Abstract
Human genetic variation plays a critical role in both spontaneous clearance of and response to interferon (IFN)-based therapies against hepatitis C virus (HCV) as shown by the success of recent genome-wide association studies (GWAS). Several GWAS and later validation studies have shown that single nucleotide polymorphisms (SNPs) at the IFNL3 (formerly IL28B) locus on chromosome 19 are involved in eliminating HCV in human patients. No doubt that this information is helping clinicians worldwide in making better clinical decisions in anti-HCV therapy, but the biological mechanisms involving the SNPs leading to differential responses to therapy and spontaneous clearance of HCV remain elusive. Recent reports including the discovery of a novel IFN (IFN-λ4) gene at the IFNL3 locus and in vitro functional studies implicating 2 SNPs as causal variants lead to novel conclusions and perhaps to new directions in research. An attempt is made in this review to summarize the major findings of the GWAS, the efforts involved in the discovery of causal SNPs; and to explain the biological basis for spontaneous clearance and response to treatment in HCV infections.
Introduction
Hepatitis C virus (HCV) is a global health problem affecting more than 3% of the world population (Lavanchy 2011). It causes chronic disease conditions of the liver, including liver cancer. The mode of transmission is by contact with contaminated blood. HCV is a flavivirus with a plus-strand RNA genome that exists as 7 genotypes (1 to 7) and 67 subtypes (a, b, c, etc.) (Smith and others 2013). Currently, there is no vaccine to prevent HCV infections. The standard of care (SOC) treatment for chronic HCV infections for several years has been a combination of Interferon-α (IFN-α) and Ribavirin (RBV) that is administered over an extended period of time (Fried and others 2002). This treatment involves severe side-effects and is poorly tolerated. Drugs targeting the different viral proteins (Direct-acting antivirals or DAAs) are being developed; some have entered into clinical practice and are showing great promise; however, the emergence of resistance mutations is a major limitation to this strategy (Alkhouri and Zein 2012; Asselah and Marcellin 2013).
Host genetic variation is a strong determinant of HCV pathogenesis and treatment-response. Genome-wide association studies (GWAS) and later validation studies conducted by several groups around the world have reported that single nucleotide polymorphisms (SNPs) around the innate immunity gene IFNL3 (formerly IL28B) encoding the cytokine IFN-λ3 on chromosome 19 have strong associations with both HCV spontaneous clearance and treatment-response (Ge and others 2009; Suppiah and others 2009; Tanaka and others 2009; Thomas and others 2009; Rauch and others 2010; Hayes and others 2012; Duggal and others 2013). However, the biological significance of these associations remains unknown, especially in in vivo situations. The objective of this review is to update the current literature on association of IFNL3 polymorphisms with HCV infections and importantly to understand the biological significance of these associations. The efforts involved in discovering the causal SNPs and the progress made so far since the initial GWAS are analyzed. The association of IFNL3 variants with treatment response in nongenotype 1 HCV infections is also addressed. The article ends with a discussion on future directions of research in light of the GWAS findings and the discovery of causal SNPs.
Genetic Studies and Innate Immunity Against HCV
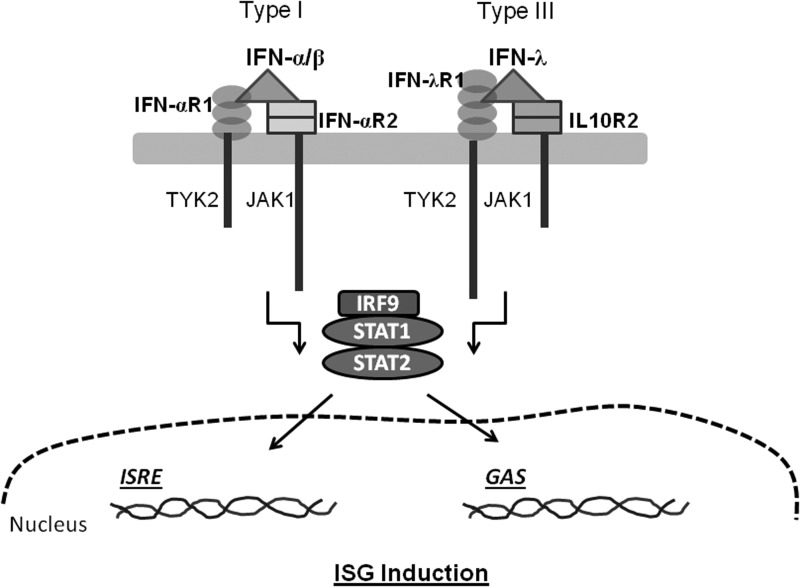
Innate immunity against RNA viruses is mediated by IFNs, which are cytokines expressed by a variety of cell types in response to viral infections. The HCV RNA replication intermediates are sensed by innate immunity pattern recognition receptors such as RIG-I (Retinoic acid inducible gene) and TLR3 (Toll-like receptor 3) (Lemon 2010). Type I IFNs (IFN-α and IFN-β) and Type III IFNs (IFN-λ) are produced in response to viral infections by several different cell types in the human body. These IFNs, on secretion by virus infected cells, act in an autocrine/paracrine manner and drive the expression of several hundreds of genes collectively called ISGs (IFN-stimulated genes) that will prepare the body/cells to fight virus infection (Fig. 1). Many of the ISGs act directly to suppress virus replication, while others act indirectly by mediating antiviral processes, including priming an optimal adaptive immune response. The disruption of RIG-I and TLR3 signaling by HCV NS3-4A protease preventing the production of type I IFNs, including IFN-α, is well recognized (Lemon 2010; Horner and Gale 2013). HCV NS3-4A is also likely to disrupt IFN-λ secretion similar to its effect on IFN-α secretion, as both cytokines are stimulated for secretion through similar pathways (Levy and others 2011; Horner and Gale 2013). In fact, a recent report confirms that HCV NS3-4A protease cleaves the adaptor protein mitochondrial anti-viral signaling protein (MAVS) which is responsible for RIG-I-dependent signaling events that lead to IFN-λ expression (Ding and others 2012).
FIG. 1.
A schematic of the signaling events that lead to production of ISGs by Type I and Type III interferons (IFNs). The IFN receptors are indicated next to their extracellular domains in different shapes. IFN molecules are shown as triangles. TYK2-Tyrosine kinase 2; JAK-Janus kinase; STAT-Signal transducer and activator of transcription; IRF9-IFN regulatory factor 9; ISRE-IFN-stimulated response element; GAS-γ activated sequence. The basic mechanisms of signal transduction are shown; for details, the readers can refer Donnelly and Kotenko (2010), and Li and others (2009).
A subset of HCV-infected individuals are able to spontaneously clear the virus within 6 months of infection (∼20% for genotype 1 HCV), while the remaining progress to chronic infection that may lead to fibrosis, cirrhosis, and even hepatocellular carcinoma (Lauer and Walker 2001; Lavanchy 2011). Among the persistent carriers when subject to SOC therapy (for a duration of 24–48 weeks depending on the HCV genotype), only a subset respond to the therapy (also dependent on HCV genotype; ∼50% for genotype 1 HCV) (Fried 2004). Response to treatment is measured by monitoring for SVR (sustained virological response, which is defined as testing negative for HCV RNA 6 months after cessation of therapy). HCV infections are known to have an ethnic bias: The African-American population is more susceptible to chronic infection and less responsive to treatment (Howell and others 2000). Clinical phenotypic variation in both natural progression and response to treatment combined with features of ethnic bias makes HCV a good candidate for genetic studies to identify host genes that are critical for viral replication and infection. Variation at several host genes has been identified mainly by candidate gene approaches to be important for spontaneous and treatment-induced viral clearance (Selvarajah and others 2010; Schlecker and others 2012). Most of these genes (such as HLA, TNF, IL10, JAK, TYK, IFNAR1, IFNAR2, IFNG, and others) belong to biological pathways involving response to virus, immune response, and inflammatory responses, based on gene ontology classification (Schlecker and others 2012). The majority of these studies have been carried out on small sample sizes and in restricted geographical regions, and, hence, the results may not be robust enough to be replicated in larger and diverse host populations (Fried and others 2006; Selvarajah and others 2010). Therefore, at the advent of the genomics era, similar to other human complex diseases HCV infections were also subject to genome-wide studies in different regions of the world.
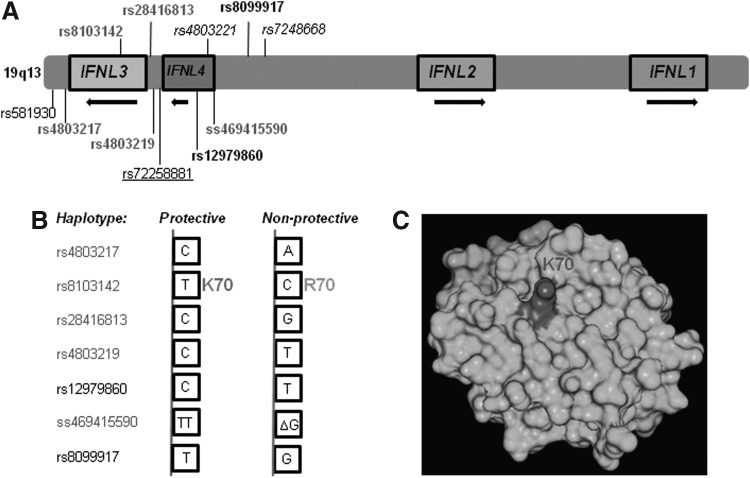
In the year 2009, 3 reports were published almost simultaneously on the results of GWAS carried out in different world populations for treatment response in patients with chronic HCV infections (Ge and others 2009; Suppiah and others 2009; Tanaka and others 2009). Another GWAS was subsequently carried out to identify SNPs that played a role in spontaneous clearance of HCV infection (Rauch and others 2010). Intriguingly, all the 4 studies had the same locus on chromosome 19q13 (with several overlapping SNPs) that showed genome-wide significance in the respective case-control studies (Ge and others 2009; Suppiah and others 2009; Tanaka and others 2009; Rauch and others 2010). The strong association seen in all 4 studies suggested a clear role for this locus that has the IFN-λ genes (IFNL2 (formerly IL28A) coding for IFN-λ2; IFNL3 (formerly IL28B) for IFN-λ3; and IFNL1 (formerly IL29) for IFN-λ1) (Fig. 2) in generating antiviral responses in patients. The majority of the strongly associated SNPs identified are within 8 kb of IFNL3 coding region at its 5′ end (Fig. 2). IFN-λ2 and IFN-λ3 share 96% identity, while IFN-λ1 shares about 81% identity with the other 2 (Gad and others 2010). The 3 genes are considered as having been evolved from a common precursor by gene duplication events (Donnelly and Kotenko 2010). Biologically, IFN-λ3 is 16-fold more potent than IFN-λ2 and about 2-fold more potent than IFN-λ1 against encephalo myocarditis virus infection in HepG2 cells (Dellgren and others 2009), while a recent report shows IFN-λ1 to be the most potent out of the 3 IFN-λs against HCV subgenomic replicons (Friborg and others 2013). More recently, Kanda and others (2013) show that IFN-λ3 induces more anti-HCV-specific ISGs (such as TLR3, MyD88, IFN-β1, and CXCL10) than the other 2 IFN-λs in HepG2 stable cell lines carrying the 3 IFN-λ genes.
FIG. 2.
SNPs at the IFNL3 locus associated with HCV infections. (A) IFNL3 locus at 19q13 has 4 genes. IFNL3 codes for IFN-λ3, IFNL2 codes for IFN-λ2, and IFNL1 codes for IFN-λ1 apart from the newly identified IFNL4 (codes for IFN-λ4) that arises due to a deletion polymorphism (ss469415590) found upstream of IFNL3 coding region. The important SNPs at the IFNL3 locus identified by several genetic studies are shown. The SNPs rs4803219, rs28416813, rs8103142, and rs4803217 were identified as causal SNPs by di Lulio and others (2011). The SNPs identified by Smith and others (2011) are in italics. The TA repeat SNP identified by Sugiyama and others (2011) is underlined. rs12979860 and rs8099917 are considered the “tag-SNPs” (in bold) that are in LD with the causal SNPs. (B) The protective and nonprotective alleles of the SNPs that were identified by different studies are shown as haplotypes. (C) Crystal structure of IFN-λ3 depicting K70 (dark shade) (PDBID: 3HHCL). The variant rs8103142 changes the amino acid K70 to R70. K70 is exposed on the outside surface of the protein.
The GWAS result is intriguing for the following reasons: (1) Genetic variation at a type III IFN locus determines the response to treatment with a type I IFN (IFN-α) in a viral infection; (2) no other gene that could be responsible for uptake, processing, or metabolism of the 2 drugs (IFN-α and RBV) showed any genome-level significance in determining the treatment response; and (3) the same region in the genome was responsible for both spontaneous clearance and response to treatment. Importantly, the GWAS result satisfactorily explained the long-standing observation of high HCV incidence and poor response rate to IFN-based therapy in the African-American population by showing that this ethnic group has a high frequency of the “nonprotective” IFNL3 allele compared with East Asians, Hispanics, or European Americans (Ge and others 2009; Thomas and others 2009). More importantly, the GWAS results reveal the significance of type III IFNs in HCV disease biology at the human patient level, even though both IFN-α and IFN-λ may have similar anti-HCV properties in vitro (Doyle and others 2006; Marcello and others 2006; Kohli and others 2012). Minor differences especially in the kinetics of ISG expression have been noted between the 2 IFNs, but overall, they seem to have similar antiviral properties in vitro (Marcello and others 2006; Kohli and others 2012). However, the target cells of IFN-λs may be more specific, as receptors for IFN-λs are mostly expressed on cells of epithelial origin; whereas those of IFN-α are expressed on most human cell types [refer Levy and others (2011) and Kotenko (2011), for a comparative review of literature on the biology of the 2 IFNs].
