Abstract
Eukaryotic translation initiation factor eIF4A is a DEAD-box helicase that resolves secondary structure elements in the 5'-UTR of mRNAs during ribosome scanning. Its RNA-stimulated ATPase and ATP-dependent helicase activities are enhanced by other translation initiation factors, but the underlying mechanisms are unclear. DEAD-box proteins alternate between open and closed conformations during RNA unwinding. The transition to the closed conformation is linked to duplex destabilization. eIF4A is a special DEAD-box protein that can adopt three different conformations, an open state in the absence of ligands, a half-open state stabilized by the translation initiation factor eIF4G and a closed state in the presence of eIF4G and eIF4B. We show here that eIF4A alone does not measurably sample the closed conformation. The translation initiation factors eIF4B and eIF4G accelerate the eIF4A conformational cycle. eIF4G increases the rate of closing more than the opening rate, and eIF4B selectively increases the closing rate. Strikingly, the rate constants and the effect of eIF4B are different for different RNAs, and are related to the presence of single-stranded regions. Modulating the kinetics of the eIF4A conformational cycle is thus central for the multi-layered regulation of its activity, and for its role as a regulatory hub in translation initiation.
INTRODUCTION
Initiation of translation is the first step in protein biosynthesis and is mediated by a set of translation initiation factors (eIFs). At the beginning of eukaryotic translation initiation, the eIF4F complex, formed by the cap-binding protein eIF4E, the scaffold protein eIF4G and the DEAD-box helicase eIF4A, binds to the m7Gppp cap at the 5'end of the messenger RNA (mRNA) (1, reviewed in 2) (Figure 1A). Together with the poly-A-binding protein (and eIF3 in mammals), the eIF4F complex recruits the 43S ribosomal pre-initiation complex that then searches for the translation start codon by scanning along the 5'-untranslated region (UTR) in 5'- to 3'-direction (reviewed in (3,4)). The DEAD-box helicase eIF4A has been ascribed a key role in removing secondary and tertiary structures and possibly also bound proteins during 5'-UTR scanning (5–7). Initiation is the rate-limiting step of translation, and is tightly regulated. Deregulation of translation initiation is often found in cancer cells, and altered expression levels of eIF4A and other translation factors are linked to transformation and cancer development (8,9).
Figure 1.
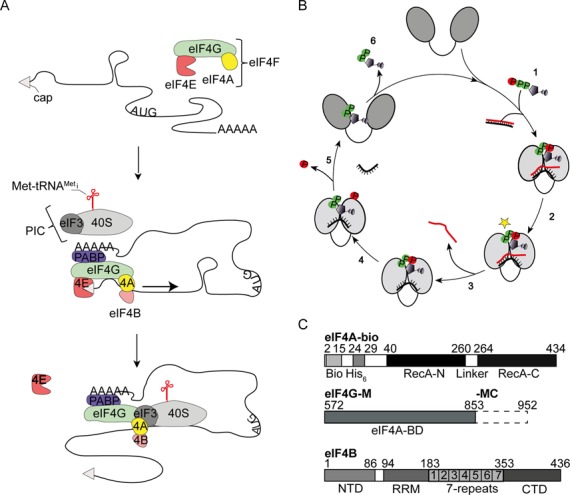
eIF4A is a DEAD-box helicase involved in translation initiation. (A) Scheme of translation initiation in eukaryotes. The DEAD-box helicase eIF4A (yellow), the cap-binding protein eIF4E (red) and the scaffold protein eIF4G (green) form the eIF4F complex that binds to the cap (gray triangle) at the 5'-end of the mRNA. The mRNA is circularized by interactions between eIF4G and the poly-A-binding protein (PABP, purple). eIF4G facilitates recruitment of the 40S ribosomal subunit (light gray, in mammals via binding to eIF3, dark gray) and to the mRNA. eIF4B (orange) interacts with eIF4A and increases its helicase activity. eIF4A then unwinds structures in the 5'-UTR of the mRNA during ribosome scanning toward the start codon (arrow). (B) Conformational cycle of DEAD-box proteins. Cooperative binding of adenosine triphosphate (ATP) and RNA (cartoons, α- and β-phosphates in green, γ-phosphate in red; (1)) to the open conformation of DEAD-box proteins leads to closure of the cleft separating the two domains of the helicase core (gray ovals). A subsequent activation step (2) populates the hydrolysis- and unwinding competent state (star), from which the first strand of the RNA substrate dissociates (3). ATP is hydrolyzed (4) and phosphate release is coupled to re-opening of the cleft in the helicase core, and dissociation of the second RNA strand (5). After dissociation of ADP (6), a subsequent cycle can be initiated. (C) Constructs used in this study. eIF4A-bio contains a 14 aa biotinylation tag and a His6-tag at the N-terminus of the helicase core (RecA_N, linker, RecA_C). eIF4G-M comprises the eIF4A-binding middle domain (eIF4A-BD) of eIF4G, eIF4G-MC in addition contains the C-terminal tail of eIF4G. eIF4B consists of an N-terminal domain (NTD), an RNA recognition motif (RRM), a 7-repeat region (1–7), and a C-terminal domain (CTD).
eIF4A (reviewed in (10)), is an RNA-dependent ATPase (11) and ATP-dependent RNA helicase (6,12–13). It consists of a helicase core that carries conserved motifs involved in RNA and nucleotide binding, ATP hydrolysis and RNA unwinding (14). A conformational cycle is central to ATP-dependent RNA unwinding by DEAD-box proteins (reviewed in (15,16)) (Figure 1B). Cooperative binding of RNA and ATP to DEAD-box proteins leads to closure of a cleft separating the two domains of the core, such that the conserved motifs from both domains jointly form the active sites for ATP hydrolysis and the RNA binding site (17,18). Formation of the closed conformer is linked to the local destabilization of a bound ribonucleic acid (RNA) duplex, facilitating release of the first RNA strand (17). Re-opening of the inter-domain cleft occurs upon phosphate release and is coupled to dissociation of the second RNA strand (18–21).
eIF4A is a very modest ATPase and RNA helicase on its own, but its activities are stimulated by the translation initiation factors eIF4B, eIF4H and eIF4G (22–27) (reviewed in (10)). eIF4G is a scaffold protein that binds to eIF4A, to the cap-binding protein eIF4E and to the poly-A-binding protein (reviewed in (2,10)). Yeast eIF4G contains five antiparallel hairpin repeats (HEAT-repeats), followed by an arginine serine (R/S)-rich domain that binds RNA (28) (Figure 1C). The eIF4G middle domain (eIF4G-M, Figure 1C) is the minimal region required for eIF4A binding and for stimulation of its ATPase activity (29). eIF4G-M stimulates the eIF4A ATPase activity by contacting both domains of the helicase core and stabilizing a ‘half-open’ conformation from which RNA and phosphate release are accelerated (20–21,29–30). Although eIF4G stimulates the eIF4A ATPase activity 13-fold, it has little effect on unwinding (20,31). Yeast eIF4B (Tif3) consists of an N-terminal domain, followed by a domain with seven repeat regions, an RNA recognition motif and a C-terminal domain (Figure 1C). eIF4B stimulates RNA unwinding by eIF4A without affecting its ATPase activity (13,26,31–32). It binds RNA (33) and has been implicated in the formation of an eIF4A-eIF4G complex (34) and of the eIF4A-43S-mRNA pre-initiation complex (35). When eIF4B and eIF4G are present at the same time, ATP hydrolysis and RNA unwinding by eIF4A are accelerated equally, by 13-fold (31), pointing to increased coupling of ATP hydrolysis to RNA unwinding when eIF4B is present.
