Abstract
The constitutive expression of the high-risk HPV E6 and E7 viral oncogenes is the major cause of cervical cancer. To comprehensively explore the composition of HPV16 early transcripts and their genomic annotation, cervical squamous epithelial tissues from 40 HPV16-infected patients were collected for analysis of papillomavirus oncogene transcripts (APOT). We observed different transcription patterns of HPV16 oncogenes in progression of cervical lesions to cervical cancer and identified one novel transcript. Multiple-integration events in the tissues of cervical carcinoma (CxCa) are significantly more often than those of low-grade squamous intraepithelial lesions (LSIL) and high-grade squamous intraepithelial lesions (HSIL). Moreover, most cellular genes within or near these integration sites are cancer-associated genes. Taken together, this study suggests that the multiple-integrations of HPV genome during persistent viral infection, which thereby alters the expression patterns of viral oncogenes and integration-related cellular genes, play a crucial role in progression of cervical lesions to cervix cancer.
Introduction
Cervical cancer is the second leading cause of cancer-related death in women worldwide. The persistent infection by high-risk human papillomavirus (HR-HPV), such as genotype 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59 are essential for the progression of cervical lesions [1], and over 50% cases are caused by HPV16 [2]. Viral oncoproteins, E6 and E7, of HR-HPVs contribute to cervical carcinogenesis by inactivating two major cellular tumor suppressor proteins, p53 and pRb, respectively [3]–[6]. These viral oncoproteins in infected cells can also result in chromosome instability and accumulation of mutation events [7].
A viral early promoter lied upstream of the E6 ORF, such as P97 in HPV16 [8], [9], P99 in HPV31 [10], [11] and P105 in HPV18 [12], [13], is responsible for almost all early gene expression, including E6 and E7. Upstream cis-elements in the LCR interact with cellular transcription factors and the viral transactivator/repressor E2 and regulate the transcription of HPV E6 and E7 genes [8], [14]. Furthermore, DNA methylation [15], alternative RNA splicing [9], [16], [17] and early poly(A) site polyadenylation signal [18], [19] also take part in the regulation of E6 and E7 gene expression [19].
To date, a full transcription map of oncogenic HPV16 and HPV18 in HPV-infected cells and raft tissues have been constructed [19], [20].
It's well known that the integration of HPV genomes is a key event in cervical carcinogenesis [21], [22]. Besides viral genome integration in activating cellular oncogenes or inactivating cellular tumor suppressive genes [23]–[25], HPV genome integration into host genome may change the transcription patterns of both viral and host genes [26]. It has been reported that the integration of HPV genomes can disrupt the viral E2 gene in cells and release its inhibition on the viral early promoter that controls the expression of E6 and E7 [27]. In addition, E6 and E7 transcripts cotranscribed with cellular sequences may be more stable, and thus enhance their expression level [28]–[30].
Transcription patterns of HPV16 in the tissues of cervical cancer have been reported [26], [31]. There were an episomal HPV early gene transcript (E7-E1∧E4) and several integrated HPV transcripts (such as E7-E1∧cellular RNA, E7-E1∧E4-cellular RNA, etc.) in HPV16-infected tissues. However, transcriptional selection in response to environmental changes is a dynamic process to achieve optimal gene expression for cell survival and carcinogenesis [32]. In this study, we applied a modified technique of amplification of papillomavirus oncogene transcripts (APOT) [26] to comprehensively explore the structure and sequences of HPV16 E7 related transcripts and their genomic annotation in 8 LSIL (low-grade squamous intraepithelial lesions), 24 HSIL (high-grade squamous intraepithelial lesions), and 8 CxCa HPV16-positive cervical biopsy samples.
Materials and Methods
Patients and specimens
Tissue samples of primary uterine cervical lesions containing dysplastic epithelium/tumor cells were collected from the Second Affiliated Hospital of Wenzhou Medical University (Zhejiang Province, China) from December 2010 to April 2012. The presence of HR-HPV was detected by HCII test, and the screening of HPV16 in HR-HPV-positive samples was done by HPV genotypes detection kit (KaiPu, Guangzhou, China) [33]. All of them did not receive radiation therapy or chemotherapy before operation and each patient underwent a colposcopically directed biopsy. The collected biopsy specimens were bisected. One portion was submitted for standard histopathologic diagnosis, while the other portion was stored in RNAlater (Ambion, Austin, Texas, USA) at −80°C for subsequent analysis. On the basis of the histopathologic diagnosis, the samples were divided into LSIL (CIN I, n = 8), HSIL (CIN II, n = 22; CIN III, n = 16) and cervix carcinoma (CxCa, n = 17). Additional 8 cervical tissues with normal cytology and HPV DNA negativity as controls were obtained from the patients who underwent hysterectomy owing to benign gynecologic diseases. The study has been approved by the Medical Ethics Committee of Second Affiliated Hospital of Wenzhou Medical University. All women were informed and gave their written consent to participate in the study.
RNA and DNA Isolation from Clinical Samples
Total RNA from biopsy samples described above was isolated using TRIzol reagent (Invitrogen, Calif., USA) according to the manufacturer's instructions. To remove the residual DNA contamination, the RNA preparation was treated with Rnase-free Dnase I (Takara, Dalian, China) according to the manufacturer's protocol. Purified total RNA was dissolved in Rnase-free water and stored at −80°C. The concentration and purity of total RNA were quantified by the ultraviolet spectrophotometer at 260 nm and 280 nm and 1% agarose gel electrophoresis. Only RNA samples with an A260/A280 ratio of 1.8–2.0 and high integrity were used for the further experiment.
Reverse Transcription and PCR Amplification of Transcripts
APOT assay reported previously was based on nested PCR reactions [26], which could only amplify the abundant transcripts and ignore the transcripts with lower levels in samples. So modified APOT assay was used to amplify the HPV oncogene transcripts. The primers for these reactions were designed according to Klaes R, et al [26]. Total RNA (1 µg) was reversely transcribed using an oligo(dT)17-primer coupled to a linker sequence RT [34] according to the manufacturer's protocol of reverse transcriptase Kit (TOYOBO, Japan). To verify first-strand cDNA quality, PCR using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) -specific primers were performed as previously described [35]. First-strand cDNA encompassing viral oncogene sequences were subsequently amplified by PCR using p1-HPV16E7 specific primer (5′-CGGACAGAGCCCATTACAAT-3′) and linker p0 (5′-GACTCGAGTCGACATCG-3′) as the reverse primer; and the PCR amplification was carried out in a reaction volume of 50 µl. Different from previous reports, the PCR cycles was increased to 35, and all specimens only performed one-round PCR reaction. To verify the specificity of this procedure, the “minus-RT” control in which reverse transcriptase was omitted from the reactions was also performed parallel.
