Abstract
BACKGROUND AND PURPOSE
The histone acetyltransferase MOF is a member of the MYST family. In mammals, MOF plays critical roles by acetylating histone H4 at K16 and non-histone substrates such as p53. Here we have investigated the role of MOF in human lung cancer and possible new substrates of hMOF.
EXPERIMENTAL APPROACH
Samples of human non-small cell lung cancer (NSCLC) were used to correlate MOF with clinicopathological parameters and NF–E2-related factor 2 (Nrf2) downstream genes. 293T-cells were used to study interactions between MOF and Nrf2, and acetylation of Nrf2 by MOF. Mouse embryonic fibroblast and A549 cells were utilized to assess involvement of MOF in antioxidative and anti-drug responses. A549 cells were used to analysis the role of MOF in anti-drug response in vitro and in vivo.
KEY RESULTS
hMOF was overexpressed in human NSCLC tissues and was associated with large tumour size, advanced disease stage and metastasis, and with poor prognosis. hMOF levels were positively correlated with Nrf2-downstream genes. MOF/hMOF physically interacted with and acetylated Nrf2 at Lys588. MOF-mediated acetylation increased nuclear retention of Nrf2 and transcription of its downstream genes. Importantly, MOF/hMOF was essential for anti-oxidative and anti-drug responses in vitro and regulated tumour growth and drug resistance in vivo in an Nrf2-dependent manner.
CONCLUSION AND IMPLICATIONS
hMOF was overexpressed in human NSCLC and was a predictor of poor survival. hMOF-mediated Nrf2 acetylation and nuclear retention are essential for anti-oxidative and anti-drug responses. hMOF may provide a therapeutic target for the treatment of NSCLC.
Keywords: hMOF, NSCLC, Nrf2, acetylation, oxidative stress, drug resistance
Introduction
The histone acetyltransferase MOF is a member of the MYST (MOS, ΚB2/Sas3, Sas2 and TIP60) family of histone acetyltransferases, and was first described in Drosophila melanogaster as an essential component of the X chromosome dosage compensation male-specific lethal (MSL) complex (Hilfiker et al., 1997; Akhtar and Becker, 2000). Acetylation of the histone H4 at K16 in higher eukaryotes is mainly carried out by MOF (Mellert and McMahon, 2009). The function of MOF in dosage compensation is mediated by its acetyltransferase activity (Bone et al., 1994; Hilfiker et al., 1997).
MOF is conserved among higher eukaryotes. Compared with functions in Drosophila dosage compensation, the roles of MOF in mammals are less well characterized. In mammals, MOF is ubiquitously expressed and is clearly targeted to all chromosomes. Loss of MOF gene in mice causes peri-implantation lethality, as a result of massive disruption of chromatin architecture in a wide range of cells (Gupta et al., 2008; Thomas et al., 2008). In addition, by maintaining normal chromatin structure, MOF is important for ataxia telangiectasia mutated-dependent cell-cycle checkpoint control (Gupta et al., 2005), and transcription activation of Hox genes in coordination with the H3K4 methyltransferase MLL (Dou et al., 2005). Loss of MOF leads to severe G2/M cell-cycle arrest, massive chromosome aberration, and defects in ionizing radiation-induced DNA damage repair by both non-homologous end-joining and homologous recombination (Li et al., 2010; Sharma et al., 2010). All these effects are mediated by acetylation activity of MOF on H4K16. MOF may also acetylate non-histone proteins. So far, two non-histone substrates of MOF have been identified.One of these, p53 is acetylated by hMOF on its DNA-binding domain (K120) and helps to distinguish the cell-cycle arrest and apoptotic functions of p53 (Sykes et al., 2006). The other non-histone substrate of MOF is the TIP5 (K633) subunit of the NoRC chromatin-remodelling complex (Zhou et al., 2009). However, the pathophysiological functions of MOF in mammals remain to be defined. Here we have investigated the role(s) of hMOF in human non-small cell lung cancer (NSCLC) and tried to identify new substrates of hMOF.
Oxidative stress is induced by a wide range of factors including xenobiotic, drugs, heavy metals and ionizing radiation. The transcription factor NF–E2-related factor 2 (Nrf2) was originally identified as a crucial factor for protecting cells from oxidative and electrophilic insults (Kaspar et al., 2009). Under homeostatic conditions, Nrf2 is retained in the cytoplasm by the anchor protein Kelch-like ECH-associated protein 1 (Keap1) and is maintained at a low level by the Keap1-dependent ubiquitination and proteasomal degradation system (Kaspar et al., 2009). When cells were exposed to oxidative or electrophonic stress, the Keap1-dependent ubiquitin ligase activity is inhibited, and Nrf2 can be phosphorylated and then translocated to the nucleus where it binds to a consensus sequence called the antioxidant response element (ARE; Jaiswal, 2004; Zhang, 2006; Kaspar et al., 2009). Nrf2 effector genes bearing ARE include many of the anti-oxidative and phase II detoxifying enzymes (Jaiswal, 2004). In addition to these enzymes, recent studies have shown that Nrf2 transactivates a wide variety of genes, including several ATP-dependent drug efflux pumps (Hayashi et al., 2003; Vollrath et al., 2006). Activation of Nrf2 in cancer cells, lung cancer cells for example, increases cell proliferation and survival from exposure to anticancer drugs (Singh et al., 2006; Ohta et al., 2008; Homma et al., 2009; Yamadori et al., 2012). Post-translational modification is important for Nrf2 activation. Current studies indicate that acetylation of Nrf2 mediates its activation. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the anti-oxidative responses (Sun et al., 2009). Moreover, Sirt1-mediated deacetylation of Nrf2 regulates its transcriptional activity and nuclear localization (Kawai et al., 2011). However, how Nrf2 is regulated in anti-oxidative and anti-drug responses is not fully understood.
Here we show that hMOF was overexpressed in NSCLC and associated with tumour size, disease stage, metastasis and patients' survival. We found that hMOF bound to Nrf2 and acetylated Nrf2 at Lys588. hMOF-mediated acetylation promoted Nrf2 nucleus maintenance and regulated anti-oxidative and drug-resistance responses by facilitating Nrf2-dependent gene expression. Furthermore, hMOF was essential for lung cancer growth and drug resistance in vivo.
Methods
Cells and cell culture
WT and Nrf2-KO mouse embryonic fibroblasts (MEF) were gifts from Xingfen Su of Nanjing University. 293T cells and NCI-H292 were purchased from ATCC (Manassas, VA, USA). Human lung cancer cell lines A549 and LC-AI were obtained from Riken BioResource Center (Tsuba, Japan). 293T, MEF, A549 and LC-AI cells were cultured in high glucose-containing DMEM supplemented with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. H292 cells were cultivated in RPMI 1640 with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. All cells were cultured at 37°C in a humidified atmosphere containing 5% CO2.
Plasmids
Lysine to arginine (K → R) substitution mutations of Nrf2 were created at Lys588 and Lys591. Nrf2 was mutated at putative acetylation sites using the QuikChange™ site-directed mutagenesis kit from Stratagene (La Jolla, CA, USA). Mutations were confirmed by DNA sequencing. HA-Nrf2WT, HA-Nrf2K588R, HA-Nrf2K591R and Flag-hMOF cDNAs were cloned into pcDNA3 vector respectively. The Nrf2 and Nrf2K588R were also cloned into pGEX-4T-1, a plasmid expressing in prokaryote.
Transfection and treatment
293T-cells were transfected with plasmids expressing HA-Nrf2, HA-Nrf2K588R or HA-Nrf2K588R with/without Flag-hMOF co-transfection with Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) according to the manufacturer's protocol. 293T, mouse embryonic fibroblasts (MEF) and lung cancer cells were transfected with the MOF/hMOF-siRNA or NC-siRNA (Santa Cruz Biotech, Dallas, TX, USA) with Lipofectamine RNAi Max (Invitrogen) according to the manufacturer's protocol.
Generation of lung cancer cell lines stably expressing hMOF short hairpin RNA
hMOF shRNA, Nrf2 shRNA and Ctrl shRNA retroviruses were packaged as described previously (Kapoor-Vazirani et al., 2008; Rachakonda et al., 2010). For transduction, A549 cells were incubated with virus-containing supernatant in the presence of 8 mg·mL−1 polybrene. After 48 h, infected cells were selected for 72 h with puromycin (2 mg·mL−1) or hygromycin (200 mg·mL−1).