In contrast to other DEAD-box proteins that alternate between open and closed conformations during their nucleotide cycle, eIF4A can adopt three different conformations, an open state in the absence of ligands (20,31,36), a half-open state when the middle domain of eIF4G is bound (20,29,30,31) and a closed state in the presence of eIF4B and eIF4G (31) (Figure 1B). We have shown previously that Saccharomyces cerevisiae (yeast) eIF4B and eIF4G jointly stimulate eIF4A ATPase and helicase activities by altering the conformational equilibrium of eIF4A (31), and proposed that the interaction partners modulate eIF4A activity by affecting the kinetics of individual steps in its nucleotide-driven conformational cycle. Here, we have followed conformational changes of yeast eIF4A in the presence of eIF4B and eIF4G in real time by single molecule (sm) total internal reflection fluorescence (TIRF) microscopy. We show that eIF4A only rarely undergoes conformational changes on its own. As soon as eIF4G is bound, however, eIF4A alternates between half-open and closed states in the presence of ATP and poly-U-RNA, with closing being the rate-limiting step in its conformational cycle. eIF4B additionally accelerates the conformational cycling of eIF4A by further accelerating closing. Interestingly, the rate constants for eIF4A conformational changes are different for different single-stranded RNAs and for double-stranded RNAs that are substrates for unwinding by eIF4A. Our results illustrate that eIF4A integrates signals from its auxiliary factors and elements in the RNA substrates and reveal the conformational cycle as a central element for the multi-faceted regulation of eIF4A activity in translation initiation.
MATERIALS AND METHODS
Purification of eIF4A-bio, eIF4B, eIF4G-M and eIF4G-MC
For in vivo biotinylation, yeast eIF4A was produced with an N-terminal 14 aa biotinylation sequence (GLNDIFEAQKIEWH, (37)) in Escherichia coli Rosetta(DE3). Lysine 10 (highlighted in bold) is biotinylated in vivo by the Escherichia coli biotin ligase BirA (37). BirA was coproduced from the plasmid pBTac-BirA, carrying the birA gene in frame with a region coding for a C-terminal His-tag (37). Cells were grown in LB medium supplemented with biotin (10 μg/ml). Protein production was induced by IPTG addition, and cells were harvested from overnight cultures at 20°C. Cells were disrupted in a microfluidizer in 50 mM Tris/HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole, 2 mM β-mercaptoethanol (BME), eIF4A-bio was bound to Ni2+-NTA sepharose and eluted with 50 mM Tris/HCl, pH 7.5, 500 mM NaCl, 200 mM imidazole, 2 mM BME. Fractions containing eIF4A-bio were further purified on a S75 size-exclusion column in 50 mM Tris/HCl, pH 7.5, 500 mM NaCl, 2 mM BME. Residual BirA ligase was removed by a final anion exchange chromatography step on MonoQ sepharose in 50 mM Tris/HCl, pH 7.5, 100 mM NaCl, 2 mM BME. eIF4A-bio was eluted in a gradient from 0.1–1 M NaCl, and shock-frozen in liquid nitrogen. The biotinylation degree was determined with a Biotin quantification kit (Thermo Scientific Pierce, USA) according to the manufacturer's instructions. Saccharomyces cerevisiae (yeast) eIF4B, eIF4G-M (comprising the eIF4A-binding middle domain, aa 572–853) and eIF4G-MC (comprising the middle domain plus the C-terminal R/S-rich tail, aa 572–952, Figure 1C) were purified from recombinant Escherichia coli Rosetta(DE3) as described (31). eIF4G-MC lacks the N-terminal eIF4E binding region and is therefore a suitable model for studying the effects of eIF4G on eIF4A in the absence of eIF4E. All proteins used in this study are >95% pure as judged from Coomassie-stained SDS-PAGE (Supplementary Figure S1).
ATPase and helicase activity
eIF4A steady state ATPase activity was measured in a coupled enzymatic assay as described previously (31) in 50 mM Tris/HCl, pH 7.5, 80 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol (DTT), 1% (v/v) glycerol with 1 μM eIF4A, 2 μM eIF4B and 2 μM eIF4G-M in the presence of 2 mM ATP and varying concentrations of poly-U-RNA. Unwinding of a 32/9mer (31,38–39) and a 50/10mer substrate (13,31) was followed in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT with 500 nM RNA and 5 μM eIF4A, eIF4B and eIF4G-MC in the presence of 3 mM ATP. To ensure single turnover conditions, unlabeled 9mer (or 10mer) was added in 10-fold molar excess. Reactions were stopped at different time points, and dsRNA substrate and ssRNA products were separated by gel electrophoresis as described previously (18).
smFRET experiments
eIF4A_186C/370C (20,31) was labeled with a two- and five-fold molar excess of AlexaFluor555 (donor) and AlexaFluor647 (acceptor) maleimides as described previously (20,31). Labeling efficiencies were calculated from absorbance spectra as described previously (18,20,31), using the following extinction coefficients: eIF4A-bio: ϵ280 nm = 17 880 M−1 cm−1; Alexa555: ϵ280 nm = 12 000 M−1 cm−1, ϵ554 nm = 150 000 M−1 cm−1; Alexa647: ϵ280 nm = 7170 M−1 cm−1, ϵ554 nm = 18 164 M-1 cm−1, ϵ650 nm = 239 000 M−1 cm−1.
For single molecule FRET (smFRET) experiments by TIRF microscopy, coverslips were cleaned with alconox (10%), KOH and plasma cleaning (Harrick Plasma PDC-3XG), amino-functionalized with 1% (v/v) 3-(2-aminoethylamino)-propyldimethoxymethylsilane in methanol with 5% (v/v) acetic acid, PEGylated with 22% (w/v) mPEG-succinimidyl valerate and 2% (w/v) biotin-PEG-succinimidyl valerate in 50 mM MOPS pH 7.5 and streptavidin-functionalized with 50 μl 0.2 mg/ml streptavidin in 10 mM Tris/HCl, pH 7.5, 50 mM NaCl. Donor/acceptor labeled eIF4A-bio in 50 mM Tris/HCl, pH 7.5, 80 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol (DTT), 1% (v/v) glycerol was immobilized on the pretreated slides. smFRET experiments were performed in the absence and presence of eIF4B, eIF4G-M or eIF4G-MC (10 μM each), 1 mM poly-U-RNA (concentration per base), 15 μM 50mer, 50/10mer or 32/9mer RNA and 3 mM ATP in 50 mM Tris/HCl, pH 7.5, 80 mM KCl, 2.5 mM MgCl2, 1% (v/v) glycerol supplemented with an oxygen scavenging system consisting of 2.5 mM protocatechuic acid, 50 nM protocatechuate-3,4-dioxygenase and with 1 mM of the triplet state quencher Trolox, 1 mM methylviologen, as well as 0.1% (w/v) bovine serum albumin. Imaging was done with an Olympus TIRF microscope equipped with a 532 nm excitation laser, a high numerical aperture temperature-controlled objective (UAPON 100x, oil immersion), a dual view beam splitter and an Andor iXON3 EM-CCD camera. The incident angle of the excitation light was adjusted via a motorized integrated TIRF illumination combiner. ZT532/638rpc (Chroma) and F37-585 (Semrock Inc., consisting of a 585/40 bandpass filter (donor), a T640lpx dichroic beam splitter and a 692/40 bandpass filter (acceptor)=) beam splitters were used to separate excitation and emission light, and donor and acceptor emission, respectively. Data acquisition, as well as mapping of donor and acceptor images was done using the Olympus xcellence software. The time resolution was 100 ms.
smFRET data analysis
Background correction, peak identification and calculation of fluorescence and FRET time traces were performed with LabVIEW programs. The apparent FRET efficiency was calculated from background-corrected donor and acceptor fluorescence intensities. Only fluorescence traces with anticorrelated changes of donor and acceptor fluorescence and single-step photobleaching of the donor (or the acceptor and the donor) were used for data analysis. The calculation of FRET efficiencies was restricted to the time before photobleaching. The numbers of traces for each experiment are summarized in Supplementary Table S1. FRET time traces were idealized by Hidden Markov Modeling (HMM) using the program vbFRET (40). The dwell times in the different FRET states and transition density plots (TDPs) were calculated using a MATLAB script.