Sequence Analysis of Transcripts
The APOT amplification products were visualized by 2.5% agarose gel electrophoresis. PCR products of interest were excised from the gel and extracted using DNA agarose gel recovery kit (TianGen, Beijing, China). The corresponding amplimeres were cloned into cloning vector (TransGen, Beijing, China) and DNA sequence analysis was executed using an ABI 3730 XL Genetic Analyser (Applied Biosystems, USA) according to standard protocols. Sequencing results were analyzed using the BLASTn program provided by the National Center for Biotechnology Information, USA. Additionally, the chromosomal integration sites were ascertained using the National Center for Biotechnology Information (BLAST) and European Molecular Biology Laboratory (EBI). Moreover, the fragile sites and genes of integration sites were defined using the NCBI fragile site map viewer and the UCSC Blat tool.
Results
Specificity of APOT assay for HPV16 oncogene transcripts
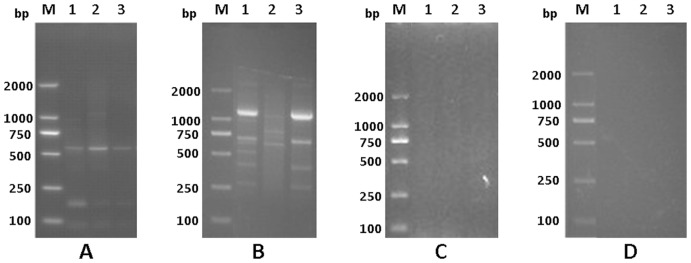
The principle of the APOT assay is a 3′ rapid amplification of cDNA ends (RACE) PCR assay that achieves amplification and cloning of the region between a single short sequence in a cDNA molecule and its unknown 3′- end [34]. In general, the integrated transcripts derived from E6 and E7 oncogenes encompass viral sequences at their 5′- ends and host genome sequences at their 3′- ends [26]. The expected size of products obtained from an episome-derived transcript is 1050 bp [26] Amplimers that displayed a size different from 1050 bp may therefore be derived from an integrated HPV genome. To testify the specificity of the modified APOT assay, cDNAs from HPV16-positive Caski cell contains the integrated HPV16 genome and HPV-negative normal cervical tissues, as well as the “minus-RT” controls in which reverse transcriptase was omitted from the reactions were used. The amplified products of the cDNAs from HPV16-positive Caski cell were similar to the previous report [26], whereas no RT product was obtained from the normal cervical tissues without HPV DNA and the “minus-RT” control (Figure 1). These data indicated the modified APOT assay can specifically detect the transcripts derived from the integrated HPV genome.
Figure 1. Specificity of APOT assay in detection of HPV16 oncogene transcripts.
Amplified products from CaSki cells (A), HPV16-positive CxCa (B), HPV negative normal cervical tissues (C) and the “minus-RT” controls of the RNA isolated from HPV 16-positive samples (D) by the APOT assay were separated on 2.5% agarose gels. A: Lane 1-3 were three different subcultured CaSki cells (as a positive controls); B: Lane 1-3 were three different CxCa samples; C: Lane 1-3 were three different normal cervical tissues (as negative controls); D: Lane 1-3 were corresponding with samples in B, respectively; M: 250-bp DNA ladders.
Characteristics of HPV16 oncogene transcripts in the tissues of cervical intraepithelial neoplasia and cervix carcinoma
To analyze the HPV16 oncogene transcripts, 40 HPV16-positive cervical specimens (LSIL, n = 8; HSIL, n = 24; CxCa, n = 8) with good quality RNA were selected among 63 collected samples in this study. Total 133 transcripts containing viral fragments were found. Among these transcripts, 64 fragments had HPV16 E7-E1* sequences at their 5′- ends and directly connected with poly A at their 3′- ends (Figure S1). Furthermore, there were four different disruptions of E1 region at nt 880, 949, 1054 and 1234 (Figure S2). The transcripts containing an E1-splice donor signal at nt 880 [36] might belong to potential episomal pattern, whereas the transcript which truncated at nt949 might be a result of internal priming by oligo dT [37]. Other transcripts which truncated at nt 1054 and 1234 neither contained poly A sequences, nor any polyadenylation site belong to viral or host, so these transcripts were viewed as potential integrated patterns. In addition, we also found another transcript which has E7 ORF spliced at nt 880 to the E4-splice acceptor site at nt 3358 and then spliced from the E4-splice donor signal at nt 3632 to the L1-splice acceptor site at 5639, and also terminated at poly A (Figure S1). In this transcript, the E4 ORF is not disrupted. Lack of a splice donor signal at nt 5815 in this transcript indicates that the HPV16 genome disrupted at nt 5815 might also take part in the virus genome integration.
In addition, there were 64 viral transcripts directly connected to host genome sequences and they were all began with the beginning of the forward primer (p1) at nt 729. These HPV16 oncogene integrated transcripts could be divided three different types (Figure 2). Among these transcripts, Type A has HPV16 E7-E1* sequences at their 5′- ends and directly connected to host genome sequences. However, there were two different integration sites of E1 region (at nt880 and nt1107) in this type (Figure S3). The site at nt880 contained an E1-splice donor signal while the site truncated at nt1107 might be more likely to linearize the viral circular genome for integration into the host genome. Transcript type B has an E2 ORF disrupted at nt2870 and the Type B sequence composes of HPV16 E7-E1∧E2* at its 5′- ends and the host genome sequence at its 3′- ends. In transcript type C, the E1∧E4 stop codon is disrupted for virus integration and an entire E1∧E4 ORF without a stop codon is fused in frame to host sequence. Among these three patterns, transcripts of Type A and C had been reported by Wentzensen N, et al. [31]. However, transcript of Type B had not previously been reported in precancerous lesions and cervical cancer.
Figure 2. Three types of HPV 16 early gene transcription pattern.
