages with the potential to become osteoclast precursors. Henkel et al. [40] reported that the majority of cells from PU.1−/− myeloid colonies were MOMA-2-positive early monocyte precursors and that the late monocytic F4/80 marker could not be detected. Taking these results together, we propose that the osteoclast precursor 4B12 cells are macrophages that may be more differentiated than osteoclast precursor cell lines such as MOCP-5 and BDM-1 [12,16]. Macrophage surface markers Mac-1 and F4/80 expressed on undifferentiated 4B12 cells disappeared during differentiation into bone-resorbing osteoclasts. This is consistent with the reports of Takahashi et al. [31] and Niida et al. [41] showing that Mac-1 expression in calcitonin receptor-positive cells disappeared during their differentiation into multinucleated osteoclasts [31] and that mouse osteoclasts are negative for Mac-1, F4/80, and MOMA-2 antigens [41]. Recently, Chang MK et al. [42] proposed that F4/80+c-Fms+ resident macrophages in the osteal microenviroment, called ‘OsteoMacs’, serve as sentinel cells in osteal tissues to regulate osteoblast function. As 4B12 cells are also F4/80 and c-Fms positive cells, these cells may also function as ‘OsteoMacs’. In future experiments, it will be important to determine whether 4B12 cells have similar capabilities as OsteoMacs to stimulate osteoblast function.
CAII and vitronectin receptor integrin αVβ3, in addition to CTK, play an important role in bone resorption as demonstrated by genetic experiments in mice [43–45]. Expression of CAII and the integrin β3 subunit of the vitronectin receptor were increased during terminal 4B12 osteoclast differentiation. We assume that the elevation of both CAII and integrin β3 gene expression is required for terminal differentiation of TRAP(+) MNCs into bone-resorbing osteoclasts. Integrin αV and CTK, however, were continuously expressed in undifferentiated 4B12 cells through to mature osteoclast formation.
The CD14 antigen expressed on undifferentiated 4B12 cells did not completely disappear during osteoclast maturation. CD14 is a membrane-anchored glycoprotein that functions as a member of the LPS receptor system [46]. Itoh et al. [47] reported that osteoclasts weakly but detectably express CD14 protein as determined by immunostaining. Most likely the difference in intensity between macrophages and osteoclasts with regards to CD14 expression may correlate with their different responses to LPS. Indeed, in bone marrow macrophages or C7-TY cells normally expressing CD14, LPS inhibits RANKL-induced osteoclastogenesis [48], while in mature osteoclasts with low CD14 expression, LPS promotes the survival of mature osteoclasts [36].
c-Fms was localized to F-actin on bone-resorbing osteoclasts formed from 4B12 and M-BMMs. It is known that c-Fms is highly expressed on mature osteoclasts [5] and it has been reported that M-CSF induces cell spreading, motility, and actin reorganization in mature osteoclasts [49]. In the future, it will be interesting to elucidate the function of c-Fms localized to F-actin in bone-resorbing osteoclasts. Another potential use of the 4B12 cells may be to analyse c-Fms signaling or the cooperation between c-Fms and RANK signals on osteoclast differentiation, in comparison with RAW264.7 or MOCP-5 cells.
In conclusion, we have generated a new osteoclast precursor cell line, 4B12, established from calvaria-derived cells of E14 mouse embryos. 4B12 cells were found to be useful for siRNA knockdown experiments before and after osteoclast differentiation making this cell line a powerful model to identify the molecular regulatory mechanisms of osteoclast differentiation and function. This osteoclast precursor cell line will be useful in dissecting specific stages during osteoclast differentiation from macrophages into mature osteoclasts and potentially for the generation of therapeutics to reduce bone loss.
Acknowledgments
Contract grant sponsor: The Ministry of Education, Culture, Sports, Science, and Technology of Japan; Contract grant number: Grant-in-Aid for Scientific Research (no. 05671513)
Contract grant sponsor:NIH NIAMS; Contract grant number: AR046798 (LFB)
We thank Dr. Yi-Ping Li (Professor, Department of Developmental Biology, Harvard School of Dental Medicine) for providing mouse osteoclast precursor cell line MOCP-5.
Footnotes
Conflict of interest: None of the authors has a conflict of interest.
Contributor Information
Shigeru Amano, Email: shigerua@dent.meikai.ac.jp.
Keisuke Sekine, Email: ksekine@dent.meikai.ac.jp.
Lynda Bonewald, Email: bonewaldl@umkc.edu.
Yoshihiro Ohmori, Email: ohmori@dent.meikai.ac.jp.
