Abstract
During Drosophila melanogaster oogenesis, the follicular epithelium that envelops the germline cyst gives rise to an elaborate eggshell, which houses the future embryo and mediates its interaction with the environment. A prominent feature of the eggshell is a pair of dorsal appendages, which are needed for embryo respiration. Morphogenesis of this structure depends on Broad, a Zinc-finger transcription factor, regulated by the EGFR pathway. While much has been learned about the mechanisms of Broad regulation by EGFR, current understanding of processes that shape the spatial pattern of Broad expression is incomplete. We propose that this pattern is defined by two different phases of EGFR activation: An early, posterior-to-anterior gradient of EGFR signaling sets the posterior boundary of Broad expression, while the anterior boundary is set by a later phase of EGFR signaling, distributed in a dorsoventral gradient. This model can explain the wild-type pattern of Broad in D. melanogaster, predicts how this pattern responds to genetic perturbations, and provides insight into the mechanisms driving diversification of eggshell patterning. The proposed model of the Broad expression pattern can be used as a starting point for the quantitative analysis of a large number of gene expression patterns in Drosophila oogenesis.
Keywords: pattern formation, Drosophila, systems biology, quantitative biology, EGFR, BMP, epithelial, morphogenesis
1. Introduction
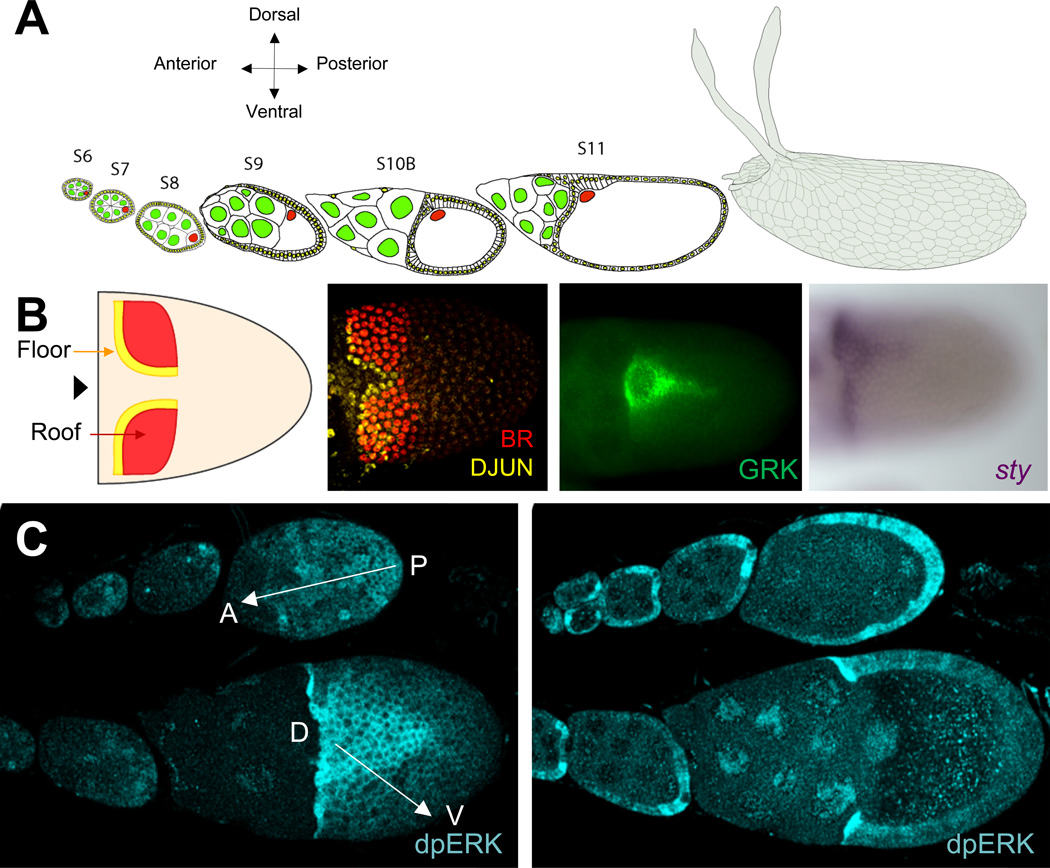
The developing germline cyst in Drosophila is encapsulated by the follicular epithelium, an epithelial sheet of approximately one thousand somatically derived follicle cells (Fig. 1A) [1, 2]. During oogenesis, this epithelium is subdivided into several domains of cells that contribute to different structures of the eggshell [1–4]. As a consequence, the dorsal-anterior region of the follicular epithelium undergoes a highly regulated sequence of cell shape changes and rearrangements that transform the epithelial sheet into an intricate three-dimensional structure, which serves as a mold for secreting eggshell proteins by the follicle cells. A prominent feature of the eggshell is a pair of dorsal appendages, tubular structures that provide enhanced gas exchange for the embryo [5]. Dorsal appendage morphogenesis depends on the specification of two adjacent groups of cells, which form the lower (“floor”) and upper (“roof”) part of the appendage (Fig. 1B) [1, 6].
Figure 1. Patterning and morphogenesis of the follicular epithelium.
(A): Cartoon schematic of the multiple stages of oogenesis starting from stage 6 to the final egg with the prominent dorsal appendages protruding from the dorsal anterior of the egg.
(B): Panel 1: During stage 10B, the floor and roof cells are specified. The dorsal midline is defined as a line equidistant from each set of floor and roof domains (marked by an arrowhead).
Panel 2: A Stage 10B egg chamber is stained for DJUN, which shows elevated levels in the floor cells (green) at stage 11, and BR (red), which marks the roof cells.
Panel 3: Antibody staining of the EGFR ligand, Gurken (GRK), which is secreted from the oocyte and is distributed in a midline pattern. The qualitative shape of GRK source is used in the model to calculate the GRK gradient.
Panel 4: The gene transcript sty is expressed in the domain of high levels of EGFR signaling (midline pattern).
(C): The level of EGFR signaling can be assayed with dpERK antibody stainings. Earlier stages show an AP gradient (marked with an arrow), whereas later stages (stage 10) show a clear DV gradient (arrow). In the second panel, a cross-section is shown. The switch in EGFR activation patterns is caused by the migration of the oocyte nucleus from the posterior half of the oocyte to the dorsal anterior during midoogenesis.