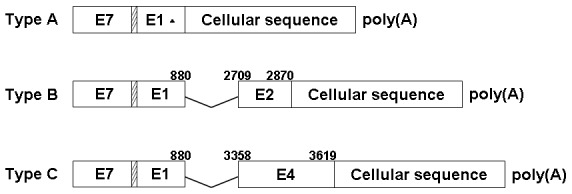
Type A shows E1 sequences spliced directly to cellular flanking sequence; Type B shows E1 spliced to E2, with E2 fused with a cellular sequence; Type C shows E1 spliced to E4, with E4 running into a cellular sequence. ▴, there are two integration sites in E1 (data shown in Figure S3). The boxes within slashes represent six nucleotides between E7 and E1gene.
Moreover, HPV16 oncogenes showed significantly different transcription patterns in the tissues of LSIL, HSIL and CxCa (Figure 3, 4 and Table 1). Among these 3 transcription patterns detected in our patients, the Type A and Type B were higher prevalence than Type C, which were observed in almost all pathological types, whereas the Type C was detected only in the samples of CxCa, with a detection frequency of 75% (Table 1 and Figure 4). All patient samples displayed the Type A, but all CxCa samples had the Type B and Type C (Figure 4). Consistent with the presumption of potential integration of the viral genome in the later stages of cancer development [38], [39], the prevalence of fusion transcripts were higher in HSIL and CxCa than LSIL.
Figure 3. Detection of HPV 16 early gene transcripts by the APOT assay.
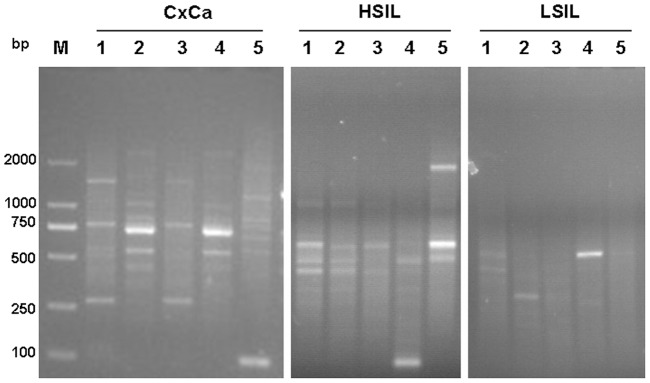
HPV16-positive clinical samples with LSIL, HSIL and CxCa were subjected to the APOT assay and separated on 2.5% agarose gels, Lane 1-5 mean five different samples in each pathological type. M: 250-bp DNA Ladders.
Figure 4. The proportion of patient samples that contain the different type of transcripts.
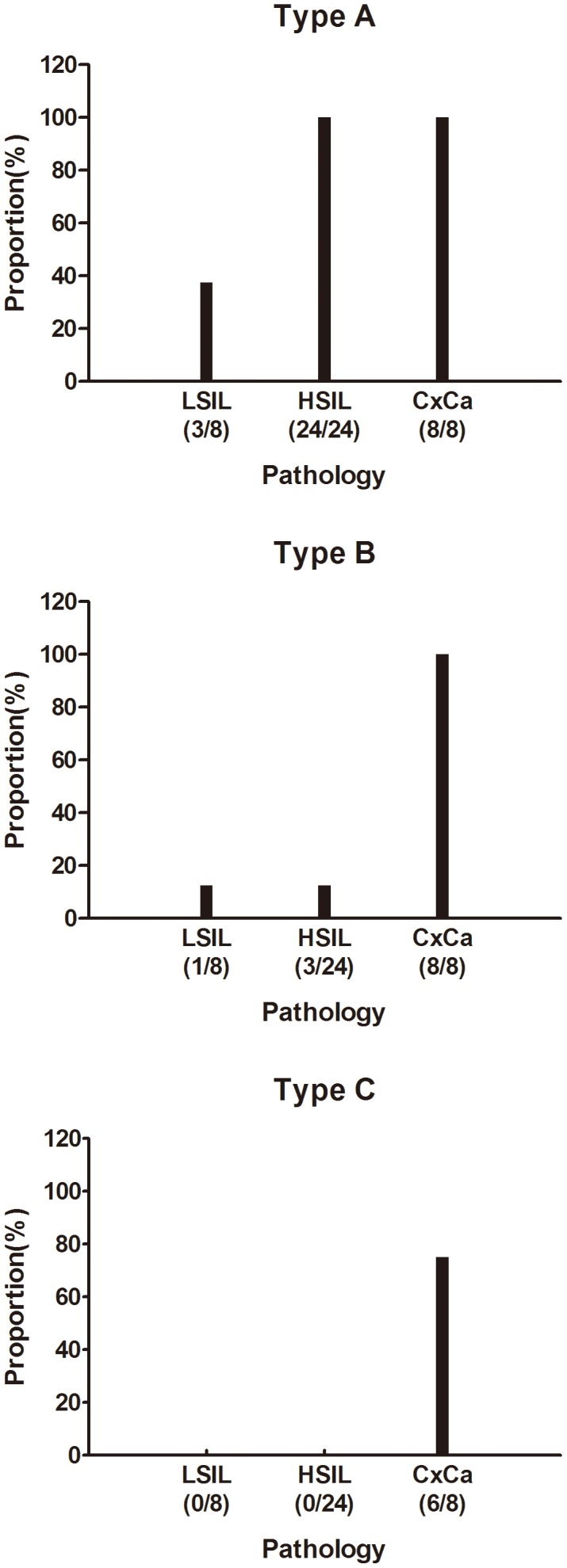
Table 1. The Transcript Number of Three Transcription Patterns in the Groups of LSIL, HSIL and CxCa.
| Different pathological types | ||||
| Pattern | Total | LSIL | HSIL | CxCa |
| Type A | 48 | 3 | 35 | 10 |
| Type B | 10 | 1 | 1 | 8 |
| Type C | 6 | - | - | 6 |
64 transcripts in total, directly connected to host genome sequences at their 3′- ends, were detected from 40 HPV 16-positive cervical specimens, including 8 LSIL, 24 HSIL and 8 CxCa. Type A and B were detected in all HPV 16-positive samples while type C was only found in CxCa. “-”, no transcripts.