References
- 1.Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2(4):389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 2.Kodama H, Nose M, Niida S, Yamasaki A. Essential role of macrophage colony-stimulating factor in the osteoclast differentiation supported by stromal cells. J Exp Med. 1991;173(5):1291–1294. doi: 10.1084/jem.173.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka S, Takahashi N, Udagawa N, Tamura T, Akatsu T, Stanley ER, Kurokawa T, Suda T. Macrophage colony-stimulating factor is indispensable for both proliferation and differentiation of osteoclast progenitors. J Clin Invest. 1993;91(1):257–263. doi: 10.1172/JCI116179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeung YG, Jubinsky PT, Sengupta A, Yeung DC, Stanley ER. Purification of the colony-stimulating factor 1 receptor and demonstration of its tyrosine kinase activity. Proc Natl Acad Sci U S A. 1987;84(5):1268–1271. doi: 10.1073/pnas.84.5.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherr CJ, Rettenmier CW, Sacca R, Roussel MF, Look AT, Stanley ER. The c-fms proto-oncogene product is related to the receptor for the mononuclear phagocyte growth factor, CSF-1. Cell. 1985;41(3):665–676. doi: 10.1016/s0092-8674(85)80047-7. [DOI] [PubMed] [Google Scholar]
- 6.Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99(1):111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 7.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93(2):165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 8.Anderson DM, Maraskovsky E, Billingsley WL, Dougall WC, Tometsko ME, Roux ER, Teepe MC, DuBose RF, Cosman D, Galibert L. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390(6656):175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 9.Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, Wang L, Xia XZ, Elliott R, Chiu L, Black T, Scully S, Capparelli C, Morony S, Shimamoto G, Bass MB, Boyle WJ. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A. 1999;96(7):3540–3545. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dougall WC, Glaccum M, Charrier K, Rohrbach K, Brasel K, De Smedt T, Daro E, Smith J, Tometsko ME, Maliszewski CR, Armstrong A, Shen V, Bain S, Cosman D, Anderson D, Morrissey PJ, Peschon JJ, Schuh J. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13(18):2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers TJ, Owens JM, Hattersley G, Jat PS, Noble MD. Generation of osteoclast-inductive and osteoclastogenic cell lines from the H-2KbtsA58 transgenic mouse. Proc Natl Acad Sci U S A. 1993;90(12):5578–5582. doi: 10.1073/pnas.90.12.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, Li YP. Generation of mouse osteoclastogenic cell lines immortalized with SV40 large T antigen. J Bone Miner Res. 1998;13(7):1112–1123. doi: 10.1359/jbmr.1998.13.7.1112. [DOI] [PubMed] [Google Scholar]
- 13.Hentunen TA, Jackson SH, Chung H, Reddy SV, Lorenzo J, Choi SJ, Roodman GD. Characterization of immortalized osteoclast precursors developed from mice transgenic for both bcl-X(L) and simian virus 40 large T antigen. Endocrinology. 1999;140(7):2954–2961. doi: 10.1210/endo.140.7.6867. [DOI] [PubMed] [Google Scholar]
- 14.Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J Bone Miner Res. 2000;15(8):1477–1488. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- 15.Gattei V, Bernabei PA, Pinto A, Bezzini R, Ringressi A, Formigli L, Tanini A, Attadia V, Brandi ML. Phorbol ester induced osteoclast-like differentiation of a novel human leukemic cell line (FLG 29.1) J Cell Biol. 1992;116(2):437–447. doi: 10.1083/jcb.116.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin JH, Kukita A, Ohki K, Katsuki T, Kohashi O. In vitro differentiation of the murine macrophage cell line BDM-1 into osteoclast-like cells. Endocrinology. 1995;136(10):4285–4292. doi: 10.1210/endo.136.10.7664646. [DOI] [PubMed] [Google Scholar]
- 17.Amano S, Hanazawa S, Kawata Y, Ohta K, Kitami H, Kitano S. An assay system utilizing devitalized bone for assessment of differentiation of osteoclast progenitors. J Bone Miner Res. 1992;7(3):321–328. doi: 10.1002/jbmr.5650070312. [DOI] [PubMed] [Google Scholar]
