Abstract
Several aspects of HIV-1 virulence and pathogenesis are mediated by the envelope protein gp41. Additionally, peptides derived from the gp41 ectodomain have been shown to induce chemotaxis in monocytes and neutrophils. Whereas this chemotactic activity has been reported, it is not known how these peptides could be produced under biological conditions. The heptad repeat 1 (HR1) region of gp41 is exposed to the extracellular environment and could therefore be susceptible to proteolytic processing into smaller peptides. Matriptase is a serine protease expressed at the surface of most epithelia, including the prostate and mucosal surfaces. Here, we present evidence that matriptase efficiently cleaves the HR1 portion of gp41 into a 22-residue chemotactic peptide MAT-1, the sequence of which is highly conserved across HIV-1 clades. We found that MAT-1 induced migration of primary neutrophils and monocytes, the latter of which act as a cellular reservoir of HIV during early stage infection. We then used formyl peptide receptor 1 (FPR1) and FPR2 inhibitors, along with HEK 293 cells, to demonstrate that MAT-1 can induce chemotaxis specifically using FPR2, a receptor found on the surface of monocytes, macrophages and neutrophils. These findings are the first to identify a proteolytic cleavage product of gp41 with chemotactic activity and highlight a potential role for matriptase in HIV-1 transmission and infection at epithelial surfaces and within tissue reservoirs of HIV-1.
Keywords: chemotaxis, formyl peptide receptor 2, gp41, matriptase
Introduction
The envelope of HIV-1 is coated in glycoproteins, gp120 and gp41, that enable the virus to bind, fuse and infect susceptible host cells. In addition to mediating viral fusion, gp41 has been implicated in immunosuppressive cytokine production and induction of apoptosis.1–3 Additionally, synthetic peptides derived from gp41 have been shown to be chemotactic agonists through the formyl peptide receptor FPR2.4–6 Despite exhibiting chemotactic properties that could affect transmission and virulence, no study has reported how these peptides could be naturally produced within the host.
The ectodomain of gp41 on the surface of the viral envelope consists predominantly of the helical heptad repeats 1 and 2 (HR1 and HR2), which are joined by a disulphide loop. HR1 is exposed to the extracellular environment, and a large number of antibodies are known to target the gp41 immunodominant domain located at the C-terminal region of HR1 at gp41 residues 68–102.7,8 This stretch of amino acids is unusually conserved and overlaps with areas of gp41 exhibiting activity other than membrane fusion.9,10 This region of gp41 is therefore of particular interest as a potential site for proteolytic digestion, which could produce peptides independent from the intact virion. These peptides may then induce chemotaxis of susceptible host cells for infection.
During infection, the majority of HIV-1 resides in host tissue reservoirs where the virus may replicate while evading antiviral immunity.11,12 One such reservoir is the prostate, where epithelial cells efficiently bind, and transfer HIV to target CCR5-expressing leucocytes.13,14 The prostate is also an important viral reservoir in terms of the dissemination of HIV, with semen being the most predominant vehicle of transmission. Within the prostate, a number of well-characterized serine proteases exist on the surface of epithelial cells or within prostatic fluid. This environment therefore represents an area where proteolytic maturation of immunomodulatory gp41 peptides is likely to occur. Synthetic HR1 peptides can be produced to represent a region of the gp41 ectodomain that would be exposed to the extracellular environment, including proteases found on the apical membranes of epithelial cells.15 In this study we determined whether an HR1-derived peptide could be cleaved by prostatic epithelial proteases into chemotaxis-inducing peptides that function through FPR2.
By examining overlapping peptides of gp41 HR1, we identified a seven-residue region of HR1 that was associated with chemotactic activity toward HL-60 neutrophils. We then evaluated the proteolytic activity of several epithelial proteases found in the prostate against an HR1 peptide using in vitro digestion assays. We determined that matriptase efficiently cleaves a region of gp41 at biochemically conserved matriptase-recognition sites located within the leucine zipper motif. A 22-residue peptide cleavage product, MAT-1, was identified and found to exhibit chemotactic activity in HL-60 neutrophils and primary monocytes. We then demonstrated that expression of the receptor FPR2 mediated the observed chemotaxis in response to MAT-1. This work demonstrates a potential mechanism by which chemotactic peptides can be produced within the host, during infection with HIV-1.
Materials and methods
Peptides and DNA constructs
The 15-mer peptide set corresponding to the HIV-1 subtype B (MN) envelope was obtained from the National Institutes of Health AIDS Research Reagent Program (Division of AIDS, NIAID, NIH). Synthetic peptides were generated using solid-phase synthesis. T21, PAP286 and MAT-1 were custom synthesized by Peptide 2.0, (Chantilly, VA). LL-37 and the peptide negative control RREQALLIYVA were synthesized by Bachem (Torrance, CA). Peptides were purified to > 99% purity by HPLC and verified by mass spectrometry. WRW4 and WKYMVm were purchased from Anaspec (Fremont, CA). N-Formylmethionyl-leucyl-phenylalanine (fMLF) was purchased from Sigma Aldrich (St Louis, MO). Cyclosporin H was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
The FPR2 gene was cloned from human monocyte cDNA using the primers FPR2_S: ATGGAAACCAACTTCTCCACTCCTCTGAAT and FPR2_AS: TCACATTGCCTGTAACTCAGTCTCTGCAGG. PCR amplicons were purified and inserted into the TOPO-TA cloning vector before being digested with EcoRI and subcloned into the mammalian expression vector pcDNA3 (Life Technologies, Carlsbad, CA). Sequences were confirmed by Sanger sequencing using FPR2 and pcDNA-specific primers FPR2_AS and T7, respectively.