Integration sites and characterization of the cellular flanking sequence
To identify the individual chromosomal locations, all 64 fusion-transcripts containing viral and cellular sequences were further analyzed by BLASTn comparisons to the whole genome database. Our data show that all chromosomes, except for Chr21 and X, were integrated with HPV16 genome, confirming the previous reports that no preferential HPV integration site was seen in selection of the human chromosome [40]. Some loci, such as 1p36.22, 1p36, 2p24, 2q33, 5q31.1, 5q31, 6p24, 8p23, 10q22.1, 13q22.1, 19q13 and 19p13.3, were reported previously [31], [41]–[45] (Table 2). Among these integration events, fourteen of 40 samples exhibited multiple integration sites (Table 2). Although local DNA rearrangements could happen frequently and rapidly after the integration [43], we found that cellular flanking sequences in 11 tissues were mapped to different chromosomes, indicating the presence of multiple independent integrations in these samples. Moreover, we found that multiple-integration events were significantly higher in CxCa tissues (75%) than in the cervical tissues of LSIL (50%) and HSIL (53.8%). Screening of all integration loci indicates that 35 of 63 mapped integration sites were located in or close to a fragile site with a distance of 26 bp to 5 Mbp (Table 2). Among the 22 mapped fragile sites, FRA13A was found in 4 independent samples. Twenty-two transcripts were not associated with any fragile site.
Table 2. Summary of All Integration Sites and Characterization of the Cellular Flanking Sequence.
| Sample ID | Pathology | Integration locus | Nearest fragile site* | Accession code* | Gene name | Nearest genes* | Integrate to |
| 79 | LSIL | 11q13 | - | PRDX5 | peroxiredoxin 5 | ESRRA CCDC88B CCDC88B |
n.a |
| 11q23.3 | - | AMICA1 | adhesion molecule, interacts with CXADR antigen 1 | MPZL3 SCN2B SCN2B |
Exon 9 | ||
| 196 | LSIL | 11q23.3 | - | AMICA1 | adhesion molecule, interacts with CXADR antigen 1 | MPZL3 SCN2B SCN2B |
Exon 9 |
| 184 | HSIL | 10q22.1 | FRA10D(10q22.1) | KIAA1279 | KIAA1279 | DDX21 ACTBP14 ACTBP14 |
Intron 7 |
| 179 | HSIL | 8p23 | - | ARHGEF10 | Rho guanine nucleotide exchange factor (GEF) 10 | MIR596 KBTBD11 KBTBD11 |
Intron 27 |
| 8p23.3 | - | LOC100131395 | LOC100131395 | - | n.a. | ||
| 8q23.1 | - | ANGPT1 | angiopoietin 1 | PGAM1P13 HMGB1P46 HMGB1P46 |
Intron 5 | ||
| 8q23 | - | EBAG9 | estrogen receptor binding site associated, antigen, 9 | PKHD1L1 SYBU SYBU |
n.a. | ||
| 120 | HSIL | 19q13.1 | FRA19A (19q13) | COX6B1 | cytochrome c oxidase subunit VIb polypeptide 1 | ETV2 UPK1A UPK1A |
n.a. |
| 19q13.13 | FRA19A (19q13) | UPK1A | uroplakin 1A | COX6B1 ZBTB32 ZBTB32 |
n.a. | ||
| 12q23.3 | FRA12E (12q24) | SLC41A2 | solute carrier family 41, member 2 | KRT18P20 CHST11 CHST11 |
Exon 10 | ||
| 55 | HSIL | 1p36.22 | FRA1A (1p36) | MIR34A | microRNA 34a | LOC727721 GPR157 GPR157 |
Intron 2 |
| 169 | HSIL | 15q21.3 | - | RFX7 | regulatory factor X, 7 | HMGB1P33 CD24P2 CD24P2 |
n.a. |
| 29 | HSIL | 13q14.11 | FRA13A(13q14) | MRPS31 | mitochondrial ribosomal protein S31 | SLC25A15 FOXO1 FOXO1 |
Exon 1 |
| 13q14.3 | FRA13A(13q14) | THSD1 | thrombospondin, type I, domain containing 1 | VPS36 TPTE2P2 TPTE2P2 |
n.a. | ||
| 17q25.1 | - | EXOC7 | exocyst complex component 7 | FOXJ1 ZACN ZACN |
Intron 6 | ||
| 203 | HSIL | 2p24 | FRA2C(2p24.2) | ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | LOC650157 PQLC3 PQLC3 |
Intron 1 |
| 2p21 | FRA2C(2p21) | MSH2 | mutS homolog 2 | EPCAM LOC644093 LOC644093 |
n.a. | ||
| 139 | HSIL | 16q22.1 | FRA16C(16q22.1) | COG4 | component of oligomeric golgi complex 4 | SF3B3 FUK FUK |
Intron 10 |
| 16q23.3 | FRA16D(16q23.2) | WWOX | WW domain containing oxidoreductase | LOC100131126 MAF MAF |
Intron 9 | ||
| 16q23 | FRA16D(16q23.2) | ADAMTS18 | ADAM metallopeptidase with thrombospondin type 1 motif, 18 | NUDT7 VN2R10P VN2R10P |
Intron 4 | ||
| 16q24.2 | FRA16D(16q24) | MAP1LC3B | microtubule-associated protein 1 light chain 3 beta | FBXO31 NR3C1P1 NR3C1P1 |
Intron 3 | ||
| 7q36 | - | LMBR1 | limb development membrane protein 1 | NOM1 RNF32 RNF32 |
Intron 5 | ||
| 7p14.3 | FRA7B(7p14) | NT5C3A | 5′-nucleotidase, cytosolic IIIA | RP9 FKBP9 FKBP9 |
Intron 7 | ||
| 9q33 | FRA9E(9q32) | TNC | tenascin C | DEC1 TNFSF8 TNFSF8 |
Intron 3 | ||
| 9q21.33 | - | DAPK1 | death-associated protein kinase 1 | C9orf170 CTSL CTSL |
Intron 3 | ||
| 68 | HSIL | 10pter-q25.3 | FRA10D(10q25) | ASCC1 | activating signal cointegrator 1 complex subunit 1 | ANAPC16 SPOCK2 SPOCK2 |
Intron 6 |