- 18.Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Luthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, Shimamoto G, DeRose M, Elliott R, Colombero A, Tan HL, Trail G, Sullivan J, Davy E, Bucay N, Renshaw-Gegg L, Hughes TM, Hill D, Pattison W, Campbell P, Sander S, Van G, Tarpley J, Derby P, Lee R, Boyle WJ. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89(2):309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 19.Walsh CA, Beresford JN, Birch MA, Boothroyd B, Gallagher JA. Application of reflected light microscopy to identify and quantitate resorption by isolated osteoclasts. J Bone Miner Res. 1991;6(7):661–671. doi: 10.1002/jbmr.5650060703. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood. 2003;101(6):2227–2234. doi: 10.1182/blood-2002-06-1740. [DOI] [PubMed] [Google Scholar]
- 22.Kamolmatyakul S, Chen W, Yang S, Abe Y, Moroi R, Ashique AM, Li YP. IL-1alpha stimulates cathepsin K expression in osteoclasts via the tyrosine kinase-NF-kappaB pathway. J Dent Res. 2004;83(10):791–796. doi: 10.1177/154405910408301011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimi E, Nakamura I, Duong LT, Ikebe T, Takahashi N, Rodan GA, Suda T. Interleukin 1 induces multinucleation and bone-resorbing activity of osteoclasts in the absence of osteoblasts/stromal cells. Exp Cell Res. 1999;247(1):84–93. doi: 10.1006/excr.1998.4320. [DOI] [PubMed] [Google Scholar]
- 24.Osdoby P, Martini MC, Caplan AI. Isolated osteoclasts and their presumed progenitor cells, the monocyte, in culture. J Exp Zool. 1982;224(3):331–344. doi: 10.1002/jez.1402240306. [DOI] [PubMed] [Google Scholar]
- 25.Mbalaviele G, Jaiswal N, Meng A, Cheng L, Van Den Bos C, Thiede M. Human mesenchymal stem cells promote human osteoclast differentiation from CD34+ bone marrow hematopoietic progenitors. Endocrinology. 1999;140(8):3736–3743. doi: 10.1210/endo.140.8.6880. [DOI] [PubMed] [Google Scholar]
- 26.Miyamoto T, Ohneda O, Arai F, Iwamoto K, Okada S, Takagi K, Anderson DM, Suda T. Bifurcation of osteoclasts and dendritic cells from common progenitors. Blood. 2001;98(8):2544–2554. doi: 10.1182/blood.v98.8.2544. [DOI] [PubMed] [Google Scholar]
- 27.Basak SK, Harui A, Stolina M, Sharma S, Mitani K, Dubinett SM, Roth MD. Increased dendritic cell number and function following continuous in vivo infusion of granulocyte macrophage-colony-stimulating factor and interleukin-4. Blood. 2002;99(8):2869–2879. doi: 10.1182/blood.v99.8.2869. [DOI] [PubMed] [Google Scholar]
- 28.Day CJ, Kim MS, Stephens SR, Simcock WE, Aitken CJ, Nicholson GC, Morrison NA. Gene array identification of osteoclast genes: differential inhibition of osteoclastogenesis by cyclosporin A and granulocyte macrophage colony stimulating factor. J Cell Biochem. 2004;91(2):303–315. doi: 10.1002/jcb.10780. [DOI] [PubMed] [Google Scholar]
- 29.Bendixen AC, Shevde NK, Dienger KM, Willson TM, Funk CD, Pike JW. IL-4 inhibits osteoclast formation through a direct action on osteoclast precursors via peroxisome proliferator-activated receptor gamma 1. Proc Natl Acad Sci U S A. 2001;98(5):2443–2448. doi: 10.1073/pnas.041493198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobrovolskaia MA, Vogel SN. Toll receptors, CD14, and macrophage activation and deactivation by LPS. Microbes Infect. 2002;4(9):903–914. doi: 10.1016/s1286-4579(02)01613-1. Review. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi N, Udagawa N, Tanaka S, Murakami H, Owan I, Tamura T, Suda T. Postmitotic osteoclast precursors are mononuclear cells which express macrophage-associated phenotypes. Dev Biol. 1994;163(1):212–221. doi: 10.1006/dbio.1994.1137. [DOI] [PubMed] [Google Scholar]
- 32.Naito A, Azuma S, Tanaka S, Miyazaki T, Takaki S, Takatsu K, Nakao K, Nakamura K, Katsuki M, Yamamoto T, Inoue J. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells. 1999;4(6):353–362. doi: 10.1046/j.1365-2443.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 33.Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, van der Heiden A, Itie A, Wakeham A, Khoo W, Sasaki T, Cao Z, Penninger JM, Paige CJ, Lacey DL, Dunstan CR, Boyle WJ, Goeddel DV, Mak TW. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13(8):1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudo O, Fujikawa Y, Itonaga I, Sabokbar A, Torisu T, Athanasou NA. Proinflammatory cytokine (TNFalpha/IL-1alpha) induction of human osteoclast formation. J Pathol. 2002;198(2):220–227. doi: 10.1002/path.1190. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura I, Kadono Y, Takayanagi H, Jimi E, Miyazaki T, Oda H, Nakamura K, Tanaka S, Rodan GA, Duong le T. IL-1 regulates cytoskeletal organization in osteoclasts via TNF receptor-associated factor 6/c-Src complex. J Immunol. 2002;168(10):5103–5109. doi: 10.4049/jimmunol.168.10.5103. [DOI] [PubMed] [Google Scholar]