RESULTS
To follow the conformational changes of eIF4A in real time, we immobilized N-terminally biotinylated, donor/acceptor-labeled eIF4A (eIF4A-bio, Figure 1C) and performed smFRET experiments by total internal reflection fluorescence microscopy. eIF4A-bio showed slightly reduced ATPase and RNA unwinding activities, but both activities were stimulated by eIF4B, eIF4G-M and eIF4G-MC (Supplementary Figure S2) to similar extents as for wild-type eIF4A (31).
Conformational states of eIF4A-bio and effect of eIF4B and eIF4G
Donor/acceptor-labeled eIF4A-bio shows intermediate donor and acceptor fluorescence (Supplementary Figure S3A). A FRET histogram reveals a unimodal distribution of FRET efficiencies. The mean FRET efficiency (EFRET) of 0.48 is virtually identical to the FRET efficiency observed for the open conformation of eIF4A in solution (EFRET = 0.45–0.47) (20,31). In the presence of eIF4G-M, comprising the middle domain of eIF4G, an increased donor and reduced acceptor fluorescence intensity is observed. The lower FRET efficiency (EFRET = 0.29; Supplementary Figure S3B) is characteristic of the half-open conformation of eIF4A (EFRET = 0.26 in solution) (20,31). Extensive rinsing of the surface with buffer leads to an increase in FRET efficiency to EFRET = 0.47 (Supplementary Figure S3C), consistent with dissociation of eIF4G-M and the return of eIF4A to the open conformation. These results show that the interaction between eIF4A and eIF4G-M is dynamic. In agreement with previous solution experiments (31), eIF4B shows no effect on the FRET efficiency and on the eIF4A-bio conformation (Supplementary Figure S3D–F).
Effects of eIF4B and eIF4G on eIF4A conformational dynamics in presence of ATP and poly-U-RNA
Poly-U-RNA is often used as a non-specific single-stranded RNA substrate for DEAD-box helicases. When we monitored FRET for donor-/acceptor-labeled eIF4A-bio in the presence of poly-U-RNA and ATP (Figure 2A), an open conformation with a medium FRET efficiency was observed. Seven out of 88 FRET time traces showed brief excursions to a high FRET state, possibly corresponding to the closed conformation. An occasional sampling of the closed state even in the absence of other translation factors is in agreement with the low, but detectable basal ATPase and RNA unwinding activities of eIF4A. FRET histograms for eIF4A-bio were unimodal (EFRET = 0.45), and did not reflect a second, high FRET state, indicating that the closed conformation is not significantly populated under these conditions. Similarly, eIF4A-bio remained in the open conformation in the presence of eIF4B (Figure 2B, EFRET = 0.44). When eIF4G-M was added, eIF4A-bio adopted the half-open conformation, with a low FRET efficiency (Figure 2C) (20,29,31). Again, only some molecules (22 out of 76) showed brief excursions to the closed conformation, and the FRET histogram remained unimodal (EFRET = 0.32; Figure 2C). In contrast to the static behavior of eIF4A-bio with eIF4B or eIF4G-M, FRET time traces in the presence of eIF4G-MC, comprising the eIF4A-binding domain and the R/S-rich C-terminal tail, show dynamic switches between low and high FRET states, with FRET efficiencies of EFRET = 0.30 and EFRET = 0.61 (Figure 2D). The high FRET state corresponds to the closed conformation of eIF4A we recently identified in solution (EFRET = 0.65) (31). Overall, immobilized eIF4A-bio (referred to as eIF4A in the following) thus recapitulates all three conformational states previously identified for eIF4A in solution, validating our experimental approach.
Figure 2.
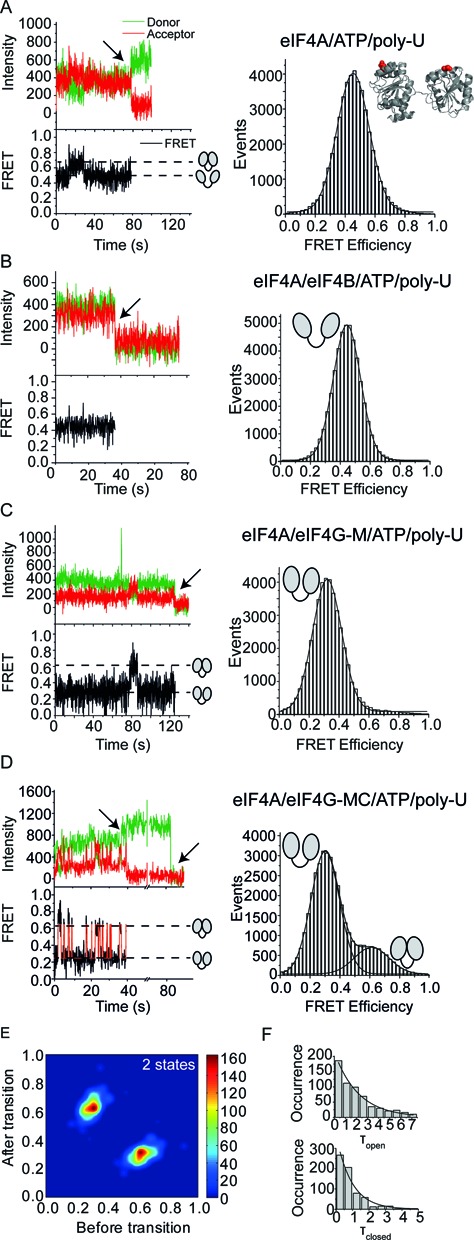
eIF4A adopts three different conformations. smTIRF experiments of donor/acceptor-labeled, immobilized eIF4A-bio in the absence and presence of its interaction partners reveals three different eIF4A conformations: open, half-open and closed. The FRET efficiencies for the three states are identical to FRET efficiencies determined with a different donor/acceptor pair in solution (31). In (A–D), the left panel shows representative donor/acceptor fluorescence and FRET time traces (top: donor fluorescence in green, acceptor fluorescence in red, bottom: FRET time traces in black). Broken horizontal lines indicate the FRET values for open, half-closed and closed conformations (see cartoons). Arrows highlight donor and acceptor photobleaching events. FRET histograms are depicted on the right. (A) eIF4A-bio in the presence of ATP and poly-U-RNA predominantly adopts an open conformation (cartoon) with a mean FRET efficiency of EFRET = 0.45. (B) eIF4A-bio in the presence of eIF4B, ATP and poly-U-RNA. eIF4B does not affect the eIF4A conformation, and eIF4A remains in the open conformation with a FRET efficiency of EFRET = 0.44. (C) eIF4A-bio in the presence of eIF4G-M, ATP and poly-U-RNA. eIF4G-M stabilized eIF4A in a half-open conformation with a FRET efficiency of EFRET = 0.32. (D) eIF4A-bio in the presence of eIF4G-MC, ATP and poly-U-RNA. The red curve in the FRET time trace is the idealized trace calculated by Hidden Markov modeling (see Methods). eIF4A switches between low and high FRET states (EFRET = 0.61, EFRET = 0.30), reflecting the half-open and closed states. From the populations of the two conformers in the FRET histogram, the equilibrium constant Kclose of 0.27 is obtained. (E) Transition density plot (TDP) for eIF4A in the presence of eIF4G-MC, poly-U-RNA and ATP shows transition probabilities between the two FRET states. The TDP was generated from 102 FRET time traces with a total of 754 closing (EFRET low → high) and 757 opening (EFRET high → low) events. (F) Dwell time histograms for the open (τopen) and closed states (τclosed) of eIF4A in the presence of eIF4G-MC, ATP and poly-U-RNA. From the exponential fit (black line), the rate constants kopen = 1.06 s−1 and kclose = 0.46 s−1 are obtained. The equilibrium constant for closing, calculated from the rate constants is 0.44.