Immunoprecipitation (IP) and immunoblotting (IB)
IP and IB analysis were done using standard protocols as described previously (Lu et al., 2011). Anti-HA and Anti-Flag antibodies were purchased from Abcam (Cambridge, MA, USA). Anti-Nrf2, anti-hMOF, anti-β-actin were purchased from Santa Cruz Biotech. Anti-ac-K (acetylated lysine) and anti-histone H3 antibodies were purchased from Cell Signaling Technology (Shanghai, China).
RNA extraction and real-time quantitative PCR (q-PCR)
Total RNA was extracted from cells or tissues with TRIzol. q-PCR was performed as previously described (Li et al., 2011). One Step RT-PCR Kit and SYBR Green were purchased from TAKARA (Dalian, China). The primers used are listed in Supporting Information Table S1.
Chromatin immunoprecipitation (ChIP)
ChIP was carried out as described previously (Lu et al., 2011) with anti-Nrf2 antibody. DNA recovered from chromatin immunoprecipitation was analysed by q-PCR as described above. The primers used in ChIP/q-PCR are listed in Supporting Table 2.
GST pull-down assay
Nrf2 or hMOF sequences were in vitro transcribed and translated according to the manufacturer's instructions (Promega, Madison, WI, USA), and glutathione-S-transferase (GST) pull-down assays were performed as previously described (Shi et al., 2006).
Cell viability assay
Cell viability was assayed with the MTT Cell Proliferation and Cytotoxicity Assay Kit (Beyotime, Beijing, China) according to the manufacturer's protocol.
Tumour xenograft experiments
All animal care and experimental procedures complied with the guidelines of Shanghai Chest Hospital, Shanghai Jiao Tong University and were approved by the animal ethics committees of Shanghai Chest Hospital, Shanghai Jiao Tong University and of Longhua Hospital, Shanghai University of Traditional Chinese Medicine. All studies involving animals are reported in accordance with the ARRIVE guidelines for reporting experiments involving animals (Kilkenny et al., 2010; McGrath et al., 2010). A total of 64 animals were used in the experiments described here.
Equal numbers of A549 cells expressing luciferase and either control or hMOF/Nrf2/Nrf2 + hMOF knockdown vectors were injected subcutaneously, within 30 min of harvesting, over the right and left flanks in male nu/nu mice between 4 and 6 weeks of age. Tumour growth was monitored using callipers. For drug-resistance analysis, tumour-bearing mice received CDPP (5 mg·kg−1, i.p.) treatment or saline (vehicle/control, i.p.) weekly since day 21 post implantation for 4 weeks. Tumour size was also quantified by bioluminescence imaging. Tumour weight was quantified at the end of experiment. Eight mice were used in each group.
Patients
This study was approved by the Clinical Research Ethics Committee of Shanghai Jiaotong University and by the Clinical Research Ethics Committee of Shanghai University of Traditional Medicine. Full informed consent was obtained from all 54 patients with NSCLC included in this study. The diagnosis of lung cancer was established using World Health Organization morphological criteria. Thirty-five patients were recruited from the Shanghai Chest Hospital (Shanghai) and the others were recruited from Longhua Hospital (Shanghai). The patients consisted of 39 (72.2%) men and 15 (27.8%) women with a mean age of 61 years. Tumours were histologically classified into 37 adenocarcinomas and 17 squamous cell carcinomas. Clinicopathological parameters including age, gender, smoking history, histological grade, tumour status, lymph node metastasis and histological type were analysed.
Immunohistochemistry
Tissues were fixed with 4% neutral formalin. Cancer or adjacent tissue sections were subjected for immunohistochemical staining with anti-hMOF antibody. Paraffin sections were subjected to high-temperature antigen retrieval, 3 min in a pressure cooker in 0.01 M citrate buffer pH 6.0. slides were treated with 3% H2O2 for 15min, blocked in 5% normal horse serum in PBS for 20min, and stained with anti-hMOF antibody in 5% normal goat serum overnight at 4°C. Secondary antibody was used according to Vectastain ABC kits, followed by DAB staining. The areas of total lung tumors and hMOF-positive areas were quantified using ImageJ. The average percentage of hMOF-positive area was 34%. This value was used to divide the subgroups of all immunohistochemical variables in our data. The patients were then divided into two groups: hMOF high expression group (≥34% hMOF-positive/total tissue cores, n = 26) and hMOF low expression group (<34% hMOF-positive/ total tissue cores, n = 28)
In vitro acetylation assay
Wide type and K588R mutant Nrf2 were expressed in E. coli and the recombinant proteins were purified. In vitro acetylation assay was performed as described (Lin et al., 2012). The reaction was performed in 40 μL of reaction mixture containing 20 mM Tris-Cl, pH 8.0, 20% glycerol, 100 mM KCl,1 mM DTT and 0.2 mM EDTA, 10 μM TSA, 10 mM nicotinamide, 100 μM acetyl-CoA, and hMOF (SignalChem, Richmond, VA, USA). After incubation at 30°C for 1 hour, the reaction was stopped by addition of 10 μL of 5 × SDS sample buffer. The samples were subjected to SDS–PAGE and Immunoblotting.
Assessment of intracellular ROS
Intracellular ROS was measured using 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) Kit (Beyotime) according to the manufacturer's protocol. After H2O2 or UV treatment, cells were washed with PBS and then incubated with 10 μM H2DCF-DA in PBS for 30 min at 37°C in darkness. Cells were then harvested and washed with PBS. After centrifugation, cells were resuspended in PBS and analysed with a FACSCalibur flow cytometer (Becton-Dickinson) with Win-MDI software at excitation and emission settings of 480 and 525 nm respectively.
Total glutathione quantification
Intracellular total glutathione was quantified using a Total Glutathione Quantification Kit (Dojindo, Rockville, MD, USA) according to the manufacturer's protocol.
Data analysis
All values were expressed as the mean ± SEM of at least three independent experiments. Statistical differences between two groups were determined using Student's t-test. The correlation of hMOF immunoreactivity with patients' clinicopathological variables was analysed by the χ2 test or Fisher's exact test. The Kaplan–Meier method was used to estimate overall and disease-free survival. Correlation of survival differences with hMOF expression was analysed by the log–rank test. Regression analysis was performed using GraphPad Prism (version 6; La Jolla, CA, USA). P < 0.05 was considered statistically significant.
Materials
The cis-diamminedichloroplatinum (cisplatin; CDDP), H2O2 and 5-fluorouracil (5-FU) were purchased from Sigma (St. Louis, MO, USA). Bleomycin was purchased from Nippon Kayaku (Chiyada-Ku, Japan). The drug and molecular target nomenclature follows Alexander et al. (2013a,2013,2013b a,b).
Results
hMOF is overexpressed in human lung cancer tissues and correlates with low survival
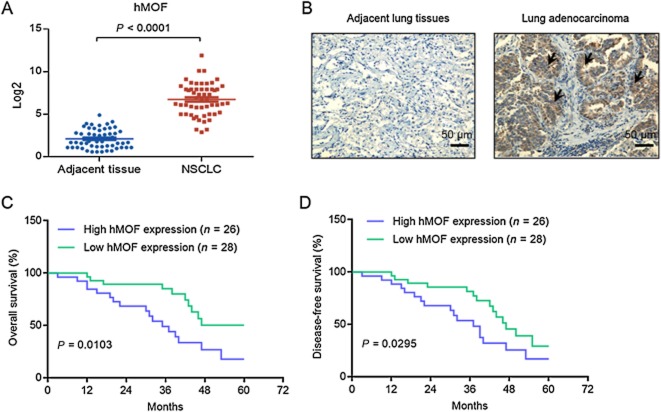
To study the role of hMOF in human NSCLC, we first analysed the pattern of expression of hMOF gene in tumour and adjacent normal tissues from 54 NSCLC patients. hMOF expression was markedly up-regulated in tumour tissues compared with their matched normal adjacent tissues (Figure 1A). To further validate these observations, we used immunohistochemistry to analyse hMOF expression in NSCLC tissues. Whereas normal adjacent lung tissues exhibited negligible hMOF staining, NSCLC tissues had a clear increase in levels of immunoreactive hMOF protein (Figure 1B). These findings indicated that hMOF may be involved in human NSCLC. We therefore analysed correlations between hMOF and clinical parameters. We found that high levels of hMOF (>34% immunopositive) were associated with metastasis and recurrence (P = 0.024), tumour size (P = 0.0113) and disease stage (P = 0.0243; Table 1). Patients with low levels of hMOF (<34% immunopositive) had a longer overall survival than those with high levels of hMOF (P = 0.0103; Figure 1C). Additionally, hMOF expression was associated with recurrence. High hMOF expression predicted adverse disease-free survival (P = 0.0295; Figure 1D).