Primary and immortalized cells
The HL-60 cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA) and was maintained in RPMI-1640 medium with penicillin, streptomycin, 10 mm HEPES and 20% fetal bovine serum (R20). For chemotaxis assays, HL-60 cells were differentiated into neutrophil-like cells as previously described.16,17 Briefly, cells were resuspended in R20 with 1·3% DMSO at 2·0 × 105 cells/ml and allowed to proliferate and differentiate into neutrophils for 6 days. Differentiation was verified morphologically using Diff-Quik stain (Dade Behring Inc., Newark, DE) with bright-field microscopy. Neutrophils were used within 3 days after differentiation. HEK 293 cells were purchased from ATCC and maintained in Dulbecco's modified Eagle's medium supplemented with penicillin, streptomycin, 10 mm HEPES and 10% fetal bovine serum. HEK 293 cells were transfected with either the pcDNA3/FPR2 construct described above or the pcDNA3 vector alone using the Effectene transfection system (Qiagen, Hilden, Germany). Stable FPR2-expressing cells were selected by passaging cells in the presence of 1·5 mg/ml G418 for 30 days. Expression of FPR2 was confirmed by RT-PCR, Western blot and flow cytometry as described below.
To obtain primary cells, blood was drawn from healthy donors following informed consent with approval from the Institutional Review Board at the University of Central Florida. Blood was collected in acid-citrate-dextrose and divided for isolation of monocytes or neutrophils. For monocyte purification by negative selection, gradient centrifugation with the RosetteSep™ Human Monocyte Enrichment Cocktail (Stem Cell Technologies, Grenoble, France) and Ficoll-Paque™ PLUS (GE Healthcare, Little Chalfont, UK) was performed following the manufacturers' instructions. This system uses tetrameric antibody complexes to achieve negative selection of monocytes. Briefly, 750 μl of antibody reagent was combined with 15 ml of whole blood and allowed to incubate for 20 min at room temperature. The blood was then diluted with an additional 15 ml PBS + 2% fetal bovine serum (FBS) and 1 mm EDTA and layered onto 15 ml of Ficoll-Paque PLUS gradient. Blood was centrifuged at 1200 g for 20 min at room temperature. Enriched cells were harvested and washed twice with PBS + 2% FBS. Monocyte purification by positive selection was carried out using cell-sorting methods. Monocytes were labelled with phycoerythrin-conjugated anti-CD14 antibody reagent (Becton Dickinson, Franklin Lakes, NJ) at 100 μl for 1·0 × 107 cells at 1·0 × 106 cells/ml peripheral blood mononuclear cells for 30 min at room temperature in Hanks' balanced salt solution (HBSS) with 1% FBS. CD14-positive monocytes were sorted (BD FACSJazz; Becton Dickinson) and maintained in RPMI-1640 with penicillin, streptomycin, 10 mm HEPES and 10% FBS before use in chemotaxis experiments. For primary neutrophil isolation, whole blood was treated with erythrocyte lysis buffer (150 mm NaCl, 10 mm NaHCO3 and 1·3 mm EDTA) at a 10 : 1 ratio of lysis buffer to blood. After 3–5 min, lysis buffer was immediately neutralized using an equal volume of HBSS. Cells were washed twice in HBSS and resuspended in HBSS with 1% FBS and then isolated by using forward scatter and side scatter (BD FACSJazz) to identify and sort neutrophils.
Chemotaxis assays
Chemotaxis was determined using a 48-well microchemotaxis chamber (Neuro Probe, Cabin John, MA). Peptides were diluted in chemotaxis medium (RPMI-1640 with penicillin/streptomycin, 10 mm HEPES and 1% BSA) and pre-warmed at 37° before being added to the lower compartment of the chemotaxis chamber. Polycarbonate filters were then positioned over the lower compartments before assembling the complete chamber. For HL-60 neutrophils, primary monocytes and HEK 293 cells, filters containing 5-μm pores were used. For primary neutrophils, filters with 3-μm pores were used. For monocyte chemotaxis, filters were purchased pre-coated with poly-vinyl-pyrollidone. For HEK 293 chemotaxis, filters were coated for 2 hr in 0·05% type-1 rat tail collagen (BD Biosciences, Bedford, MA) in RPMI-1640 with 70 mm HEPES. Before chemotaxis, cells were washed once in PBS, resuspended in chemotaxis media (5·0 × 105 cells/ml for HL-60 neutrophils and 1·0 × 106 cells/ml for primary monocytes, primary neutrophils and HEK 293 cells), and applied to the upper compartment of the chamber. The microchemotaxis chamber was then incubated for 1 hr (neutrophils and monocytes) or 5 hr (HEK 293 cells) at 37° and 5% CO2. Filters were scraped to remove cells that did not migrate and fixed in methanol for 1 min. Filters were stained using Diff-Quik (Dade Behring Inc.) and mounted to glass slides with coverslips for imaging. Cells from five high-powered fields (400 ×) were quantified using the fiji package of imagej software and averaged for each repeat. The chemotaxis index was calculated for each condition as the fold-increase in migrated cells versus media alone.