The specification of the roof and floor cell domains is initiated by the EGF ligand Gurken (GRK), which is secreted from the dorsal-anterior cortex of the oocyte and signals through the Epidermal Growth Factor Receptor (EGFR) in the follicle cells, establishing a dorsoventral (DV) gradient of EGFR activation (Fig. 1C, 2A) [7, 8]. The roof cells are marked by high expression levels of Broad (BR), a Zinc-finger transcription factor [9]. EGFR signaling controls BR expression through an incoherent feedforward loop, a network where an input activates both a target gene and its repressor. As a consequence, BR is expressed in the regions of the follicular epithelium that correspond to the intermediate levels of GRK [10–12]. In addition to its regulation by EGFR, BR is controlled by the Decapentaplegic (DPP) pathway [9]. A BMP2/4 type ligand, DPP is secreted at the anterior boundary of the follicular epithelium and signals through the DPP receptors in the follicle cells [13, 14]. During the early stages of DV patterning, the DPP receptors are expressed throughout the follicular epithelium [10, 14, 15]. A combination of DPP produced in the anterior and its uniformly expressed receptors results in anteroposterior (AP) gradient of DPP signaling, which represses BR in a narrow band of the anterior follicle cells (Fig. 2A) [10].
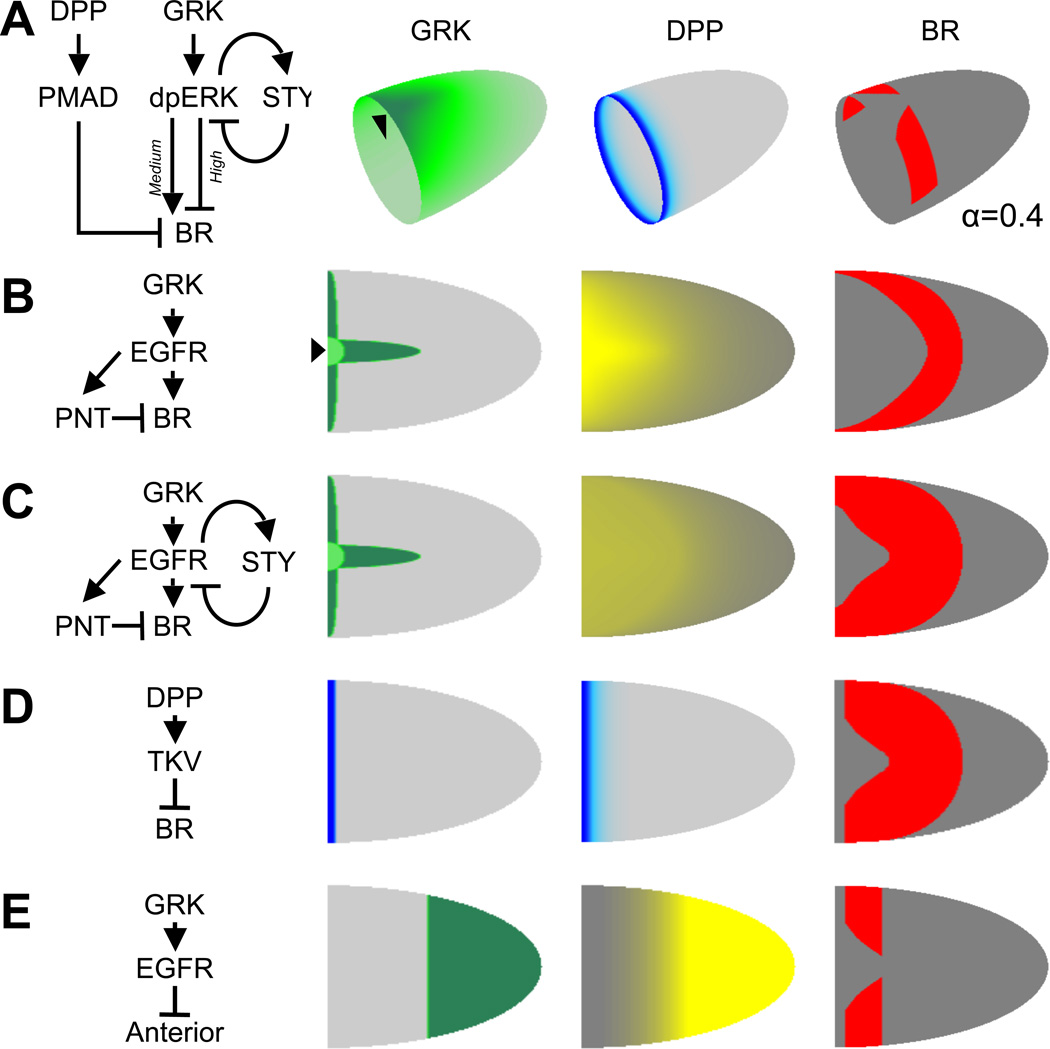
Figure 2. Model of the full 2D pattern of BR.
(A): Panel 1: A simplified network model describing the patterning of BR. DPP promotes high levels of PMAD signaling required to repress BR in the anterior cells. GRK activates high levels of dpERK, which also repress BR, whereas medium levels of dpERK upregulates BR. STY forms a negative feedback loop that modulates EGFR signaling levels. Arrowheads in this and subsequent figures denote the dorsal midline.
Panel 2/3: EGFR/DPP signaling gradients.
Panel 4: Two-dimensional BR pattern based on the defined regulatory network shown in Panel 1.
(B–E): Panel 1: Network motifs (see text) contributing to the overall regulatory network.
Panel 2: Spatial distribution of the morphogen source: circles denote locations of the oocyte nuclei at different developmental time points.
Panel 3: Corresponding signaling gradients for the ligand sources in Panel 2.
Panel 4: Predicted shape of the BR pattern with each motif added to the pattern above.
(C): The dorsal boundary is set by repression through PNT (this is effect is most likely indirect). A “horseshoe” expression pattern is predicted which is inconsistent with experimental observation.
(D): Including a negative feedback inhibitor, STY, provides a dorsal-anterior boundary of BR that is closer to the observed pattern.
(E): DPP represses BR along the anterior boundary.
(F): The posterior boundary is set at an early phase of EGFR signaling. Only the anterior cells are competent to receive the activation and repression signals at later stages. The poster domain is refractive to later EGFR signaling.
Genetic studies of BR regulation led to a model whereby the pattern of BR is established by the EGFR and DPP signaling gradients that act through an intricate network of feedforward and feedback loops [9, 12, 16–18]. While this model is consistent with a large number of experimental observations in the wild type and mutant backgrounds, it cannot fully explain the two-dimensional pattern of BR expression: The model predicts that BR should be expressed in a simply connected horseshoe-like pattern, which is different from the experimentally observed pattern with two dorsolateral patches (Fig. 2B). To explain the wild-type pattern, we propose that the posterior boundary of the BR domain is established by an earlier phase of EGFR signaling, when the oocyte nucleus is located at the posterior end and directs local secretion of GRK, resulting in the posterior-to-anterior gradient of the EGFR activation in the follicular epithelium [19, 20]. This model can explain the wild-type pattern of Broad in D. melanogaster, predicts how this pattern responds to genetic perturbations, and generates hypotheses regarding the diversification of the eggshell patterning network in species with alternate numbers of dorsal appendages [5, 21, 22]. The proposed model of the Broad pattern provides a starting point for the quantitative analysis of a large number of two-dimensional gene expression patterns established by the EGFR and DPP pathways in oogenesis [23].