Figure 1.
hMOF was overexpressed in human lung cancer tissues and correlated with lower survival. (A) hMOF mRNA increases in human lung cancer. The mRNA levels of hMOF in adjacent and cancer tissues from 54 NSCLC patients were analysed with q-PCR. (B) hMOF protein level is up-regulated in human lung cancer. Paraffin sections of adjacent and cancer tissues from 54 NSCLC patients were subjected to immunohistochemical analysis with anti-hMOF antibody. (C–D) Kaplan–Meier curve showing high hMOF expression correlates with lower overall (C) and disease-free (D) survival.
Table 1.
Clinical characteristic of the patients with NSCLC
| Variables | Total | hMOF low expression | hMOF high expression | P value |
|---|---|---|---|---|
| Age (years) | ||||
| ≥60 | 30 | 15 | 15 | 0.7905 |
| <60 | 24 | 13 | 11 | |
| Gender | ||||
| Male | 39 | 19 | 20 | 0.55 |
| Female | 15 | 9 | 6 | |
| Smoking history | ||||
| Never | 22 | 15 | 7 | 0.5756 |
| Ever | 32 | 19 | 13 | |
| Histology type | ||||
| Adenocarcinomas | 37 | 17 | 20 | 0.1429 |
| Squamous cell carcinomas | 17 | 4 | 13 | |
| Tumour size (cm) | ||||
| ≥3 | 21 | 6 | 15 | 0.0113 |
| <3 | 33 | 22 | 11 | |
| Metastasis and recurrence | ||||
| Present | 19 | 14 | 5 | 0.024 |
| Absent | 35 | 14 | 21 | |
| Stage | ||||
| I | 18 | 14 | 4 | 0.0243 |
| II | 19 | 8 | 11 | |
| III | 17 | 6 | 11 |
Low expression of hMOF is <34% MOF-positive cells and high expression represents >34 % MOF-positive cells. The P-values less than 0.05 are in bold.
hMOF is correlated with Nrf2-downstream genes
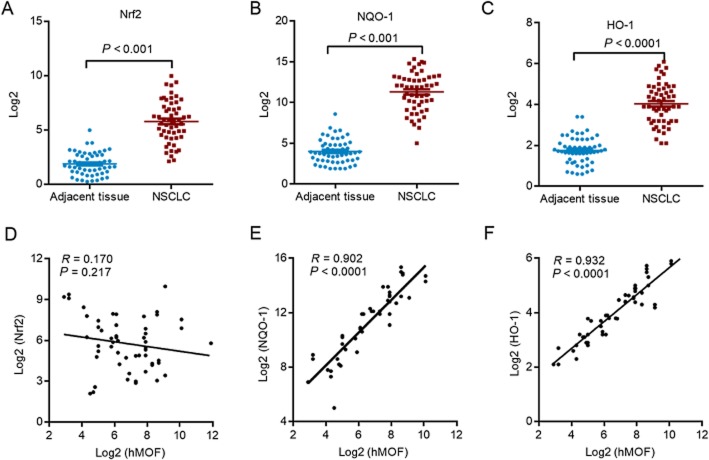
We next investigated how hMOF might mediate human NSCLC. The transcription factor, Nrf2 activated NSCLC in humans and facilitated lung cancer cell proliferation and drug resistance (Singh et al., 2006; Ohta et al., 2008; Homma et al., 2009). Further, Nrf2 acetylation and deacetylation are important for its translocation and activation (Sun et al., 2009; Kawai et al., 2011). Therefore, we evaluated acetylation of Nrf2 by MOF and the consequent effects on genes downstream of Nrf2, samples of human NSCLC. First, we assessed the expression of Nrf2 and its downstream genes in lung cancer and adjacent normal tissues from human NSCLC. The expression of Nrf2 and its target genes haem oxygenase 1 (HO-1) and NAD(P)H quinone oxidoreductase 1 (NQO1) were significantly up-regulated in tumour samples compared with adjacent normal samples (Figure 2A–C). Furthermore, linear regression analysis indicated that HO-1 and NQO1 levels were significantly and positively correlated with the level of hMOF. However, the level of Nrf2 was not related with hMOF level (Figure 2D–F). These findings suggested that hMOF might regulate Nrf2 at a posttranslational level. One likely possibility is that hMOF can acetylate Nrf2, increasing nuclear retention of Nrf2 and thus facilitate the transcriptional activity of Nrf2 (Kawai et al., 2011).
Figure 2.
hMOF correlates with NQO1 and HO-1 levels in human lung cancer tissues. q-PCR was performed to analysis the mRNA levels of Nrf2 (A), NQO1 (B) or HO-1 (C) in adjacent and cancer tissues from human NSCLC. The relation between the levels of hMOF and Nrf2 (D), NQO1 (E) or HO-1 (F) in lung cancer tissues was analysed with linear regression analysis.
hMOF physically interacts with Nrf2
If hMOF did acetylate Nrf2 there should be a physical interaction between the two proteins. Flag-tagged hMOF (Flag-hMOF) and HA-tagged Nrf2 (HA-Nrf2) were expressed or co-expressed in 293T-cells and cell lysates were subjected to IP and IB analysis. The results revealed that HA-Nrf2 was associated with Flag-hMOF (Figure 3A) and IB analysis of endogenous Nrf2 also revealed a specific interaction with exogenous hMOF and vice versa (Figure 3B). Notably, the hMOF-Nrf2 interaction was augmented by H2O2 treatment (Figure 3B), suggesting that H2O2-dependent translocation of Nrf2 to the nucleus facilitates its interaction with hMOF. However, we also found that hMOF could bind to Nrf2 in lung cancer tissues (Supporting Information Fig. S1). Next, we performed in vitro GST pull-down assay to test whether hMOF can directly interact with Nrf2. The results showed that hMOF directly interacted with Nrf2 (Figure 3C).
Figure 3.
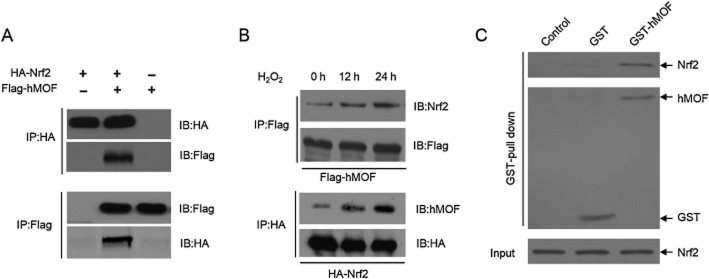
hMOF interacts with Nrf2. (A) hMOF can interact with Nrf2. Plasmids expressing HA-Nrf2 or Flag-hMOF were transfected or co-transfected into 293T-cells. 48 h after transfection, total proteins were subjected to IP and IB analysis with indicated antibodies. (B) H2O2 promotes the interaction between hMOF and Nrf2. 293T-cells were transfected with plasmids expressing HA-Nrf2 or Flag-hMOF. Thirty-six hours later, the cells were left untreated or were treated with 100 μM H2O2 for additional 12 or 24 h. Cell lysates were subjected to IP and IB analysis with the indicated antibodies. (C) GST pull-down assay showing Nrf2 binds directly to hMOF. In vitro Nrf2 bound purified recombinant GST-hMOF. GST served as the pull-down control. Equal amounts of input Nrf2 were measured by CBB staining, while the pull-down products were detected with anti-Nrf2 antibody.