Analysis of FPR2 expression
FPR2-specific primers for real-time RT-PCR were designed using beacon design software version 7.5 (PREMIER Biosoft, Palo Alto, CA). The following primer pairs were used for RT-PCR, S: CACATTACCATTCCTCAT and AS: AACCAATCAAGAAGACAC for FPR2 and S: TGGTATCGTGGAAGGACTC and AS: AGTAGAGGCAGGGATGATG for glyceraldehye 3-phosphate dehydrogenase (GAPDH). HEK 293 cells transfected with pcDNA3, pcDNA3/FPR2, or untransfected cells were grown to confluence in six-well plates. After removal of media, cells were washed once with PBS and treated with 0·5 ml of TRIzol Reagent (Life Technologies) following the manufacturer's instructions. After isolation of RNA, cDNA was produced by incubation with reverse transcriptase using the iScript system (Bio-Rad, Hercules, CA). Each 20-μl reaction contained 1 μg of template RNA. Relative levels of cDNA were measured using the SYBR Green RT-PCR Reagents Kit (Bio-Rad). Briefly, FPR2 and GAPDH cDNA were amplified using one cycle of melting at 95° for 5 min followed by 40 cycles of amplification with annealing and extension at 55·7°. Melt curve analysis confirmed the absence of non-specific products and curve-fitted relative fluorescence unit values were baseline subtracted. Cycle thresholds were normalized to GAPDH and fold-expression was calculated using the ΔΔCT method.18
For confirmation of FPR2 protein expression, HEK 293 cells were grown to confluence in six-well plates and lysed to isolate membrane fractions as shown previously.19 Protein concentrations were determined using the DC Protein Assay (Bio-Rad) with a BSA standard curve. For each cell type, 15 μg of total protein was resolved on pre-cast 4–20% gradient polyacrylamide gels (Thermo-Fisher Pierce Protein Biology Products, Rockford, IL). Proteins were transferred to PVDF membranes and blotted for FPR2 using rabbit polyclonal anti-FPR2 (ab63022, Abcam, Cambridge, UK) following an established Western blot protocol.20 PVDF membranes were blotted using secondary goat anti-rabbit horseradish peroxidase-conjugated antibodies and developed using Protein Simple chemiluminescent substrate (GE Healthcare). Developed membranes were visualized with a Chemi-Doc imaging system (Bio-Rad)
FPR2 expression in HEK 293 cells was determined using a FACSJazz flow cytometer. Phycoerythrin-conjugated anti-FPR2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was incubated with cells for 30 min at room temperature in HBSS with 1% FBS. A 2-ml suspension of HEK 293 cells at 1·0 × 106 cells/ml was labelled following the manufacturer's instructions at 1 μg of antibody/1·0 × 106 cells. Analysis was performed using flowjo Version X software.
Identification of proteolytic cleavage products
Prostate-specific antigen (PSA)/KLK3 purified from human semen was obtained commercially (Sigma Aldrich). Recombinant prostasin was purified as described previously21 and matriptase was obtained commercially (R&D systems, Minneapolis, MN). All peptide digestions were prepared in Dulbecco's modified PBS (DPBS). T21 and PAP286 peptides were used at 50 μm concentrations. Prostasin and matriptase were used at final concentrations of 2 nm and 1 nm, respectively. Prostate-specific antigen was used at 50 nm as previously described.22 Digestions were carried out in 10-μl reactions in DPBS and incubated for 24 hr at 37°, shaking at 300 rpm. Digestion products were electrophoresed on Tricine-SDS 16% polyacrylamide gels for 3 hr at 70 V. The remaining digestion products were brought to 50% acetonitrile and 0·1% trifluoroacetic acid before mixing 1 : 1 with 10 mg/ml α-cyano-4-hydroxycinnamic acid matrix dissolved in 50% acetonitrile and 0·1% trifluoroacetic acid. Samples were analysed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) on a Microflex mass spectrometer (Bruker Daltonics, Billerica, MA) in positive reflectron mode. External calibration was performed using a peptide-range mass standards kit (Bruker Daltonics). Mass spectra results were analysed using flex analysis software (Bruker Daltonics).
Protein and DNA sequence analysis
The HIV and SIV Env sequences were obtained from the 2012 HIV Sequence Compendium (Los Alamos National Laboratory, Los Alamos, NM). Representative protein alignments were prepared using the MUSCLE multiple sequence alignment software in MEGA version 5.23 Shannon Entropy for Group M Subtype B sequences was calculated using the Entropy-One tool provided by the HIV Sequence Database (Los Alamos National Laboratory): http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy_one.html. DNA alignments were prepared from representative gp41 ectodomain sequences from diverse HIV clades. Maximum likelihood trees with bootstrapping were generated from alignments using MEGA version 5.
Statistical analysis
Chemotactic activity for each peptide in the initial HR1 screen was compared with the media control using Student's t-test. For experiments with multiple peptide concentrations, a two-way analysis of variance with Bonferroni post tests was used to compare every concentration of unknown or positive control to its corresponding equal concentration of negative control peptide. D'Agostino normality and Mann–Whitney U-tests were used to analyse non-parametric median Shannon Entropy data.
Results
Chemotactic activity of gp41 HR1 is localized to two distinct regions
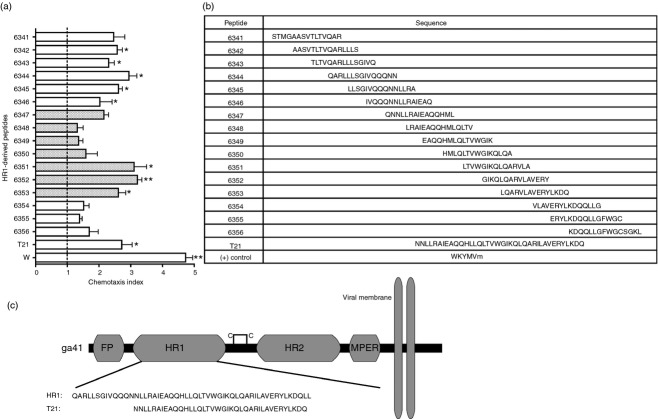
To determine the regions of gp41 HR1 associated with chemotaxis, overlapping 15-mer peptides derived from HIV LAI HR1 were individually tested for chemotactic activity in HL-60-derived neutrophils. These cells have been shown in earlier studies to express FPR2,24,25 a receptor previously implicated in the chemotactic activity of gp41,5,6 which we confirmed (not shown). Of the 16 HR1 peptides we tested, 6342–6346 and 6351–6353 were shown to significantly induce chemotaxis, exhibiting twofold to threefold greater activity than the vehicle control (Fig. 1a,b). The peptide WKYMVm was used as a positive control for HL-60 neutrophil chemotaxis through FPR2.26 The region of HR1 represented by peptides 6351–6353 overlaps with the synthetic peptide T21/DP-107 (Fig. 1c). T21 has been previously characterized as a chemotactic peptide for primary monocytes and neutrophils.6 Peptides 6351–6353 also share the LQARVLA sequence, which potentially imparts chemotactic activity.