| 10q21.2 | - | ZNF365 | zinc finger protein 365 | RTKN2 ADO ADO |
Intron 5 | ||
| 18q21.1 | FRA18B(18q21.3) | SETBP1 | SET binding protein 1 | KRT8P5 LOC10013166 LOC10013166 |
Intron 3 | ||
| 155 | HSIL | 6p24 | - | MAK | male germ cell-associated kinase | GCM2 LOC100506379 LOC100506379 |
Intron 8 |
| C15 | HSIL | 19q13 | FRA19A(19q13) | ACTN4 | actinin, alpha 4 | EIF3K CAPN12 CAPN12 |
Intron 1 |
| 51 | HSIL | 4q32-q33 | FRA4C(4q33) | TLL1 | tolloid-like 1 | LOC646995 SPOCK3 SPOCK3 |
Intron 2 |
| 5q31 | FRA5C(5q31.1) | PCDHA@ | protocadherin alpha cluster, complex locus | LOC100421074 PCDHB1 PCDHB1 |
Intron 1 | ||
| 5q31.1 | FRA5C(5q31.1) | Egr1 | early growth response 1 | Reep2 RPL7P19 RPL7P19 |
Intron 1 | ||
| 28 | HSIL | 14q21.2 | FRA14A(14q21) | FANCM | Fanconi anemia, complementation group M | SNORD127 MIS18BP1 MIS18BP1 |
Intron 4 |
| 42 | CxCa | 11q13 | - | PRDX5 | peroxiredoxin 5 | ESRRA CCDC88B CCDC88B |
n.a. |
| 13q22.1 | FRA13C(13q21.2) | PIBF1 | progesterone immunomodulatory binding factor 1 | BORA PSMD10P3 PSMD10P3 |
n.a. | ||
| 22q12.3 | FRA22B (22q12.2) | TIMP3 | TIMP metallopeptidase inhibitor 3 | FBXO7 LARGE-AS1 LARGE-AS1 |
Intron 2 | ||
| 97 | CxCa | 11q13 | - | PRDX5 | peroxiredoxin 5 | ESRRA CCDC88B CCDC88B |
n.a. |
| 2q37 | FRA2J(2q37.3) | AGAP1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 | Loc642692 Gbx2 Gbx2 |
Intron 14 | ||
| 13q14.11 | FRA13A(13q14) | MRPS31 | mitochondrial ribosomal protein S31 | SLC25A15 FOXO1 FOXO1 |
Exon 1 | ||
| 6 | CxCa | 2q33 | FRA2I(2q33) | CD28 | CD28 molecule | LOC100287498 KRT18P39 KRT18P39 |
n.a. |
| 2p22-p21 | FRA2C(2p21) | EIF2AK2 | eukaryotic translation initiation factor 2-alpha kinase 2 | SULT6B1 HEATR5B HEATR5B |
Intron 2 | ||
| 1q25.3 | FRA1G(1q25) | CACNA1E | calcium channel, voltage-dependent, R type, alpha 1E subunit | GM140 ZNF648 ZNF648 |
n.a. | ||
| 11q13 | - | PRDX5 | peroxiredoxin 5 | ESRRA CCDC88B CCDC88B |
n.a. | ||
| 206 | CxCa | 3p14.3 | FRA3B(3p14.2) | DENND6A | DENN/MADD domain containing 6A | PDHA1P1 ARF4 ARF4 |
n.a. |
| 7q21.11 | FRA7E(7q21.2) | SEMA3E | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E | SEMA3A PCLO PCLO |
Intron 2 | ||
| 94 | CxCa | 11q13 | - | PRDX5 | peroxiredoxin 5 | ESRRA CCDC88B CCDC88B |
n.a. |
| 213 | CxCa | 8q23 | - | EBAG9 | estrogen receptor binding site associated, antigen, 9 | PKHD1L1 SYBU SYBU |
n.a. |
| 4p13 | - | RHOH | ras homolog family member H | N4BP2 CHRNA9 CHRNA9 |
Intron 9 | ||
| 131 | CxCa | 13q22.1 | FRA13C(13q21.2) | PIBF1 | progesterone immunomodulatory binding factor 1 | BORA PSMD10P3 PSMD10P3 |
Exon 18 |
| 13q14.11 | FRA13A(13q14) | MRPS31 | mitochondrial ribosomal protein S31 | SLC25A15 FOXO1 FOXO1 |
Exon 1 | ||
| 19p13.3-p13.2 | FRA19B(19p13) | INSR | insulin receptor | LOC100996504 MBD3L3 MBD3L3 |
Intron 3 | ||
| 19q12 | - | CCNE1 | cyclin E1 | PLEKHF1 LOC126170 LOC126170 |
n.a. | ||
| 11q13 | - | PRDX5 | peroxiredoxin 5 | ESRRA CCDC88B CCDC88B |
n.a. | ||
| 190 | CxCa | 13q14.11 | FRA13A(13q14) | MRPS31 | mitochondrial ribosomal protein S31 | SLC25A15 FOXO1 FOXO1 |
Exon 1 |
| 9q21.33 | - | DAPK1 | death-associated protein kinase 1 | C9orf170 CTSL CTSL |
Intron 3 |
*“-”, no entry of fragile sites or nearest genes; n.a., not applicable because fusion transcript is in antisense orientation.
Genes highly relevant to cervix cancer which located in integration sites are indicated in italics, and genes indicated by an underline shows they are related to tumor.
The cellular flanking sequences of viral-cellular fusion transcripts were further examined for known genes. Most of these fused transcripts had a cellular sequence from the coding orientation of known genes and thirty transcripts had the cellular sequence from an intron region, and 8 transcripts were fused with a sense exon sequence of the predicted genes (Table 2). Among these predicted genes, AMICA1, DAPK1, EBAG9, PIBF1 were affected twice, MRPS31 four times and PRDX5 even six times by the viral integration. At the same time, the nearest host genes to each integration site in the direction of transcription were also analyzed (Table 2). Among these predicted genes integrated or closed to the integration site, we identified several tumor-associated genes, including PRDX5, CD28, ROCK2, RHOH, TIMP3 and DAPK1, etc. As shown in Table 1, the transcripts type D and E were only detected in CxCa and most of their integration loci were located in or close to the fragile sites of FRA13C, FRA22B, FRA2I and FRA13A. The genes associated with the transcripts type D and E were oncogenes (CD28 and EBAG9), tumor suppressor genes (TIMP3), or tumor-related genes (PIBF1and MRPS31).