hMOF acetylates Nrf2 and promotes Nrf2 nucleus retention
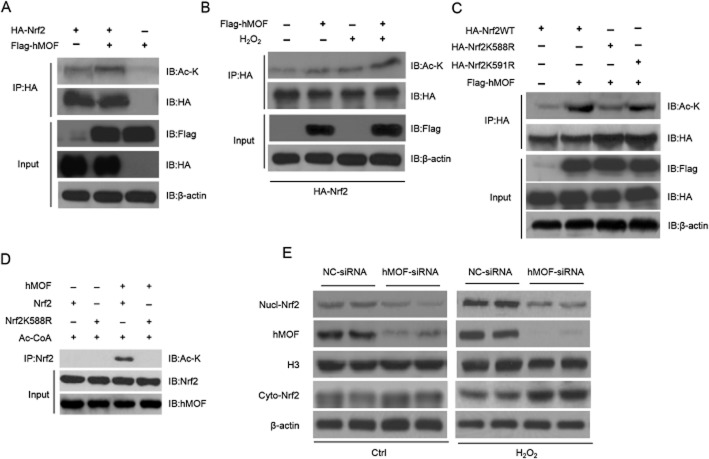
We next explored whether Nrf2 is a non-histone substrate of hMOF. We found that hMOF enhanced the acetylation of Nrf2 (Figure 4A). In addition, H2O2 promoted the acetylation of Nrf2, and hMOF overexpression significantly up-regulated Nrf2 acetylation induced by H2O2 (Figure 4B). Conversely, when hMOF was knocked down, H2O2 was unable to up-regulate the acetylation of Nrf2 (Supporting Information Fig. S2a).
Figure 4.
hMOF acetylates Nrf2 and promotes its nuclear retention. (A) hMOF acetylates Nrf2. Plasmids expressing HA-Nrf2 and Flag-hMOF were transfected respectively or together into 293T-cells for 48 h. Cell lysates were subjected to IP and IB analysis with indicated antibodies. (B) H2O2 promotes the acetylation of Nrf2 by hMOF. 293T-cells were transfected with plasmids expressing HA-Nrf2 with/without Flag-hMOF. Thirty-six hours later, the cells were left untreated or were treated with 100 μM H2O2 for additional 12 h. Then, cell lysates were subjected to IP and IB analysis with indicated antibodies. (C–D) hMOF acetylates Nrf2 at lysine 588. (C) 293T-cells were transfected with the plasmids expressing the indicated proteins. Forty-eight hours post-transfection, cells lysates were subjected to IP and IB analysis with indicated antibodies. (D) Recombinant wide-type Nrf2 and Nrf2K588R proteins were expressed in Escherichia coli, and the purified proteins were subjected to in vitro acetylation with/without recombinant hMOF. The products were subjected to IB analysis with indicated antibodies. (E) hMOF promotes Nrf2 nuclears retention. 293T-cells were transfected with control siRNA (NC-siRNA) or siRNA targeting hMOF (hMOF-siRNA) for 36 h. Then the cells were treated with or without 100 μM H2O2 for 12 h. The nuclear and cytoplasmic proteins were subjected to IB analysis with the indicated antibodies.
hMOF and Sirt1 exert opposing effects on H4K16, p53 and TIP5 acetylation (Zhou et al., 2009; Dai and Gu, 2010; Hajji et al., 2010), as Sirt1 deacetylated Nrf2 on Lys588/591 in the Neh3 region (Kawai et al., 2011). We therefore We created two mutant Nrf2 structures which could not be acetylated by MOF, by changing lysine to arginine at Lys588 or Lys591. As shown in Figure 4C, the K588R mutant, but not the K591R mutant, was totally resistant to acetylation by hMOF, indicating that hMOF acetylates Nrf2 at Lys588. To exclude the possibility that other lysine residues could be acetylated by hMOF, we generated the K588R Nrf2 mutant and cloned the gene to a prokaryotic vector. We expressed wild-type Nrf2 and the Nrf2 mutant in Escherichia coli and purified the proteins. Further, an in vitro acetylation assay was performed. We found that hMOF acetylated the wild-type Nrf2, while the Nrf2K588R mutant could not be acetylated (Figure 4D). This finding indicates that only the Lys588 of Nrf2 can be acetylated by hMOF.
Acetylation of Nrf2 regulates its nuclear retention (Sun et al., 2009; Kawai et al., 2011). We therefore measured the nuclear retention of Nrf2 in our system. 293T-cells were transfected with control siRNA (si-Ctrl) or siRNA targeting hMOF (si-hMOF) followed by H2O2 treatment. As shown in Figure 4E, hMOF knockdown markedly reduced the nuclear content of Nrf2, under basal and oxidative conditions. Conversely, hMOF overexpression up-regulated Nrf2 levels in the nucleus (Supporting Information Fig. S2b). Importantly, hMOF overexpression was not able to increase the nuclear abundance of the mutant Nrf2K588R (Supporting Information Fig. S2c).
MOF regulates Nrf2-dependent oxidative stress response
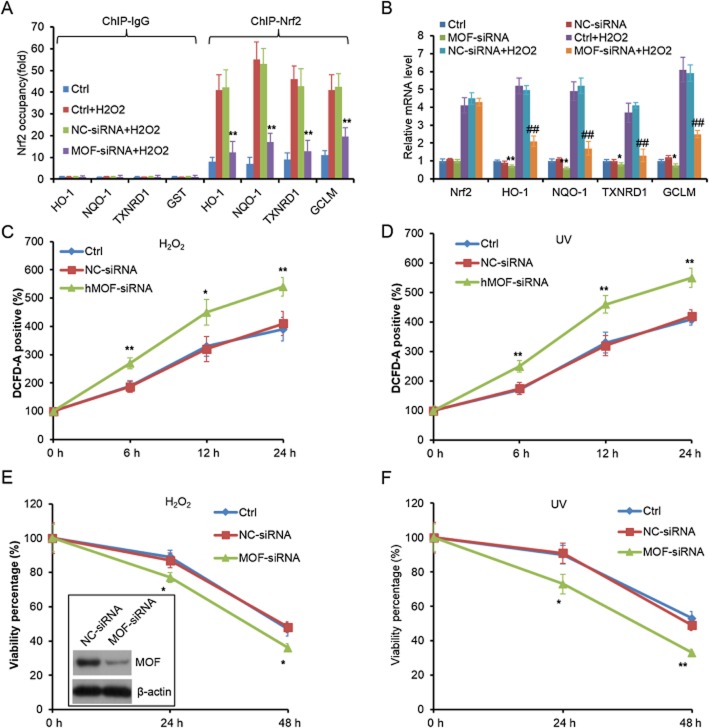
The acetylation of Nrf2 is important for its transcriptional activity, which prompted us to explore whether hMOF facilitates the binding of Nrf2 to the promoters of its target genes and mediates their transcription, and thus regulates oxidative stress response. As Figure 5A shows, MOF knockdown significantly down-regulated H2O2-induced Nrf2 binding to the promoters of HO-1, NQO1, thioredoxin reductase 1 and glutamate-cysteine ligase modifier subunit (GCLM). Similarly, Nrf2-dependent ARE genes were down-regulated under basal and H2O2-induced conditions when MOF was knocked down (Figure 5B and data not shown). However, MOF knockdown did not change the recruitment of HA-Nrf2K588R to ARE regions, and did not change the expression of anti-oxidative stress genes in Nrf2−/− MEFs (data not shown), indicating that the effects of MOF in conditions of oxidative stress were totally dependent on the presence of Lys588 in Nrf2. As our results show, the levels of MOF did not affect the mRNA and protein levels of Nrf2 itself (Figures 7F). Those results were consistent with the results in Figure 2D, suggesting that MOF regulates Nrf2 activity, but not its expression.
Figure 5.