2. Materials and Methods
2.1 Fly stocks and clonal analysis
The FLP/FRT technique was used to generate loss-of-function clones, null clones of which are marked by the loss of a GFP marker [24]. The sty−/− mosaic clones have the following genotype: e22c>flp; styΔ5 FRT2A/hv-GFP FRT2A [25]. GAL4 driver lines include: CY2-GAL4 [26] and GMR-GAL4 [27]. Oregon R was used as the wild-type control. D. yakuba was a gift from D. Stern), and D. virilis, D. phalerta are gifts from J. Duffy).
siRNAs targeting the sty mRNA were generated by the Dharmacon siDesign center (http://www.dharmacon.com/designcenter/designcenterpage.aspx). 71-nt DNA oligonucleotides (Integrated DNA Technologies, IDT) containing sty siRNAs were designed and inserted into either the pHB or pNE3 vectors as described in [28, 29], followed by concatenation to produce a tandem styshmiR expression vector. Multiple, independent UAS-styshmiR fly lines were produced by Best-Gene Inc. (Chino Hills, CA) using standard methods. Oligonucleotide sequences used for UAS-styshmiR vector construction are below:
Sty siRNA #1 (pHB): TTTCTCGCATAGCTTCATGCA
pHB Top: agcttagtTGCATGAAGCAATGCGAGATAtagttatattcaagcataTTTCTCGCATAGCTTCATGCAgcg
pHB Bottom: gatccgcTGCATGAAGCTATGCGAGAAAtatgcttgaatataactaTATCTCGCATTGCTTCATGCAacta
sty siRNA #2 (pNE3): TTTCGCAGTATGCTCCTGGCG
pNE3 Top: ctagcagtCGCCAGGAGCTTACTGCGATAtagttatattcaagcataTTTCGCAGTATGCTCCTGGCGgcg
pNE3 Bot: aattcgcCGCCAGGAGCATACTGCGAAAtatgcttgaatataactaTATCGCAGTAAGCTCCTGGCGactg
2.2 Immunostaining and microscopy
Antibodies used in this study include mouse anti-BR core (1:100, DSHB), rabbit or sheep anti-GFP (1:1000, Chemicon International and Biogenesis, respectively) and rabbit anti-dpERK (1:100, Cell Signaling). DAPI (VECTASHIELD Mounting Medium for Fluorescence with DAPI, Vector Laboratories) was used to stain for nuclei. Alexa Fluor- and Oregon Green-conjugated secondary antibodies (1:1000, Molecular Probes) were used. The immunostaining protocols are described elsewhere [10, 11]. The BR antibody showed significant cross-reactivity in other species; this allowed us to visualize the expression patterns of BR in multiple species.
2.3 Mathematical modeling
The model of the GRK gradient is an extension of an earlier 2D model of the GRK gradient [8]. Equations for calculating the ligand distribution function, g, can be found in the previous work [8]. The previous model assumed a symmetrical hemispherical shape of GRK secretion that is convex with respect to the dorsal midline. In this study, the two dimensional geometry of GRK is an important feature required for explaining the 2D shape. As shown in Fig. 1B the GRK gradient shows a clear T-shape pattern which is used in the current model (Fig. 1B., Fig. 2B, second panel).
The analytical solution of Equation 3 is given by the following expression:
where θI ≡ Θ / a and δ ≡ Δ / a ≪ 1. In the limit of no inhibitory feedback (α=0), using L’Hopital’s rule, leads to i = 0.5 (1 + (g − θI) / δ) when θI − δ < g < θI + δ, as expected.
Parameter values for the computational model are as follows: The shape of the gradients are given by the Thiele moduli, FGRK = 2.7 and FDPP=20, and were calculated through modeling and experiments in previous studies [8, 14, 16]. The thresholds for the AP patterning are: qR,AP =qI,AP =0.27 and qAnterior=0.1. For DV patterning, the thresholds are: qR=0.7, qI=0.7, qBR =0.25, qBR,DPP=0.6. For the D. phaleratera pattern in Fig. 4, the following perturbations to threshold values were used: qR=0.5, qI=0.5, qBR,DPP =1 (i.e., DPP was chosen to not repress BR). No AP patterning was added. For the D. virilis pattern: qR,AP =qI,AP =0.29, and qBR,DPP =1.
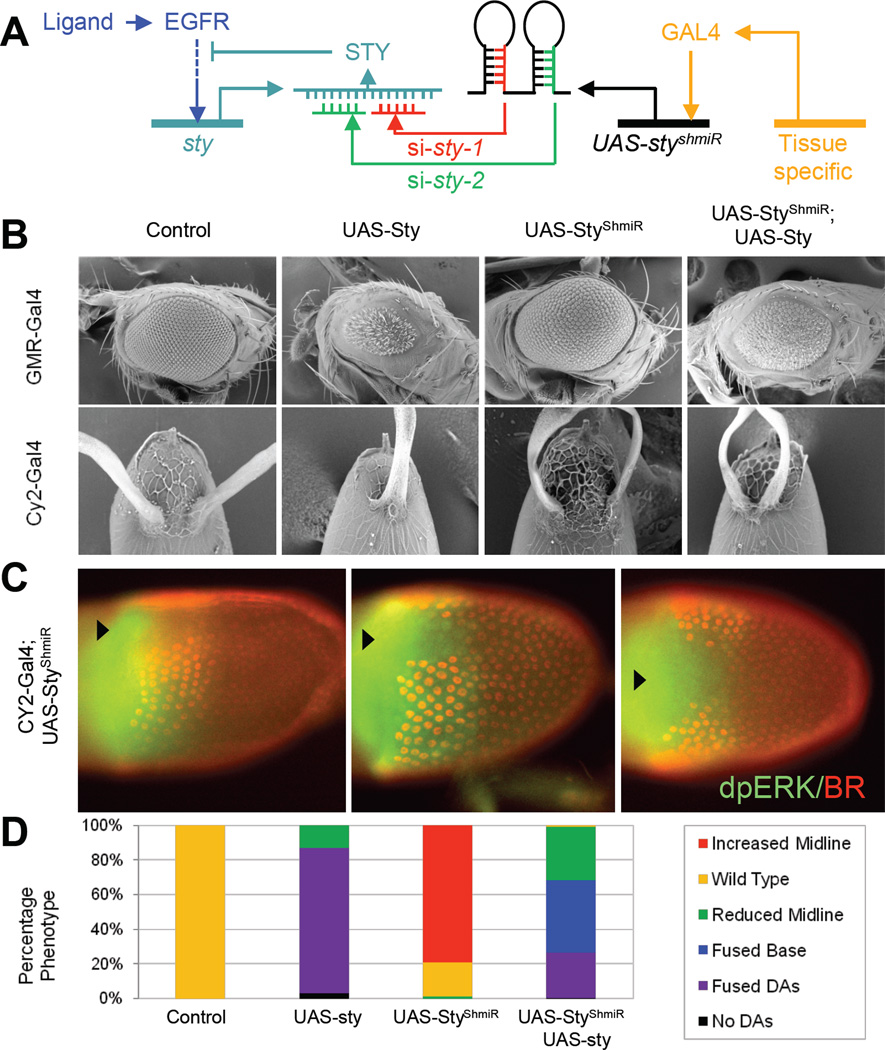
Figure 4. Perturbing later phases of EGFR signaling.