A review of the GWAS in HCV infections
The initial GWAS carried out on treatment response to SOC therapy and on spontaneous clearance have been extensively reviewed elsewhere (Balagopal and others 2010; Kelly and others 2011; Clark and Thompson 2012; Hayes and others 2012). Briefly, Ge and others (2009) identified SNP rs12979860 about 3 kb upstream of the IFNL3 gene on chromosome 19 to be strongly associated in European Americans with the achievement of SVR (C allele-protective; T allele-nonprotective, Fig. 2B). After this report, Thomas and others (2009) showed that rs12979860 was also involved in spontaneous clearance of HCV in a candidate gene study. Tanaka and others (2009) and Suppiah and others (2009) saw that rs8099917 lying ∼7.5 kb upstream of IFNL3 coding region showed the strongest association with virological responses against SOC therapy from a Japanese and Australian cohort, respectively (T allele-protective; G allele-nonprotective, Fig. 2B). In a subsequent GWAS, Rauch and others (2010) also saw that rs8099917 was associated with spontaneous clearance of HCV at genome-wide significance. This study also did resequencing and recombinant mapping at the IFNL3 region in a cohort containing spontaneous clearers and persistently infected patients and identified a 5-SNP containing haplotype that had 2 promoter SNPs (rs4803219 and rs28416813), one exonic SNP (rs8103142), and two 3′ UTR SNPs (rs4803217 and rs581930) to have strong association with spontaneous clearance (Fig. 2) (Rauch and others 2010). Apart from this study, a recent study reports on a GWAS carried out on a large number of individuals in 13 international study centers, to identify SNPs responsible for spontaneous clearance of HCV (Duggal and others 2013). This study involving 919 patients who had spontaneously cleared HCV and 1482 chronic HCV patients, using the Illumina Human Omni-Quad array with more than a million SNPs, showed 2 loci to be significantly associated with spontaneous clearance (with P values below the threshold of 1×10−8). The strongest association was, however, with rs12979860 at the IFNL3 locus (P=2.17×10−30), while rs4273729 near the class II HLA locus on chromosome 6 also showed strong genome-wide significance (P=1.71×10−16). The latter SNP lies in between DQA2 and DQB1 (coding for the α and β chains of MHC class II, respectively) genes, and the region shows strong linkage disequilibrium (LD).
Several follow-up studies have been carried out since the original GWAS data from different regions of the world were published (Kelly and others 2011; Clark and Thompson 2012; Hayes and others 2012). In all of the studies with genotype 1 HCV, the association has been replicated, suggesting that the polymorphisms reported around the IFNL3 gene are critical to HCV replication, irrespective of the ethnicity of the hosts. Several studies have showed a clear correlation of HCV viremia with the IFNL3 genotype post-treatment (Howell and others 2012; Lindh and others 2011a, 2011b). For example, Lindh and others (2011a, 2011b) report that IFNL3 genotype at the SNP rs12979860 correlates with both earlier and later phases of viral decline in genotype 1, 2, and 3 HCV. Genotype CC at the SNP site shows more viral clearance from serum when compared with the CT or TT genotype. This shows that similar to the results from in vitro studies (Marcello and others 2006), IFN-λs may be directly responsible for viral clearance in affected patients.
Polymorphisms at IFNL3 Locus and Association with Nongenotype 1 HCV Infections
Even though the initial GWA studies were carried out in genotype 1 HCV infections, the association with response to SOC therapy was later replicated in other HCV genotypes such as genotypes 4, 5, and 6 (Bitetto and others 2011; Asselah and others 2012; Seto and others 2013). However, the association with response to treatment in genotype 2 and 3 HCV-infected patients is controversial. In genotype 2 and 3 HCV infections, the majority of the studies have shown a significant association with early virological responses but not with sustained responses (Moghaddam and others 2011; Sarrazin and others 2011; Bucci and others 2013); whereas some studies have shown an association with both early and sustained responses in genotype 2 or genotype 2/3 HCV infections (Sakamoto and others 2011; Cavalcante and others 2013). However, recent studies (Schreiber and others 2012) show that when data are combined from several studies involving genotype 2 and 3 HCV infections, the association of treatment responses with IFNL3 SNPs is replicated even with these HCV genotypes. This indicates that due to high treatment-response rates in these easy-to-treat HCV genotypes (Tapper and Afdhal 2013), the studies that showed no association may have suffered from a lack of power. Furthermore, relapse rates are known to be higher in genotype 3 HCV infections (possibly due to shorter durations of therapy, Zeuzem and others 2009) and may be a factor affecting the results of association tests (Moghaddam and others 2011; Bucci and others 2013). Therefore, it is likely that all genotypes of HCV may have to deal with similar, if not identical, innate immune responses that are mediated by IFNL3, even though the outcome may differ due to viral-genotype effects (Bellecave and others 2010; Horner and Gale 2013). However, the predictive value of IFNL3 genotypes in treatment response involving genotype 2 and 3 HCV infections is low (Schreiber and others 2012). Comparative studies between the easy-to-treat genotypes 2 and 3 and the difficult-to-treat genotypes 1 and 4 HCV in terms of viral load, IFNL3 genotypes and especially of ISG expression are lacking. Such studies can delineate the differences that lead to different treatment-response rates between different HCV genotypes.
Discovery of the Causal SNP(s) and IFN-λ4
Although the GWAS for HCV infections has met with unprecedented success, the progress on discovery of the causal SNP(s) has been very slow. One of the reasons for this is the limitations associated with a large-scale genetic study such as GWAS (Klein and others 2012). In a typical GWAS, a million or more SNPs of the human genome are spotted on chip-arrays along with their flanking sequences as “probes.” Next, sheared genomic DNA from cases and controls is allowed to hybridize, after which the call for a particular variant (depending on base-pair hybridization) is obtained by fluorescence-assisted methods (For a review of basic study design refer Dube and Hegele 2013). Then, each SNP is interrogated for its association with case versus control using an appropriate statistical method. Multiple testing correction is imparted to compensate for the large number of tests carried out to avoid false positives, and after the correction, the SNPs that have P values below a fixed threshold (usually 1×10−8) are considered genome-wide significant. However, the oligonucleotides that are arrayed on the chip do not represent the complete set of SNPs which are identified (more than 40 million submitted so far (dbSNP build138, NCBI) and several of them are yet to be identified) on the human genome but a sample of SNPs that are chosen based on how strongly they are correlated to nearby SNPs by LD (Bush and Moore 2012). These “tag-SNPs” are selected from available datasets such as Hapmap or the 1000 genomes project. Therefore, rarely does a GWAS directly identify the “causal” variant, as it may not have been chosen as a “tag-SNP” to be arrayed on to the SNP array. In the simplest of situations, the causal variant may be one or a very few critical SNPs that affect the expression of the causal gene or function of the causal protein and lie close to the “tag-SNP” within the LD block (region of the genome that has strong LD and co-segregates in successive generations). In such situations, fine-mapping by sequencing the DNA around the GWA signal region within the LD block will identify the causal SNP, which usually is a common SNP (the minor allele occurring in>5% of the population). However, it is possible that a large number of rare variants (that are not picked up by most of the available GWAS platforms) which are individually or in small combinations are causative of the phenotype in a nonoverlapping manner could be giving rise to “synthetic associations” in GWAS (Cirulli and Goldstein 2010). These rare variants are usually not in LD with the “tag-SNP” and, hence, occur at a distant location (several megabases) from the “tag-SNP”. Identifying such rare variants that give rise to synthetic associations in a GWAS will require whole-genome sequencing that will have coverage of the genome beyond the LD block which defined the GWA signal. In view of the intrinsic complexities involved in such studies, it is well recognized that identification of the causal SNP(s) post-GWAS is the bottle-neck in complex disease genetics (Goldstein 2009; Chakravarti and others 2013). It is also imperative that genetic studies are followed up with appropriate functional studies to validate the causal SNPs to gain a mechanistic understanding of the underlying biology (Chakravarti and others 2013). After the initial success of GWAS in HCV infections, several groups have been engaged in this venture, and have reported varying success.
Suppiah and co-workers have followed up their GWA studies and sequenced about 100 kb of DNA sequence around the IFNL3 gene in about 200 responder and nonresponder (to SOC therapy) HCV-infected patients by using massively parallel sequencing to identify the causal variants (Smith and others 2011). However, even though some new SNPs (rs4803221 and rs7248668, Fig. 2) were discovered with better predictive values for treatment response than their GWAS SNP rs8099917, they did not come across any definitive causal variant(s) that drastically increased the odds ratios. The authors did an in silico analysis and showed that certain SNPs may give rise to transcription factor binding sites or may change potential methylation sites in the promoter DNA but no experiments were carried out to investigate these findings. The Mizokami group tried to functionally characterize several of the SNPs that their group identified from the initial GWAS on HCV infections but did not find any significant result apart from identifying another new SNP which affected transcription in in vitro reporter assays (Sugiyama and others 2011). This SNP, rs72258881 that is present ∼750 bp upstream of the first exon of IFNL3 (Fig. 2) had multiple copies of TA repeats when genotyped in different healthy Japanese individuals. The number of repeats (11 or 13) correlated with levels of transcription of reporter gene in vitro. However, the association of this novel SNP with response to treatment or spontaneous clearance of HCV remains to be established. SNP rs1188122 that was initially identified in their GWAS and thought to be occurring near a splice-site junction was tested for function and was not found to be affecting splicing. SNP rs28416813 was also tested for influence on splicing and found to have no effect. The number of exons present in the IFNL3 gene is not clear as Kotenko and others (2003) (Donnelly and Kotenko 2010) predicted 5 exons, while Sheppard and others (2003) claimed IFNL3 to have 6 exons (with a small, 4-amino-acid coding 1st exon being extra in the latter report). Sugiyama and others (2011) carried out 5′ RACE and showed that IFNL3 has 6 exons and that rs28416813 is located in the first intron. While rs28416813 may not have affected splicing, there is a possibility that it could affect transcription due to its proximity to the transcription start site (Chinnaswamy and others 2013). This possibility was not tested by Sugiyama and others (2011). Instead, they tested rs4803219 that is located in the promoter region (∼300 bp upstream of the 1st exon of IFNL3, Fig. 2) (Kotenko and others 2003) for its role in reporter gene expression, and found no significant influence of the alternate alleles on transcription.
In a report from Egypt, a 20 kb region at the IFNL3 locus bounded by rs12980275 and rs8099917 was speculated to include the causal SNP(s) for spontaneous clearance of (genotype 4) HCV (Pedergnana and others 2012). The LD block in the European population comprised a much larger fragment that included DNA all the way up till IFNL2 coding region, while genotype data from the Egyptian population reduced the size of the LD block to ∼20 kb in size. Within this 20 kb region, the strongest association with spontaneous clearance was with rs12979860 and rs8103142 (Pedergnana and others 2012). Rauch and others (2010) and di Lulio and others (2011) undertook a comprehensive genetic analysis approach in identifying the causal SNP involved in spontaneous clearance of HCV. They examined the distribution of haplotypes in patients with “extreme” genotype-phenotype (discordant) combinations that is, persistently infected but carrying protective alleles at rs12979860 and rs8099917 and spontaneously cleared but carrying the nonprotective alleles at the 2 SNP positions and compared them with concordant genotype-phenotype combination groups (Rauch and others 2010). This recombinant mapping analysis pointed to 4 SNPs as prime candidates to be causal (Fig. 2): rs4803219, rs28416813, rs8103142, and rs4803217 (di Lulio and others 2011). The 4 SNPs were found to be in high LD between each other and with the tag-SNP rs12979860. Further, di Lulio and others (2011) tested the association of these 4 SNPs with spontaneous clearance in both a multiple-source and a single-source cohort. The multiple-source cohort had patients who were infected with multiple HCV genotypes and were not homogeneous for age or sex or other viral (HIV) coinfections. On the other hand, the single-source cohort included 27 spontaneous clearers and 44 persistently infected women who had acquired genotype 1b HCV infection from a single source several years ago and had no HIV coinfection. The haplotype containing protective alleles of the 4 SNPs (Fig. 2) were highly predictive of spontaneous clearance of HCV in both cohorts but more strongly with the single-source cohort. The study with the single-source cohort had no confounders such as differences in viral genotype, age, and sex of the patients and, hence, was a test of the net contribution of host genetics in spontaneous clearance of HCV. Therefore, the strong association of these 4 SNPs with spontaneous clearance of HCV in this cohort is an indication of the presence of causal SNP(s) within the 4 studied SNPs or the haplotype block. The study also examined for occurrence of copy number variants (CNVs) in the IFNL3 region. An analysis of the available data from 1,000 genomes project and also by q-PCR analysis from their patient samples failed to detect any CNVs and it was concluded that CNVs at IFNL3 region may be unlikely to have any role in their association with HCV infection outcome (di Lulio and others 2011). This study also noted the technical challenges involved in sequencing the IFNL3 region due to the presence of the homologous IFNL2 in the vicinity, an observation also made by others (Smith and others 2011; Sugiyama and others 2011; Reynolds and others 2012; Chinnaswamy and others 2013). In view of these difficulties, extreme care is needed in designing primers for PCR-based sequencing/genotyping methods and interpretation of genotyping results, especially with the SNPs closer toward the 5′ end, on the coding region, and at 3′ ends of IFNL3. In a separate study, the 4 SNPs identified by di Lulio and others (2011) were tested in a cohort of HCV-infected patients receiving SOC therapy for association with SVR and 3 of them (rs28416813, rs8103142 and rs4803217, Fig. 2) showed the strongest association (de Castellarnau and others 2012), suggesting that the predicted causal SNPs for spontaneous clearance of HCV were also important in response to treatment. Interestingly, 3 out of the 4 potential causal SNPs identified by di Lulio and others (2011) (viz. the promoter SNP rs28416813, the exonic SNP rs8103142, and the 3′UTR SNP rs4803217, see Fig. 2) along with the tag-SNP rs12979860 were found to be under strong positive selection during evolution in the European and Asian population (Manry and others 2011). This suggests that these ethnic groups may have evolved to deal with HCV infections better than other world populations due to evolutionary pressures. The protective allele frequency at rs12979860 is known to be higher in East Asians and European Americans than the African or Hispanic populations (Ge and others 2009). Furthermore, evolutionary selection has not operated uniformly across all the 3 IFN-λ genes, as low LD was noted between them in all populations (Manry and others 2011).