MOF knockdown reduces Nrf2-dependent gene expression and anti-oxidative response. (A) MOF knockdown decreases Nrf2 binding to the promoters of its downstream genes. MEF cells were transfected with negative control siRNA (NC-siRNA) or MOF-siRNA for 36 h (knockdown efficiency was shown in Figure 3E) and followed by 100 μM H2O2 treatment for 12 h. Then cells were subjected to chromatin IP (ChIP) and q-PCR analysis with indicated antibodies and primers, respectively. **P < 0.01; significantly different from Ctrl + H2O2. (B) MOF knockdown reduces the expression of Nrf2-downstream genes. The total RNA of the cells in (A) was subjected to q-PCR analysis with indicated primers. *P < 0.05, **P < 0.01; significantly different from Ctrl; #P < 0.05, ##P < 0.01; significantly different from Ctrl + H2O2. (C–D) MOF knockdown increases intracellular ROS induced by H2O2 (C) and UV (D). MEF cells were transfected with NC-siRNA or MOF-siRNA for 24 h and then the cells were treated with 100 μM H2O2 or 2 Jm−2 UV-C irradiation. Intracellular ROS was measured with DCFH-DA at the indicated time points. (E–F) MOF knockdown decreases cell survival following treatment with H2O2 (E) or UV (F). MEF cells were transfected with NC-siRNA or MOF-siRNA for 24 h and then the cells were treated with 500 μM H2O2 or 10 Jm−2 UV-C. The cell survival was measured with MTT method at the indicated time points. *P < 0.05, **P < 0.01; significantly different from Ctrl.
Figure 7.
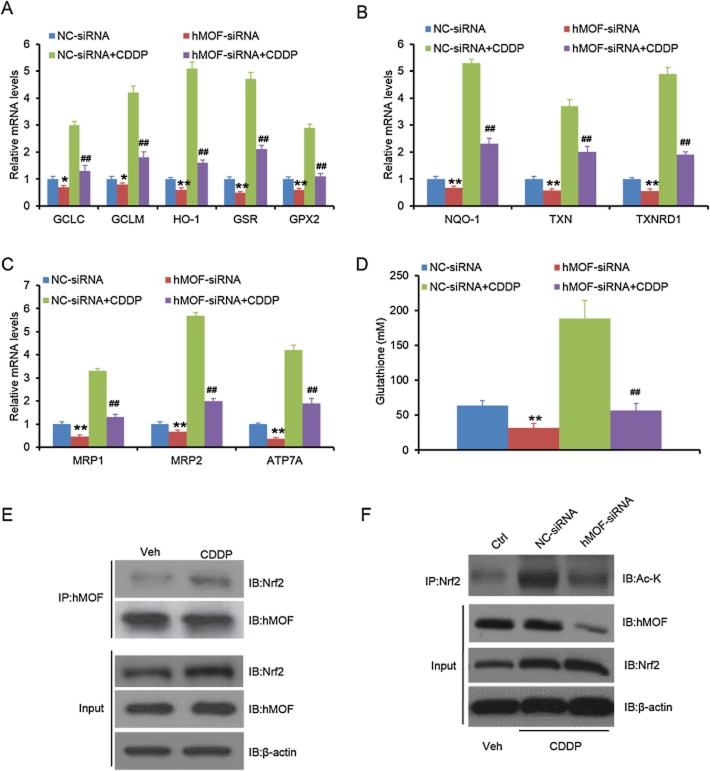
hMOF regulates the expression of drug-defence genes. (A–C) hMOF knockdown reduces expression of self-defence genes. A549 cells were transfected with NC-siRNA or hMOF-siRNA for 24 h, followed by 5 μΜ CDDP treatment for 48 h. Total RNA was subjected to q-PCR analysis for the indicated antioxidant genes (A), drug metabolising enzymes (B) and ATP-dependent drug efflux pumps (C). (D) hMOF knockdown reduces glutathione production. The intracellular total glutathione of the cells in (A) was quantified. (E) CDPP enhances Nrf2 interaction with hMOF. (F) hMOF knockdown reduces CDDP-induced acetylation of Nrf2. The total proteins in (A) were subjected to IP and IB analysis with the indicated antibodies. *P < 0.05, **P < 0.01; significantly different fromNC-siRNA; ##P < 0.01, significantly different from NC-siRNA + CDDP.
To further investigate the function of MOF in the environmental stress response, we analysed the effects of MOF on the cellular response to oxidative (H2O2) or DNA damage (UV irradiation). Firstly, MOF knockdown increased the production of intracellular ROS induced by H2O2 and UV irradiation (Figure 5C and D). In addition, MOF knockdown increased the susceptibility of MEFs to H2O2 and UV treatment (Figure 5E and F). However, MOF knockdown was not sufficient to change intracellular ROS production and sensitivity of Nrf2–/– MEFs to H2O2 and UV (Supporting Information Fig. S3). Taken together, these results demonstrated that MOF participated in stress responses in an Nrf2-dependent manner.
hMOF regulates drug resistance and cell proliferation of human lung cancer cells
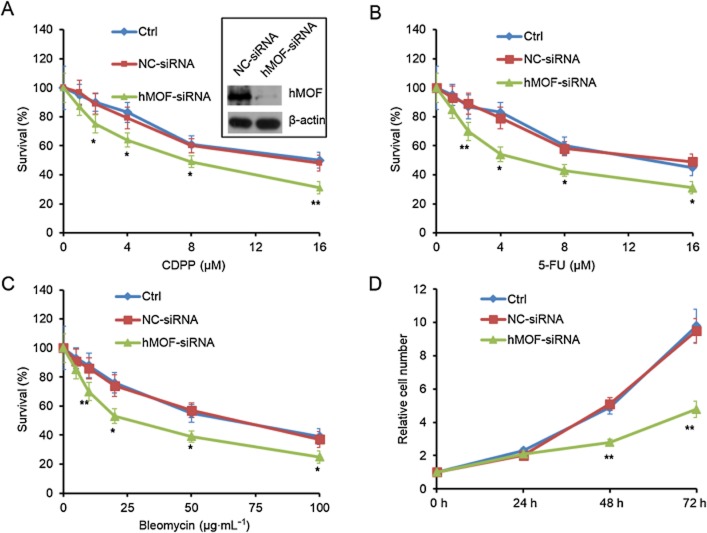
Overexpression and hyperactivation of Nrf2 contribute to carcinogenesis and chemo-resistance in a large number of solid cancers and leukaemia (Ganan-Gomez et al., 2013). Nrf2 has been extensively studied in NSCLC, which is the most common type of lung cancer and is characterized by a low sensitivity to chemotherapy. To assess the effects of hMOF on drug resistance and proliferation of human NSCLC, the expression of hMOF was knocked down in A549 cells. hMOF knockdown significantly increased the sensitivity of these cells to CDDP (Figure 6A) and to two other types of cytotoxic agent, 5-FU (Figure 6B) or bleomycin (Figure6C). We also assessed the effects of hMOF knockdown on the growth of A549 cells in the absence of any cytotoxic agent and found that the proliferation of these cells was markedly decreased (Figure 6D). Those results are paralleled with Nrf2-knockdown (Homma et al., 2009).
Figure 6.
hMOF regulates drug resistance and cell proliferation in human lung cancer cells. (A–C) hMOF knockdown enhances drug-induced decrease of survival. Human lung cancer A549 cells were transfected with NC-siRNA or hMOF-siRNA for 24 h. Then the cells were treated with CDDP (A), 5-FU (B) or Bleomycin (C) of the indicated concentration for 48 h. Cell survival was monitored by MTT method. (D) hMOF knockdown decreases cell proliferation. A549 cells were transfected with NC-siRNA or hMOF-siRNA. Cell proliferation was monitored by MTT method at the indicated time points post-transfection. *P < 0.05, **P < 0.01; significantly different from Ctrl.
To explore whether Nrf2 is essential for the effect of hMOF, we assessed the effects of hMOF on LC-AI and NCI-H292, two lung cancer cell lines expressing low levels of Nrf2 and less resistant to CDDP treatment, compared with A549 cells (Homma et al., 2009), but their expression of hMOF is as high as in A549 cells (Homma et al., 2009 and Supporting Information Fig. S4e). The results showed that hMOF knockdown did not change the sensitivity of LC-AI and NCI-H292 cells to CDDP (Supporting Information Fig. S4a and b) or their basal proliferation, i.e., in the absence of cytotoxic agent (Supporting Information Fig. S4c and d).
hMOF regulates expression of self-defence genes in lung cancer cells
Several studies have shown that the development of drug resistance is related to the expression of self-defence genes such as those encoding drug metabolizing enzymes, drug efflux transporters and antioxidant enzymes. Nrf2 regulated the expression of such self-defence genes in human NSCLC (Homma et al., 2009). In our experiments, we found that CDDP treatment of A549 cells induced expression of gens for antioxidant enzymes (heavy and light subunits of γ-glutamyl cysteine synthetase (GCLC and GCLM), HO-1, glutathione reductase and glutathione peroxidase 2), xenobiotic metabolism enzymes (NQO1, thioredoxin and thioredoxin reductase 1) and ATP-dependent drug efflux pumps (multidrug resistance-associated protein 1 and 2 and ATP7A; Figure 7A–C). When hMOF was knocked down, the basal and CDDP-induced expression of those genes were markedly reduced (Figure 7A–C). We also found that CDDP enhanced Nrf2 binding to hMOF and thus increased Nrf2 acetylation, which was reduced by hMOF knockdown (Figure 7E–F). However, we did not observe any changes in expression of self-defence genes when hMOF was knocked down in LC-AI and NCI-H292 cells (data not shown).