Discussion
Integration of HPV genome into host chromosomes represents an early clonal event to provide an additional selective advantage for the expansion of the neoplasm. Viral transcripts have been detected by the APOT assay [26], [31], [42]–[45]. Although APOT assay has some advantages in detection transcripts from each chromosome integration site, there are several limitations. First, it is difficult to amplify very long integration-derived transcripts, which will underestimate the number of tumors with integrated HPV DNA [45]. Second, APOT is one type of nested PCR, which may tend to amplify the transcripts with higher levels and ignore those with lower levels. Third, It has been reported that the internal poly A priming could replace the oligo(dT) primer within certain limits, and generating a set of anchored oligo(dT) primers for cDNA synthesis. These sequences caused by internal priming interrupted the generating of full-length cDNA and confused the analysis of alternative splicing [37]. With our modified APOT assay to detect the transcription pattern of the cervical tissues, we did find many viral transcripts connected with poly A or host genome sequences in HPV16-infected cervical squamous epithelial tissues. We noticed that there were a lot of viral transcripts directly ended with poly A at their 3′- ends. Except for the reported E1-splice donor signal site (nt 880), the truncation sites at nt1054, 1234 and 5815 neither contained internal poly A sequences nor any polyadenylation signals should be potential novel integrated sites and need for further analysis. The viral-cellular fusion transcript of type A and C has been reported previously [26], [31], [41]. In the Type C transcript, the integration disruption of E4 termination codon would result in the E4 to use a host termination codon. In this study, we also noticed that some cervical cancer samples contained all three types of transcripts were viral-cellular fusion transcripts.
HPV16 transcription patterns in LSIL, HSIL, and CxCa were significantly different. We found that the Type C transcript was only detected in the samples of CxCa and more random integration sites existed in our tissue samples. Similar to previous reports [31], [38]–[42], [46], [47], our study indicates that HPV integration has no preferential site in the human genome. Except for chromosome 21 and X, other chromosomes are all susceptible to HPV16 integration. Approximately 55% integrations are located in or close to a fragile site. Different from previous reports [42], [45], we noticed that integration events often occur multiple times significantly more in cervical cancer than in LSIL and HSIL. These data not only provide biological support to the epidemiologic observation that persistent infection by specific types of HR-HPV is the important cause of cervical carcinoma [1], but also indicate that subsequent selection for and accumulation of mutations in yet-to-be-identified key cellular regulatory genes promotes further progression to cervical cancer.
The integration not only changes the transcription pattern relevant for the dysregulated expression of the viral oncogenes, but also affects the expression of the host gene with virus genome integration. The integration alters the expression of host genes in integration sites, even if this occurs within the intron sequences [43], [45]. In our study, we identified a broad spectrum of cancer-associated genes in the integration sites and flanking sequence regions. Most of genes in the integration sites were associated with tumor development, and nineteen genes were strongly related to cervical cancer. Some of them act as tumor suppressors (such as, miR-34a, MSH2, WWOX and TIMP3, et al) or oncogenes (such as, ROCK2, CD28, EBAG9 and ANGPT1, et al). Interestingly, most of them were not reported in previous documents [31], [45]. MiR-34a, an important tumor suppressor, is down-regulated in cervical cancer [48], [49]. It has been reported that oncoprotein E6 of HPV16 and HPV18 can inhibit the expression of tumor-suppressive miR-34a by destabilization of p53 and resulted in cell proliferation [50]. The disruption of miR-34a gene might further interpret the phenomenon of reduced expression of miR-34a in cervical cancer. MSH2 is a DNA mismatch repair protein, and associated with DNA repair pathway [51], [52]. Decreased expression of MSH2 might be a risk factor in the early stage cervical cancer [53]. ROCK2, an important signaling molecule, can promote cervical cancer metastasis by upregulating and activating the expression and function of moesin protein through RhoA/ROCK2 pathway [54]. Besides the cancer-associated genes, the genes in integration sites and flanking sequence regions might be also beneficial for viral genome integration. FANCM which is a DNA translocase and highly related to DNA replication regulates checkpoint signaling and replication fork progression [55], [56]. Other genes, such as COX6B1 is related to cell apoptosis [57] and ESRRA also have been reported associated with cervical cancer [58]. In addition, among 45 integration events, 13 events led to antisense transcription of the coding sequences, such as PRDX5, EBAG9 and CD28, etc. These integrations were generally deemed of no interest. However, their sense sequences were associated with DNA restoration or tumor development and might affect both host and viral gene expression during the development of cervical cancer. The most integration in the antisense orientation was the gene encoding peroxiredoxin 5 (PRDX5), a protective emzyme against oxidative stress [59], [60]. Its altered expression due to HPV16 integration could have significant virological consequence, along with the integration into DNA repair genes, such as FANCM and MSH2. Upregulation of EBAG9 expression has been observed in several malignant tumors [61]. The synergistic stimulation factor of CD28 which maintains immune homeostasis plays a role in increasing susceptibility to cervical cancer [47].
In conclusion, changes of the transcription patterns of HPV 16 early genes go along with the progression from cervical intraepithelial neoplasia to cervix carcinoma and viral genome integration into host chromosome. The change or selection of transcription patterns and the integration on the expression of host genes in the integration sites and flanking cellular sequence regions might all take part in oncogenesis of HPV16-induced cancers.
Supporting Information
The types of viral sequences connected with poly A at their 3′- ends. The type of Class I shows E1 sequences directly ended with poly A; the type of Class II shows E1 spliced to E4 and then to L1 and also ended with poly A sequences. ▴, there are several truncation sites in E1 (data shown in Figure S2).
(TIF)
Different truncation sites in E1 region in the type of Class I. There are four truncated sites in E1, 880, 949, 1054 and 1234, respectively.
(TIF)
Several spliced donor sites in E1. In Type A there are two integration sites in E1, 880, and 1107, respectively. The solid boxes mean cellular sequences.
(TIF)
Acknowledgments
We thank Zhi-Ming Zheng of NIH and Kong-Nan Zhao for helpful comments and revisions.