(A): Two unique shmiRs targeting sty were expressed from a single transgene to enhance gene silencing efficiency. Tissue specific drivers can be utilized for selective knock-down of STY’s inhibitory effect of the EGFR pathway.
(B): The miRNA-based RNA silencer (UAS-styShmiR) rescues the overexpression of UAS-sty phenotype in the eye. GMR-Gal4 is an eye-specific driver that drives expression of UAS-transgenes. Similarly, the UAS-styShmiR line rescues the single appendage phenotype of CY2>sty. The CY2-GAL4 driver is active during the mid-stages of oogenesis. Controls correspond to siblings carrying balancer chromosomes.
(C): When STY levels are knocked down, the levels of high dpERK signaling can transition from a concave shape to a convex shape. The dorsal, but not posterior, boundaries of BR are affected, consisted with the interpretation that the early stages of EGFR signaling are not perturbed by a loss of negative feedback through STY. Arrowheads give the approximate location of the dorsal midline.
(D): Scoring of DA phenotypes for eggshell images in B.
3. Results and discussion
In the wild-type follicular epithelium, BR is expressed in the cells that are exposed to intermediate levels of the EGFR signaling. This pattern and its changes in mutants with altered levels of the EGFR activation are consistent with an incoherent feedforward loop model, in which the activation of BR is overridden by a repressor that is induced by high levels of the EGFR signaling [10, 12, 16]. The output of this circuit is modulated by negative feedback loops [11, 12, 17, 30–32]. In particular, Sprouty (STY), an intracellular inhibitor of EGFR signaling that is activated by EGFR, controls the shape and size of the BR expression domains (Fig. 1B, 2C). In the absence of STY, the distance between the two BR patches is increased and their anteroposterior extent is reduced (Fig. 3) [11, 12]. Below we argue that these observations reveal a missing layer of regulation in the eggshell patterning network. To illustrate this point, we use a mathematical model that captures what we believe to be the essential features of BR regulation.
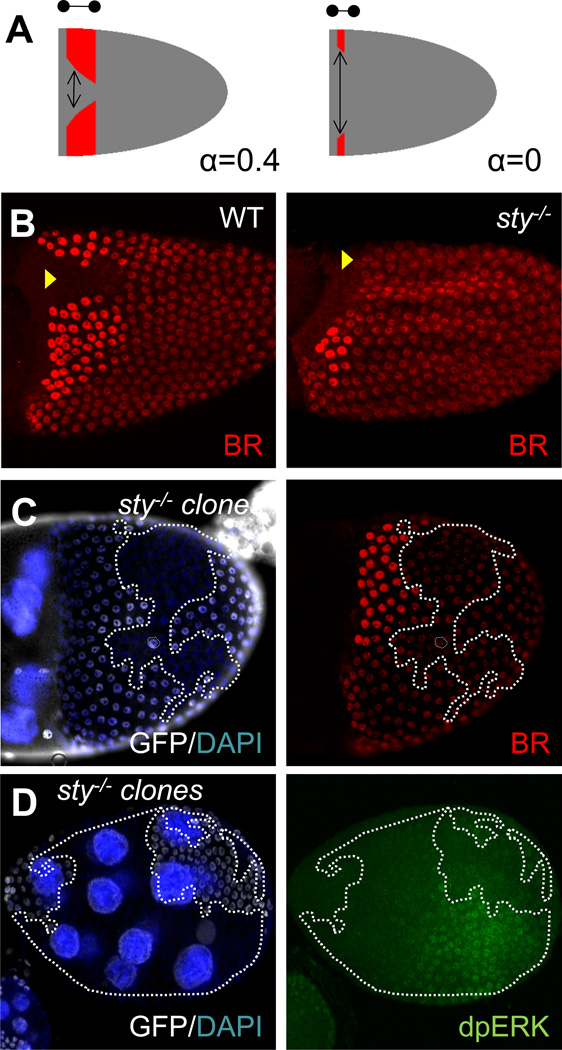
Figure 3. Experimental support for the 2D model.
(A): A model prediction: removing negative feedback at all time points will affect both the dorsal and posterior boundaries while removing negative feedback at later time points will principally affect only the dorsal boundary of the BR expression domain.
(B): Experimentally observed BR patterns in WT and sty−/− FCs. Yellow arrowheads give the approximate location of the dorsal midline.
(C): Posterior clones that remove sty lead to repression of BR, consistent with the model’s predictions.
(D): Levels of dpERK staining are higher in sty−/− clones compared to wild-type cells at earlier stages of oogenesis. The sty−/− cells are marked by a loss of GFP.
3.1 Mathematical model of cell autonomous negative feedback
We assume that the level of the EGFR signaling, denoted by S, is proportional to the local concentration of the oocyte-derived morphogen, denoted by G, which is assumed to form a steady-state concentration gradient across the follicle cells. The morphogen induces an inhibitor (such as STY), denoted by I, which lowers the net signaling level according to the following expression:
| (1) |
where A is the amplification of signaling and B is the strength of inhibitory feedback.