Of the 4 predicted causal SNPs, the most amenable to functional analysis is the exonic SNP rs8103142. The polymorphism that resides on the 70th codon in the second exon of IFN-λ3 changes the amino acid from Lysine to Arginine. K70 is not a part of the receptor binding site of IFN-λ3 (Gad and others 2009), but it is located on the surface of the protein (Fig. 2) and, hence, could be important in its function by affecting interaction with other factors. However, 3 independent studies have ruled this possibility out by showing that the conservative substitution does not affect the activity of the protein, at least in vitro. Urban and others (2010) used both Escherichia coli-expressed and HEK293T-cell-expressed recombinant IFN-λ3 variants and tested for anti-HCV replicon activity in Huh7.5 cells and found no difference. Similarly, Sugiyama and others (2011) used 293-F-cell-expressed and purified variants of IFN-λ3 and saw no difference in the stimulation of ISRE activity in different human liver cell lines. Friborg and others (2013) used synthetic human recombinant IFN-λ3 wt and IFN-λ3K70R against HCV subgenomic replicons from different genotypes and found no difference in their potencies. These results suggest that rs8103142 may itself not be a causal SNP but is in LD with some other causal SNP(s) in the IFNL3 region.
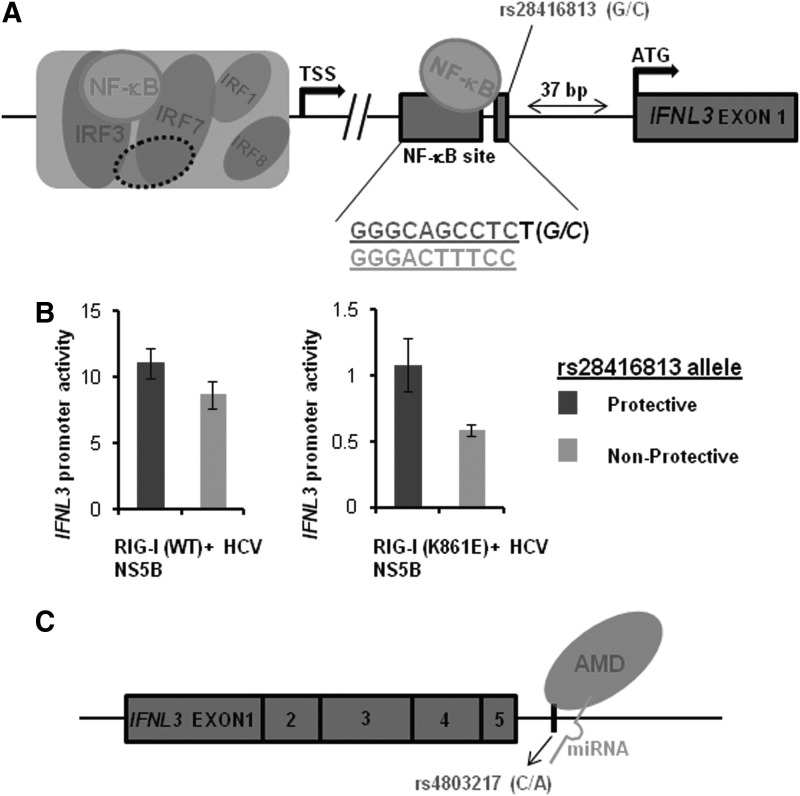
The first evidence of functional roles for IFNL3 SNPs associated with HCV infections has come recently, where 2 SNPs-rs28416813 and rs4803217-have been shown to influence reporter gene expression by utilizing different mechanisms (Chinnaswamy and others 2013; McFarland and others 2013). In the first report, Chinnaswamy and others (2013) found that rs28416813 due to its close proximity to an NF-κB-binding site (Fig. 3A) in the downstream promoter region of IFNL3, influenced gene expression in luciferase reporter assays. This SNP lies 37 bp upstream of start codon of IFNL3 (Kotenko and others 2003; Osterlund and others 2007) and one nucleotide away from a nonconsensus NF-κB binding site (Fig. 3A). After confirming for binding to the transcription factor NF-κB, the authors showed by molecular modeling that rs28416813 lies in a position that can contact the residue R41 in p65 component of NF-κB in solved co-crystal structures. The nonprotective allele (Tanaka and others 2009; Rauch and others 2010; di Lulio and others 2011) at this SNP position was responsible for decreasing transcription from a 1.4 kb promoter construct by 25%–50% compared with the protective allele when either the NF-κB was overexpressed or the endogenous NF-κB was stimulated by physiological inducers such as dsRNA and TNF-α or by an active HCV RNA-dependent RNA polymerase in HEK293T cells (Fig. 3B). Deletion of the nonconsensus NF-κB-binding site increased the transcription activity, suggesting that this region of the promoter may be acting as a silencer; while scrambling the site to eliminate NF-κB binding had a modest reduction of transcription, which was similar to the effect produced by the nonprotective allele (Chinnaswamy and others 2013). The SNP rs28416813 seems to be ideally poised to have a subtle effect on the weak binding of NF-κB to this site. A stronger interaction resulting from the binding of NF-κB to a consensus binding element would have weakened the effect of the nearby SNP, hence a nonconsensus site may have evolved so that the alternate alleles at the nearby site would give the host additional control for transcriptional regulation. It is also possible that rs28416813 may be affecting the binding affinity of different NF-κB functional dimers (such as p50 homodimers, p65 homodimers, or p50+p65 heterodimers) to the DNA that can result in the differential regulation of transcription (Udalova and others 2000). Although rs28416813 is outside the binding motif of NF-κB by 2 bases, based on X-ray crystal data, such flanking sequences can affect affinity of p50 homodimers to the NF-κB element (Chen and others 1998). Interestingly, the promoter region of the homologous IFNL2 also has this nonconsensus NF-κB site but no SNP is reported to occur around the site. In a separate study, Ding and others (2012) have shown that a 261 bp distal region of the IFNL3 promoter, including this nonconsensus NF-κB site, is not enough for transcription. This suggests that a major part of the IFN-λ3 enhanceosome that will include transcription factors such as NF-κB, IRF1, 3, 7, and 8 (Osterlund and others 2007) seems to be farther away from this distal region that has the only known functional SNP (affecting transcription) associated with HCV infections (Fig. 3). A major future thrust area of research should be to structurally and functionally characterize the IFN-λ3 enhanceosome complex, similar to the efforts that were put in for the IFN-β enhanceosome (Balachandran and Beg 2011).
FIG. 3.
rs28416813 and rs4803217 are functional SNPs. (A) A schematic of the putative enhanceosome complex of IFNL3. The enhanceosome (large open box) comprises of the factors identified by Osterlund and others (2007) that bind to the promoter region of IFNL3. The dotted oval represents factors that are still to be identified in the enhanceosome. The nonconsensus NF-κB-binding site at +115 to +124 (in relation to transcription start site TSS) and the IFNL3 exon 1at +163 are shown as closed rectangles. The start codon ATG on exon 1 (Kotenko and others 2003; Osterlund and others 2007) is shown. rs28416813 at +126 is depicted as a small rectangle adjacent to the nonconsensus NF-κB-binding site. The weak NF-κB-DNA interaction is depicted on the nonconsensus site as well as its interaction with the alleles at rs28416813. The nucleotides forming the nonconsensus NF-κB-binding site are shown (top). In contrast, the nucleotides in a consensus NF-κB-binding site are shown below (bottom). rs28416813 occurs as G or C (in italics) in the population and is just one nucleotide away from the nonconsensus site. (B) This panel is adapted from Chinnaswamy and others (2013). The nonprotective allele at rs28416813 reduces downstream gene expression in a reporter assay. Firefly luciferase reporter activity (after normalizing with Renilla luciferase activity) of a 1.4 kb IFNL3 promoter construct that includes the nonconsensus NF-κB-binding site. The construct with the alternate alleles at rs28416813 was cotransfected with Renilla luciferase controls after expressing the HCV NS5B and RIG-I (WT or mutant) proteins in HEK293T cells. HCV NS5B will generate ligands for RIG-I (exogenous or endogenous) that can lead to stimulation of IFNL3 promoter. A WT RIG-I can increase the promoter activity by 10-fold (compared with an RNA-binding mutant of RIG-IK861E). The nonprotective allele at rs28416813 can decrease the promoter activity ether in the presence of a WT RIG-I or with the endogenous RIG-I (as in RIG-IK861E). (C) rs4803217 affects mRNA stability. The mRNA transcript of IFNL3 with 5 exons is shown along with 5′ and 3′ UTRs. rs4803217 in the DNA occurs close to the last exon of IFNL3 (53 bases downstream) and affects mRNA stability by affecting binding/functioning of AMD factors and HCV-induced miRNA binding to the 3′ UTR (McFarland and others 2013).
McFarland and others (2013) report on the second functional SNP rs4803217 in the 3′UTR region to be affecting IFNL3 mRNA stability resulting from effects on AU-rich element (ARE) mediated decay (AMD) and binding of HCV-induced miRNAs. The authors performed in vitro luciferase reporter assays using constructs with the 3′UTR region of IFNL3 cloned downstream of the luciferase gene. They found that the nonprotective allele at rs4803217 was responsible for increased mRNA degradation, resulting in 30%–40% less reporter activity in HepG2 and Huh7 cells compared to constructs that had the protective allele in the 3′UTR. Further, HCV-induced miRNAs (hsa-mir-208b and hsa-mir-499) differentially bound to the region involving the SNP rs4803217 in the luciferase transcripts with IFNL3 3′UTR. This led to loss of degradation of mRNA by miRNA-induced silencing complex (miRISC) preferentially from transcripts originating from constructs that had the protective allele at rs4803217 (Fig. 3C). This finding highlights the role of 2 HCV-induced miRNAs in HCV pathogenesis. Along with the previously known miR-122, these HCV-induced miRNAs could be exploited for future therapeutic purposes (McFarland and others 2013).
All 4 causal SNPs predicted by di Lulio and others (2011) (Fig. 2A, B) have, therefore, been functionally analyzed and 2 of them have been demonstrated to be functionally relevant [rs28416813 in the promoter (Chinnaswamy and others 2013) and rs4803217 in the 3′UTR (McFarland and others 2013)] while the other 2 have not been found to have functional roles [rs4803219 in the promoter region (Sugiyama and others 2011) and rs8103142 in the coding region (Urban and others 2010; Sugiyama and others 2011; Friborg and others 2013)]. The 2 functional SNPs influence transcription and post-transcriptional events that may lead to increased IFNL3 expression and production of IFN-λ3 in patients carrying the protective alleles.
The effect of the 2 functional SNPs on expression of reporter genes in in vitro systems was not very high (average effect of inhibitory alleles on expression, less than 50%) (Chinnaswamy and others 2013; McFarland and others 2013). Presuming that these in vitro findings may also be true in vivo in HCV-infected patients, the modest effect of the functional SNPs on IFNL3 transcription may be significant because IFNs are known to function by feed-forward amplification loops (Balachandran and Beg 2011; Choubey and Moudgil 2011). For example, IRF-7 that is required for the transcription of IFNs is also an ISG that will further contribute to IFN expression in a loop (Levy and others 2002). NF-κB that is required for IFN induction is also induced by IFNs (Pfeffer 2011). Due to this phenomenon, modest changes in IFN expression can cause large downstream effects in ISG expression. Therefore, it is important to appreciate small differences in IFN-λ3 mRNA or protein levels in future clinical studies by having sufficiently powered sample sizes to avoid false-negatives in order to ascertain the effect of IFNL3 genotypes on IFN-λ3 expression. The 2 functional SNPs rs28416813 and rs4803217 are in strong LD (r2=0.99) with each other and with rs12979860 (r2=0.94) (di Lulio and others 2011). Interestingly, neither rs28416813 nor rs4803217 was arrayed on any of the GWAS chips that were used in the 4 studies conducted in 2009-10 (Balagopal and others 2010). rs28416813 was identified by fine-mapping by Ge and others (2009), by Tanaka and others (2009), and by Rauch and others (2010); while rs4803217 was identified by Rauch and others (2010) by recombinant mapping. It is important to note here that Rauch and others (2010) and di Lulio and others (2011) had identified and predicted rs28416813 and rs4803217 as 2 of the 4 potential causal SNPs for spontaneous clearance of HCV by using solely genetic methods (Fig. 2). Hence, the discovery of rs28416813 and rs4803217as 2 functional SNPs associated with HCV infections is a vindication of the value of well-performed genetic tests in human complex disease research. The in vivo significance of transcriptional regulation by both rs28416813 and rs4803217 in HCV infections still needs to be determined, although their association with HCV infections is well validated (Suppiah and others 2009; Tanaka and others 2009; Rauch and others 2010; Chen and others 2011; di Lulio and others 2011).