Idealization of FRET time traces by Hidden Markov modelling (HMM) and generation of dwell time histograms (Figure 2D and F) allows for the determination of the underlying rate constants of the observed conformational transitions. In the presence of eIF4G-MC, the two eIF4A conformations inter-convert with rate constants of kopen = 1.06 s−1 and kclose = 0.46 s−1 (Table 1), corresponding to an equilibrium constant of closing, Kclose, of 0.43. This value is within 1.5-fold of the equilibrium constant determined from populations of half-open and closed states in FRET histograms (Kclose = 0.27; Figure 2D), and in agreement with the equilibrium constant determined previously from smFRET data in solution (Kclose = 0.54) (31). Strikingly, FRET time traces (down to integration times of 35 ms) do not reveal any excursions of eIF4A to the open state. Transition density plots (TDPs) support that only two FRET states are populated (Figure 2E, Supplementary Figure S4). Thus, eIF4A directly alternates between half-open and closed states when eIF4G is present.
Table 1.
Rate constants of eIF4A opening and closing and effect of eIF4B, eIF4G and different RNAs
| kopen (s−1) | kclose (s−1) | |
|---|---|---|
| eIF4A/eIF4G-M/eIF4B/polyU-RNA/ATP | 0.23 | 0.13 |
| eIF4A/eIF4G-MC/polyU-RNA/ATP | 1.06 | 0.46 |
| eIF4A/eIF4G-MC/eIF4B/polyU-RNA/ATP | 1.69 | 1.61 |
| eIF4A/eIF4G-MC/50mer/ATP | 0.34 | 0.24 |
| eIF4A/eIF4G-MC/eIF4B/50mer/ATP | 0.42 | 1.49 |
| eIF4A/eIF4G-MC/50–10mer/ATP | 0.36 | 0.44 |
| eIF4A/eIF4G-MC/eIF4B/50–10mer/ATP | 0.50 | 1.64 |
| eIF4A/eIF4G-MC/32–9mer/ATP | 0.90 | 0.20 |
| eIF4A/eIF4G-MC/eIF4B/32–9mer/ATP | 0.66 | 0.29 |
We have previously shown that eIF4B promotes the population of the closed eIF4A conformer when either eIF4G-M or eIF4G-MC is bound (31). Now, we investigated the effect of eIF4B on the conformational dynamics of eIF4A when eIF4G-M is present. FRET time traces for eIF4A in presence of eIF4G-M, eIF4B, ATP and poly-U-RNA show frequent switches between the low FRET half-open and the high FRET closed conformation (EFRET = 0.31 and EFRET = 0.61; Figure 3A). The corresponding equilibrium constant for closing of Kclose = 0.57 (0.18 according to the populations from FRET histograms) is within 1.5-fold of the equilibrium constant from previous data in solution (ca. 0.39) (31). The rate constants of eIF4A opening and closing in the presence of eIF4B and eIF4G-M are kopen = 0.23 s−1, and kclose = 0.13 s−1 (Table 1). Thus, eIF4B induces rapid conformational cycling of eIF4A in the formerly static eIF4A/eIF4G-M complex.
Figure 3.
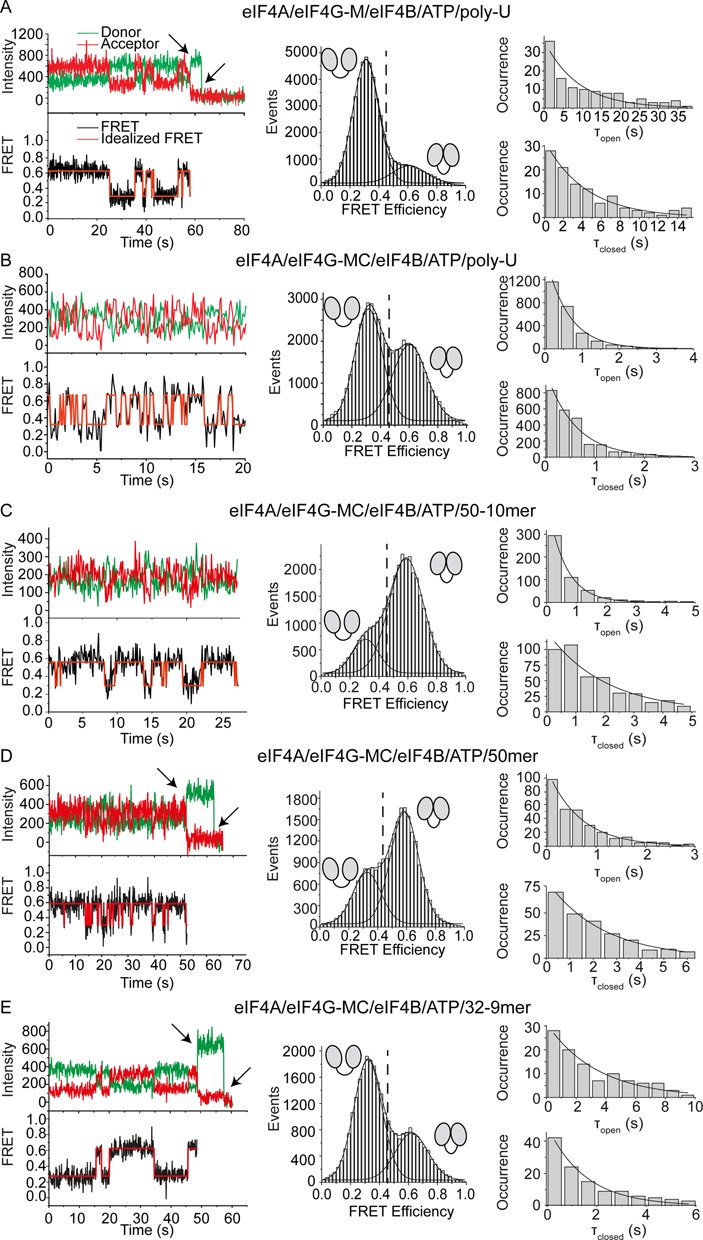
eIF4B, eIF4G and the bound RNA modulate the kinetics of the eIF4A conformational cycle. smTIRF experiments of donor/acceptor-labeled, immobilized eIF4A-bio in the absence and presence of its interaction partners and different RNAs reveal different rate constants for opening and closing. The left panel shows representative donor/acceptor fluorescence and FRET time traces (top: donor fluorescence in green, acceptor fluorescence in red, bottom: FRET time traces in black, idealized traces obtained by HMM in red). FRET histograms are depicted in the center. The vertical broken line indicates the FRET value for the open eIF4A conformer, and half-open and closed conformers are indicated by the cartoons. Dwell time histograms are shown on the right. The corresponding experiments in the absence of eIF4B are shown in Figure 2 C and D and Supplementary Figure S5. All rate constants are summarized in Table 1. (A) eIF4A in the presence of eIF4B, eIF4G-M, ATP and poly-U-RNA switches between low and high FRET states (EFRET = 0.29, EFRET = 0.59), reflecting the half-open and closed states, with rate constants kopen and kclose of 0.23 s−1 and 0.13 s−1. The equilibrium constant Kclose for closing, calculated from the rate constants or from populations of the two conformers in the FRET histogram, is 0.57 (only molecules that show transitions, 22 out of 76) or 0.18 (all molecules). (B) eIF4A in the presence of eIF4B, eIF4G-MC, ATP and poly-U-RNA switches between half-open and closed states with kopen and kclose of 1.69 s−1 and 1.61 s−1. Kclose is 0.95 (rate constants) or 0.84 (histogram), respectively. (C) eIF4A in the presence of eIF4B, eIF4G-MC, ATP and 50/10mer RNA switches between half-open and closed states with rate constants kopen and kclose of 0.50 s−1 and 1.64 s−1. Kclose is 3.3 (rate constants) or 4.5 (histogram). (D) eIF4A in the presence of eIF4B, eIF4G-MC, ATP and 50mer RNA switches between half-open and closed states with kopen and kclose of 0.42 s−1 and 1.49 s−1, respectively. Kclose is 3.5 (rate constants) or 2.2 (histogram). (E) eIF4A in the presence of eIF4B, eIF4G-MC, ATP and 32/9mer RNA switches between half-open and closed states with kopen and kclose of 0.66 s−1 and 0.29 s−1. Kclose is 0.44 (rate constants) or 0.45 (histogram).