The distribution of the morphogen, G, is calculated based on our previous model of the GRK gradient (see Materials and Methods). According to this model, the steady-state ligand concentration is given by G = Gmaxg, where Gmax is the concentration that would be observed if the spatial pattern of ligand secretion was uniform along the surface of the oocyte. g is a function of position on the surface of the follicular epithelium; this function characterizes the shape of the steady-state distribution of secreted ligand, and depends on the pattern of ligand release, the size of the egg, and the characteristic distance of ligand degradation (the distance to which the ligand diffuses before it is internalized by the follicle cells). Briefly, the model assumes a constant flux, Q, of the morphogen G from a local area over the oocyte nucleus, reversible binding of G with its receptor, and that the system is in the limited-ligand regime (receptor is a constant). The level of pathway activation depends on the rate of internalization of the bound ligand-receptor complex. Additionally, a prolate spheroid geometry for the egg chamber is assumed. Complete equations and estimation of the parameters for the morphogen gradient can be found in our earlier work on quantifying the GRK gradient [8]. In this work, we have modified the 2D shape of the source to be T-shaped pattern based on antibody staining of GRK (Fig. 1B., Fig. 2B, second panel). With the ligand field as the input, the modified signaling levels of EGFR can be calculated as discussed below.
We assume that the level of inhibitor expression is an algebraic function of the EGFR signaling: I = Imaxf (S), where Imax is the maximal level of inhibitor expression, and f (S) increases from zero to one as a function of S. For simplicity, we use the following piecewise linear function:
| (2) |
In this equation, Θ is the threshold for the induction of the inhibitor and Δ characterizes the sharpness of the activation of the expression of inhibitor by the EGFR signaling. Combining Equations 1 and 2, we get an implicit equation for the level of inhibitor expression:
| (3) |
where i ≡ I / Imax, a ≡ AGmax, α = BImax. Without the loss of generality, we take a = 1. Equation (3) can be solved analytically (see Materials and Methods), leading to an explicit relation between the level of the morphogen, and the strength of the negative feedback:
| (4) |
where θI ≡ Δ / a. This equation can be used to analyze the effect of the strength of the negative feedback and the threshold for the induction of inhibitor on the local signaling level.
3.2 Feedback control of the incoherent feedforward loop
To explore how negative feedback affects the pattern of BR, we use a threshold-based model, in which BR is expressed when the level of the EGFR signaling is between two threshold levels, which correspond to the level of the EGFR signaling necessary for the induction of BR and the induction of its repressor [16]. The values of the thresholds are denoted by Sa and Sr, respectively. When combining this threshold-based model with the models of the gradient and negative feedback, we first calculate the steady-state GRK gradient, denoted by g. Next, based on Equation 4, we calculate the spatial pattern of inhibitor expression. Finally, based on Equation 1, we calculate the spatial pattern of EGFR signaling. Combining the results, we find that the BR domain is defined by the following inequalities:
| (5) |
Thus, the BR expression pattern is delineated by the level-sets, i.e. the lines of constant concentration, of the GRK gradient. When the strength of the feedback is set to zero, the boundaries of BR expression are predicted to move ventrally, because the levels of signaling that correspond to each of the thresholds are now realized at a lower value of morphogen concentration.
Since the level sets (locations of constant concentration) of the steady state morphogen concentration become progressively convex (with respect to the dorsal midline) as a function of the threshold (Fig. 2B, C), this model predicts that the boundaries of BR expression domain should become more convex when the strength of the negative feedback is reduced (Fig. 2B). According to this model, the dorsal boundary of BR expression, which is defined by its repression by an intracellular repressor, should be more sensitive to the reduction of the feedback strength than the ventral boundary. This is due to the fact that the level of inhibitor expression decreases from dorsal to ventral regions. Hence, removal of inhibitor (setting a=0 in the model) does not affect the ventral boundary of the BR pattern (Fig. 2C).
Both of these predictions are supported by the results of previous genetic and imaging experiments in which we analyzed BR expression in the egg chambers with large clones of sty−/− follicle cells, which results in a cell autonomous loss of negative feedback and should reduce the value of α in the model (Fig. 3B) [11]. Thus, a model that relies on threshold-based signaling and negative feedback can explain why the dorsal boundary of the BR domain moves to a more ventral position in response to removal of STY.
3.3 Problems of a simple model and proposed resolution
The same model (Fig. 2C) predicts that, in response to removal of negative feedback inhibitors, the posterior boundary of BR expression domain should also shift in a direction opposing the EGFR signaling gradient. In contrast to this prediction, however, the posterior boundary moves in an anterior direction (Fig. 3C). The pattern of BR expression becomes thinner, as a consequence of the dorsal movement of the anterior boundary and the anterior shift of the posterior boundary. Thus, the presented model does not fully account for the experimentally observed effects of cell-autonomous negative feedback on the BR expression pattern. The second problem of the presented model has to do with the wild-type pattern. According to the model described above, the BR expression domain should look like a “horseshoe” between the two level sets of the GRK gradient (Fig. 2C). Instead, it appears as two patches straddling the dorsal midline with a separation of about four cells.
The anterior repression of BR can be attributed to the DPP signaling gradient, which is generated due to a combination of anteriorly-produced DPP ligand and uniformly-expressed DPP receptors (Fig. 2A, D). In the past, the DPP signaling gradient was also proposed to mediate the posterior splitting of the BR expression domain [33]. Specifically, if DPP acts as activator of BR expression, then, by “intersecting” with the dorsoventral gradient of the EGFR signaling, the anterior DPP gradient could generate a posterior split in the pattern of BR. While this model potentially can account for the wild-type pattern of BR, experiments in our and other groups rule out the activating role of DPP in BR regulation [9, 10, 34].
What splits the posterior boundary of the BR pattern is currently unclear. It is certainly possible that some yet to be defined molecular component regulates the posterior domain of BR expression. In the meantime, we propose a plausible alternative that explains the emergence of the wild-type pattern and behavior of this pattern in mutants. Specifically, we propose that the posterior boundary of the BR pattern is specified by an earlier phase of the EGFR signaling, when the EGFR pathway is activated in a posterior-to-anterior gradient (Fig. 2E). At this stage of oogenesis, the oocyte nucleus is at its posterior position, which directs GRK secretion towards the posterior follicle cells [19, 20]. We hypothesize that this results in the posterior-to-anterior signaling gradient that pre-patterns the follicular epithelium. As a consequence, only the anterior follicle cells can assume dorsal cell fates and express BR. By separating the patterning of EGFR signaling in time, the level sets specified at two distinct developmental time points can now “intersect” to define the dorsal, ventral and posterior boundaries, almost exclusively by a single signaling pathway.
3.4. Experimental tests of the temporal EGFR gradient model
The proposed model makes several predictions regarding the effect of reduced negative feedback on the pattern of BR expression. First, high levels of the EGFR signaling should be detected in the posterior follicle cells at earlier stages of oogenesis. Second, removing the action of the negative inhibitor STY during both early and later patterning steps should lead to higher levels of EGFR signaling along the AP axis at earlier stages. At later stages, removal of STY should push the two roof domains away from the dorsal midline, while at the same time the posterior boundary should be shifted in the anterior direction (Fig. 3A).