Because GCLC and GCLM are rate-limiting enzymes for glutathione biosynthesis, we then quantified total intracellular glutathione levels in A549 cells. hMOF knockdown significantly reduced basal and CDDP-induced synthesis of glutathione (Figure 7D).
hMOF regulates tumour growth and drug resistance in vivo
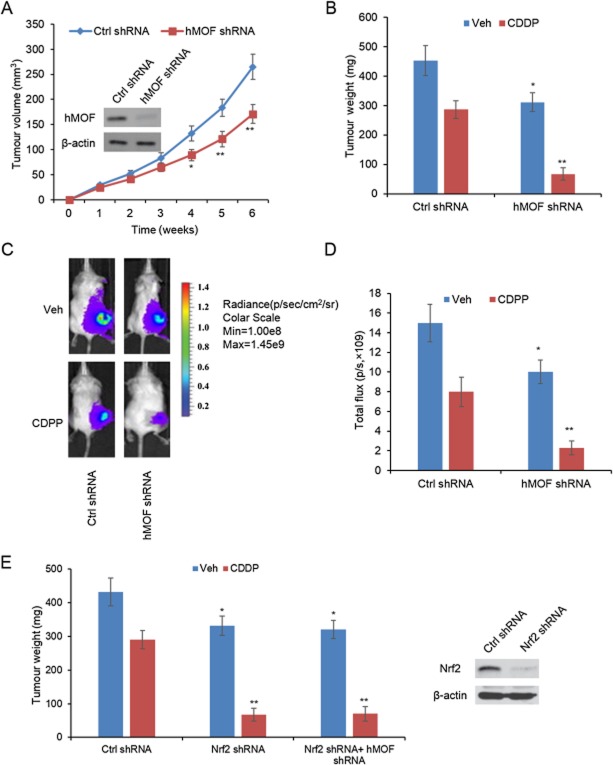
Because hMOF can regulate stress response and drug resistance in vitro, and the high level of hMOF predicts poor survival, we wanted to know whether hMOF can regulate drug resistance in vivo. We therefore generated A549 cells constitutively expressing shRNA targeting hMOF or Ctrl shRNA and injected them subcutaneously over the right and left flanks in male nu/nu mice between 4 and 6 weeks of age. Tumour growth 4 weeks after implantation was less in the mice bearing A549 cells with hMOF knockdown (Figure 8A). In tumour-bearing mice treated with CDDP, the mean tumour weight in the Ctrl shRNA group decreased to 63%, whereas the mean tumour weight of hMOF shRNA group decreased to 22% (Figure 8B). Similar results were obtained in bioluminescence imaging analysis (Figure 8C and D). Finally, we showed that hMOF knockdown did not induce additional reduction of cell growth either with or without CDPP treatment when Nrf2 was silenced (Figure 8E), indicating that Nrf2 is an essential component of the response to hMOF in tumour cells.
Figure 8.
hMOF regulates lung cancer growth and drug resistance in vivo. (A) A549 cells constructively expressing sh-RNA targeting hMOF (hMOF shRNA) were injected in the flank of male athymic nude mice (n = 8). A549 cells expressing Ctrl shRNA were used as control (n = 8). Weekly measurements were made of the tumours. *P < 0.05, **P < 0.01; significantly different from Ctrl shRNA. (B) Mice bearing hMOF shRNA- or Ctrl shRNA-expressing A549 cells were treated with CDPP (5 mg·kg−1, i.p.) or saline (vehicle/control, i.p.) weekly after day 21 for 4 weeks. Each treatment group involved eight tumour-bearing mice. Weight of the tumour was recorded at the termination of the experiment. (C) Tumour size was quantified by in vivo bioluminescence imaging. (D) Quantitative analysis of total photon flux values. (E) Mice bearing Nrf2-shRNA, Nrf2-shRNA + hMOF shRNA- or Ctrl shRNA-expressing A549 cells were treated with CDPP (5 mg·kg−1, i.p.) or saline (vehicle/control, i.p.) weekly after day 21 for 4 weeks. Each group involved eight tumour-bearing mice. Weight of the tumour was recorded at the termination of the experiment. *P < 0.05 significantly different from Ctrl shRNA + Veh; **P < 0.01, significantly different from Ctrl shRNA + CDPP.
Discussion and conclusions
In this study we found that hMOF was overexpressed in human NSCLC and was correlted with a poor prognosis. We identified Nrf2 as a novel non-histone substrate of hMOF. MOF acetylated Nrf2 at Lys588. We showed that MOF promoted Nrf2 nucleus retention and transcriptional activation by acetylating Nrf2. MOF facilitated anti-oxidative responses and drug resistance via Nrf2-dependent manners in vitro and regulated tumour growth and drug resistance in vivo.
MOF plays an essential role in mammals during carcinogenesis. In comparison with primary cells or normal tissues, all immortalized human normal and tumour cell lines and primary tumours showed similar or elevated hMOF level. Accordingly, MOF overexpression correlates with increased cellular proliferation, oncogenic transformation and tumour growth (Gupta et al., 2008). Abnormal gene expression of hMOF is involved in certain primary cancers, including breast cancer (Kapoor-Vazirani et al., 2011), renal cell carcinoma (Wang et al., 2013), ovarian cancer (Liu et al., 2013), medulloblastoma (Pfister et al., 2008), as well as lung cancers (Song et al., 2011; Zhao et al., 2013). Song et al. (2011) found that hMOF mRNA was frequently overexpressed in lung cancer tissues and that hMOF knockdown led to a significant inhibition of growth in the Calu-6 cell line. However, the mechanism underlying the involvement of hMOF inlung cancer remained unknown. When the present paper was under preparation, Zhao et al. (2013) reported that hMOF promoted proliferation, migration and adhesion of NSCLC cells in vivo, and attributed the effects of hMOF to acetylation of H4 at K16 and S-phase entry. However, they did not confirm their conclusions in vivo and in human lung cancer samples.
Here, we have shown that hMOF was markedly overexpressed in human NSCLC tissues compared with normal adjacent lung tissues, and that hMOF regulated tumour growth and drug resistance in vitro and in vivo. High levels of hMOF were significantly correlated with tumour size, disease stage, metastasis and recurrence. In addition, hMOF overexpression predicted decreased overall and disease-free survival. Importantly, we demonstrated that hMOF expression was significantly correlated with the expression of Nrf2-downstream genes, NQO1 and HO-1, two important genes in carcinogenesis and drug resistance (Dinkova-Kostova and Talalay, 2010; Ganan-Gomez et al., 2013). Our in vitro results strongly suggested that hMOF increased expression of self-defence genes and facilitated drug resistance and proliferation of human lung cancer cells. These effects of MOF/hMOF were largely Nrf2-dependent. Therefore, hMOF may facilitate the development and drug resistance of NSCLC, through regulating Nrf2-dependent genes. Our findings in lung cancer cell lines and in NSCLC patients have allowed us to identify a novel mechanism by which hMOF could support the development of lung cancer.