We next investigated the effect of eIF4B on the conformational changes of eIF4A bound to eIF4G-MC. Compared to the experiments in presence of eIF4G-M, FRET time traces for eIF4A in presence of eIF4G-MC, eIF4B, ATP and poly-U-RNA show even more frequent switching events between the two FRET states (EFRET = 0.32, EFRET = 0.60; Figure 3B, Supplementary Figure S4A). The increased conformational dynamics point to significant contributions of the eIF4G C-terminal tail to acceleration of the eIF4A conformational cycle. The rate constants for eIF4A conformational changes in the presence of eIF4B and eIF4G-MC are kopen = 1.69 s−1 and kclose = 1.61 s−1 (Table 1). These rate constants are 1.6-fold and 3.5-fold higher than in the absence of eIF4B (kopen = 1.06 s−1, kclose = 0.46 s−1, Figure 2D–F, Table 1). eIF4B thus promotes an additional shift of the eIF4A conformational equilibrium toward the closed conformation by increasing kclose more than kopen. The equilibrium constant Kclose is 0.95 (0.84 from FRET histogram), similar to the equilibrium constant from previous data in solution (ca. 1.17) (31).
Effect of the non-hydrolyzable ATP analog ADPNP on eIF4A conformational changes
ADPNP (5'-adenylyl-β,γ-imidotriphosphate) has been used successfully to trap a number of DEAD-box helicases in their active, closed state in crystallographic (17,21,41–43) and smFRET studies (18,19). Yet we have previously reported that ADPNP does not lead to an increased population of the high-FRET eIF4A species in equilibrium (31). We now monitored the effect of ADPNP on eIF4A conformation in real time. FRET time traces for eIF4A in the presence of eIF4B, eIF4G-MC, poly-U-RNA and ADPNP show occasional, short excursions to a high FRET state (Figure 4). However, no trapping of eIF4A in the closed state is observed (Figure 4), in agreement with our equilibrium studies (31). The absence of a stable closed conformer is consistent with previous studies that reported a failure to cross-link eIF4A to RNA in the presence of non-hydrolyzable ATP analogs (44,45). The requirement for cofactors to adopt the closed conformation implies that the closed state of eIF4A is less stable than for other DEAD-box proteins. The energy of nucleotide binding to DEAD-box proteins is lower for ADPNP than for ATP (19), and ADPNP may therefore simply not provide enough binding energy to be coupled to the conformational change of eIF4A.
Figure 4.
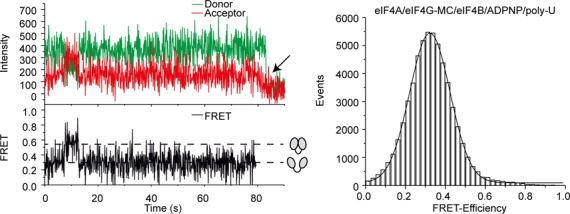
eIF4A is not trapped in the closed conformation in the presence of a non-hydrolyzable ATP analog. smTIRF experiments of donor/acceptor-labeled, immobilized eIF4A-bio in the presence of eIF4B, eIF4G-MC, poly-U-RNA and ADPNP. A selected donor/acceptor fluorescence time trace (left, top, green: donor, red: acceptor), and the resulting FRET time trace (left, bottom) and FRET histogram (right) are shown. eIF4A occasionally (in 14 out of 70 traces) switches from the low FRET (half-open) to a high FRET (closed) state. However, no stable trapping of eIF4A in the closed state is observed in the presence of the non-hydrolyzable ATP analog.
Effect of different RNAs containing single-stranded regions on eIF4A conformational dynamics
Although poly-U-RNA has frequently been used for the characterization of DEAD-box proteins, it is not a substrate for unwinding. We therefore tested the effect of a 50/10mer RNA that has previously been used as an unwinding substrate for eIF4A (12) on the conformational dynamics of eIF4A. In the presence of eIF4B, eIF4G-MC, ATP and the 50/10mer RNA, eIF4A showed rapid changes of donor and acceptor signals, and thus of the FRET efficiency (EFRET = 0.31 and 0.59) (Figure 3C), in agreement with rapid conformational cycling of eIF4A during the unwinding reaction. The TDP reveals transitions between two species, the half-open and closed states (Supplementary Figure S4C). From dwell time histograms, the rate constants kopen = 0.50 s−1 and kclose = 1.64 s−1 are obtained (Figure 3C, Table 1). The rate constant of opening is 3.3-fold lower than with poly-U-RNA, whereas the rate constant for closing is identical with both RNAs. As a result of the reduced opening rate, opening of the cleft in the helicase core becomes rate-limiting for the eIF4A conformational cycle under these conditions. The equilibrium constant for closing is Kclose = 3.3, in agreement with Kclose = 4.5 from populations in the FRET histograms (Figure 3C). The conformational cycle of eIF4A is thus substantially different in the presence of poly-U-RNA and this unwinding substrate.
We next investigated the conformational dynamics of eIF4A in presence of only the single-stranded 50mer, the product of the unwinding reaction. In the presence of this RNA, eIF4A also undergoes rapid conformational cycles (Figure 3D, Supplementary Figure S4B). The rate constants kopen and kclose (0.42 s−1 and 1.49 s−1, respectively; Table 1) are very close to the values in presence of the 50/10mer RNA (kopen = 0.50 s−1 and kclose 1.64 s−1; see above). Hence, eIF4A undergoes a similar conformational cycle when bound to the 50mer or the 50/10mer RNA. Possibly, the effects of the 50/10mer unwinding substrate on the eIF4A conformational cycle are dominated by the single-stranded region 5' of the duplex (see below).
More insight into the effect of single-stranded RNAs on eIF4A conformational dynamics can be gained from comparing kopen and kclose of eIF4A in presence of poly-U-RNA and the 50mer. When all factors are present (eIF4B, eIF4G-MC and ATP), the rate constants for eIF4A closing are similar (1.49 s−1 for the 50mer and 1.61 s−1 for poly-U-RNA; see above). Thus, kclose seems to show little dependence of the identity of the bound single-stranded RNA. In contrast, the rate constant for eIF4A opening is substantially different, and 4-fold lower when the 50mer is bound than for poly-U-RNA (kopen = 0.42 s−1 versus 1.69 s−1). Comparing eIF4A conformational dynamics with the 50mer, poly-U-RNA, or the 50/10mer RNA bound, the highest rate constants (ca. 1.6 s−1 for kopen and kclose) are observed with poly-U-RNA, again indicating that poly-U-RNA elicits very special behavior of eIF4A.