We have tested these predictions experimentally. First, we found that the spatial pattern of double phosphorylated Extracellular Signal Regulated Mitogen Activated Protein Kinase (dpERK), an intracellular mediator of the EGFR signaling [35, 36], forms a posterior-to-anterior gradient in the follicular epithelium during earlier stages of oogenesis (Fig. 1C). Second, somatic clones homozygous for a mutant allele of sty−/− generated at early stages of development lead to higher levels of dpERK in the posterior FCs (Fig. 3D), and affect the posterior boundary of the BR patches, as mentioned previously (Fig. 3C). This demonstrates that prior to the anterior migration of the oocyte nucleus, the EGFR pathway is activated in a posterior-to-anterior gradient and that this gradient is sensitive to the removal of the intracellular negative feedback provided by STY.
In experiments with genetic mosaics, removal of STY affects both the early and late phases of the EGFR signaling, and has an effect on both the anterior and posterior boundaries of the BR pattern. According to our model, the ventral shift of the dorsal boundary in response to genetic removal of STY reflects the action of STY during the dorsoventral phase of EGFR activation by GRK. On the other hand, the anterior shift of the posterior boundary reflects the inhibitory action of STY during the earlier, posterior phase of the EGFR signaling.
To summarize, the DV and posterior boundaries of the BR pattern are specified by the same pathway, but at different points in time. Furthermore, the positions of both boundaries are controlled by cell-autonomous negative feedback, which positions the expression boundary closer to the source of ligand production. This model can readily explain both the posterior split in the wild-type pattern of BR and the effects of sty−/− clones, since removal of STY early in oogenesis would affect both the AP and DV phases of EGFR signaling.
To test whether the posterior boundary of the BR pattern is specified during the AP stage of EGFR activation, we used conditional knock-down experiments, in which we inhibited the action of STY only during the dorsoventral phase of the EGFR signaling. Our revised model predicts that this should affect mainly the anterior boundary the BR pattern and not influence the AP extent of the BR patches. To remove the action of sty during the DV phase of EGFR signaling, we made use of artificial microRNAs (shmiRs) expressed at mid-stages of oogenesis using the GAL4/UAS binary expression system [28, 29], (Fig. 4A). The GAL4/UAS system is a tool that enables the experimentalist to selectively drive expression of a transgene construct in a specific tissue (Duffy, 2002). The driver includes the tissue-specific enhancer that regulates the expression of GAL4, a transcription factor that binds to UAS sites to direct expression of the UAS-transgene. In our experiment, the CY2-GAL4 driver drives expression in the FCs in contact with the oocyte from stage 8 onwards (Queenan et al. 1997). Here, the follicle cell driver, CY2-GAL4 [26, 37] is active in the FCs contacting the oocyte from stage 8 onwards and was used to express a tandem shmiR cassette (UAS-styshmiR), which produces two, unique small interfering RNAs (siRNAs) targeting sty mRNA (Fig. 4A).
While ectopic expression of UAS-sty with CY2-GAL4 induced fusion of the DAs and reduced midline, UAS-styshmiR expression produced an opposing phenotype. Co-expression of UAS-sty and UAS-styshmiR was able to rescue the phenotypes generated by both constructs (during both eye and egg development), demonstrating the specificity of action for each transgene (Fig. 4B). Scoring the phenotypes reveals that a distribution of morphological outcomes in which patterning robustness is lost, suggesting stochastic expression of the knockdown cassette (Fig. 4D). However, we found multiple egg chambers that had a convex pattern of dpERK (high EGFR signaling) and increased separation between the two BR domains (Fig. 4C).
As described above, the model predicts that only the shape of the anterior boundary of the BR pattern will be affected by inhibitory modulation of the late EGFR activation. In accordance with this prediction, we did not observe a dramatic effect on the posterior border characteristic of the sty−/− clones (Fig. 4C), confirming that later stages of EGFR signaling have no effect on setting the position of the posterior boundary of the BR pattern. These results suggest that EGFR activation controls the posterior boundary of BR expression through pre-patterning at an earlier stage. Loss of the negative feedback at both early and late stages affects both the dorsal-anterior and posterior boundaries of BR. Selectively modulating the strength of negative feedback at later stages will principally affect only the dorsal boundary, as was observed. Thus, the revised model provides a mechanistic explanation of the wild type pattern of BR and can be used to predict how this pattern responds to perturbations of the EGFR signaling pathway.
4. Conclusion and outlook
The EGFR-mediated patterning of the follicular epithelium provides a striking example of how complex gene expression domains can be specified parsimoniously by a single pathway. We present a simple model that explains the two-dimensional pattern of BR, a transcription factor that marks the cells contributing to the roof part of the dorsal appendages in Drosophila oogenesis. Our model can be summarized as follows: The two BR domains are limited to the anterior domain by the earlier phase of EGFR signaling, which defines an anterior band of cells competent to express high levels of BR necessary for the formation of dorsal appendages. At a later stage of oogenesis, the level sets (lines of constant concentration) of the dorsoventral EGFR signaling gradient intersect with this competence zone, splitting the BR expression domain into two patches (Fig. 2).
In addition to rationalizing the wild-type pattern of BR, our model can be used to generate hypotheses regarding the patterning of eggshell morphologies in other Drosophila species. The number and size of dorsal appendages varies greatly across the phylogenetic spectrum, providing a mechanism of adaptation to the nature of the oviposition substrate [5]. Since BR is a key regulator of dorsal appendage morphogenesis, one can expect that changes in the expression pattern of BR can provide a mechanism for the diversification of dorsal appendages. As a first step towards testing this hypothesis, we analyzed the expression of BR in mid-oogenesis, when the domains of elevated BR are first established (Fig. 5).