Plasticity in transcription regulation can be achieved by dynamic regulation of the chromatin structure, involving ATP-dependent changes in nucleosome positions, modification of histone tails or DNA methylation. Notably, the modification of transcriptional regulators and factors is also important. In Drosophila, MOF acetylates H4K16 and other proteins, including MSL3, a subunit of the dosage compensation complex. Acetylation modulates the interaction of MSL3 with roX2 RNA (Hilfiker et al., 1997; Akhtar and Becker, 2000; Buscaino et al., 2003). Posttranslational modification of the DNA-binding domain of p53 is important for its role in apoptosis. p53 is rapidly acetylated after DNA damage and this is catalysed by hMOF and TIP60 (Sykes et al., 2006; 2009). In addition, acetylation regulates the interaction of NoRC with promoter-associated RNA (pRNA), which in turn affects heterochromatin formation, nucleosome positioning and rDNA silencing. The acetylation of NoRC is regulated by MOF (Zhou et al., 2009). Here, we present data showing that Nrf2 is another non-histone substrate of MOF. Importantly, Nrf2 is the second transcriptional factor identified to be a substrate of hMOF. Such evidence strongly indicates that MOF regulates transcription through acetylating both histone (H4K16), and non-histone proteins, especially transcriptional factors (p53, Nrf2) and regulators (TIP5, NoRC).
MOF plays important roles in DNA damage repair (DDR) by modulating recruitment of the DDR protein, Mdc1 (Li et al., 2010). Several earlier studies have shown MOF to be involved in many steps of the DDR process (Gupta et al., 2005; Taipale et al., 2005; Sharma et al., 2010). Also, MOF interacts with proteins such as the MORF-related gene on chromosome 15 (Pardo et al., 2002; Garcia et al., 2007), Ruvb1/2 (Dou et al., 2005; Jha et al., 2008), and DNA-dependent protein kinase catalytic subunit (Sharma et al., 2010) that are important for DDR. Acetylation of p53 occurs rapidly after DNA damage and is catalysed by the MYST family acetyltransferases, hMOF and TIP60 (Sykes et al., 2006). In addition, hMOF is important for G2/M cell-cycle arrest (Gupta et al., 2005; Sykes et al., 2006; Zhao et al., 2013). Our present evidence showed that hMOF reduced oxidative stress and induced expression of self-defence genes to prevent apoptosis and promote survival. Taken together, those data indicate that MOF is involved in several steps of oxidative stress, DNA damage repair and apoptosis to promote cell survival. MOF not only acetylates H4K16, changes the local chromatin structure and recruits DDR proteins, but acetylates non-histone proteins such as p53 to regulate cell arrest and apoptosis, and orchestrates responses to oxidative stress that induces DNA damage by acetylating Nrf2.
In summary, our present findings uncovered novel mechanisms in which MOF dynamically interacts with and activates Nrf2 through acetylation at Lys588. We have provided additional mechanistic insights into the processes underlying regulation by MOF of responses to oxidative damage and of drug resistance in vitro and in vivo. Our results have also explained how Nrf2 is regulated during cellular responses to oxidative stress and cytotoxic drug treatment. Finally, our data suggest that hMOF may serve as a potential target in developing new drugs for the treatment of NSCLC.
Acknowledgments
This study was supported by grants from the Youth Foundation of Shanghai Municipal Public Health Bureau (2010Y043); International S&T Cooperation Program of China (2012DFG31320); Shanghai Foundation for Leaders of Disciplines in Science, China (13XD1403300); and the Science and Technology Commission of Shanghai (06DZ19501).
Glossary
- ARE
antioxidant response element
- CDDP
cis-diamminedichloroplatinum
- GCLM
glutamate-cysteine ligase modifier subunit
- HO-1
haem oxygenase 1
- Keap1
Kelch-like ECH-associated protein 1
- MEF
mouse embryonic fibroblast
- MOF
males absent on the first
- MSL
male-specific lethal
- Nrf2
NF–E2-related factor 2
- NQO1
NAD(P)H quinone oxidoreductase 1
- NSCLC
non-small cell lung cancer
- 5-FU
5-fluorouracil
Author contributions
Zhiwei Chen and Xiangyun Ye performed most experiments. Naiwang Tang and Shengping Shen collected human samples. Ziming Li performed clinical analysis. Xiaomin Niu performed bio-information and statistical analysis. Shun Lu and Ling Xu designed the study and wrote the paper.
Conflict of interests
None.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
http://dx.doi.org/10.1111/bph.12661
Figure S1 hMOF interacts with Nrf2 in human lung cancer tissues. Proteins from human lung cancer tissues were immunoprecipitated with anti-IgG or anti-Nrf2 antibodies. The immunoprecipitates and input were subjected to IB analysis with anti-hMOF, anti-Nrf2 and anti-β-actin antibodies.
Figure S2 MOF promotes Nrf2 acetylation and nuclear retention. (A) hMOF knockdown reduces H2O2-induced acetylation of Nrf2. 293T-cells were transfected with negative control siRNA (NC-siRNA) or siRNA targeting hMOF (hMOF-siRNA) for 36 h and then treated with/without 100 μM H2O2 for additional 12 h. Cells lysates were subjected to IP analysis and the immunoprecipitates and input were analysed by IB with indicated antibodies. (B) hMOF overexpression promotes Nrf2 nuclear retention. 293T-cells were transfected with plasmids expressing Flag-hMOF for 48 h. Nuclear and cytoplasmic proteins were extracted and subjected to IB analysis with indicated antibodies. (C) hMOF-mediated Nrf2 nucleus retention depends on K588 of Nrf2. Plasmids expressing HA-Nrf2WT or HA-Nrf2K588R were transfected to Nrf2-KO MEF cells with/without Flag-hMOF co-transfection. 48 h post transfection, nuclear and cytoplasmic proteins were extracted respectively and subjected to IB analysis with indicated antibodies.
Figure S3 MOF does not affect the intracellular ROS production and survival of Nrf2-KO MEFs. (A–B) Nrf2-KO MEF cells were transfected with NC-siRNA or MOF-siRNA for 24 h followed by 100 μM H2O2 (A) or UV (B) treatment for the indicated time. DCFH-DA method was used to analyse ROS level. (C-D) Nrf2-KO MEF cells were transfected with NC-siRNA or MOF-siRNA for 24 h followed by 500 μM H2O2 (C) or UV (D) treatment for 24 or 48 h. MTT method was used to analyse survival. (E) IB showing the knockout efficacy of Nrf2. (F) IB showing the knockdown efficacy of MOF. Values are expressed as mean ± SEM of three independent experiments.
Figure S4 hMOF does not affect survival and proliferation of LC-AI and NCI-H292 cells. (A–B) hMOF knockdown does not affect LC-AI and NCI-H292 cell survival. NC-siRNA or hMOF-siRNA were transfected to LC-AI (A) or NCI-H292 (B) cells for 24 h, and then the cells were treated with CDDP of the indicated concentration for 48 h. (C–D) hMOF knockdown does not affect LC-AI and NCI-H292 cell proliferation. NC-siRNA or hMOF-siRNA were transfected to LC-AI (C) or NCI-H292 (D) cells. MTT was used to analysis cell survival and proliferation. (E) IB result showing Nrf2 and hMOF protein levels in A549, LC-AI and NCI-H292 lung cancer cells. Values are expressed as mean ± SEM of three independent experiments.
Table S1 Primers used for q-PCR analysis.
Table S2 Primers used for CHIP/q-PCR analysis.