Figure 1.
Chemotactic activity of gp41 heptad repeat 1 (HR1) is localized in two distinct regions. (a) Chemotactic activity of gp41-derived peptides was quantified to determine chemotactic regions of the gp41 HR1 helix. HL-60 neutrophil migration was assessed in response to 15 overlapping 15-mer peptides, T21, and the positive control WKYMVm. Shaded bars represent peptides that overlap with T21. Error bars represent SEM. Average cells/field from unknown and positive control conditions were compared with vehicle control to test for significant chemotaxis (n = 3, *P < 0·05, **P < 0·01). (b) Sequences for each peptide are listed in this table. (c) The regions of the gp41 ectodomain are shown with amino acid sequences of HR1 and T21 displayed beneath.
An HR1-derived peptide is specifically cleaved by matriptase to a lower molecular weight product
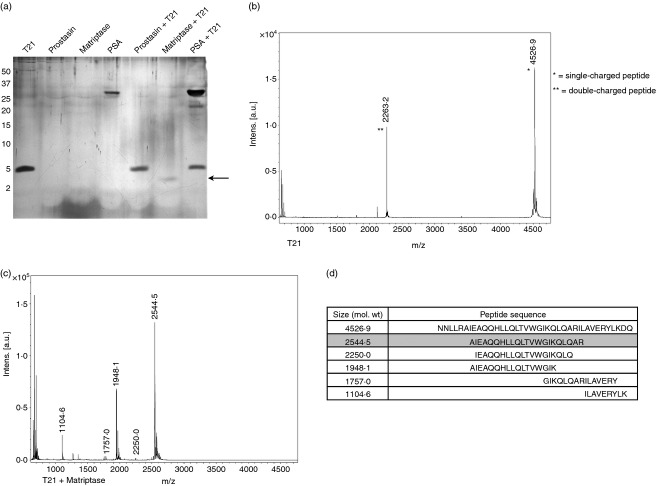
To determine if peptides corresponding to chemotactic regions of gp41 may be naturally produced via proteolytic processing, the HR1-derived T21 peptide was incubated with several prostate-derived proteases. Prostate-specific antigen (PSA), prostasin and matriptase were incubated with T21 and digestions were analysed for cleavage by SDS–PAGE. We verified that our commercially-obtained PSA maintained activity by incubating it with a PSA-sensitive peptide, PAP28620 derived from prostatic acid phosphatase (data not shown). While both PSA and prostasin failed to cleave T21, matriptase was found to effectively cleave T21 to a lower molecular weight product (Fig. 2a). A potential matriptase cleavage product was observed at approximately 2500 molecular weight (MW). Proteolytic digestion products were then identified using MALDI-TOF mass spectrometry. Peaks corresponding to the mass of T21 remained present after treatment with PSA and prostasin (data not shown). After matriptase digestion, the T21 peak (Fig. 2b) was completely absent and peaks corresponding to several cleavage products were present (Fig. 2c). Sequences of cleavage products were determined based on the mass of the digested parent peptide sequence (Fig. 2d). Based on the specificity of cleavage and peak intensity, we continued to pursue the peptide with mass 2544·5 (MAT-1). This peptide partially overlaps with the previously characterized chemotactic sequence from our peptide library, LQARVLA, while possessing LLR/AI and QAR/IL cleavage sites at the N and C termini, respectively. The LLR and QAR residues represent the matriptase P1–P3 protease recognition sites whereas the hydrophobic AI and IL residues represent the P1'–P2' sites. These residues are at positions consistent with substrates commonly recognized by matriptase.27
Figure 2.
A heptad repeat 1 (HR1) -derived peptide is specifically cleaved by matriptase to a lower molecular weight product. (a) T21 digestion reactions were resolved using SDS–PAGE and silver stained. T21 is visible as a ∼5000 molecular weight (MW) band while prostate-specific antigen is also visible as a ∼ 29 000 MW band. The arrow indicates the mass of the major matriptase cleavage product of T21. (b) Mass spectra in the peptide range (500–5000 MW) are shown for T21 alone and (c) matriptase + T21 conditions. Peak masses are labelled with mass [M] values reflecting the true mass of peptides. (d) The ExPASy FindPept tool was used to identify the sequence of peptides represented by mass peaks based on the parent T21 mass. MAT-1 is shown shaded in grey. Peaks corresponding to matriptase alone were neglected in the analysis.