Funding Statement
This work is supported by grants from the National Natural Science Foundation of China (81001343, 81172463), the Department of Education of Zhejiang Province (Y200907403) and Wenzhou Science and Technology Bureau (Y20090103, Y20100175 and H20100063). These sponsors provided the funding for the experiments and the collection of specimens. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. zur Hausen H (1991) Human papillomaviruses in the pathogenesis of anogenital cancer. Virology 184: 9–13. [DOI] [PubMed] [Google Scholar]
- 2. Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, et al. (2003) Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 348: 518–527. [DOI] [PubMed] [Google Scholar]
- 3. Kelley ML, Keiger KE, Lee CJ, Huibregtse JM (2005) The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J Virol 79: 3737–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63: 1129–1136. [DOI] [PubMed] [Google Scholar]
- 5. Wang J, Sampath A, Raychaudhuri P, Bagchi S (2001) Both Rb and E7 are regulated by the ubiquitin proteasome pathway in HPV-containing cervical tumor cells. Oncogene 20: 4740–4749. [DOI] [PubMed] [Google Scholar]
- 6. Ying H, Xiao ZX (2006) Targeting retinoblastoma protein for degradation by proteasomes. Cell Cycle 5: 506–508. [DOI] [PubMed] [Google Scholar]
- 7. Incassati A, Patel D, McCance DJ (2006) Induction of tetraploidy through loss of p53 and upregulation of Plk1 by human papillomavirus type-16 E6. Oncogene 25: 2444–2451. [DOI] [PubMed] [Google Scholar]
- 8. Romanczuk H, Thierry F, Howley PM (1990) Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol 64: 2849–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smotkin D, Wettstein FO (1986) Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc Natl Acad Sci U S A 83: 4680–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hummel M, Hudson JB, Laimins LA (1992) Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol 66: 6070–6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ozbun MA, Meyers C (1997) Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol 71: 5161–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schneider-Gadicke A, Schwarz E (1986) Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J 5: 2285–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thierry F, Heard JM, Dartmann K, Yaniv M (1987) Characterization of a transcriptional promoter of human papillomavirus 18 and modulation of its expression by simian virus 40 and adenovirus early antigens. J Virol 61: 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steger G, Corbach S (1997) Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J Virol 71: 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galvan SC, Martinez-Salazar M, Galvan VM, Mendez R, Diaz-Contreras GT, et al. (2011) Analysis of CpG methylation sites and CGI among human papillomavirus DNA genomes. BMC Genomics 12: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cornelissen MT, Smits HL, Briet MA, van den Tweel JG, Struyk AP, et al. (1990) Uniformity of the splicing pattern of the E6/E7 transcripts in human papillomavirus type 16-transformed human fibroblasts, human cervical premalignant lesions and carcinomas. J Gen Virol 71 (Pt 5): 1243–1246. [DOI] [PubMed] [Google Scholar]
- 17. Smotkin D, Prokoph H, Wettstein FO (1989) Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J Virol 63: 1441–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schwartz S (2008) HPV-16 RNA processing. Front Biosci 13: 5880–5891. [DOI] [PubMed] [Google Scholar]
- 19. Zheng ZM, Baker CC (2006) Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci 11: 2286–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, Meyers C, Wang HK, Chow LT, Zheng ZM (2011) Construction of a full transcription map of human papillomavirus type 18 during productive viral infection. J Virol 85: 8080–8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Badaracco G, Venuti A, Sedati A, Marcante ML (2002) HPV16 and HPV18 in genital tumors: Significantly different levels of viral integration and correlation to tumor invasiveness. J Med Virol 67: 574–582. [DOI] [PubMed] [Google Scholar]
- 22. Hopman AH, Smedts F, Dignef W, Ummelen M, Sonke G, et al. (2004) Transition of high-grade cervical intraepithelial neoplasia to micro-invasive carcinoma is characterized by integration of HPV 16/18 and numerical chromosome abnormalities. J Pathol 202: 23–33. [DOI] [PubMed] [Google Scholar]
- 23. Ferber MJ, Thorland EC, Brink AA, Rapp AK, Phillips LA, et al. (2003) Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene 22: 7233–7242. [DOI] [PubMed] [Google Scholar]
- 24. Peter M, Rosty C, Couturier J, Radvanyi F, Teshima H, et al. (2006) MYC activation associated with the integration of HPV DNA at the MYC locus in genital tumors. Oncogene 25: 5985–5993. [DOI] [PubMed] [Google Scholar]
- 25. Reuter S, Bartelmann M, Vogt M, Geisen C, Napierski I, et al. (1998) APM-1, a novel human gene, identified by aberrant co-transcription with papillomavirus oncogenes in a cervical carcinoma cell line, encodes a BTB/POZ-zinc finger protein with growth inhibitory activity. EMBO J 17: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, et al. (1999) Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res 59: 6132–6136. [PubMed] [Google Scholar]
- 27. Nishimura A, Ono T, Ishimoto A, Dowhanick JJ, Frizzell MA, et al. (2000) Mechanisms of human papillomavirus E2-mediated repression of viral oncogene expression and cervical cancer cell growth inhibition. J Virol 74: 3752–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jeon S, Allen-Hoffmann BL, Lambert PF (1995) Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol 69: 2989–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeon S, Lambert PF (1995) Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci U S A 92: 1654–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. von Knebel Doeberitz M, Bauknecht T, Bartsch D, zur Hausen H (1991) Influence of chromosomal integration on glucocorticoid-regulated transcription of growth-stimulating papillomavirus genes E6 and E7 in cervical carcinoma cells. Proc Natl Acad Sci U S A 88: 1411–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wentzensen N, Ridder R, Klaes R, Vinokurova S, Schaefer U, et al. (2002) Characterization of viral-cellular fusion transcripts in a large series of HPV16 and 18 positive anogenital lesions. Oncogene 21: 419–426. [DOI] [PubMed] [Google Scholar]