References
- Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- Alexander SPH, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, et al. The Concise Guide to PHARMACOLOGY 2013/14: Enzymes. British Journal of Pharmacology. 2013a;170:1797–1867. doi: 10.1111/bph.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, et al. The Concise Guide to PHARMACOLOGY 2013/14: Transporters. British Journal of Pharmacology. 2013;170:1706–1796. doi: 10.1111/bph.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, et al. The Concise Guide to PHARMACOLOGY 2013/14: Enzymes. British Journal of Pharmacology. 2013b;170:1797–1867. doi: 10.1111/bph.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone JR, Lavender J, Richman R, Palmer MJ, Turner BM, Kuroda MI. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- Buscaino A, Kocher T, Kind JH, Holz H, Taipale M, Wagner K, et al. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol Cell. 2003;11:1265–1277. doi: 10.1016/s1097-2765(03)00140-0. [DOI] [PubMed] [Google Scholar]
- Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Talalay P. NAD(P)H: quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys. 2010;501:116–123. doi: 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Ganan-Gomez I, Wei Y, Yang H, Boyano-Adanez MC, Garcia-Manero G. Oncogenic functions of the transcription factor Nrf2. Free Radic Biol Med. 2013;65:750–764. doi: 10.1016/j.freeradbiomed.2013.06.041. [DOI] [PubMed] [Google Scholar]
- Garcia SN, Kirtane BM, Podlutsky AJ, Pereira-Smith OM, Tominaga K. Mrg15 null and heterozygous mouse embryonic fibroblasts exhibit DNA-repair defects post exposure to gamma ionizing radiation. FEBS Lett. 2007;581:5275–5281. doi: 10.1016/j.febslet.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajji N, Wallenborg K, Vlachos P, Fullgrabe J, Hermanson O, Joseph B. Opposing effects of hMOF and SIRT1 on H4K16 acetylation and the sensitivity to the topoisomerase II inhibitor etoposide. Oncogene. 2010;29:2192–2204. doi: 10.1038/onc.2009.505. [DOI] [PubMed] [Google Scholar]
- Hayashi A, Suzuki H, Itoh K, Yamamoto M, Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem Biophys Res Commun. 2003;310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N, et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15:3423–3432. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor-Vazirani P, Kagey JD, Powell DR, Vertino PM. Role of hMOF-dependent histone H4 lysine 16 acetylation in the maintenance of TMS1/ASC gene activity. Cancer Res. 2008;68:6810–6821. doi: 10.1158/0008-5472.CAN-08-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor-Vazirani P, Kagey JD, Vertino PM. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol. 2011;31:1594–1609. doi: 10.1128/MCB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Garduno L, Theodore M, Yang J, Arinze IJ. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J Biol Chem. 2011;286:7629–7640. doi: 10.1074/jbc.M110.208173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG. Animal research: Reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, He Y, Wang L, Mo C, Zhang J, Zhang W, et al. D-galactose induces necroptotic cell death in neuroblastoma cell lines. J Cell Biochem. 2011;112:3834–3844. doi: 10.1002/jcb.23314. [DOI] [PubMed] [Google Scholar]
- Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-Y, Li TY, Liu Q, Zhang C, Li X, Chen Y, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- Liu N, Zhang R, Zhao X, Su J, Bian X, Ni J, et al. A potential diagnostic marker for ovarian cancer: involvement of the histone acetyltransferase, human males absent on the first. Oncol Lett. 2013;6:393–400. doi: 10.3892/ol.2013.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Li L, Lv X, Wu X-S, Liu D-P, Liang C-C. Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin. Cell Res. 2011;21:1182–1195. doi: 10.1038/cr.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Drummond G, McLachlan E, Kilkenny C, Wainwright C. Guidelines for reporting experiments involving animals: the ARRIVE guidelines. Br J Pharmacol. 2010;160:1573–1576. doi: 10.1111/j.1476-5381.2010.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert HS, McMahon SB. hMOF, a KAT(8) with many lives. Mol Cell. 2009;36:174–175. doi: 10.1016/j.molcel.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, et al. Loss of keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J Biol Chem. 2002;277:50860–50866. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- Pfister S, Rea S, Taipale M, Mendrzyk F, Straub B, Ittrich C, et al. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int J Cancer. 2008;122:1207–1213. doi: 10.1002/ijc.23283. [DOI] [PubMed] [Google Scholar]
- Rachakonda G, Sekhar KR, Jowhar D, Samson PC, Wikswo JP, Beauchamp RD, et al. Increased cell migration and plasticity in Nrf2-deficient cancer cell lines. Oncogene. 2010;29:3703–3714. doi: 10.1038/onc.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JS, Chun S-M, Lee JY, Kim DK, Kim YH, Jang SJ. The histone acetyltransferase hMOF is overexpressed in non-small cell lung carcinoma. Korean J Pathol. 2011;45:386–396. [Google Scholar]
- Sun Z, Chin YE, Zhang DD. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol Cell Biol. 2009;29:2658–2672. doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Stanek TJ, Frank A, Murphy ME, McMahon SB. Acetylation of the DNA binding domain regulates transcription-independent apoptosis by p53. J Biol Chem. 2009;284:20197–20205. doi: 10.1074/jbc.M109.026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol. 2008;28:5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath V, Wielandt AM, Iruretagoyena M, Chianale J. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang R, Wu D, Lu Z, Sun W, Cai Y, et al. Epigenetic change in kidney tumor: downregulation of histone acetyltransferase MYST1 in human renal cell carcinoma. J Exp Clin Cancer Res. 2013;32:1–8. doi: 10.1186/1756-9966-32-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamadori T, Ishii Y, Homma S, Morishima Y, Kurishima K, Itoh K, et al. Molecular mechanisms for the regulation of Nrf2-mediated cell proliferation in non-small-cell lung cancers. Oncogene. 2012;31:4768–4777. doi: 10.1038/onc.2011.628. [DOI] [PubMed] [Google Scholar]
- Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- Zhao L, Wang D-L, Liu Y, Chen S, Sun F-L. Histone acetyltransferase hMOF promotes S phase entry and tumorigenesis in lung cancer. Cell Signal. 2013;25:1689–1698. doi: 10.1016/j.cellsig.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Schmitz KM, Mayer C, Yuan X, Akhtar A, Grummt I. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol. 2009;11:1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 hMOF interacts with Nrf2 in human lung cancer tissues. Proteins from human lung cancer tissues were immunoprecipitated with anti-IgG or anti-Nrf2 antibodies. The immunoprecipitates and input were subjected to IB analysis with anti-hMOF, anti-Nrf2 and anti-β-actin antibodies.
Figure S2 MOF promotes Nrf2 acetylation and nuclear retention. (A) hMOF knockdown reduces H2O2-induced acetylation of Nrf2. 293T-cells were transfected with negative control siRNA (NC-siRNA) or siRNA targeting hMOF (hMOF-siRNA) for 36 h and then treated with/without 100 μM H2O2 for additional 12 h. Cells lysates were subjected to IP analysis and the immunoprecipitates and input were analysed by IB with indicated antibodies. (B) hMOF overexpression promotes Nrf2 nuclear retention. 293T-cells were transfected with plasmids expressing Flag-hMOF for 48 h. Nuclear and cytoplasmic proteins were extracted and subjected to IB analysis with indicated antibodies. (C) hMOF-mediated Nrf2 nucleus retention depends on K588 of Nrf2. Plasmids expressing HA-Nrf2WT or HA-Nrf2K588R were transfected to Nrf2-KO MEF cells with/without Flag-hMOF co-transfection. 48 h post transfection, nuclear and cytoplasmic proteins were extracted respectively and subjected to IB analysis with indicated antibodies.
Figure S3 MOF does not affect the intracellular ROS production and survival of Nrf2-KO MEFs. (A–B) Nrf2-KO MEF cells were transfected with NC-siRNA or MOF-siRNA for 24 h followed by 100 μM H2O2 (A) or UV (B) treatment for the indicated time. DCFH-DA method was used to analyse ROS level. (C-D) Nrf2-KO MEF cells were transfected with NC-siRNA or MOF-siRNA for 24 h followed by 500 μM H2O2 (C) or UV (D) treatment for 24 or 48 h. MTT method was used to analyse survival. (E) IB showing the knockout efficacy of Nrf2. (F) IB showing the knockdown efficacy of MOF. Values are expressed as mean ± SEM of three independent experiments.
Figure S4 hMOF does not affect survival and proliferation of LC-AI and NCI-H292 cells. (A–B) hMOF knockdown does not affect LC-AI and NCI-H292 cell survival. NC-siRNA or hMOF-siRNA were transfected to LC-AI (A) or NCI-H292 (B) cells for 24 h, and then the cells were treated with CDDP of the indicated concentration for 48 h. (C–D) hMOF knockdown does not affect LC-AI and NCI-H292 cell proliferation. NC-siRNA or hMOF-siRNA were transfected to LC-AI (C) or NCI-H292 (D) cells. MTT was used to analysis cell survival and proliferation. (E) IB result showing Nrf2 and hMOF protein levels in A549, LC-AI and NCI-H292 lung cancer cells. Values are expressed as mean ± SEM of three independent experiments.
Table S1 Primers used for q-PCR analysis.
Table S2 Primers used for CHIP/q-PCR analysis.