Matriptase cleavage sites in gp41 are conserved across diverse HIV clades
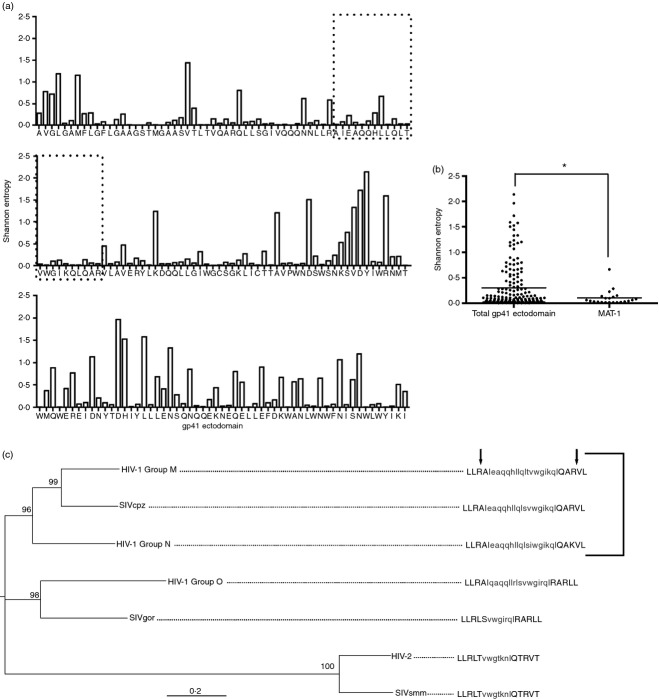
Due to the high genetic and amino acid variation across HIV, we next assessed whether diverse HIV isolates also contained matriptase recognition sites in T21. This information would then indicate whether matriptase cleavage is only relevant in a minority of strains, or if this is a conserved phenomenon across multiple clades of HIV. Using the most recent genetic data available from the HIV sequence compendium,28 we generated alignments of HIV-1 Group M subtype B containing 173 taxa. We then evaluated amino acid diversity across gp41 using Shannon Entropy (Fig. 3a), which determines the variation in sequence at each amino acid position in a given alignment. This method was implemented to measure amino acid diversity, due to overlapping coding regions within gp41 that would interfere with tests for nucleic acid diversity. Amino acids recognized by matriptase, as well as the MAT-1 peptide, were significantly conserved when compared with the entire gp41 subtype B ectodomain (Fig. 3b).
Figure 3.
Matriptase cleavage sites in gp41 are conserved across diverse HIV clades. HIV-1 Group M subtype B amino acid sequences from 173 taxa were aligned using MUSCLE. (a) Shannon Entropy was calculated for each amino acid spanning the gp41 ectodomain. Low values correspond to low variation and so more conserved residues. The area enclosed in dotted boxes represents the MAT-1 peptide. (b) Average Entropy values for each amino acid were plotted and the median values for MAT-1 and the entire gp41 ectodomain were compared (*P < 0·05). (c) DNA sequences from representative strains of diverse HIV and SIV clades were aligned with ClustalW. Maximum likelihood was used to generate a phylogeny. Nodal support is represented as bootstrap values. Consensus amino acid sequences were generated for each taxa and matriptase cut sites are shown for the N and C terminals of MAT-1 as indicated by arrows. The bracketed taxa represent those containing conserved matriptase recognition sites.
Alignments were generated using representative DNA sequences of the gp41 ectodomain from diverse HIV clades. The phylogeny was prepared using the maximum likelihood method to show relationships between clades (Fig. 3c). Matriptase recognition sites were found to be biochemically conserved in HIV-1 Group M, SIVcpz and HIV-1 Group N, with a substitution of arginine 68 of gp41 with lysine in Group N. This polymorphism would be unlikely to affect the proteolytic activity of matriptase because of the preference of matriptase for both arginine and lysine residues at the P1 site.27 These findings suggest that the majority of HIV-1 strains would be susceptible to matriptase cleavage, and that our observations are not confounded by infrequent polymorphisms in the matriptase-digested peptide.
The peptide MAT-1 induces chemotaxis in neutrophils and primary monocytes and acts through FPR2
To assess the chemotactic activity of the MAT-1 cleavage product, we synthesized MAT-1 and designed experiments in primary neutrophils and monocytes using a range of peptide concentrations. The FPR2 agonist WKYMVm was used as a positive control along with a negative control peptide, RREQALLIYVA, identified by scrambling the sequence within a chemotactic region of HR1. Neutrophils were sorted from whole blood using forward and side scatter. To control for unintended stimulation, monocytes were isolated by either positive or negative selection. Positively selected monocytes were sorted from donor-derived peripheral blood mononuclear cells based on CD14 expression, whereas negatively sorted cells were purified by gradient centrifugation with tetrameric antibodies targeting lymphocytes. Induction of chemotaxis by MAT-1 was determined to exhibit peak activity at a concentration of 100 nm, with an average chemotaxis index of 5·4 in neutrophils (Fig. 4a) and 2·0 and 4·4 for positively and negatively selected monocytes, respectively (Fig. 4b,c). A similar pattern of concentration-dependent activity was observed for WKYMVm. The negative peptide control exhibited no significant chemotactic activity. To assess the chemotactic activity of MAT-1 against a naturally occurring, known FPR2 agonist, equimolar concentrations cathelicidin LL-37 were compared. LL-37 has previously been shown to induce migration of several FPR2-expressing cells, including primary neutrophils and monocytes.29 At 100 nm, we observed that LL-37 and MAT-1 were equally chemotactic using primary neutrophils (Fig. 4d).
Figure 4.