We also determined the rate constants for eIF4A conformational changes in the presence of poly-U-RNA, 50mer and 50/10mer, but in the absence of eIF4B (Figure 2D, Supplementary Figure S5A and B, Table 1). Without eIF4B, the rate constant for eIF4A opening shows a somewhat larger dependence on the nature of the bound RNA than the rate for closing: While kopen varies by a factor of three (1.06 s−1 for poly-U-RNA, 0.34 s−1 for the 50mer, 0.36 s−1 for the 50/10mer), kclose solely varies by a factor of two (0.46 s−1 for poly-U-RNA, 0.24 s−1 for the 50mer, 0.44 s−1 for the 50/10mer). When the opening rates are compared, kopen for the 50mer and 50/10mer is identical. Poly-U-RNA elicits different behavior, and kopen is threefold higher. On the other hand, kclose is identical for the poly-U-RNA and the 50/10mer, but now the 50mer induces different behavior of eIF4A, with a kclose that is nearly halved. In the presence of the single-stranded RNAs, kopen is larger than kclose, and closing is thus the rate-limiting step in the eIF4A conformational cycle. Only when the 50/10mer is bound does kclose become larger than kopen, and the rate-limiting step shifts to opening. It thus appears that, in the absence of eIF4B, the 50/10mer unwinding substrate can to some degree trap itself by accelerating closure of the eIF4A inter-domain cleft.
The effect of eIF4B on the eIF4A conformational cycle can be deduced from a comparison of the rate constants of opening and closing in the presence of the different RNAs with and without eIF4B (Table 1). Notably, the effect of eIF4B on these rate constants is very similar for all three RNAs: In all cases, eIF4B has little effect on the rate of opening (less than 1.6-fold), but accelerates closing (by 3.5- to 6.2-fold). As a result, eIF4B shifts the eIF4A conformational equilibrium toward the closed state by 2.2- to 5-fold (Table 1). eIF4A thus exhibits different conformational dynamics depending on the RNA bound, both in the absence and presence of eIF4B.
Overall, our findings suggest that poly-U-RNA should not be considered a representative and general model substrate for eIF4A, and underline the importance of studying more than one (model) RNA substrate to arrive at general mechanistic conclusions.
Effect of an RNA substrate lacking single-stranded regions on eIF4A conformational dynamics
We finally tested the effect of single-stranded regions in the RNA substrate on the conformational cycle of eIF4A by investigating the effect of a different unwinding substrate that lacks single-stranded regions. We have previously studied unwinding of a 32/9mer (46) by eIF4A (31). This RNA consists of a 9 bp duplex that is unwound by eIF4A, flanked by a hairpin formed by a five-base pair stem and a five-base loop (31). In contrast to the 50/10mer, the 32/9mer RNA contains no extended single-stranded region. In the presence of eIF4B, eIF4G-MC, ATP and the 32/9mer, eIF4A switched between half-open and closed states (Figure 3E, Supplementary Figure S4D) with rate constants of 0.66 s−1 (kopen) and 0.29 s−1 (kclose; Table 1). The rate constant kclose is 5.7-fold smaller for the 32/9mer than in the presence of the 50/10mer unwinding substrate. In contrast, kopen is very similar for both RNAs. As a result, the equilibrium constant for closing is only 0.44 in the presence of the 32/9mer RNA (0.45 from the FRET histogram), compared to 3.3 for the 50/10mer RNA.
In the absence of eIF4B (Supplementary Figure S5C), the rate constant of opening is 1.5-fold higher (kopen = 0.90 s−1), and kclose is 1.5-fold lower than when eIF4B is present. Thus, the accelerating effect of eIF4B on the eIF4A conformational cycle is very moderate for this RNA. In fact, the rate constant kopen even decreases slightly in the presence of eIF4B. The 32/9mer is also the only RNA where the rate constant for closing still remains rate-limiting when eIF4B is present. Overall, our results clearly demonstrate that different RNA substrates have very different impacts on the conformational dynamics of eIF4A.
DISCUSSION
Conformational cycling of eIF4A is regulated differently by eIF4B and eIF4G
We have shown here that conformational cycling by eIF4A is slow in the absence of interaction partners, with only very few switches between open and closed conformations, in agreement with the low intrinsic ATP hydrolysis and RNA unwinding activities. From the previously reported effects of eIF4B and eIF4G on the conformational equilibrium of eIF4A, we have suggested that the interaction partners differentially affect the kinetics of conformational transitions (31). The kinetic data we present here unravel the underlying mechanism of eIF4A stimulation by eIF4B and eIF4G.
In the presence of eIF4G, eIF4A alternates directly between half-open and closed states, without detectable excursions to the open state. The half-open and closed conformations are hence the functionally relevant states of eIF4A during RNA unwinding. As we can only determine rate constants for the eIF4A conformational changes in the presence, but not in the absence of eIF4G, we cannot quantify the effect of eIF4G on the conformational dynamics of eIF4A. However, in the presence of eIF4B and either eIF4G-M or eIF4G-MC, eIF4A also shows rapid conformational cycling, and the rate constants for opening and closing can be determined. From a comparison, the effect of the eIF4G C-terminal tail on the kinetics of eIF4A conformational changes can be deduced: In the presence of eIF4G-MC, both closing and opening are accelerated compared to when eIF4G-M is bound. This accelerated passage of eIF4A through its conformational cycle is in agreement with the stimulatory effect of the C-tail on ATP hydrolysis (31). eIF4G-MC accelerates closing of eIF4A more than opening, in agreement with the effects of the C-tail on the conformational equilibrium, i.e. a shift to the closed state. The shift in equilibrium toward the closed eIF4A conformer in the presence of eIF4G-MC is related to a reduction in the energetic difference between the functionally relevant conformational states (thermodynamic effect). The increase in rate constants is linked to the activation barrier separating these states (kinetic effect). A reduction of the energetic difference between the two states can be brought about either by stabilization (reduction in energy) of the closed state, or by destabilization (increase in energy) of the open state. There is no data available that points to stabilization of the closed state of eIF4A by eIF4G. On the other hand, it is likely that the half-open state in the presence of eIF4G is an energetically higher state than the open state in the absence of eIF4G. A higher energy level of the half-open state would reduce the energetic difference between the two states. At the same time, it would reduce the activation barrier for closing, but not for opening (Figure 5A). The effect of eIF4G-MC on opening is not explained yet, but can be rationalized by postulating a reduced transition state energy when eIF4G-MC is present (Figure 5A). A lower energy transition state reduces the activation energy for both opening and closing. This scenario is thus in agreement with the thermodynamic and kinetic effects we observed.
Figure 5.