Among the studies that were initiated to identify the causal SNP(s), the study from the NIH which reported the discovery of IFN-λ4 has provided evidence to understand a major part of the puzzle (see next section). In this study by Prokunina-Olsson and others (2013), RNA sequencing was performed from primary human hepatocyte cultures after stimulating with polyI:C (a dsRNA mimic) to identify the regions in the genome that are stimulated for expression in response to a dsRNA mimic. Surprisingly, it was seen that a transcript was consistently produced which mapped to a novel gene near the promoter region of IFNL3 (Fig. 2). This novel protein resembled IFN-λ3 but was smaller in size and due to its similarities in structure and function to IFN-λ3, it was christened by human genome organization (HUGO) as IFN-λ4. Several other smaller transcripts were also produced from the same transcription start site but had premature stop codons and some of the transcripts encoded proteins that did not align with any other known proteins using BLAST. Importantly, a simple nucleotide polymorphism (a single nucleotide deletion, dbSNP) ss469415590 (ΔG) was responsible for a frame-shift that gave rise to the open reading frame for the 179-amino-acid IFN-λ4 protein (which is 29% similar to IFN-λ3 protein at amino-acid level) and this variant was present in a higher frequency in nonresponders than in responders to SOC therapy. The association seen with this SNP with response to treatment is stronger than that seen with rs12979860 (Kramer and others 2013; Prokunina-Olsson and others 2013). Interestingly, the authors noted that IFN-λ4 was not secreted in to the media, but showed the expression of similar ISGs to IFN-λ3 and IFN-α when overexpressed within the cells and was able to suppress a subgenomic HCV replicon in cells. Moreover, cells that were transfected with an IFN-λ4 construct did not respond to IFN-λ3 and IFN-α by further up-regulation of their ISGs. The study found that IFN-λ4 does not have sequence similarity to IFN-λ3 at the region that binds to one of the receptors (IL10R2, Fig. 1). However, a recent report found that IFN-λ4 signals by using the same receptors as IFN-λ3 and is equally potent as the latter in in vitro study systems (Hamming and others 2013); therefore, the mechanism of in vivo action of IFN-λ4 is not clear. The TT/-G polymorphism giving rise to IFN-λ4 was also shown by Bibert and others (2013) to be associated with response to treatment in HCV-infected patients.
Biological Basis of a Differential Response to IFN Treatment Among Patients with Chronic HCV Infections
The biological basis for a subset of the individuals not responding to SOC therapy remained a conundrum thus far, but recent studies are showing great promise in reaching a clearer understanding about this phenomenon (Clark and Thompson 2012; Hayes and others 2012; Horner and Gale 2013). However, several questions still remain as explained next. A high level of ISG induction and a low level of basal (pretreatment) viremia have been consistently seen in patients who will eventually not respond to the SOC therapy as reported by many studies (Chen and others 2005; Sarasin-Filipowicz and others 2008; Chen and others 2010; Honda and others 2010; Urban and others 2010; Dill and others 2011; Onomoto and others 2011). This observation, although seemingly counterintuitive, suggests that a saturation of the IFN response pathways by endogenously expressed IFNs in some patients leads to no further induction of ISG stimulation by exogenously provided IFN and, therefore, these patients have a higher chance of being nonresponders.
IFN-λs were discovered a decade ago (Kotenko and others 2003; Sheppard and others 2003) but were largely neglected for their role as antiviral cytokines in HCV research until the GWAS reports showed their significance in HCV infections even though IFN-λ3 was known to have anti-HCV activity in vitro (Marcello and others 2006). Since then, studies have started emerging that clearly demonstrate the anti-HCV properties of IFN-λs both in vitro (Diegelmann and others 2010; Friborg and others 2013; Shindo and others 2013) and in vivo (Muir and others 2010). In fact, IFN-λ1 is being developed as a promising alternative to IFN-α in anti-HCV therapy (Baugh and others 2013). IFN-α and IFN-λ have several similarities in their biological activities beginning from receptor-mediated signaling to stimulation of specific ISGs (Fig. 1) (Levy and others 2011; Horner and Gale 2013). Several of the important anti-HCV proteins such as OAS-1, ISG-15, MX-1, RNase-L, PKR, and others are induced by both these IFNs after stimulation by dsRNA (Marcello and others 2006; Kohli and others 2012). However, the kinetics of induction of ISGs differs between the 2 cytokines. IFN-λ is known to act slowly but leads to a steady increase in ISG induction, whereas IFN-α not only induces ISGs rapidly but also leads to their rapid decline (Marcello and others 2006; Kohli and others 2012; Shindo and others 2013). The less ubiquitous distribution of receptors for IFN-λ in humans may be the reason for lesser side-effects observed with IFN-λ1 anti-HCV therapy compared with IFN-α therapy (Muir and others 2010). The strategic locations of the different SNPs that have been identified to be associated with HCV infections suggest that the alternate alleles may be involved in the differential regulation of the transcription of some of the IFN-λ genes (Hayes and others 2012; Horner and Gale 2013). This is confirmed by recent reports which show that SNP rs28416813 due to its proximity to a nonconsensus NF-κB site, and SNP rs4803217 by interfering with mRNA stability, affect reporter gene expression in vitro and, therefore, can potentially affect IFNL3 gene expression in vivo (Chinnaswamy and others 2013; McFarland and others 2013).
The best possible explanation for the biological basis of the GWAS findings involving a differential response to IFN-α–based therapy is that there is an IL28B genotype-dependent difference in the level of expression of IFN-λs in potential responders versus nonresponders (Barth 2011; Horner and Gale 2013) (Note: Since several of the reports cited in this section use the old nomenclature for the IFN-λ genes, the same is used in this section of the article to assist readers interested in examining data from these references). The same would hold true to explain spontaneous clearance versus persistent HCV infection. However, clinical data seem conflicting in support of the explanation, especially when intra-hepatic expression of IFN-λs is taken into consideration. Urban and others (2010) did not see a statistically significant difference in IL28A/B mRNA levels between the different genotypes at rs12979860 in HCV-infected liver tissue. Similarly, Honda and others (2010) did not see a significant difference in IL28B expression (mRNA) between the genotypes at rs8099917 in HCV-infected liver biopsy samples. Asahina and others (2012) failed to observe a statistically significant difference in intra-hepatic expression of IL28A/B (mRNA) in patients with chronic HCV infection based on genotypes at rs8099917 and rs12979860. However, several previous and recent studies show that the IL28B genotypes/alleles are associated with IFN-λ expression levels in blood or ex vivo stimulated peripheral blood mononuclear cells (PBMCs) with the favorable alleles showing higher expression (Suppiah and others 2009; Tanaka and others 2009; Langhans and others 2011; Rallon and others 2012; Shi and others 2012; Bibert and others 2013; Murata and others 2013; Torres and others 2013). In support of this association, Dill and others (2011) show data from 92 patients where intra-hepatic mRNA levels of IL28B correlate with genotypes at rs12979860 with the C allele responsible for higher expression. In addition, Abe and others (2011) showed that the protective TT genotype at rs8099917 showed a significantly higher expression of IL28 mRNA in HCV-infected liver tissue but not in blood; while Fukuhara and others (2010) showed that the protective T allele was responsible for a decrease in intra-hepatic IL28B mRNA levels. Therefore, a consensus cannot be reached on the correlation between IL28B genotype and intra-hepatic expression of IL28B gene product. The reason for the discrepancies could be the difficulty to design primers to exclude the homologous IL28A transcripts from IL28B transcripts and the inherent heterogeneous nature of the hepatic cell population in liver biopsies. However, available data from several studies seems convincing in agreement with the association of IL28B genotype on its own expression in blood or ex vivo stimulated PBMCs (Suppiah and others 2009; Tanaka and others 2009; Langhans and others 2011; Rallon and others 2012; Shi and others 2012; Bibert and others 2013; Murata and others 2013; Torres and others 2013). However, there are a few exceptions to this generalization (Ge and others 2009; O'Connor and others 2013). Ge and others (2009) used the SNPExpress database and saw no correlation of IL28B genotypes (at rs12979860) with IL28B mRNA levels in PBMCs from 80 individuals without HCV infection. A recent report showed no association of rs12979860 with IL28B mRNA expression from unstimulated or IFN-α-stimulated PBMCs (O'Connor and others 2013) isolated from 20 healthy patients. The limitation of the latter reports was that they did not include HCV-infected patients (Ge and others 2009) nor were the isolated PBMCs from healthy patients stimulated with appropriate ligands that are known inducers of IL28B expression (O'Connor and others 2013) (IFN-α is reported to induce expression of IFN-λ by some but not others; Ank and others 2006; Ding and others 2012; Yin and others 2012). It is likely that experiments with PBMCs optimally stimulated for IL28B expression or from HCV-infected patients will be more informative on the influence of IL28B genotype on its expression. In line with this, Shi and others (2012) determined IFN-λ3 expression by measuring the protein levels instead of mRNA levels by using enzyme-linked immunosorbent assay (ELISA) from a large number of Chinese individuals. They saw that no differences indeed existed in IFN-λ3 protein levels in healthy and spontaneously cleared individuals, but the levels were significantly lower in persistently infected patients compared with the healthy and spontaneous clearance group (Shi and others 2012). When stratified by genotype at rs12979860 of 227 patients, the patients with the protective CC genotype had significantly higher expression of IFN-λ3 protein compared with the CT and TT genotypes (P=0.02). The difference in expression between the genotypes was more significant (P=0.013) within the spontaneously cleared group of patients, suggesting a role for IFN-λ3 protein in spontaneous clearance of HCV. The inconsistencies observed between the reports discussed earlier could be due to technical issues such as the need for specific primers and antibodies against IL28B gene and protein to exclude the homologous counterparts of IL28A; larger sample sizes to have sufficient power to avoid false negative results and appropriate conditions that will induce IL28B gene expression.
The association of IL28B genotypes and expression of ISGs in liver tissue or PBMCs with response to treatment is also not clear. Some studies reported that Il28B genotype does not independently associate with the expression of intra-hepatic ISGs (Dill and others 2011; McGilvray and others 2012) or ISGs in PBMCs (Naggie and others 2012) but is a covariate along with response to treatment. Other studies have showed that the intra-hepatic ISG levels correlate with IL28B gene polymorphisms in chronic HCV patients (Honda and others 2010; Abe and others 2011) and in one study, the correlation is independent of SVR status (Urban and others 2010). All of them show that the protective alleles are responsible for a lower intra-hepatic ISG expression (Honda and others 2010; Urban and others 2010; Abe and others 2011), although one study (Abe and others 2011) noted that ISG expression in PBMCs did not have significant differences between IL28B genotypes. Interestingly, a recent study shows that this trend is opposite in normal liver, where the protective IL28B alleles associate with higher levels of ISG induction (Raglow and others 2013) whereas a previous study on a large number of individuals with no HCV infection had not seen such a relationship (Shebl and others 2010). However, no conflict seems to exist at intra-hepatic levels of ISG expression when patients are stratified based on response to SOC therapy as several studies showed that potential nonresponders have high levels of ISG induction compared with responders who have significantly lower levels of ISG induction (Chen and others 2005; Sarasin-Filipowicz and others 2008; Chen and others 2010; Honda and others 2010; Urban and others 2010; Dill and others 2011; Onomoto and others 2011).
The conflicting results on the involvement of IL28B genotypes with the expression of both IL28B and intra-hepatic ISGs has led to speculation that IL28B polymorphisms may not have a significant role in expression or activity of IFN-λ3 (Park and others 2012; Booth and George 2013), which would, in turn, affect ISG expression. This was also prompted by a lack of association of type III IFN levels with the clearance of HCV at the acute stage of infection in chimpanzee models (Park and others 2012) and by reports that ruled out the involvement of the exonic SNP (rs8103142) in affecting the function of IFN-λ3 (Urban and others 2010). It was thought that some other mechanisms with or without the involvement of IL28B SNPs was functional in HCV infections. The argument gained credence with the discovery of IFN-λ4 (Prokunina-Olsson and others 2013). The study by Prokunina-Olsson and others (2013) concluded that more of nonresponders than responders expressed IFN-λ4 (due to a novel SNP located ∼3.2 kb from IL28B coding region) in response to chronic HCV infection and, hence, explained the observed differences of ISG expression between the 2 groups. However, the study also showed that rs12979860 is in high LD with the novel SNP that gives rise to IFN-λ4. Therefore, it fails to explain the apparent nonassociation of intra-hepatic ISG expression with the IL28B SNP rs12979860 reported by others (Dill and others 2011; McGilvray and others 2012), even though the discrepancies could be due to varying LDs at the IFN-λ locus between populations.
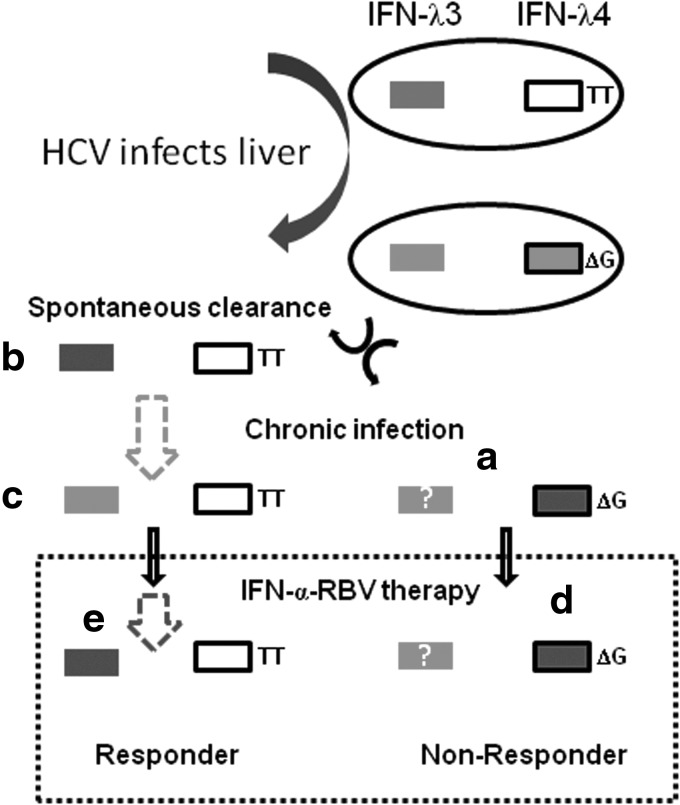
The discovery of IFN-λ4 is a big stride in improving our understanding of the conundrum involving differential treatment response in chronic HCV infections, but some questions remain and the puzzle is only partially complete (Fig. 4). First, the question remains: How do the nonresponder patients, having high levels of ISG induction, fail to reduce viral replication? It is possible that the HCV uses certain mechanisms such as shielding its replication sites within the cytoplasm from the action of any of the direct-acting ISG products by concentrating their replication complexes in lipid-rich membranes called “membranous webs” (Romero-Brey and others 2013). These lipid-rich webs may provide protection from direct-acting anti-viral proteins generated by the IFN response. Furthermore, if expression of IFN-λ4 can explain most of the variance with expression of ISGs in the liver, it is likely that IFN-λ4 is a weak IFN in that it does not express certain critical ISGs needed to inhibit HCV replication. However, a recent study has examined the functioning of a recombinant IFN-λ4 in in vitro cell culture and found no differences in the mode of signaling of IFN-λ4 and IFN-λ3 even though the former is not secreted out of the cells as efficiently (Hamming and others 2013) but has similar anti-HCV potency as the latter. This needs to be validated in vivo and also it is unclear whether other IFN-λs are also expressed at higher levels (or lower levels) in intra-hepatic tissue with chronic HCV infection (Fig. 4). The puzzle is only partially complete, because if the favorable CC genotype at rs12979860 co-segregates with the non-IFN-λ4 generating TT genotype at ss469415590 due to high LD between them as shown by Prokunina-Olsson and others (2013) and confirmed by others (Bibert and others 2013), then which IFN-λ is responsible for clearing HCV during the acute stage in cases of spontaneous clearance? It is well validated that CC genotype at rs12979860 is responsible for spontaneous clearance of HCV at the acute stage (Rauch and others 2010; di Lulio and others 2011). In this context, the roles of the other IFN-λs, especially IFN-λ3 encoded by IL28B, remain unclear in HCV infections. The relevant questions that emerge are as follows:
1) Is there a role for IFN-λ3 during treatment response?