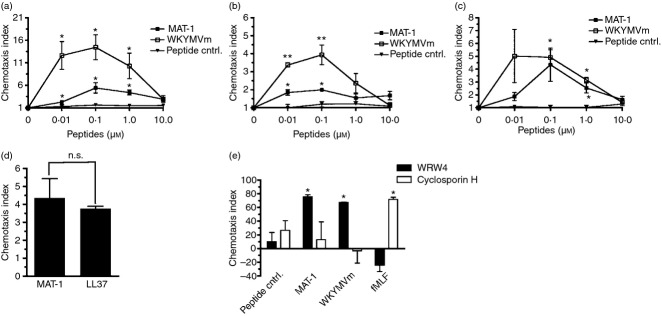
The peptide MAT-1 induces chemotaxis in primary neutrophils and monocytes. Chemotaxis was assessed using increasing concentrations of peptides in primary neutrophils (a) and monocytes were obtained by either positive selection (b) or negative selection (c). WKYMVm was used as a positive control for chemotaxis in both conditions. A negative peptide control was also used to compare with activity of MAT-1 and WKYMVm. The chemotaxis index is defined as fold change in average cells/field from media alone. Chemotaxis indices were compared at each concentration with a corresponding concentration of peptide control. Error bars represent SEM (n = 3, *P < 0·05, **P < 0·01). (d) The chemotactic activity of MAT-1 was compared with the known formyl peptide receptor 2 (FPR2) agonist LL-37 using primary neutrophils. Both MAT-1 and LL-37 were used at 100 nm concentrations (n.s. denotes no significant difference). (e) Chemotaxis inhibitors WRW4 and cyclosporin H were incubated for 15 min with neutrophils before determining chemotaxis in response to peptides. Chemotactic peptides were diluted to 100 nm for the control peptide, MAT-1, and WKYMVm, and 500 nm for fMLF. Per cent inhibition was calculated as reduction in chemotaxis index compared with pre-incubation with a vehicle control. Error bars represent SEM (n = 3, *P < 0·05).
Specificity for the receptors FPR1 and FPR2 was investigated using the peptide chemotaxis inhibitors cyclosporin H (CSH) and WRW4, respectively (Fig. 4e). Inhibitors were incubated with neutrophils for 15 min before chemotaxis using 50 μm for WRW4 and 10 μm for CSH. MAT-1, WKYMVm, and the negative peptide control were all used at 100 nm while the FPR1-specific peptide agonist fMLF was used at 500 nm. In vehicle pre-treatment conditions, chemotaxis was observed for MAT-1, WKYMVm and fMLF. No significant chemotaxis was observed using the peptide control. After pre-treatment with WRW4, chemotaxis was significantly inhibited for both MAT-1 and WKYMVm conditions. WRW4 had no significant effect on fMLF-induced neutrophil chemotaxis. Alternatively, CSH pre-treatment had no significant effect on chemotaxis stimulated by MAT-1 and WKYMVm, but significantly inhibited chemotaxis in response to fMLF. Taken together, these results indicate that FPR2, but not FPR1, is necessary for the chemotactic response to MAT-1 in neutrophils.
To determine if the matriptase cleavage product MAT-1 induces chemotaxis through FPR2 alone, we generated HEK 293 cells that stably express FPR2. FPR2 expression was determined by RT-PCR (not shown), and confirmed by Western analysis and flow cytometry (Fig. 5a,b). Although untransfected HEK 293 cells do not produce FPR2, the FPR2-transfected cells express high levels of FPR2 under the cytomegalovirus promoter. Functional FPR2 activity in FPR2-HEK 293 cells was initially assessed and compared to untransfected and pcDNA3 vector control cells (Fig. 5c). Furthermore, the MAT-1 peptide induced significant chemotaxis in FPR2-HEK 293 cells, with concentration-dependent activity (Fig. 5e), which was absent in the scrambled peptide control. We then confirmed specificity through FPR2 by testing MAT-1 against untransfected and pcDNA-transfected cells for chemotactic activity (Fig. 5d). These findings reveal that the matriptase cleavage product MAT-1 possesses chemotactic activity directed toward FPR2-expressing cells, including primary neutrophils and monocytes.
Figure 5.
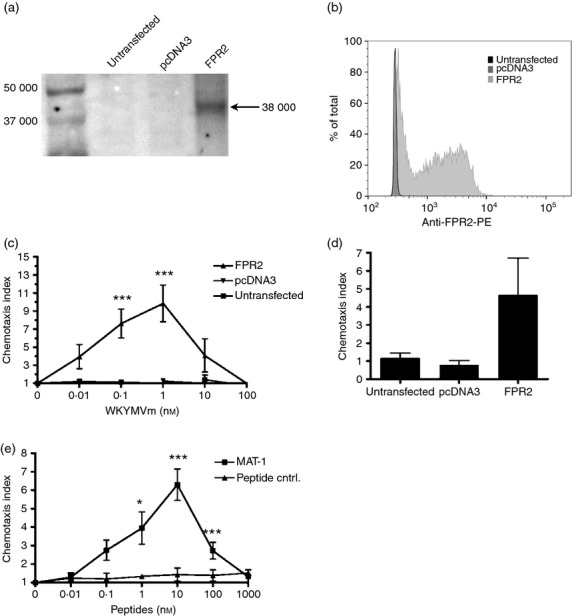
The peptide MAT-1 induces chemotaxis using the formyl peptide receptor 2 (FPR2) receptor. (a) Western blot using anti-FPR2 antibody with different HEK 293 cells shows FPR2 as a 38 000 molecular weight (MW) band. (b) Cytometric analysis of FPR2 expression confirms expression in FPR2-transfected cells. (c) Chemotaxis was assessed using increasing concentrations of WKYMVm in FPR2-transfected, pcDNA3-transfected, and untransfected HEK 293 cells. Chemotaxis indices at each WKYMVm concentration with FPR2-expressing cells were compared with a corresponding concentration with pcDNA3 and untransfected cells. (d) Chemotaxis was assessed using 10 nm MAT-1 with FPR2-transfected, pcDNA3-transfected, and untransfected HEK 293 cells to confirm specificity for FPR2. (e) Chemotaxis assays with FPR2-transfected cells compared MAT-1 with the negative peptide control at the indicated concentrations. Error bars represent SEM (n = 3, *P < 0·05, **P < 0·01, ***P < 0·001).