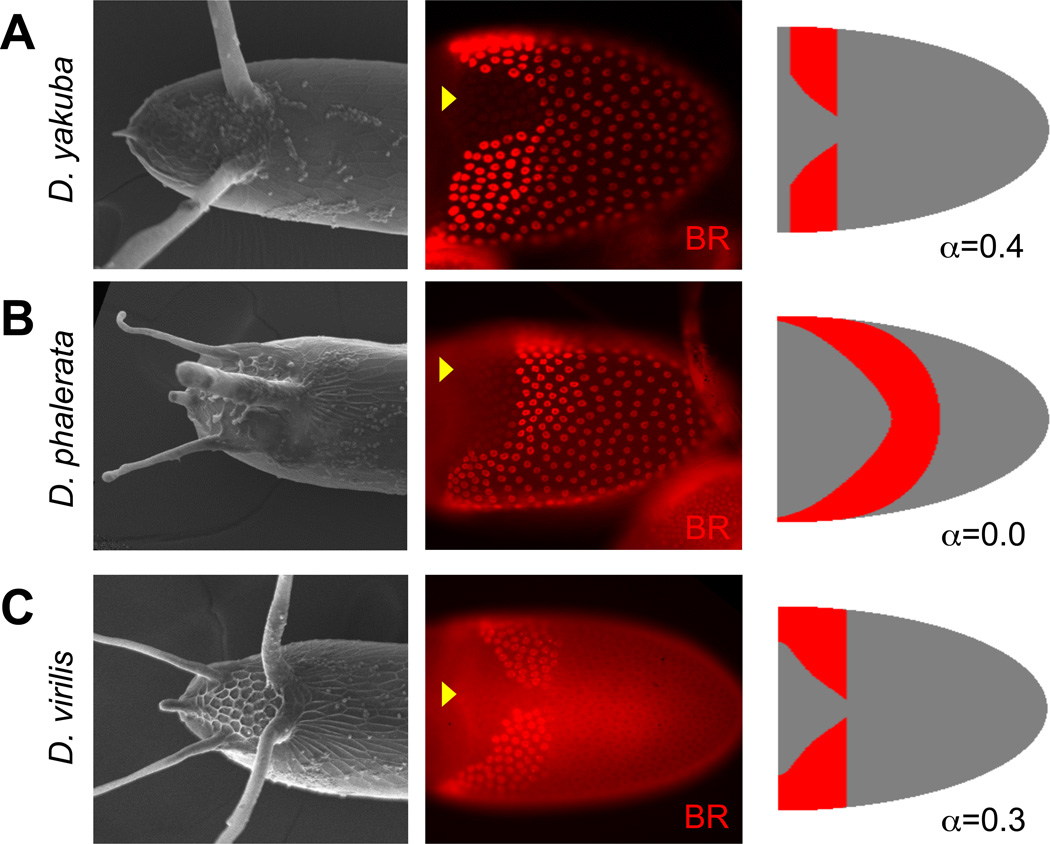
Figure 5. Concavity of BR expression across Drosophila species.
(A): Examples of eggshells with 2 (D. yakuba), 3 (D. phalerata) , or 4 (D. virilis) appendages.
(B): BR expression patterns in each species at stage 10, when elevated BR domains are first specified, but before tube morphogenesis is initiated. The dorsal midline is indicated with yellow arrowheads. In addition to the shape of the roof domain that has high levels of BR, the posterior FCs also have a basal level of BR – which the current model does not include.
(C): Model predictions of BR patterns with different values of inhibitory feedback strength. In D. phalerata and D. virilis, any regulation from BR was excluded, and relative threshold values were modified to more closely fit each expression pattern, but the shapes of the GRK and DPP gradients were held constant. Model parameters are specified in the Materials and Methods chapter.
We found that the BR pattern is dynamic and shows significant transitions during development; however, several stereotypic patterns at stage 10B of oogenesis emerge for species that have 2, 3 or 4 dorsal appendages (Fig. 5A–C). The number of contiguous BR patches is not equal to the number of dorsal appendages. At the same time, a clear pattern emerges regarding changes in concavity of the BR domain. For species with two appendages, the BR patches are split along the dorsal midline (along the DV axis) and each of the two patches has a dorsal anterior boundary that is concave relative to the DV/AP coordinate system (Fig. 5A). Species with three dorsal appendages show a continuous BR patch that is convex (Fig. 5B). For four appendages, curvature of the boundary appears to switch between concave to convex and then back to concave (Fig. 5C).
Thus, the shape of the boundary of the BR expression domain diverges across species and may alternate between convex and concave (D. vir.) or may be simply convex (D. phal.). With the change in concavity, the number of appendages also changes from 2 for D. mel. to 3 for D. phal. and 4 for D. vir. Our mathematical model can recapitulate qualitatively some aspects of the transition in BR expression, simply by varying the strength of negative feedback and thresholds. One mechanism for converting a D. mel. pattern into a D. phal. pattern involves reducing the strength of inhibition (or the shape of the GRK source) and shifting the anterior/posterior pre-pattern in the posterior direction. A comprehensive comparison of the shape of the GRK source and the function of the feedback inhibitors across species will provide a further test of this model. At the same time, investigation of BR patterning in other species can establish the limits of the model.
One of the most important questions for future work is how the quantitative changes in the expression pattern of BR give rise to discrete changes in the number of dorsal appendages. We speculate the local concavity of the BR pattern drives the temporal order of cell intercalations and specifies where the floor cells form a hinge that closes the forming tube. As such, changes in concavity could lead to mechanical “instabilities” that further subdivide the BR cells into smaller domains to form extra tubular appendages. Interestingly, recent work by Celeste Berg and colleagues published while this work was in preparation provides possible support for this model: they found that genetic perturbations or laser ablation of the cells along the dorsal anterior boundary of the roof domain blocks tube formation [38]. The next steps in increasing the scale of our understanding regarding patterning and morphogenesis will require models that integrate geometry and mechanics with signaling dynamics, as well as quantitative approaches to validating model predictions.
Acknowledgements
We thank S. Leffler, C. Watson, E. Oeffinger for technical assistance, T. Schüpbach, J. Duffy, M. Krasnow for advice and stocks. J. Z. was supported by the Fannie and John Hertz Foundation and the Princeton Wu fellowship. B. H. was supported by an American Cancer Society Postdoctoral Fellowship. The following NIH grants have supported this research: P50 GM071508 and RO1 GM078079 to S.Y.S.