2) If there is a role, then what is the mechanism of inhibition of its expression in the responders group when infection takes a chronic course? (Fig. 4)
3) What is the role of IFN-λ3 in spontaneous clearance of HCV infection at the acute phase?
4) Are the causal SNPs in the 2 cases (spontaneous clearance and response to treatment) same or different?
FIG. 4.
Transcription activity at the IFNL3 locus determines the natural progression and response to treatment in HCV infections. The figure is a summary and projection of the likely scenario in HCV infection with regard to transcription of IFN-λ3 and IFN-λ4 coding genes. Two groups of people are represented by 2 ellipses showing the transcription activity at IFNL3 locus. IFNL3 is shown as an open rectangle, while IFNL4 is depicted as a rectangle with a black border (a white box represents absence of IFNL4 gene due to absence of the deletion mutant ΔG at ss469415590 that will give rise to IFN-λ4 while TT at ss469415590 will give rise to the IFNL4 gene). The polymorphisms relevant to IFNL3 like rs28416813 are presumed to be in high LD with the IFN-λ4-generating SNP ss469415590. When HCV infects these 2 groups of people: (a) the nonprotective alleles that will have lower levels of expression of IFN-λ3 combined with a simultaneous expression of IFN-λ4 that will saturate the interferon-responsive pathways cannot clear the virus spontaneously (Shi and others 2012; Aka and others 2013; Prokunina-Olsson and others 2013) and go on to have chronic infection; (b) the protective alleles that will increase the expression of IFN-λ3 (due to LD the protective allele TT at ss469415590 will be present in this group) will clear the HCV (Shi and others 2012); (c) unknown mechanisms operate that will progress some patients with the protective alleles to have a chronic course of disease (dashed arrow between b and c). The mechanisms that will suppress IFN-λ3 expression (Rallon and others 2012) in these patients are unknown; (d) when the chronic HCV-infected patients are subject to therapy with IFN-α-ribavirin the patients that carry nonprotective alleles/genotypes will have no or lower levels of IFN-λ3 and high levels of IFN-λ4, a feature characteristic of nonresponse; (e) whereas the patients with the protective alleles/genotypes will show an increase in IFN-λ3 expression (dashed arrow between c and e) (Rallon and others 2012) and will help the patients eliminate the virus. The question marks are used when experimental data for gene expression are not available or clear. All shades in the rectangles within the two ellipses (white, light gray, and dark gray) denote very low or no gene expression. From (a) to (e): White shade in rectangle denotes “no” expression, light gray denotes “no” or “low” expression and dark gray denotes “high” expression.
The answer to the first question comes from a recent report which shows that ss469415590 is associated with differential expression of IL28B (Bibert and others 2013). The polyI:C stimulated PBMCs from (IFN-λ4-generating) ΔG genotype patients show low levels of IL28B mRNA that corresponds to nonresponse to therapy, while the TT genotype shows a high IL28B mRNA expression that corresponds to response to therapy (SVR). This will suggest that ss469415590 even though it is responsible for generating IFN-λ4 may have a significant role in the transcription of IL28B gene either directly or indirectly by being in LD with a functional SNP in the IL28B promoter (such as rs28416813 and rs4803217). ss469415590 lies on a CpG island and it is likely that this SNP may affect epigenetic regulation of IFN-λ3 expression as proposed by Bibert and others (2013). This may also answer the second question, as epigenetic regulation may play a role in regulating expression of IFN-λ3 in potential responder patients. It is known that plasma IL28B protein levels increase in patients with chronic HCV infection when administered with exogenous IFN-α in an IL28B genotype-dependent manner (Rallon and others 2012). Rallon and others (2012) have showed significant increases in IFN-λ3 protein levels in serum in HCV-HIV co-infected patients undergoing SOC therapy from day 0 to 4th week of therapy. This difference was significant only in CC genotype carriers at rs12979860. Interestingly, the difference depended on the genotype at rs12979860 and not on whether the patients would eventually be responders or nonresponders to SOC therapy. In contrast to this study, a recent study did not see a difference in IL28A/B and IL29 protein levels in serum between responders and nonresponders either at baseline or at 12 weeks of therapy (Torres and others 2013). However, the study notes a trend of higher IL29 protein levels in the SVR group both at baseline and at 12 weeks of therapy but with no statistical significance, while rs8099917 was significantly associated with IL29 protein levels in the pooled samples. This study did not differentiate between IFN-λ3 and IFN-λ2 proteins as the antibody used in the ELISA was against IFN-λ2 that also cross-reacted with IFN-λ3, whereas the study by Rallon and others (2012) included a much larger number of patients (112 versus 45 in the study by Torres and others 2013) and used antibodies that were specific for IFN-λ3 with no significant cross-reactivity with the other 2 IFN-λs. Furthermore, measuring the IFN-λ3 levels before or around 4th week may be more informative than at 12th week due to higher virus titers being present before the 4th week of SOC therapy. It is known that potential responders who have eliminated the virus (from blood) at the 4th week (referred to as RVR or rapid virological response) are more likely to remain so at the 12th week and thereon (Thompson and others 2010); hence, optimal induction of IL28B expression by HCV may be lacking at later time points during SOC therapy.
The third question becomes even more relevant with the recent report which showed that TT genotype at ss469415590 is over-represented in patients who show spontaneous clearance of HCV (Aka and others 2013). So, if TT genotype that fails to produce IFN-λ4 is responsible for spontaneous clearance of HCV, then it is likely that other IFN-λs may have roles to play in eliminating HCV at the acute stage. Bibert and others 2013 showed that the ΔG genotype is responsible for lower expression of IFN-λ3 and IP-10 levels from ex vivo stimulated PBMCs of HCV-infected patients. Chinnaswamy and others (2013) and McFarland and others (2013) show that a SNP in the distal promoter region and another SNP in the 3′UTR of IL28B can affect expression of the downstream gene although by different mechanisms and that the protective alleles at both SNP sites are responsible for higher levels of gene expression. Although the data are from in vitro assays, it has given convincing evidence to extrapolate and understand the role of transcriptional regulation by the functional SNPs at the IL28B locus in human patients. The LD status of rs28416813, rs4803217, and ss469415590 is not known. Data so far point to the role of IFN-λ3 in spontaneous clearance of HCV (Shi and others 2012; Horner and Gale 2013), although the roles of IFN-λ1 and IFN-λ2 remain to be determined. Langhans and others (2011) reported that IL29 (IFN-λ1) protein serum levels are lower in patients with a chronic HCV infection than those who show spontaneous clearance, although the levels of IL28A/B protein also showed a similar trend but was not statistically significant. The report by Shi and others (2012) from China confirms this finding where Il28B protein levels were higher in patients who cleared HCV spontaneously than those with chronic infection with their IL28B genotype significantly affecting expression levels. Murata and others (2013) show that ex vivo stimulated PBMCs from patients undergoing SOC therapy produce IFN-λ3 in an IL28B genotype-dependent manner (protective genotypes causing higher expression) that also correlates with the treatment-response in the respective patients. Yoshio and others (2013) showed more IL28B protein levels expressed from (BDAC3) (blood dendritic cell antigen 3) (+) dendritic cells derived from healthy subjects with the protective TT genotype at rs8099917 than from those with the TG genotype when infected with HCV in cell culture.
Finally, the answer to the fourth question will depend on a further dissection of in vivo mechanism(s) involving the causal SNPs at the IL28B locus that will help in completing the puzzle.
Toward a Complete Understanding of HCV Disease Biology
Even after identifying all the causal SNPs at the IFNL3 locus, we may be far from deciphering the host-virus interactions that can completely explain the phenotypic variance seen in natural progression and treatment response in HCV infections. Chronic HCV infection, similar to any other complex disease, will involve a set of major and minor genes interacting in a network that will contribute to the phenotype and, therefore, will have to deal with all the limitations and challenges of complex-trait genetic analysis such as pleiotropy, epistasis, genetic and allelic heterogeneity, and others (Schork 1997; Stranger and others 2011). The genetic risk and the environmental risk together contribute to the observed phenotypic variance in a typical complex disease of humans. The common small-effect genes that contribute to a network of gene–gene and gene–environment interactions which lead to an anti-viral state detrimental to HCV infection and replication (Fig. 5) will remain difficult to identify. Due to stringent testing criteria involved in a hypothesis-free study such as GWAS, the small-effect genes (SNPs) involved in a complex network will generally fail to emerge as “GWAS-hits”; therefore, large sample sizes in case-control studies and alternate genetic approaches are needed to identify these small-effect genes. In research involving human complex disease genetics, the scope for experimentation is very limited due to ethical concerns, and the researcher has to make objective observations and draw conclusions from “experiments” that have already taken place in nature. To make confident observations of events that are very subtle but still true at the population level will require a large sample size. Formation of consortiums between different research groups both within and between different countries is a positive trend in this direction, where samples can be pooled to increase the power of the study but it should include appropriate measures to check for phenotypic heterogeneity. One distinct advantage with an infectious disease such as HCV infection is that the major phenotype (which in this case is presence of the virus in blood) is easy to test, quantitative, and highly reliable. This may be one reason for the vastly acknowledged success of GWAS in HCV infections. Therefore, in order to utilize this distinct advantage (which happens to be the major limitation in other human complex diseases such as cardiovascular diseases), it is important to continue with large-scale genetic studies in HCV infections. Apart from this approach, newer and creative strategies are required to identify small-effect genes. Prior knowledge of the biology involving the genes can be used to treat some genes more uniquely than others (thereby relaxing the stringency criteria) to define their association with phenotypes in large-scale genetic studies (Goldstein and others 2013) and subsequently rigorous functional studies can be used to ascertain their true positive nature. In other words, compromises in allowing for false-positive outcomes are needed to make sure that small-effect genes are not “lost” in association studies. In compensation, a combination of rigorous functional characterization of the identified genes and replication studies will filter in the true positives. Fortunately, in HCV research due to relentless efforts from researchers around the world, a lot of progress is made and continues to be made in defining the involvement of several host genes needed for HCV replication. However, the significance of these genes at the human patient level and the variation in their coding regions or in regions affecting their expression and their association with HCV infections needs to be evaluated in vivo. A recent study incorporated a large-scale candidate gene approach where 112 candidate genes were tested at the genome-wide level for SNPs in a consortium cohort for spontaneous clearance of HCV (Mosbruger and others 2010). The study identified SNPs in 4 gene regions (TNFSF18, HAVCR1, TANK, and IL18BP) to be significant at P<0.01 in 2 ethnic populations. More of such studies and their validation studies are required to identify the “other” genes in HCV infections. This information can be used to build networks that will have weighted contributions from each gene in the pathway (Chakravarti and others 2013; Goldstein and others 2013). The challenge is to identify those genes significantly contributing to the antiviral state, their interactions with each other and with the “major gene” IFNL3, and in combination their interactions with the environment, which in this case includes the pathogen itself (Fig. 5). Suppiah and others (2011) combined IFNL3 genotypes with HLA-C and KIR variation in their cohort and saw that the positive predictive value for failure to respond to treatment increased from 66% to 80%. By logistic regression analysis, they found that the 3 genes interacted physiologically in raising an anti-viral state against HCV. Neukam and others (2013) also found that along with variants at IFNL3, those at other genes such as TGF-β, AQP-2, and VLDLR were also independently associated with SVR in HIV-HCV (genotype 1 or 4) coinfected patients. These variants modified the predictive value of IFNL3 variants, suggesting a genetic interaction between them.
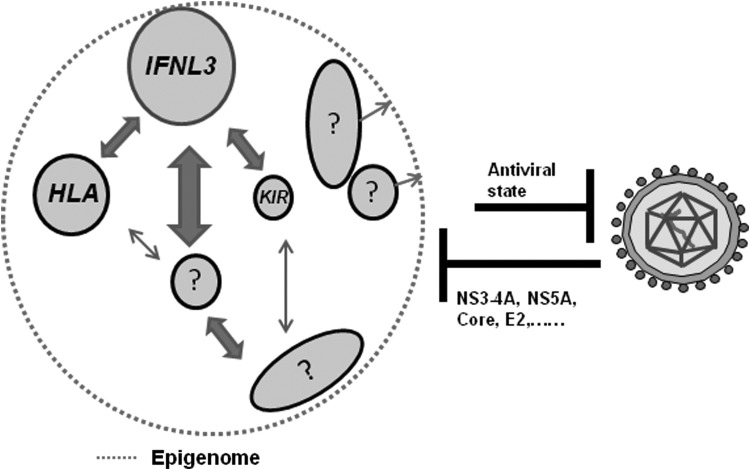
FIG. 5.