Discussion
Our study presents the first evidence that chemotactic gp41 peptides are the products of proteolytic cleavage. This finding provides a physiological basis for a mechanism by which HIV can exploit the host's immunity through chemotactic regions of gp41. Monocytes and macrophages are known to express FPR2 in addition to the viral co-receptor CCR5.30 R5-tropic HIV, which uses CCR5 for entry, is predominantly responsible for infection of a new host during transmission.31 Additionally, more recent evidence suggests that some clinical strains of R5-tropic HIV can use FPR2 as a co-receptor.32–34 Hence, either by inducing migration of CCR5+ cells at the prostate epithelium or by producing chemotactic peptides that could be delivered in semen to the new host, the matriptase cleavage product identified here would be expected to promote viral transmission.
While this work demonstrates that gp41 contains conserved amino acids that are recognized as substrates by matriptase, this activity remains to be observed using intact virions. We focused our experiments on using a peptide representing the gp41 ectodomain found at the surface of the viral envelope. Therefore, the activity we observed with matriptase would be expected to be exerted upon the exposed gp41 under biological conditions. This is especially the case for gp120 shedding, an event that further exposes the gp41 ectodomain to the extracellular environment as ‘gp41 stubs', which are believed to function in immune evasion.35,36 These exposed gp41 regions can act as antigens for the production of antibodies shown to enhance HIV infection.7 Cleavage and dissemination of gp41 peptides could therefore potentially act to induce expression of these immunoevasive antibodies.
MAT-1, the most abundant matriptase cleavage product that we detected, was cleaved at reported matriptase-recognition sites that we determined to be conserved across HIV-1 subtype B and represent the consensus sequence across all of HIV-1 Group M. This is significant as Group M represents > 98% of all HIV-1 infections.37 The prevalence of residues surrounding, and within, MAT-1 across diverse viral clades reflects the necessity of this region for overall viral replication. This is supported by MAT-1 being part of the leucine zipper motif of gp41, a region known to be critical for viral fusion.9 The role of MAT-1 in transmission and infection would therefore be relevant in all but the most divergent HIV strains, accounting for < 2% of infections.
This finding provides a basis for future research into how chemotactic gp41-derived peptides can affect infection while identifying matriptase as a potential target for preventing gp41-induced chemotaxis. We have also identified FPR2 as the receptor for MAT-1, which could have far-reaching effects on both pathogenesis and transmission considering the important roles of FPR2-expressing monocytes and macrophages in HIV-1 infection. Combined with previous studies with gp41-induced chemotaxis, and the evidence that FPR2 can act as a co-receptor for both laboratory-adapted and clinically isolated HIV-1 strains, our work supports FPR2 as a potential target for future drug development. By identifying a mechanism by which gp41 can be degraded into peptides within the host, these results promote further research into how MAT-1 and other gp41 peptide cleavage products can be produced and how they may modify host immunity.
Acknowledgments
The authors would like to thank Dorilyn Hitchcock and Jeanette Vance for their assistance with donor venepuncture and sample collection. We would also like to acknowledge Justin Price for providing expertise with fiji software. This work was supported by the National Institutes of Health [grant numbers AI052017, AI082623, AI082693 to AMC].
Author contributions
MPW, CRE and ALC performed the experiments; MPW, AMC, ALC, LMC, and KXC designed the study; MPW, AMC, ALC, LMC, and KXC analysed the data; and, MPW and AMC wrote the paper.
Disclosures
The authors report no conflicts of interest.
References
- 1.Bloch I, Quintana F, Gerber D, Cohen T, Cohen I, Shai Y. T-cell inactivation and immunosuppressive activity induced by HIV gp41 via novel interacting motif. FASEB J. 2007;21:393–401. doi: 10.1096/fj.06-7061com. [DOI] [PubMed] [Google Scholar]
- 2.Garg H, Blumenthal R. Role of HIV Gp41 mediated fusion/hemifusion in bystander apoptosis. Cell Mol Life Sci. 2008;65:3134–44. doi: 10.1007/s00018-008-8147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morozov V, Morozov A, Semaan M, Denner J. Single mutations in the transmembrane envelope protein abrogate the immunosuppressive property of HIV-1. Retrovirology. 2012;9:67. doi: 10.1186/1742-4690-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Y, Jiang S, Hu J, et al. N36, a synthetic N-terminal heptad repeat domain of the HIV-1 envelope protein gp41, is an activator of human phagocytes. Clin Immunol. 2000;96:236–42. doi: 10.1006/clim.2000.4896. [DOI] [PubMed] [Google Scholar]
- 5.de Paulis A, Florio G, Prevete N, et al. HIV-1 envelope gp41 peptides promote migration of human Fc epsilon RI+ cells and inhibit IL-13 synthesis through interaction with formyl peptide receptors. J Immunol. 2002;169:4559–67. doi: 10.4049/jimmunol.169.8.4559. [DOI] [PubMed] [Google Scholar]
- 6.Su S, Gao J, Gong W, et al. T21/DP107, A synthetic leucine zipper-like domain of the HIV-1 envelope gp41, attracts and activates human phagocytes by using G-protein-coupled formyl peptide receptors. J Immunol. 1999;162:5924–30. [PubMed] [Google Scholar]