Abbreviations list
- BR
Broad
- DV
dorsal- ventral
- AP
anterior-posterior
References
- 1.Berg CA. The Drosophila shell game: patterning genes and morphological change. Trends Genet. 2005;21(6):346–355. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Horne-Badovinac S, Bilder D. Mass transit: Epithelial morphogenesis in the Drosophila egg chamber. Developmental Dynamics. 2005;232(3):559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- 3.Zartman JJ, Shvartsman SY. Unit operations in tissue engineering: epithelial folding. Annual Review of Chemical and Biomolecular Engineering. 2010;1:231–246. doi: 10.1146/annurev-chembioeng-073009-100919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X, Tanwar PS, Raftery LA. Drosophila follicle cells: morphogenesis in an eggshell. Semin Cell Dev Biol. 2008;19(3):271–282. doi: 10.1016/j.semcdb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinton HE. Biology of Insect Eggs. Vol. 1. Oxford; New York: Pergamon Press; 1981. [Google Scholar]
- 6.Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA. bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Developmental Biology. 2004;267(2):320–341. doi: 10.1016/j.ydbio.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75(1):165–174. [PubMed] [Google Scholar]
- 8.Goentoro LA, Reeves GT, Kowal CP, Martinelli L, Schupbach T, Shvartsman SY. Quantifying the gurken morphogen gradient in Drosophila oogenesis. Dev Cell. 2006;11(2):263–272. doi: 10.1016/j.devcel.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng WM, Bownes M. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development. 1997;124(22):4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- 10.Yakoby N, Lembong J, Schupbach T, Shvartsman SY. Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development. 2008;135(2):343–351. doi: 10.1242/dev.008920. [DOI] [PubMed] [Google Scholar]
- 11.Zartman JJ, Kanodia JS, Cheung LS, Shvartsman SY. Feedback control of the EGFR signaling gradient: superposition of domain splitting events in Drosophila oogenesis. Development. 2009;136(17):2903–2911. doi: 10.1242/dev.039545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boisclair Lachance JF, Fregoso Lomas M, Eleiche A, Bouchard Kerr P, Nilson LA. Graded Egfr activity patterns the Drosophila eggshell independently of autocrine feedback. Development. 2009;136(17):2893–2902. doi: 10.1242/dev.036103. [DOI] [PubMed] [Google Scholar]
- 13.Twombly V, Blackman RK, Jin H, Graff JM, Padgett RW, Gelbart WM. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development. 1996;122(5):1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- 14.Lembong J, Yakoby N, Shvartsman SY. Spatial regulation of BMP signaling by patterned receptor expression. Tissue Eng Part A. 2008;14(9):1469–1477. doi: 10.1089/ten.tea.2008.0098. [DOI] [PubMed] [Google Scholar]
- 15.Jekely G, Rorth P. Hrs mediates downregulation of multiple signalling receptors in Drosophila. Embo Reports. 2003;4(12):1163–1168. doi: 10.1038/sj.embor.7400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lembong J, Yakoby N, Shvartsman SY. Pattern formation by dynamically interacting network motifs. Proc Natl Acad Sci. 2009;106(9):3213–3218. doi: 10.1073/pnas.0810728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peri F, Bokel C, Roth S. Local Gurken signaling and dynamic MAPK activation during Drosophila oogenesis. Mech Dev. 1999;81:75–88. doi: 10.1016/s0925-4773(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 18.Atkey MR, Lachance JF, Walczak M, Rebello T, Nilson LA. Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror. Development. 2006;133(11):2115–2123. doi: 10.1242/dev.02369. [DOI] [PubMed] [Google Scholar]
- 19.Nilson LA, Schupbach T. EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol. 1999;44:203–243. doi: 10.1016/s0070-2153(08)60471-8. [DOI] [PubMed] [Google Scholar]
- 20.González-Reyes A, St Johnston D. Patterning of the follicle cell epithelium along the anterior-posterior axis during Drosophila oogenesis. Development. 1998;125(15):2837–2846. doi: 10.1242/dev.125.15.2837. [DOI] [PubMed] [Google Scholar]
- 21.Kagesawa T, Nakamura Y, Nishikawa M, Akiyama Y, Kajiwara M, Matsuno K. Distinct activation patterns of EGF receptor signaling in the homoplastic evolution of eggshell morphology in genus Drosophila. Mech Dev. 2008;125(11–22):1020–1032. doi: 10.1016/j.mod.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 22.James KE, Berg CA. Temporal comparison of Broad-Complex expression during eggshell-appendage patterning and morphogenesis in two Drosophila species with different eggshell-appendage numbers. Gene Expr Patterns. 2003;3(5):629–634. doi: 10.1016/s1567-133x(03)00136-4. [DOI] [PubMed] [Google Scholar]
- 23.Yakoby N, Bristow CA, Gong D, Schafer X, Lembong J, Zartman JJ, Halfon MS, Schüpbach T, Shvartsman SY. A combinatorial code for pattern formation in Drosophila oogenesis. Dev Cell. 2008;15(5):725–737. doi: 10.1016/j.devcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu T, Rubin GM. Analysis Of Genetic Mosaics In Developing And Adult Drosophila Tissues. Development. 1993;117(4):1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 25.Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92(2):253–263. doi: 10.1016/s0092-8674(00)80919-8. (92): p. 253–63. [DOI] [PubMed] [Google Scholar]
- 26.Queenan AM, Ghabrial A, Schupbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124(19):3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- 27.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87(4):651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 28.Haley B, Foys B, Levine M. Vectors and parameters that enhance the efficacy of RNAi-mediated gene disruption in transgenic Drosophila. Proc Natl Acad Sci. 2010 Jun 4; doi: 10.1073/pnas.1006689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haley B, Hendrix D, Trang V, Levine M. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Developmental Biology. 2008;321(2):482–490. doi: 10.1016/j.ydbio.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reich A, Sapir A, Shilo BZ. Sprouty, a general inhibitor of receptor tyrosine kinase signaling. Development. 1999;126(18):413–447. doi: 10.1242/dev.126.18.4139. [DOI] [PubMed] [Google Scholar]
- 31.Ghiglione C, Carraway KL, Amundadottir LT, Boswell RE, Perrimon N, Duffy JB. The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell. 1999;96(6):847–856. doi: 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 32.Wasserman JD, Freeman M. An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell. 1998;95:355–364. doi: 10.1016/s0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- 33.Peri F, Roth S. Combined activities of Gurken and Decapentaplegic specify dorsal chorion structures of the Drosophila egg. Development. 2000;127:841–850. doi: 10.1242/dev.127.4.841. [DOI] [PubMed] [Google Scholar]
- 34.Dequier E, Souid S, Pal M, Maroy P, Lepesant JA, Yanicostas C. Top-DER- and Dpp-dependent requirements for the Drosophila fos/kayak gene in follicular epithelium morphogenesis. Mech Dev. 2001;106(1–2):47–60. doi: 10.1016/s0925-4773(01)00418-x. [DOI] [PubMed] [Google Scholar]
- 35.Gabay L, Seger R, Shilo B. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- 36.Gabay L, Seger R, Shilo BZ. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development. 1997;124(18):3535–3541. doi: 10.1242/dev.124.18.3535. [DOI] [PubMed] [Google Scholar]
- 37.Goentoro LA, Yakoby N, Goodhouse J, Schupbach T, Shvartsman SY. Quantitative analysis of the GAL4/UAS system in Drosophila oogenesis. Genesis. 2006;44(2):66–74. doi: 10.1002/gene.20184. [DOI] [PubMed] [Google Scholar]
- 38.Boyle MJ, French RL, Cosand KA, Dorman JB, Kiehart DP, Berg CA. Division of labor: subsets of dorsal-appendage-forming cells control the shape of the entire tube. Dev Biol. 2010;346(1):68–79. doi: 10.1016/j.ydbio.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]