Host-virus-environment interactions determine the outcome of HCV infections. On the left is shown the immunome that is a subset of the larger host genome and on the right is shown HCV with its genome enclosed in a protein cover. The components of the immunome are shown as genes (circles) or pathways (ellipses). The size of the circle or ellipse is representative of the relative importance in the immunome. The arrows in between genes and pathways refer to genetic and physiological interactions between the components. The size and length of the arrows represent the relative importance and physical location on the genome respectively. The identified genes relevant to clearance of HCV infection are shown (such as IFNL3, HLA, and KIR, Smith and others 2011) and other yet unidentified or not validated ones are shown with question marks. The epigenome shown as a dotted line denotes the pervading influence of epigenetic (mostly transcriptional) regulation by means of alterations in the chromatin state throughout the immunome. The genes and pathways that influence (arrows pointing toward dotted line) the epigenome relevant to “HCV immunome” are not known, hence shown with question marks. The combined effect of the immunome (including innate and adaptive components) will lead to an antiviral state that will inhibit HCV replication and eliminate it from the host. On the other hand, HCV can influence the host immunome, including the epigenome. The viral factors that are known to affect the immunome (such as NS3-4A cleaving MAVS and TRIF and inhibiting innate immune signaling, Lemon 2010; core affecting DNA methylation, Lee and others 2013) are shown.
Epigenetic regulation can inflate or deflate the effect of the host genetic variation, therefore posing a limit to attaining a complete understanding of the genetic risk involved in the phenotypic variance. Environmental effects are an important consideration in the biology of complex diseases. These effects are usually heterogeneous in the population and significantly contribute to the observed variance in the phenotypes. While some environmental agents can directly interact with human DNA and change the genetic sequence leading to the phenotype (eg, nicotine in lung cancer), others could affect the epigenetic regulation of host gene expression (eg, alcohol in alcoholic liver disease, Osna 2011). Low estimates of heritability and sibling relative risks have been noted for both spontaneous and treatment-induced clearance of HCV, suggesting a major role for an “environmental effect” in HCV infections (Fried and others 2006). In HCV infections, it is important to consider the virus to be a part of the environment that interacts with the host. Although it can be argued that the virus is a common factor affecting all the individuals and hence should not contribute to the variance, one should not forget the interaction of the virus with the environment and its differential outcome. For instance, a favorable interaction of the virus with the host micro-environment (which is a reflection of the host macro-environment such as nutrient intake or alcohol consumption) can significantly change the latter and lead to an outcome different than the one expected from an unfavorable virus and a host micro-environment interaction. In other words, the “virus-effect” can significantly change/increase the otherwise subtle “environmental effect.” This enhanced “environmental effect” will impinge on the host regulatory mechanisms (such as transcriptional regulation of host genes) and will expose the underlying host genetic variation in the form of explainable phenotypic variance. For example, about 15% of the variance observed in spontaneous clearance of HCV is explained by IFNL3 and HLA variants (Duggal and others 2013). If this exaggerated “environmental effect” in HCV infections is true, then eliminating the “virus-effect” from the overall “environmental effect” should have dramatic consequences on the observed phenotypic variance. In support of this argument, SVR rates have increased to as high as 70%–80% (from<10%) in difficult-to-treat genotype 1 HCV infections (Ahlenstiel and others 2012; Lange and others 2013) by including DAA protease inhibitors bocepravir and telaprevir in SOC protocols (Triple therapy) in recent years. This is suggestive of the substantial contribution of the virus in the overall “environmental effect” that contributes to phenotypic variance in HCV infections. If the virus did not contribute to the “environmental effect” substantially and if the “environmental effect” minus the “virus-effect” was significant enough to account for a large part of phenotypic variance, then merely decreasing the viral replication by DAAs would have only led to a transient virological response and high relapse rates and, therefore, no net change in SVR rates. Instead, we have seen large increases in SVRs with triple therapy (Lange and others 2013; Tapper and Afdhal 2013) in the last couple of years.
This argument is not in conflict with high response rates prevailing in easy-to-treat HCV genotypes such as genotype 2 and 3. These easy-to-treat genotypes of HCV may have a lesser contribution in the overall “environmental effect.” It is necessary to distinguish the “virus-effect” that will and will not contribute to phenotypic variance. For instance, genotype 3 HCV is known to cause steatosis (Tapper and Afdhal 2013) but steatosis status is not known to impact SVR in these infections (Asselah and others 2006). Therefore, in these easy-to-treat infections, the “environmental effect” may not alter the host features (such as expression of relevant genes) sufficiently so that the effect of the underlying host genetic variation on the phenotypic variation becomes apparent. In agreement with this, a lesser impact of IFNL3 polymorphisms in treatment response in genotype 2 and 3 HCV infections has been observed (Lange and Zeuzem 2011; Schreiber and others 2012). In addition, a weaker association of IFNL3 genotypes with virological responses (Holmes and others 2012) in triple therapy is also suggestive of a clear role for the virus in exposing the genetic variation of the host in response to therapy. A chronic infection by HCV is necessary to appreciate a stronger effect of IFNL3 variation in treatment response, as SVR rates from SOC therapy are less dependent on IFNL3 polymorphisms in acute HCV-HIV coinfections than in chronic HCV-HIV coinfections (Nattermann and others 2011). In addition, even though the achievement of RVR is dependent on IFNL3 polymorphisms, the subsequent achievement of SVR in these patients does not strongly depend on the variation at IFNL3 (Thompson and others 2010). On the contrary, IFNL3-effect is stronger on the achievement of SVR in patients who fail to achieve RVR.
These observations underlie a phenomenon that needs further attention—“HCV does not passively utilize the available micro-environment within the host but has an active role in remodeling this micro-environment, which is reflected as a dependence of the host on IFNL3 polymorphisms in countering the infection”. Possibly due to this active role, HCV elicits a stronger effect from IFNL3 polymorphisms in response to SOC (IFN-α and RBV) therapy only during the following conditions: (1) where HCV has replicated to higher levels in the human patients (ie, before initiation of therapy; Thompson and others 2010); (2) in therapy that does not include DAAs which directly interfere with its replication (Holmes and others 2012); or (3) when HCV was able to establish a chronic infection (Nattermann and others 2011). The viral genotype is, of course, a determining factor in this role of HCV in remodeling the host micro-environment: The difficult-to-treat genotypes (1 and 4) are likely to have bigger roles than the easy-to-treat genotypes (2 and 3) (presuming that infections with all HCV genotypes have similar, if not identical, innate immunity mechanisms and that IFNL3 is the “major gene” in the antiviral pathway). The “environmental effect” that includes the “virus-effect” is a highly dynamic rather than a static entity in HCV infections which can alter the genetic risk contribution to the phenotypic variance. In view of this, an additional requirement in the genetics of HCV infections apart from identifying all the genetic risk factors is to define the contribution of the virus to the environment. To summarize, in a chronic complex disease with the involvement of a pathogen, the combined effect of the environment and pathogen will exert pressure on the host to bring its genetic factors in to play. In HCV infections, the pressure appears to be on regulation of transcription and chromatin state at the IFNL3 locus (Bibert and others 2013; Chinnaswamy and others 2013; McFarland and others 2013). In this context, it is tempting to speculate that the HCV would use its resources to alter the chromatin state and, therefore, transcription at the IFNL3 locus at some point during the process of establishing a chronic infection. Further research is needed to confirm this speculation.
HCV could be interfering with the host at the genome, epigenome, transcriptome, proteome, or metabolome levels. Much progress is made in understanding how HCV proteins subvert host immune responses by directly interacting with several host proteins (Horner and Gale 2009; Horner and Gale 2013). HCV proteins are also known to down-regulate transcription of some host genes (Ke and Chen 2012; Matsui and others 2012). However, much remains to be known especially at the patient level if and how HCV would interfere with the host epigenome to regulate the host immunome transcription (Fig. 5) to make it conducive to establish a chronic infection. From the data available so far and as elaborated earlier, it seems that the host genetic variation alone would not explain a large portion of the observed variance in chronic HCV infections. For example, how will patients with protective alleles at the IFNL3 locus fail to generate an immune response at the acute stage while the same response will free them from infection after receiving treatment when the disease takes a chronic course? Therefore, virus-mediated epigenetic regulation of the immune response is crucial to understand chronic HCV infection. Identifying the genes and pathways that are critical in the epigenetic landscape of the HCV immunome can open up avenues for interfering in the host-virus interactions to tilt the scale toward eliminating the pathogen.
Conclusions
The genetic variation at the IFNL3 locus especially the recent discovery of the variation that gives rise to IFN-λ4 and its association with HCV infections has enabled us to gain a better understanding of the biological basis of response to treatment in chronic HCV infections. The finding of functional SNPs rs28416813 and rs4803217 that can potentially affect the expression of IFNL3 gene in vivo has further added to our understanding of how the levels of IFN-λs may be important in controlling HCV replication at the patient level. However, questions still remain on how the variation at this important innate immunity gene locus guides the progression of disease from acute stage to chronic infection. The role of epigenetic regulation at the IFNL3 locus may be a fertile area of research that can help us to a better understanding of the disease biology and host-virus interactions.
Acknowledgments
The author thanks Prof. Partha Majumder, Director, National Institute of Biomedical Genomics (NIBMG), Kalyani for support and encouragement. The author also thanks Dr. Tapojyoti Das for help with literature search. The funding for this work was provided by NIBMG, Kalyani.
Author Disclosure Statement
No competing financial interests exist.