- 32. Van Tine BA, Kappes JC, Banerjee NS, Knops J, Lai L, et al. (2004) Clonal selection for transcriptionally active viral oncogenes during progression to cancer. J Virol 78: 11172–11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dictor M, Warenholt J (2011) Single-tube multiplex PCR using type-specific E6/E7 primers and capillary electrophoresis genotypes 21 human papillomaviruses in neoplasia. Infect Agent Cancer 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frohman MA, Dush MK, Martin GR (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A 85: 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hsu EM, McNicol PJ, Guijon FB, Paraskevas M (1993) Quantification of HPV-16 E6-E7 transcription in cervical intraepithelial neoplasia by reverse transcriptase polymerase chain reaction. Int J Cancer 55: 397–401. [DOI] [PubMed] [Google Scholar]
- 36. Ordonez RM, Espinosa AM, Sanchez-Gonzalez DJ, Armendariz-Borunda J, Berumen J (2004) Enhanced oncogenicity of Asian-American human papillomavirus 16 is associated with impaired E2 repression of E6/E7 oncogene transcription. J Gen Virol 85: 1433–1444. [DOI] [PubMed] [Google Scholar]
- 37. Nam DK, Lee S, Zhou G, Cao X, Wang C, et al. (2002) Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci U S A 99: 6152–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu SS, Tsang PC, Chan KY, Cheung AN, Chan KK, et al. (2008) Distribution of six oncogenic types of human papillomavirus and type 16 integration analysis in Chinese women with cervical precancerous lesions and carcinomas. Tumour Biol 29: 105–113. [DOI] [PubMed] [Google Scholar]
- 39. Arias-Pulido H, Peyton CL, Joste NE, Vargas H, Wheeler CM (2006) Human papillomavirus type 16 integration in cervical carcinoma in situ and in invasive cervical cancer. J Clin Microbiol 44: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu B, Chotewutmontri S, Wolf S, Klos U, Schmitz M, et al. (2013) Multiplex Identification of Human Papillomavirus 16 DNA Integration Sites in Cervical Carcinomas. PLoS ONE 8: e66693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thorland EC, Myers SL, Gostout BS, Smith DI (2003) Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene 22: 1225–1237. [DOI] [PubMed] [Google Scholar]
- 42. Ziegert C, Wentzensen N, Vinokurova S, Kisseljov F, Einenkel J, et al. (2003) A comprehensive analysis of HPV integration loci in anogenital lesions combining transcript and genome-based amplification techniques. Oncogene 22: 3977–3984. [DOI] [PubMed] [Google Scholar]
- 43. Dall KL, Scarpini CG, Roberts I, Winder DM, Stanley MA, et al. (2008) Characterization of naturally occurring HPV16 integration sites isolated from cervical keratinocytes under noncompetitive conditions. Cancer Res 68: 8249–8259. [DOI] [PubMed] [Google Scholar]
- 44. Kraus I, Driesch C, Vinokurova S, Hovig E, Schneider A, et al. (2008) The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res 68: 2514–2522. [DOI] [PubMed] [Google Scholar]
- 45. Schmitz M, Driesch C, Jansen L, Runnebaum IB, Durst M (2012) Non-random integration of the HPV genome in cervical cancer. PLoS ONE 7: e39632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng ZM, Wang X (2011) Regulation of cellular miRNA expression by human papillomaviruses. Biochim Biophys Acta 1809: 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guzman VB, Yambartsev A, Goncalves-Primo A, Silva ID, Carvalho CR, et al. (2008) New approach reveals CD28 and IFNG gene interaction in the susceptibility to cervical cancer. Hum Mol Genet 17: 1838–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X, Meyers C, Guo M, Zheng ZM (2011) Upregulation of p18Ink4c expression by oncogenic HPV E6 via p53-miR-34a pathway. Int J Cancer 129: 1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li B, Hu Y, Ye F, Li Y, Lv W, et al. (2010) Reduced miR-34a expression in normal cervical tissues and cervical lesions with high-risk human papillomavirus infection. Int J Gynecol Cancer 20: 597–604. [DOI] [PubMed] [Google Scholar]
- 50. Wang X, Wang HK, McCoy JP, Banerjee NS, Rader JS, et al. (2009) Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA 15: 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Borelli I, Barberis MA, Spina F, Casalis Cavalchini GC, Vivanet C, et al. (2013) A unique MSH2 exon 8 deletion accounts for a major portion of all mismatch repair gene mutations in Lynch syndrome families of Sardinian origin. Eur J Hum Genet 21: 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martin LM, Marples B, Davies AM, Atzberger A, Edwards C, et al. (2013) DNA mismatch repair protein MSH2 dictates cellular survival in response to low dose radiation in endometrial carcinoma cells. Cancer Lett 335: 19–25. [DOI] [PubMed] [Google Scholar]
- 53. Nijhuis ER, Nijman HW, Oien KA, Bell A, ten Hoor KA, et al. (2007) Loss of MSH2 protein expression is a risk factor in early stage cervical cancer. J Clin Pathol 60: 824–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He M, Cheng Y, Li W, Liu Q, Liu J, et al. (2010) Vascular endothelial growth factor C promotes cervical cancer metastasis via up-regulation and activation of RhoA/ROCK-2/moesin cascade. BMC Cancer 10: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Luke-Glaser S, Luke B, Grossi S, Constantinou A (2010) FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J 29: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schwab RA, Blackford AN, Niedzwiedz W (2010) ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J 29: 806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Al-Mahrouki AA, Karshafian R, Giles A, Czarnota GJ (2012) Bioeffects of ultrasound-stimulated microbubbles on endothelial cells: gene expression changes associated with radiation enhancement in vitro. Ultrasound Med Biol 38: 1958–1969. [DOI] [PubMed] [Google Scholar]
- 58. Choi YW, Bae SM, Kim YW, Lee HN, Park TC, et al. (2007) Gene expression profiles in squamous cell cervical carcinoma using array-based comparative genomic hybridization analysis. Int J Gynecol Cancer 17: 687–696. [DOI] [PubMed] [Google Scholar]
- 59. Flohe L, Harris JR (2007) Introduction. History of the peroxiredoxins and topical perspectives. Subcell Biochem 44: 1–25. [PubMed] [Google Scholar]
- 60. Poynton RA, Hampton MB (2014) Peroxiredoxins as biomarkers of oxidative stress. Biochim Biophys Acta 1840: 906–912. [DOI] [PubMed] [Google Scholar]
- 61. Giaginis C, Giagini A, Theocharis S (2009) Receptor-binding cancer antigen expressed on SiSo cells (RCAS1): a novel biomarker in the diagnosis and prognosis of human neoplasia. Histol Histopathol 24: 761–776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The types of viral sequences connected with poly A at their 3′- ends. The type of Class I shows E1 sequences directly ended with poly A; the type of Class II shows E1 spliced to E4 and then to L1 and also ended with poly A sequences. ▴, there are several truncation sites in E1 (data shown in Figure S2).
(TIF)
Different truncation sites in E1 region in the type of Class I. There are four truncated sites in E1, 880, 949, 1054 and 1234, respectively.
(TIF)
Several spliced donor sites in E1. In Type A there are two integration sites in E1, 880, and 1107, respectively. The solid boxes mean cellular sequences.
(TIF)