- 36.Yamane T, Kunisada T, Yamazaki H, Era T, Nakano T, Hayashi SI. Development of osteoclasts from embryonic stem cells through a pathway that is c-fms but not c-kit dependent. Blood. 1997;90(9):3516–3523. [PubMed] [Google Scholar]
- 37.Arai F, Miyamoto T, Ohneda O, Inada T, Sudo T, Brasel K, Miyata T, Anderson DM, Suda T. Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors. J Exp Med. 1999;190(12):1741–1754. doi: 10.1084/jem.190.12.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udagawa N, Takahashi N, Akatsu T, Tanaka H, Sasaki T, Nishihara T, Koga T, Martin TJ, Suda T. Origin of osteoclasts: mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells. Proc Natl Acad Sci U S A. 1990;87(18):7260–7264. doi: 10.1073/pnas.87.18.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraal G, Rep M, Janse M. Macrophages in T and B cell compartments and other tissue macrophages recognized by monoclonal antibody MOMA-2. An immunohistochemical study. Scand J Immunol. 1987;26(6):653–661. doi: 10.1111/j.1365-3083.1987.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 40.Henkel GW, McKercher SR, Leenen PJ, Maki RA. Commitment to the monocytic lineage occurs in the absence of the transcription factor PU.1. Blood. 1999;93(9):2849–2858. [PubMed] [Google Scholar]
- 41.Niida S, Amizuka N, Hara F, Ozawa H, Kodama H. Expression of Mac-2 antigen in the preosteoclast and osteoclast identified in the op/op mouse injected with macrophage colony-stimulating factor. J Bone Miner Res. 1994;9(6):873–881. doi: 10.1002/jbmr.5650090613. [DOI] [PubMed] [Google Scholar]
- 42.Chang MK, Raggatt LJ, Alexander KA, Kuliwaba JS, Fazzalari NL, Schroder K, Maylin ER, Ripoll VM, Hume DA, Pettit AR. Osteal tissue macrophages are intercalated throughout human and mouse bone lining tissues and regulate osteoblast function in vitro and in vivo. J Immunol. 2008;181(2):1232–1244. doi: 10.4049/jimmunol.181.2.1232. [DOI] [PubMed] [Google Scholar]
- 43.McMahon C, Will A, Hu P, Shah GN, Sly WS, Smith OP. Bone marrow transplantation corrects osteopetrosis in the carbonic anhydrase II deficiency syndrome. Blood. 2001;97(7):1947–1950. doi: 10.1182/blood.v97.7.1947. [DOI] [PubMed] [Google Scholar]
- 44.McHugh KP, Hodivala-Dilke K, Zheng MH, Namba N, Lam J, Novack D, Feng X, Ross FP, Hynes RO, Teitelbaum SL. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. J Clin Invest. 2000;105(4):433–440. doi: 10.1172/JCI8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saftig P, Hunziker E, Wehmeyer O, Jones S, Boyde A, Rommerskirch W, Moritz JD, Schu P, von Figura K. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc Natl Acad Sci U S A. 1998;95(23):13453–13458. doi: 10.1073/pnas.95.23.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249(4975):1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 47.Itoh K, Udagawa N, Kobayashi K, Suda K, Li X, Takami M, Okahashi N, Nishihara T, Takahashi N. Lipopolysaccharide promotes the survival of osteoclasts via Toll-like receptor 4, but cytokine production of osteoclasts in response to lipopolysaccharide is different from that of macrophages. J Immunol. 2003;170(7):3688–3695. doi: 10.4049/jimmunol.170.7.3688. [DOI] [PubMed] [Google Scholar]
- 48.Hayashi S, Yamada T, Tsuneto M, Yamane T, Takahashi M, Shultz LD, Yamazaki H. Distinct osteoclast precursors in the bone marrow and extramedullary organs characterized by responsiveness to Toll-like receptor ligands and TNF-alpha. J Immunol. 2003;171(10):5130–5139. doi: 10.4049/jimmunol.171.10.5130. [DOI] [PubMed] [Google Scholar]
- 49.Insogna KL, Sahni M, Grey AB, Tanaka S, Horne WC, Neff L, Mitnick M, Levy JB, Baron R. Colony-stimulating factor-1 induces cytoskeletal reorganization and c-src-dependent tyrosine phosphorylation of selected cellular proteins in rodent osteoclasts. J Clin Invest. 1997;100(10):2476–2485. doi: 10.1172/JCI119790. [DOI] [PMC free article] [PubMed] [Google Scholar]