References
- Abe H, Hayes CN, Ochi H, Maekawa T, Tsuge M, Miki D, Mtsui F, Hiraga N, Imamura M, Takahashi S, Kubo M, Nakamura Y, Chayama K. 2011. IL28 variation affects expression of interferon stimulated genes and peg-interferon and ribavirin therapy. J Hepatol 54:1094–1101 [DOI] [PubMed] [Google Scholar]
- Ahlenstiel G, Booth DR, George J. 2012. Will IL28B polymorphisms remain relevant to direct-acting antiviral treatment paradigms? Antiviral Res 17:1163–1170 [DOI] [PubMed] [Google Scholar]
- Aka P, Kuniholm MH, Pfeiffer RM, Wang AS, Tang W, Chen S, Astemborski J, Plankey M, Villacres MC, Paters MG, et al. 2013. Association of the IFNL4-ΔG allele with impaired spontaneous clearance of hepatitis C virus. J Infect Dis. [Epub ahead of print]; DOI: 10.1093/infdis/jit433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhouri N, Zein NN. 2012. Protease inhibitors: silver bullets for hepatitis C virus infection? Cleveland Clin J Med 79:213–222 [DOI] [PubMed] [Google Scholar]
- Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. 2006. Lambda interferon (IFN-lambda), a type III IFN is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol 80:4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina Y, Tsuchiya K, Muraoka M, Tanaka K, Suzuki Y, Tamaki N, Hoshioka Y, Yasui Y, Katoh T, Hosokawa T, et al. 2012. Association of gene expression involving innate immunity and genetic variation in interleukin 28B with antiviral response. Hepatology 55:20–29 [DOI] [PubMed] [Google Scholar]
- Asselah T, De Muynck S, Broet P, Masliah-Planchon J, Blanluet M, Bieche I, Lapalus M, Martinet-Peignoux M, Lada O, Estrabaud E, Zhang Q, El Ray A, Vidaud D, Ripault MP, Boyer N, Bedossa P, Valla D, Vidaud M, Marcellin P. 2012. IL28B polymorphism is associated with treatment response in patients with genotype 4 chronic hepatitis C. J Hepatol 56:527–532 [DOI] [PubMed] [Google Scholar]
- Asselah T, Marcellin P. 2013. Interferon free therapy with direct acting antivirals for HCV. Liver Int 33 (Suppl. 1):93–104 [DOI] [PubMed] [Google Scholar]
- Asselah T, Rubbia-Brandt L, Marcellin P, Negro F. 2006. Steatosis in chronic hepatitis C: why does it really matter? Gut 55:123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Beg AA. 2011. Defining emerging roles of NF-κB in antivirus responses: revising the interferon-β enhanceosome paradigm. PLoS Pathogens 7(10):e1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal A, Thomas DL, Thio CL. 2010. IL28B and the control of hepatitis C virus infection. Gastroenterology 139:1865–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H. 2011. Insights in to the role of interferon lambda in hepatitis C virus infection. J Hepatol 54:844–847 [DOI] [PubMed] [Google Scholar]
- Baugh JM, Garcia-Rivera JA, Gallay PA. 2013. Host-targeting agents in the treatment of hepatitis C: a beginning and an end? Antiviral Res 100(2):555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellecave P, Sarasin-Filipowicz M, Donze O, Kennel A, Goutternoire J, Meylan E, Terracciano L, Tschopp J, Sarrazin C, Berg T, Moradpour D, Heim MH. 2010. Cleavage of mitochondrial antiviral signaling protein in the liver of patients with chronic hepatitis C correlates with a reduced activation of the endogenous interferon system. Hepatology 5:1127–1136 [DOI] [PubMed] [Google Scholar]
- Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, Duong FHT, Gerlach T, Malinverni R, Moradpour D, Negro F, Mullhaupt B, Bochud P-Y; the Swiss hepatitis C coort study. 2013. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med 210: 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitetto D, Fattovich G, Fabris C, Ceriani E, Falleti E, Fornasiere E, Pasino M, Ieluzzi D, Cussigh A, Cmet S, Pirisi M, Toniutto P. 2011. Complementary role of vitamin D deficiency and the interleukin-IL28B rs12979860C/T polymorphism in predicting antiviral response in chronic hepatitis C. Hepatology 53:1118–1126 [DOI] [PubMed] [Google Scholar]
- Booth D, George J. 2013. Loss of function of the new IFN-λ4 may confer protection from hepatitis C. Nat Genet 45:119–120 [DOI] [PubMed] [Google Scholar]
- Bucci C, Delft AV, Christian A, Flemming VM, Harrison A, Halliday J, Collier J, Manganis C, Klenerman P, Irving W, Barnes E. 2013. “Favorable” IL28B polymorphisms are associated with a marked increase in baseline viral load inhepatitis C virus subtype 3a infection and do not predict a sustained virological response after 24 weeks of therapy. J Gen Virol 94:1259–1265 [DOI] [PubMed] [Google Scholar]
- Bush WS, Moore JH. 2012. Genome-Wide Association Studies. PLoS Comput Biol 8(12):e1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcante LN, Abe-Sandes K, Angelo ALD, Machado TMB, Lemaire DC, Mendes CMC, Pinho JR, Malta F, Lyra LGC, Lyra AC. 2013. IL28B polymorphisms are markers of therapy response and are influenced by genetic ancestry in chronic hepatitis C patients from an admixed population. Liver Int 32(3):476–486 [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Clark AG, Mootha VK. 2013. Distilling pathophysiology from complex disease genetics. Cell 155:21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FE, Huang DB, Chen YQ, Ghosh G. 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 391:410–413 [DOI] [PubMed] [Google Scholar]
- Chen JY, Lin CY, Wang CM, Lin YT, Kuo SN, Shiu CF, Chang SW, Wu J, Sheen IS. 2011. IL28B genetic variations are associated with high sustained virological response (SVR) of interferon-α plus ribavirin therapy in Taiwanese chronic HCV infection. Gene Immun 12:300–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Borozan I, Feld J, Sun J, Tannis L-L, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. 2005. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology 128:1437–1444 [DOI] [PubMed] [Google Scholar]
- Chen L, Borozan I, Sun J, Guindi M, Fischer S, Feld J, Anand N, Heathcote J, Edwards AM, McGilvray ID. 2010. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology 138:1123–1133 [DOI] [PubMed] [Google Scholar]
- Chinnaswamy S, Chatterjee S, Boopathi R, Mukherjee S, Bhattacharjee S, Kundu TK. 2013. A single nucleotide polymorphism associated with hepatitis C virus infections located in the distal region of the IL28B promoter influences NF-κB-mediated gene transcription. PLoS One 8(10):e75495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D, Moudgil KD. 2011. Interferons in autoimmune and inflammatory diseases: regulation and roles. J Interferon Cytokine Res 31:857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB. 2010. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev 11:415–425 [DOI] [PubMed] [Google Scholar]
- Clark PJ, Thompson AJ. 2012. Host genomics and HCV treatment response. J Gastro Hep 27:212–222 [DOI] [PubMed] [Google Scholar]
- de Castellarnau M, Aparicio E, Parera M, Franco S, Tural C, Clotet B, Martinez MA. 2012. Deciphering the interleukin 28B variants that better predict response to pegylated interferon-a and ribavirin therapy in HCV/HIV-1 coinfected patients. PLoS One 7:e31016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. 2009. Human interferon-λ3 is a potent member of the type III interferon family. Genes Immun 10:125–131 [DOI] [PubMed] [Google Scholar]
- Diegelmann J, Beigel F, Zitzmann K, Kaul A, Goke B, Auernhammer CJ, Bartenschlager R, Diepolder HM, Brand S. 2010. Comparative analysis of the lambda-interferons IL-28A and IL-29 regarding their transcriptome and their antiviral properties against hepatitis C virus. PLoS One 5(12):e15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Lulio J, Ciuffi A, Fitzmaurice K, Kelleher D, Rotger M, Fellay J, Martinez R, Pulit S, Furrer H, Gunthard HF, Battegay M, Bernasconi E, Schmid P, Hirschel B, Barnes E, Klenerman P, Telenti A, Rauch A; Swiss HIV cohort study. 2011. Estimating the net contribution of interleukin-28B variationto spontaneous hepatitis C virus clearance. Hepatology 53:1446–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill MT, Duong FHT, Vogt JE, Bibert S, Bochud P-Y, Terracciano L, Papassotiropoulos A, Roth V, Heim MH. 2011. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology 140:1021–1031 [DOI] [PubMed] [Google Scholar]
- Ding Q, Huang B, Lu J, Liu Y-J, Zhong J. 2012. Hepatitis C virus NS3/4A protease blocks IL-28 production. Eur J Immunol 42:2374–2382 [DOI] [PubMed] [Google Scholar]
- Donnelly RP, Kotenko SV. 2010. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res 30:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, Yao L, Liu H, Barahmand-Pour F, Sivakumar P, Chan C, Birks C, Foster D, Clegg CH, Wietzke-Braun P, Mihm S, Klucher KM. 2006. Interleukin-29 uses a type 1 interferonlike program to promote antiviral responses in human hepatocytes. Hepatology 44(4):896–906 [DOI] [PubMed] [Google Scholar]
- Dube JB, Hegele RA. 2013. Genetics 100 for cardiologists: basics of genome-wide association studies. Can J Cardiol 29:10–17 [DOI] [PubMed] [Google Scholar]
- Duggal P, Thio CL, Wojcik GL, Goedert JJ, Mangia A, Latanich R, Kim AY, Lauer GM, Chung RT, Peters MG, et al. 2013. Genome-wide association study of spontaneous resolution of hepatitis C virus infection. Ann Intern Med 158:235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friborg J, Levine S, Chen CQ, Sheaffer AK, Chaniewski S, Voss S, Lemm JA, McPhee F. 2013. Combinations of lambda interferon with direct-acting antiviral agents are highly efficient in suppressing hepatitis C virus replication. Antimicrob Agents Chemo 57:1312–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried MW. 2004. Viral factors affecting the outcome of therapy for chronic hepatitis C. Rev Gastroentrol Disord 4:S8–S13 [PubMed] [Google Scholar]
- Fried MW, Kroner BL, Preiss LR, Wilhelmsen K, Goedert JJ. And the multicenter hemophilia cohort study. 2006. Hemophillic siblings with chronic hepatitis C: familial aggregation of spontaneous and treatment-related viral clearance. Gastroenterology 131:757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, Haussinger D, Diago M, Carosi G, Dhumeax D, Craxi A, Lin A, Hoffman J, Yu J. 2002. Peginterferon alpha-2a plus ribavirin for chronic hepatitis C virus infection. N Eng J Med 347:975–982 [DOI] [PubMed] [Google Scholar]
- Fukuhara T, Taketomi A, Motomura T, Okano S, Ninomiya A, Abe T, Uchiyama H, Soejima Y, Shirabe K, Matsuura Y, Maehara Y. 2010. Variants in IL28B in liver recepients and donorscorrelate with response to peg-interferon and ribavirin therapy for recurrent hepatitis C. Gastroenterology 139:1577–1585 [DOI] [PubMed] [Google Scholar]
- Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. 2009. Interferon-λ is functionally an interferon but structurally related to the interleukin-10 family. J Biol Chem 284:20869–20875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad HH, Hamming OJ, Hartmann R. 2010. The structure of human interferon lambda and what it has taught us. J Interferon Cytokine Res 30:565–571 [DOI] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment induced viral clearance. Nature 461:399–401 [DOI] [PubMed] [Google Scholar]
- Goldstein DB. 2009. Common genetic variation and human traits. N Eng J Med 360:1696–1698 [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Allen A, Keebler J, Margulies EH, Petrou S, Petrovski S, Sunyaev S. 2013. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet 14:460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming OJ, Terczynsca-Dyla E, Vieyres G, Dijkman R, Jorgensen SE, Akhtar H, Siupka P, Pietschmann T, Thiel V, Hartmann R. 2013. Interferon lambda signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J 32: 3055–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CN, Imamura M, Aikata H, Chayama K. 2012. Genetics of IL28B and HCV-response to infection and treatment. Nat Rev Gastro Hep 9:406–417 [DOI] [PubMed] [Google Scholar]
- Holmes JA, Desmond PV, Thompson AJ. 2012. Does IL28B genotyping still have a role in the era of direct-acting antiviral therapy for chronic hepatitis C infection? J Viral Hepat 19:677–684 [DOI] [PubMed] [Google Scholar]
- Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizokoshi E, Sakai Y, Yamashita T, Nakamura M, Shirasaki T, Horimoto K, Tanaka Y, Tokunaga K, Mizokami M, Kaneko S; the Hokuriku liver study group. 2010. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology 139:499–509 [DOI] [PubMed] [Google Scholar]
- Horner SM, Gale M, Jr., 2009. Intracellular immune cascades and interferon defenses that controls hepatitis C virus. J Interferon Cytokine Res 29:489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Gale M, Jr., 2013. Regulation of hepatic immunity by hepatitis C virus. Nat Med. [Epub ahead of print]; DOI: 10.1038/nm.3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell C, Jeffers L, Hoofnagle JH. 2000. Hepatitis C in African Americans: summary of a workshop. Gastroenterology.119:1385–1396 [DOI] [PubMed] [Google Scholar]
- Howell CD, Gorden A, Ryan KA, Thompson AJ, Ibrahim C, Fried M, Afdhal NH, McHutchison JG, Shianna KV, Goldstein DB, Shuldiner AR, Mitchell BD. 2012. Single nucleotide polymorphism upstream of interleukin 28B associated with phase 1 and phase 2 of early viral kinetics in patients infected with HCV genotype 1. J Hepatol 56:557–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Jiang X, Nakamoto S, Nakamura M, Miyamura T, Wu S. 2013. Different effects of three interferons L on toll-like receptor-related gene expression in HepG2 cells. Cytokine 64:577–583 [DOI] [PubMed] [Google Scholar]
- Kelly C, Klenerman P, Barnes E. 2011Interferon lambdas: the next cytokine storm. Gut 60:1284–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke P-Y, Chen SS-L. 2012. Hepatitis C virus and cellular stress response: implications to molecular pathogenesis liver diseases. Viruses 4:2251–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Lohmann K, Ziegler A. 2012. The promise and limitations of genome-wide association studies. JAMA 308:1867–1868 [Google Scholar]
- Kohli A, Zhang X, Yang J, Russell RS, Donnelly RP, Sheikh F, Sherman A, Young H, Imamichi T, Lempicki RA, Masur H, Kottilil S. 2012. Distinct and overlapping genomic profiles and antiviral effects of Interferon lambda and alpha on HCV-infected and non infected hepatoma cells. J Viral Hepat 19:843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV. 2011. IFN-λs. Curr Opin Immunol 23:583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. 2003. IFN-λs mediate antiviral protection through a distincr class II cytokine receptor complex. Nat Immunol 4:69–77 [DOI] [PubMed] [Google Scholar]
- Kramer B, Nischalke HD, Boeseche C, Ingiliz P, Voigt E, Mauss S, Stellbrink H-J, Baumgarten A, Rockstroh JK, Spengler U, Nattermann J. 2013. Variation in IFNL4 genotype and response to interferon-based therapy of hepatitis C in HIV-positive patients with acute and chronic hepatitis C. AIDS. [Epub ahead of print]; DOI: 10.1097/01.aids.0000433234.78807.54 [DOI] [PubMed] [Google Scholar]
- Lange CM, Jacobson IR, Rice CM, Zeuzem S. 2013. Emerging therapies for the treatment of hepatitis C. EMBO Mol Med. [Epub ahead of print]; DOI 10.1002/emmm.201303131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange CM, Zeuzem S. 2011. IL28B single nucleotide polymorphisms in the treatment of hepatitis C. J Hepatol 55:692–701 [DOI] [PubMed] [Google Scholar]