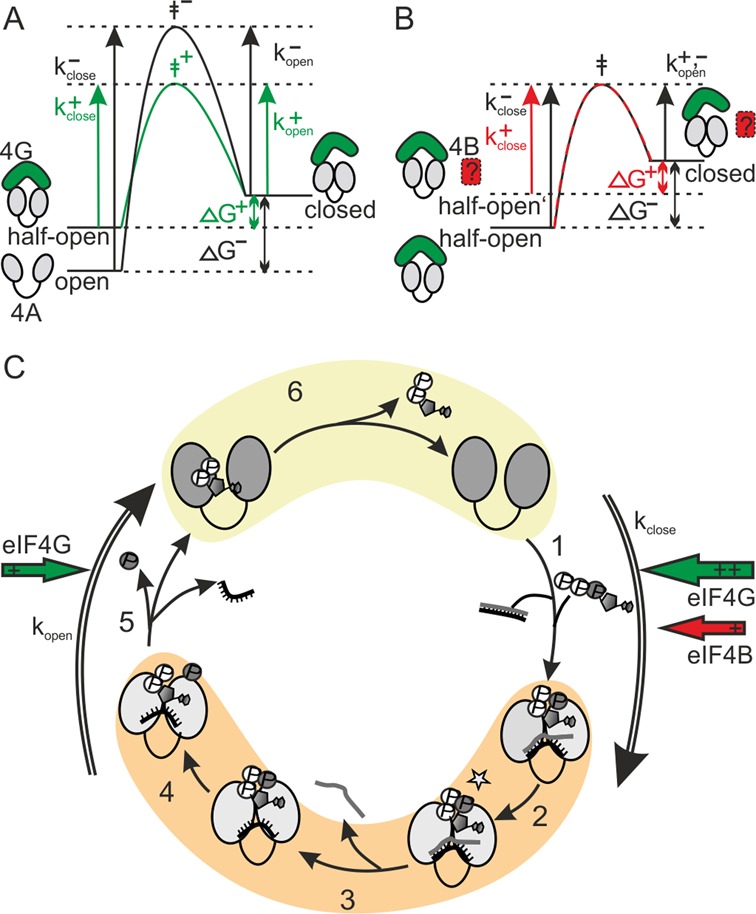
Model for the effect of eIF4G and eIF4B on the eIF4A conformational cycle. (A) In the presence of (poly-U) RNA and ATP, eIF4G (green) accelerates closing of eIF4A (gray) more than opening, and shifts the conformational equilibrium to the closed state. This effect is caused by altering the reference state (half-open), leading to a reduction in ΔG between (half-)open and closed state, and an increase in kclose. A concomitant reduction of the activation energy (green: with eIF4G, black: without eIF4G) increases kclose and kopen. (B) In the presence of (poly-U) RNA and ATP, eIF4B (red, in presence of eIF4G, green) predominantly accelerates closing, and shifts the conformational equilibrium to the closed state. The selective effect on kclose can be explained by an increased energy level of the half-open state that reduces the activation barrier (red: with eIF4B, black: without eIF4B) for closing only, but not for opening. The ΔG for closing is reduced, and the conformational equilibrium is altered by an increase in Kclose. (C) Individual steps in a productive conformational cycle of eIF4A in the presence of eIF4G and eIF4B, and rate constants for opening and closing determined from dwell time histograms. In the nucleotide-free and ADP states, at the beginning and the end of the conformational cycle, eIF4A is in the half-open state (yellow) when eIF4G is bound. In the ATP-bound state, the hydrolysis- and unwinding competent activated state (ATP*), and the ADP·Pi-state, eIF4A is in the closed conformation (orange). The rate constants determined from smFRET dwell times, kopen and kclose, are the rate constants for processes leading away from the closed and open states, respectively, and are dominated by the rate-limiting step in the relevant sequence of the catalytic cycle. The red and green arrows indicate the effects of eIF4B (red) and eIF4G (green) on opening and closing rates. For simplification, unproductive cycles with ATP hydrolysis in the absence of unwinding are not shown.
eIF4B leads to an additional shift of the eIF4A conformational equilibrium toward the closed state, brought about by the selective increase of the eIF4A closing rate. This selective effect on the closing rate without affecting opening can be explained by a further increase in the energy level of the half-open state (Figure 5B). eIF4B may increase the energy of the half-open state by interacting with the linker region of eIF4A, and facilitating closure by pre-ordering it. Such a restructuring effect is in agreement with the previously proposed role of eIF4B in assembling the eIF4A/eIF4G-complex (34). eIF4B accelerates the overall conformational cycling of eIF4A by less than 3-fold, in agreement with its modest effect on the eIF4A ATPase rate (31). These considerations are based on the experiments in the presence of poly-U-RNA. The conformational dynamics of eIF4A, and thus the energy landscape, depend on the structural features of the RNA substrate (see ‘Different RNAs affect the eIF4A conformational cycle differently’ section).
Coupling of the conformational cycle to ATP hydrolysis and duplex unwinding
Overall, rapid conformational cycling of eIF4A in the presence of its interaction partners rationalizes the stimulation of ATPase and duplex unwinding activities (Figure 5C). However, conformational cycling is four-fold faster than ATP hydrolysis (in presence of eIF4B, eIF4G-M and poly-U-RNA; Table S2), demonstrating that only every four conformational cycles an ATP molecule is hydrolyzed (Table S3). The coupling is reduced when eIF4G-MC is present, and 11 cycles (in the absence of eIF4B) or 30 cycles (in the presence of eIF4B) lead to one hydrolysis event. Thus, eIF4A does not strictly couple its conformational cycle to ATP hydrolysis, even when eIF4B and eIF4G are present.
Coupling of the conformational cycle to duplex unwinding is even less favorable (Table S3). In the presence of eIF4B and eIF4G-MC, 240 conformational cycles are required to unwind one 50/10mer and 320 cycles lead to unwinding of one 32/9mer. The number of conformational cycles required for unwinding of one 32/9mer is independent of the presence of eIF4B (317 cycles with eIF4B versus 320 cycles without eIF4B; Table S3). The unwinding rates for the 32/9mer are also independent of the presence of eIF4B (31). Together, these data suggest that coupling of conformational cycling to RNA unwinding is not altered by eIF4B.
From the individual coupling of ATP hydrolysis or duplex unwinding to the conformational cycle, we can now directly calculate the number of ATP molecules hydrolyzed per duplex unwound. In the presence of eIF4B and eIF4G-MC, 8 ATP molecules are hydrolyzed for unwinding one 50/10mer, and 11 ATP molecules are required for unwinding one 32/9mer (Table S3). In the absence of eIF4B, 27 ATP molecules are hydrolyzed per unwinding of one 32/9mer (Table S3). These numbers support a role of eIF4B in coupling ATP hydrolysis to duplex unwinding (26,31).
Overall, eIF4A thus undergoes futile conformational cycles that do not lead to ATP hydrolysis. Other futile cycles do lead to ATP hydrolysis, but not to duplex unwinding. Futile cycles occur both in the absence and presence of eIF4G and eIF4B, but the conformational transitions are accelerated when eIF4B and eIF4G are present. This is the first study on the conformational dynamics of a DEAD-box helicase. It will be interesting to see how other DEAD-box proteins couple their conformational cycle to ATP hydrolysis and duplex unwinding.
Different RNAs affect the eIF4A conformational cycle differently
In the presence of all RNAs we studied here, eIF4B shifts the conformational equilibrium of eIF4A toward the closed state by its dominant effect on the rate constant of closing. Altogether, eIF4B significantly reduces the total cycling time of eIF4A when poly-U-RNA, the 50mer or the 50/10mer are bound (2.6- to 1.9-fold), but very little when 32/9mer RNA (1.2-fold) is bound. The consequences are different for different RNAs, however: When poly-U-RNA is bound, closing is rate-limiting in the absence of eIF4B, and occurs with a similar rate constant to opening when eIF4B is present. For eIF4A bound to 50mer or 50/10mer RNA, closing and opening occur at similar rates in the absence of eIF4B, but opening becomes rate-limiting in the presence of eIF4B. Finally, for the 32/9mer, closing is rate-limiting in the absence and also in the presence of eIF4B. As a result, the dwell times of eIF4A in the open state are nearly identical for poly-U-RNA, the 50mer or the 50/10mer (0.61–0.67 s), but much higher (3.5 s) when the 32/9mer is bound. The time eIF4A spends in the open state thus may be considered as a waiting time for unwinding to occur.
From the different lengths of the duplexes in the 50/10mer and the 32/9mer (10 bp versus 9 bp), we would have predicted a faster unwinding of the 32/9mer. However, unwinding of the 32/9mer is slower than unwinding of the 50/10mer, in agreement with the longer overall passage time of eIF4A through its conformational cycle when this RNA is present. The 32/9mer has no extended single-stranded regions, whereas the 50/10mer contains a long single-stranded overhang. A preferential unwinding of RNAs containing single-stranded regions 5' of the duplex in the presence of eIF4G has been demonstrated previously (25), and may explain why unwinding of the 50/10mer is (slightly) faster than for the 32/9mer, despite the longer duplex region. These results hence suggest that single-stranded regions in the RNA substrate that are naturally encountered by eIF4A in the 5'-UTRs during scanning are important for the capability of eIF4B to accelerate eIF4A closure. Through the presence of single-stranded regions, RNAs can therefore facilitate their own trapping, and their unwinding by eIF4A.