- 7.Robinson W, Gorny M, Xu J, Mitchell W, Zolla-Pazner S. Two immunodominant domains of gp41 bind antibodies which enhance human immunodeficiency virus type 1 infection in vitro. J Virol. 1991;65:4169–76. doi: 10.1128/jvi.65.8.4169-4176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrer R, Haessig-Einius S, Aubertin A, Moog C. Neutralizing as well as non-neutralizing polyclonal immunoglobulin (Ig)G from infected patients capture HIV-1 via antibodies directed against the principal immunodominant domain of gp41. Virology. 2005;333:102–13. doi: 10.1016/j.virol.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 9.Chan D, Fass D, Berger J, Kim P. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–73. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 10.Yang O. Candidate vaccine sequences to represent intra- and inter-clade HIV-1 variation. PLoS ONE. 2009;4:e7388. doi: 10.1371/journal.pone.0007388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisele E, Siliciano R. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity. 2012;37:377–88. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun T, Fauci A. HIV reservoirs: pathogenesis and obstacles to viral eradication and cure. AIDS. 2012;26:1261–8. doi: 10.1097/QAD.0b013e328353f3f1. [DOI] [PubMed] [Google Scholar]
- 13.Le Tortorec A, Satie A, Denis H, et al. Human prostate supports more efficient replication of HIV-1 R5 than X4 strains ex vivo. Retrovirology. 2008;5:119. doi: 10.1186/1742-4690-5-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith D, Kingery J, Wong J, Ignacio C, Richman D, Little S. The prostate as a reservoir for HIV-1. AIDS. 2004;18:1600–2. doi: 10.1097/01.aids.0000131364.60081.01. [DOI] [PubMed] [Google Scholar]
- 15.Lawless MK, Barney S, Guthrie KI, Bucy TB, Petteway SR, Jr, Merutka G. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry. 1996;35:13697–708. doi: 10.1021/bi9606962. [DOI] [PubMed] [Google Scholar]
- 16.Millius A, Weiner O. Chemotaxis in neutrophil-like HL-60 cells. Methods Mol Biol. 2009;571:167–77. doi: 10.1007/978-1-60761-198-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins SJ, Ruscetti FW, Gallagher RE, Gallo RC. Normal functional characteristics of cultured human promyelocytic leukemia cells (HL-60) after induction of differentiation by dimethylsulfoxide. J Exp Med. 1979;149:969–74. doi: 10.1084/jem.149.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔCT) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Pryde J. Partitioning of proteins in Triton X-114. Methods Mol Biol. 1998;88:23–33. doi: 10.1385/0-89603-487-9:23. [DOI] [PubMed] [Google Scholar]
- 20.Martellini JA, Cole AL, Venkataraman N, et al. Cationic polypeptides contribute to the anti-HIV-1 activity of human seminal plasma. FASEB J. 2009;23:3609–18. doi: 10.1096/fj.09-131961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen LM, Skinner ML, Kauffman SW, et al. Prostasin is a glycosylphosphatidylinositol-anchored active serine protease. J Biol Chem. 2001;276:21434–42. doi: 10.1074/jbc.M011423200. [DOI] [PubMed] [Google Scholar]
- 22.Martellini J, Cole A, Svoboda P, et al. HIV-1 enhancing effect of prostatic acid phosphatase peptides is reduced in human seminal plasma. PLoS ONE. 2011;6:e16285. doi: 10.1371/journal.pone.0016285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulay F, Tardif M, Brouchon L, Vignais P. The human N-formylpeptide receptor. Characterization of two cDNA isolates and evidence for a new subfamily of G-protein-coupled receptors. Biochemistry. 1990;29:11123–33. doi: 10.1021/bi00502a016. [DOI] [PubMed] [Google Scholar]
- 25.Murphy P, Ozçelik T, Kenney R, Tiffany H, McDermott D, Francke U. A structural homologue of the N-formyl peptide receptor. Characterization and chromosome mapping of a peptide chemoattractant receptor family. J Biol Chem. 1992;267:7637–43. [PubMed] [Google Scholar]
- 26.Le Y, Gong W, Li B, et al. Utilization of two seven-transmembrane, G protein-coupled receptors, formyl peptide receptor-like 1 and formyl peptide receptor, by the synthetic hexapeptide WKYMVm for human phagocyte activation. J Immunol. 1999;163:6777–84. [PubMed] [Google Scholar]
- 27.Beliveau F, Desilets A, Leduc R. Probing the substrate specificities of matriptase, matriptase-2, hepsin and DESC1 with internally quenched fluorescent peptides. FEBS J. 2009;276:2213–26. doi: 10.1111/j.1742-4658.2009.06950.x. [DOI] [PubMed] [Google Scholar]
- 28.Kuiken C, Foley B, Leitner T, et al. HIV Sequence Compendium. Los Alamos, NM: Los Alamos National Laboratory, Theoretical Biology and Biophysics Group; 2012. [Google Scholar]
- 29.De Y, Chen Q, Schmidt AP, et al. LL-37, the neutrophil granule- and epithelial cell-derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J Exp Med. 2000;192:1069–74. doi: 10.1084/jem.192.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Y, Yang Y, Cui Y, et al. Receptors for chemotactic formyl peptides as pharmacological targets. Int Immunopharmacol. 2002;2:1–13. doi: 10.1016/s1567-5769(01)00150-3. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen C, Pedersen C, Lundgren JD, Gerstoft J. Biological properties of HIV isolates in primary HIV infection: consequences for the subsequent course of infection. AIDS. 1993;7:1035–40. doi: 10.1097/00002030-199308000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Nedellec R, Coetzer M, Shimizu N, et al. Virus entry via the alternative coreceptors CCR3 and FPRL1 differs by human immunodeficiency virus type 1 subtype. J Virol. 2009;83:8353–63. doi: 10.1128/JVI.00780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu N, Tanaka A, Mori T, et al. A formylpeptide receptor, FPRL1, acts as an efficient coreceptor for primary isolates of human immunodeficiency virus. Retrovirology. 2008;5:52. doi: 10.1186/1742-4690-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimizu N, Tanaka A, Oue A, et al. Broad usage spectrum of G protein-coupled receptors as coreceptors by primary isolates of HIV. AIDS. 2009;27:761–9. doi: 10.1097/QAD.0b013e328326cc0d. [DOI] [PubMed] [Google Scholar]
- 35.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–42. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 36.Crooks ET, Tong T, Osawa K, Binley JM. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol. 2011;85:5825–39. doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]