- Langhans B, Kupfer B, Braunschweiger I, Arndt S, Schulte W, Nischalke HD, Nattermann J, Oldenburg J, Sauerbruch T, Spengler U. 2011. Interferon-lambda serum levels in hepatitis C. J Hepatol 54:859–865 [DOI] [PubMed] [Google Scholar]
- Lauer GM, Walker BD. 2001. Hepatitis C virus infection. N Eng J Med 345:41–52 [DOI] [PubMed] [Google Scholar]
- Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17:17–115 [DOI] [PubMed] [Google Scholar]
- Lee H, Woo YJ, Kim SS, Kim SH, Park BJ, Choi D, Jang KL. 2013. Hepatitis C virus core protein overcomes all-trans retinoic acid-induced cell growth arrest by inhibiting retinoic acid receptor-β2 expression via DNA methylation. Cance Lett 335:372–379 [DOI] [PubMed] [Google Scholar]
- Lemon SM. 2010. Induction and evasion of innate antiviral responses by hepatitis C virus. J Biol Chem 285:22741–22747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Marie I, Smith E, Prakash A. 2002. Enhancement and diversification of IFN induction by IRF-7 mediated positive feedback. J Interferon Cytokine Res 22:87–93 [DOI] [PubMed] [Google Scholar]
- Levy DE, Marie IJ, Durbin JE. 2011. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol 1:476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Liu X, Zhou Y, Su SB. 2009. Interferon-λs: the modulators of antivirus, antitumor, and immune responses. J Leuk Biol 86:23–32 [DOI] [PubMed] [Google Scholar]
- Lindh M, Lagging M, Arnholm B, Eilard A, Nilsson S, Norkrans G, Soderholm J, Wahlberg T, Wejstal R, Westin J, Hellstrand K. 2011a. IL28B polymorphisms determine early viral kinetics and treatment outcome in patients receiving peginterferon/ribavirin for chronic hepatitis C genotype 1. J Viral Hepat 18:e325–e331 [DOI] [PubMed] [Google Scholar]
- Lindh M, Lagging M, Farkkila M, Langeland N, Morch K, Nilsson S, Norkrans G, Pedersen G, Buhl MR, Westin J, Hellstrand K. 2011b. Interleuking 28B gene variation at rs12979860 determines early viral kinetics during treatment in patients carrying genotypes 2 and 3 of hepatitis C virus. J Infectious Dis 203:1748–1752 [DOI] [PubMed] [Google Scholar]
- Manry J, Laval G, Patin E, Fornarino S, Itan Y, Fumagalli M, Sironi M, Tichit M, Bouchier C, Casanova J-L, Barreiro LB, Quintana-Murci L. 2011. Evolutionary genetic dissection of human interferons. J Exp Med 208:2747–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. 2006. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 131:1887–1898 [DOI] [PubMed] [Google Scholar]
- Matsui C, Shoji I, Kaneda S, Sianipar IR, Deng L, Hotta H. 2012. Hepatitis C virus infection suppresses GLUT2 gene expression via downregulation of hepatocyte nuclear factor 1α. J Virol 86:12903–12911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA, Delker DA, Hagedorn CH, Carrington M, Gale M, Savan R. 2013. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol. [Epub ahead of print]; DOI: 10.1038/ni.2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGilvray I, Feld JJ, Chen L, Pattullo V, Guindi M, Fischer S, Borozan I, Xie G, Selzner N, Heathcote EJ, Siminovitch K. 2012. Hepatic cell-specific gene expression better predicts HCV treatment outcome than IL28B genotype. Gastroenterology 142:1122–1131 [DOI] [PubMed] [Google Scholar]
- Moghaddam A, Melum E, Reinton N, Ring-Larsen H, Verbaan H, Bjoro K, Dalgard O. 2011. IL28B genetic variation and treatment response in patients with hepatitis C virus genotype 3 infection. Hepatology 53:746–754 [DOI] [PubMed] [Google Scholar]
- Mosbruger TL, Duggal P, Goedert JJ, Kirk GD, Hoots WK, Tobler LH, Busch M, Peters MG, Rosen HR, Thomas DL, Thio CL. 2010. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. J Infec Dis 201:1371–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir AJ, Shiffinan ML, Zaman A, Yoffe B, de la Torre A, Flamm S, Gordon SC, Marotta P, Vierling JM, Lopez-Talavera JC, Byrnes-Blake K, Fontana D, Freeman J, Gray T, Hausman D, Hunder NN, Lawitz E. 2010. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology 52:822–832 [DOI] [PubMed] [Google Scholar]
- Murata K, Sugiyama M, Kimura T, Yoshio S, Kanto T, Kirikae I, Saito H, Aoki Y, Hiramine S, Matsui T, Ito K, Korenaga M, Imamura M, Masaki N, Mizokami M. 2013. Exvivo induction of IFN-λ3 by a TLR7 agonist determines response to peg-IFN/ribavirin therapy in chronic hepatitis C patients. J Gastroenterol [Epub ahead of print]; DOI 10.1007/s00535-013-0814-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naggie S, Osinusi A, Katsounas A, Lempicki R, Herrmann E, Thompson AJ, Clark PJ, Patel K, Muir AJ, McHutchison JG, et al. 2012. Dysregulation of innate immunity in hepatitis C virus genotype 1 IL28B-unfavorable genotype patients: impaired viral kinetics and therapeutic response. Hepatology 56:444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natterman J, Vogel M, Nischalke HD, Danta M, Mauss S, Stellbrink H-J, Baumgarten A, Mayr C, Bruno R, Tural C, et al. 2011. Genetic variation in IL28B and treatment-induced clearance of hepatitis C virus in HIV-positive patients with acute and chronic hepatitis C. J Infec Dis 203:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukam K, Caruz A, Rivero-Juarez A, Barreiro P, Merino D, Real LM, Herrero R, Camacho A, Soriano V, Di Lello FA, Macias J, Rivero A, Pineda JA. 2013. Variations at multiple genes improve interleukin 28b genotype predictive capacity for response to therapy against hepatitis C genotype 1 or 4 infection. AIDS. [Epub ahead of print]; DOI: 10.1097/01.aids.0000432459.36970.a9 [DOI] [PubMed] [Google Scholar]
- O'Connor KS, Ahlenstiel G, Suppiah V, Schibeci S, Ong A, Leung R, van der Poorten D, Douglas MW, Weltman MD, Stewart GJ, Liddle C, George J, Booth DR. 2013. IFNL3 mediates interaction between innate immune cells: implications for hepatitis C virus pathogenesis. Innate Immun. [Epub ahead of print]; DOI: 10.1177/1753425913503385 [DOI] [PubMed] [Google Scholar]
- Onomoto K, Morimoto S, Kawaguchi T, Toyoda H, Tanaka M, Kuroda M, Uno K, Kumada T, Matsuda F, Shimotohno K, Fujita T, Murakami Y. 2011Dysregulation of IFN system can lead to poor response to pegylated interferon and ribavirin therapy in chronic hepatitis C. PLoS One 6:e19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osna NA. 2011. Epigenetic regulation in alcoholic liver disease. World J Gastroenterol 17:2456–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. 2007. IFN regulatory family members differentially regulate the expression of type III IFN (IFN lambda) genes. J Immunol 179:3434–3442 [DOI] [PubMed] [Google Scholar]
- Park H, Serti E, Eke O, Muchmore B, Prokunina-Olsson L, Capone S, Folgori A, Rehermann B. 2012. IL29 is the dominant type III interferon produced by hepatocytes during acute hepatitis C virus infection. Hepatology 56:2060–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedergnana V, Abdel-Hameed M, Guergnon J, Mohsen A, Le Fouler L, Theodorou I, Mohamed MK, Fontanet A, Plancoulaine S, Abel L. 2012. Alalysis of IL28B variants in an Egyptian population defines the 20 kilobases minimal region involved in spontaneous clearance of HCV. PLoS One 7(6):e38578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer LM. 2011. The role of nuclear factor κB in the interferon response. J Interferon Cytokine Res 31:553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, et al. 2013. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 45:164–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raglow Z, Thoma-Perry C, Gilroy R, Wan Y-JY. 2013. IL28B genotype and the expression of ISGs in normal liver. Liver Int 33(7):991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallon NI, Soriano V, Naggie S, Restrepo C, McHutchison J, Vispo E, Benito JM. 2012. Impact of IL28B gene polymorphisms on interferon-l3 plasma levels during pegylated interferon-α/ribavirin therapy for chronic hepatitis C in patientscoinfected with HIV. J Antimicrob Chemother 67(5):1246–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Kutalik Z, Descombes P, Cai T, di Lulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, et al. 2010. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138:1338–1345 [DOI] [PubMed] [Google Scholar]
- Reynolds JM, Paciga SA, Sanders FA, Hyde CL, Loomis AK, Johnston GI. 2012. Sequence analysis of the IL28A/IL28B inverted gene duplication that contains polymorphisms associated with treatment response in hepatitis C patients. PLoS One 7:e29983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I, Merz A, Chiramel A, Lee J-Y, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Locker-Krijnse J, Barthensclager R. 2013. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8(12):e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto N, Nakagawa M, Tanaka Y, Sekine-Osajima Y, Ueyama M, Kurosaki M, Nishida N, Tamori A, Yuki N-S, Itsui Y, et al. 2011. Association of IL28B variants with response to pegylated-interferon alpha plus ribavirin combination therapy reveals intersubgenotypic differences between genotypes 2a and 2b. J Med Virol 83:871–878 [DOI] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terraciano L, Filipowicz W, Heim MH. 2008. Interferon signaling and treatment outcome in chronic hepatitis C. Pro Natl Acad Sci USA 105:7034–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin C, Susser S, Doehring A, Lange CM, Muller T, Schlecker C, Hermann E, Lotsch J, Berg T. 2011. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatology 54:415–421 [DOI] [PubMed] [Google Scholar]
- Schlecker C, Ultsch A, Geisslinger G, Lotsch J. 2012. The pharmacogenetic background of hepatitis C treatment. Mutat Res 751:36–48 [DOI] [PubMed] [Google Scholar]
- Schork NJ. 1997. Genetics of complex disease: approaches, problems and, solutions. Am J Respir Crit Care Med 156:s103–s109 [DOI] [PubMed] [Google Scholar]
- Schreiber J, Moreno C, Garcia BG, Louvet A, Trepo E, Henrion J, Thabut D, Mathurin P, Deltenre P. 2012. Meta-analysis: the impact of IL28B polymorphisms on rapid and sustained virological response in HCV-2 and -3 patients. Aliment Pharmacol Ther 36:353–362 [DOI] [PubMed] [Google Scholar]
- Selvarajah S, Tobler LH, Simmons G, Busch MP. 2010. Host genetic basis for HCV clearance: a role for blood collection centers. Curr Opin Hematol 17:550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto WK, Tsang OT, Liu K, Chan JM, Wong DK, Fung J, Lai CL, Yuen MF. 2013. Role of IL28B and inosine triphosphatase polymorphisms in the treatment of chronic hepatitis C virus genotype 6 infection. J Viral Hepat 20:470–477 [DOI] [PubMed] [Google Scholar]
- Shebl FM, Maeder D, Shao Y, Prokunina-Olsson L, Schadt EE, O'Brien TR. 2010. In the absence of HCV infection, interferon stimulated gene expression in liver is not associated with IL28B genotype. Gastroenterology 139:1422–1424 [DOI] [PubMed] [Google Scholar]
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. 2003. Nat Immunol 4:63–68 [DOI] [PubMed] [Google Scholar]
- Shindo H, Maekawa S, Komase K, Miura M, Kadokura M, Sueki R, Komatsu N, Shindo K, Amemiya F, Nakayama Y, Inoue T, Sakamoto M, Yamashita A, Moriishi K, Enomoto N. 2013. IL28B (IFN-λ3) and IFN-α synergistically inhibit HCV replication. J Viral Hepat 20:281–289 [DOI] [PubMed] [Google Scholar]
- Shi X, Pan Y, Wang M, Wang D, Li W, Jiang T, Zhang P, Chi X, Jiang Y, Gao Y, Zhong J, Sun B, Xu D, Jiang J, Niu J. 2012. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in chinese population. PLoS One 7(5):e37054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2013. Expanded classification of hepatitis C virus in to 7 genotypes and 67 subtypes: updated criteria and updated web resource. Hepatology. [Epub ahead of print]; DOI: 10.1002/hep.26744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KR, Suppiah V, O'Connor K, Berg T, Weltman M, Abate ML, Spengler U, Bassendine M, Matthews G, Irving WL, Powell E, Riordan S, Ahlenstiel G, Stewart GJ, Bahlo M, George J, Booth DR; Int. HCV genetics Consortium. 2011. Identification of improved Il28B SNPs and haplotypes for prediction of drug response in treatment of hepatitis C using massively parallel sequencing in a cross-sectional European cohort. Genome Med 3:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Stahl EA, Raj T. 2011. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics 187:367–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Tanaka Y, Wakita T, Nakanishi M, Mizokami M. 2011. Genetic variation of the IL-28B promoter affecting gene expression. PLoS One 6:e26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V, Gaudieri S, Armstrong NJ, O'Connor KS, Berg T, Weltman M, Abate ML, Spengler U, Bassendine M, Dore GJ, et al. ; (International Hepatitis C Genetics Consortium). 2011. IL28B, HLA-C, and KIR variants additively predict response to therapy in chronic hepatitis C virus infection in a european cohort: a cross-sectional study. PLoS Med 8:e1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, et al. 2009. IL28B is associated with response to chronic hepatitis C interferon-a and ribavirin therapy. Nat Genet 41:1100–1104 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, et al. 2009. Genome-wide association of IL28B with response to pegylated interferon-a and ribavirin therapy for chronic hepatitis C. Nat Genet 41:1105–1109 [DOI] [PubMed] [Google Scholar]
- Tapper EB, Afdhal NH. 2013. Is 3 the new 1: perspectives on virology, natural history and treatment for hepatitis C genotype 3. J Viral Hepat 20:669–677 [DOI] [PubMed] [Google Scholar]
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, Urban T, Afdhal NH, Jacobson IM, Esteban R, et al. 2010. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology 139:120–129 [DOI] [PubMed] [Google Scholar]
- Torres C, Brahm J, Venegas M. 2013. Lambda interferon serum levels in patients with chronic hepatitis C virus infection according to their response to therapy with pegylated interferon and ribavirin. J Interferon Cytokine Res. [Epub ahead of print]; DOI: 10.1089/jir.2013.0005 [DOI] [PubMed] [Google Scholar]
- Udalova IA, Richardson A, Denys A, Smith C, Ackerman H, Foxwell B, Kwiatkowski D. 2000. Functional consequences of a polymorphism affecting NF-κB p50-p50 binding to the TNF promoter region. Mol Cel Biol 20:9113–9119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV, McHutchison JG, Goldstein DB, Afdhal N. 2010. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology 52:1888–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, Lewis-Antes A, Amrute SB, Garrigues U, Doyle S, Donnelly RP, Kotenko SV, Fitzgerald-Bocarsly P. 2012. J Immunol 189:2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N, Ishida H, Hiramatsu N, Nagano H, Sugiyama M, Murata K, Fukuhara T, Matsuura Y, Hayashi N, Mizokami M, Takehara T. 2013. Human blood dendritic cell antigen 3 (BDCA3)(+) dendritic cells are a potent producer of interferon-lambda in response to hepatitis C virus. Hepatology 57:1705–1715 [DOI] [PubMed] [Google Scholar]
- Zeuzem S, Berg T, Moeller B, Hinrichsen H, Mauss S, Wedemeyer H, Sarrazin C, Hueppe D, Zehnter E, Manns MP. 2009. Expert opinion on the treatment of patients with chronic hepatitis C. J Viral Hepat 16:75–90 [DOI] [PMC free article] [PubMed] [Google Scholar]