Altogether, the translation initiation factors eIF4B and eIF4G, as well as different structures in the 5'-UTR, modulate the energy landscape underlying the eIF4A conformational cycle. By changing the energetic differences and the energy barriers between functionally relevant conformational states, rate constants for their inter-conversion and their equilibrium distributions are altered, affecting the (loosely) coupled ATP hydrolysis and duplex unwinding reactions. Modulation of the conformational cycle is most likely a common principle for the regulation of ATP hydrolysis and RNA unwinding activities of yeast and mammalian eIF4A, and of other DEAD-box proteins, although the mechanistic details may vary. The dependence of eIF4A on auxiliary factors to adopt the closed conformation provides a large dynamic range for regulatory mechanisms. As a conformational sensor, eIF4A can integrate a wide variety of inter-dependent stimuli and functions as regulatory hub in translation initiation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Jacqueline Butter and Bruno Stephanidis for their contributions to LabVIEW programs, Dorothy Beckett for providing the pBtac-BirA plasmid, Daniela Bremann for excellent technical assistance.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) (KL1153/7-1). Funding for open access charge: DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grifo J.A., Tahara S.M., Morgan M.A., Shatkin A.J., Merrick W.C. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 1983;258:5804–5810. [PubMed] [Google Scholar]
- 2.Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinnebusch A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W.C., Sonenberg N. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 5.Svitkin Y.V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G.J., Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5' secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozen F., Edery I., Meerovitch K., Dever T.E., Merrick W.C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pause A., Methot N., Svitkin Y., Merrick W.C., Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindqvist L., Oberer M., Reibarkh M., Cencic R., Bordeleau M.E., Vogt E., Marintchev A., Tanaka J., Fagotto F., Altmann M., et al. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One. 2008;3:e1583. doi: 10.1371/journal.pone.0001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindqvist L., Pelletier J. Inhibitors of translation initiation as cancer therapeutics. Future Med. Chem. 2009;1:1709–1722. doi: 10.4155/fmc.09.122. [DOI] [PubMed] [Google Scholar]
- 10.Andreou A.Z., Klostermeier D. The DEAD-box helicase eIF4A: paradigm or the odd one out? RNA Biol. 2012;10:19–32. doi: 10.4161/rna.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grifo J.A., Abramson R.D., Satler C.A., Merrick W.C. RNA-stimulated ATPase activity of eukaryotic initiation factors. J. Biol. Chem. 1984;259:8648–8654. [PubMed] [Google Scholar]
- 12.Rogers G.W., Jr, Komar A.A., Merrick W.C. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 13.Rogers G.W., Jr, Richter N.J., Merrick W.C. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 14.Linder P., Lasko P.F., Ashburner M., Leroy P., Nielsen P.J., Nishi K., Schnier J., Slonimski P.P. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 15.Putnam A.A., Jankowsky E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. 2013;1829:884–893. doi: 10.1016/j.bbagrm.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilbert M., Karow A.R., Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol. Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 17.Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 18.Theissen B., Karow A.R., Kohler J., Gubaev A., Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aregger R., Klostermeier D. The DEAD-box helicase YxiN maintains a closed conformation during ATP hydrolysis. Biochemistry. 2009;48:10679–10681. doi: 10.1021/bi901278p. [DOI] [PubMed] [Google Scholar]
- 20.Hilbert M., Kebbel F., Gubaev A., Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2011;39:2260–2270. doi: 10.1093/nar/gkq1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montpetit B., Thomsen N.D., Helmke K.J., Seeliger M.A., Berger J.M., Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers G.W., Jr, Richter N.J., Lima W.F., Merrick W.C. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 23.Korneeva N.L., First E.A., Benoit C.A., Rhoads R.E. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J. Biol. Chem. 2005;280:1872–1881. doi: 10.1074/jbc.M406168200. [DOI] [PubMed] [Google Scholar]
- 24.Rozovsky N., Butterworth A.C., Moore M.J. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA. 2008;14:2136–2148. doi: 10.1261/rna.1049608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajagopal V., Park E.H., Hinnebusch A.G., Lorsch J.R. Specific domains in yeast eIF4G strongly bias the RNA unwinding activity of the eIF4F complex towards duplexes with 5'-overhangs. J. Biol. Chem. 2012;287:20301–20312. doi: 10.1074/jbc.M112.347278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozes A.R., Feoktistova K., Avanzino B.C., Fraser C.S. Duplex unwinding and ATPase activities of the DEAD-Box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 2011;412:674–687. doi: 10.1016/j.jmb.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altmann M., Muller P.P., Wittmer B., Ruchti F., Lanker S., Trachsel H. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 1993;12:3997–4003. doi: 10.1002/j.1460-2075.1993.tb06077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berset C., Zurbriggen A., Djafarzadeh S., Altmann M., Trachsel H. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA. 2003;9:871–880. doi: 10.1261/rna.5380903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schutz P., Bumann M., Oberholzer A.E., Bieniossek C., Trachsel H., Altmann M., Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberer M., Marintchev A., Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andreou A.Z., Klostermeier D. eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. J. Mol. Biol. 2014;426:51–61. doi: 10.1016/j.jmb.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 32.Bi X., Goss D.J. Wheat germ poly(A)-binding protein increases the ATPase and the RNA helicase activity of translation initiation factors eIF4A, eIF4B, and eIF-iso4F. J. Biol. Chem. 2000;275:17740–17746. doi: 10.1074/jbc.M909464199. [DOI] [PubMed] [Google Scholar]
- 33.Methot N., Pickett G., Keene J.D., Sonenberg N. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA remotif. RNA. 1996;2:38–50. [PMC free article] [PubMed] [Google Scholar]
- 34.Park E.H., Walker S.E., Zhou F., Lee J.M., Rajagopal V., Lorsch J.R., Hinnebusch A.G. Yeast eukaryotic initiation factor (eIF) 4B enhances complex assembly between eIF4A and eIF4G in vivo. J. Biol. Chem. 2013;288:2340–2354. doi: 10.1074/jbc.M112.398537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker S.E., Zhou F., Mitchell S.F., Larson V.S., Valasek L., Hinnebusch A.G., Lorsch J.R. Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA. 2013;19:191–207. doi: 10.1261/rna.035881.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caruthers J.M., Johnson E.R., McKay D.B. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckett D., Kovaleva E., Schatz P.J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karow A.R., Theissen B., Klostermeier D. Authentic interdomain communication in an RNA helicase reconstituted by expressed protein ligation of two helicase domains. FEBS J. 2007;274:463–473. doi: 10.1111/j.1742-4658.2006.05593.x. [DOI] [PubMed] [Google Scholar]
- 39.Linden M.H., Hartmann R.K., Klostermeier D. The putative RNase P motif in the DEAD box helicase Hera is dispensable for efficient interaction with RNA and helicase activity. Nucleic Acids Res. 2008;36:5800–5811. doi: 10.1093/nar/gkn581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronson J.E., Fei J., Hofman J.M., Gonzalez R.L., Jr., Wiggins C.H. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersen C.B., Ballut L., Johansen J.S., Chamieh H., Nielsen K.H., Oliveira C.L., Pedersen J.S., Seraphin B., Le Hir H., Andersen G.R. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 42.Bono F., Ebert J., Lorentzen E., Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Del Campo M., Lambowitz A.M. Structure of the yeast DEAD-box protein Mss116p reveals two wedges that crimp RNA. Mol. Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorsch J.R., Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 45.Pause A., Methot N., Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diges C.M., Uhlenbeck O.C. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


