Abstract
Werner syndrome (WS), caused by loss of function of the RecQ helicase WRN, is a hereditary disease characterized by premature aging and elevated cancer incidence. WRN has DNA binding, exonuclease, ATPase, helicase and strand annealing activities, suggesting possible roles in recombination-related processes. Evidence indicates that WRN deficiency causes telomeric abnormalities that likely underlie early onset of aging phenotypes in WS. Furthermore, TRF2, a protein essential for telomere protection, interacts with WRN and influences its basic helicase and exonuclease activities. However, these studies provided little insight into WRN's specific function at telomeres. Here, we explored the possibility that WRN and TRF2 cooperate during telomeric recombination processes. Our results indicate that TRF2, through its interactions with both WRN and telomeric DNA, stimulates WRN-mediated strand exchange specifically between telomeric substrates; TRF2's basic domain is particularly important for this stimulation. Although TRF1 binds telomeric DNA with similar affinity, it has minimal effects on WRN-mediated strand exchange of telomeric DNA. Moreover, TRF2 is displaced from telomeric DNA by WRN, independent of its ATPase and helicase activities. Together, these results suggest that TRF2 and WRN act coordinately during telomeric recombination processes, consistent with certain telomeric abnormalities associated with alteration of WRN function.
INTRODUCTION
Patients with the autosomal recessive disease Werner syndrome (WS) prematurely develop many of the characteristics of normal aging, including graying and loss of hair, cataracts, atherosclerosis and cancer following relatively normal childhood development (1–3). WS results from defects in a single gene product, Werner syndrome protein (WRN) (4,5), that possesses 3′ to 5′ helicase, 3′ to 5′ exonuclease and strand annealing activities (6–9). WRN is a member of the RecQ helicase family that also includes the human proteins BLM and RecQ4, deficient in the premature-aging and cancer-prone diseases Bloom syndrome and Rothmund–Thomson syndrome, respectively (4,10,11). Cells derived from WS patients are characterized by genomic instability including large-scale chromosomal insertions, deletions and translocations (12,13). Compared to normal cells, WS cells also have a reduced replicative lifespan that is extended by the expression of telomerase (14), suggesting that the premature senescence of WS cells may be due to telomere dysfunction. The notion that WRN participates in telomere maintenance was further strengthened by the findings of premature aging phenotypes reminiscent of WS and telomere anomalies in mouse models deficient for Wrn and telomerase (15,16) and WRN co-localization and co-immunoprecipitation (predominantly during S phase) with telomeric DNA and telomeric factors (17–20).
Telomeres are the protective structures present at the ends of linear chromosomes and are composed of 5–15 kb of the repeating sequence, TTAGGG/AATCCC, ending in a 3′ overhang of the G-rich strand. A complex of proteins (including TRF1, TRF2, TIN2, POT1, TPP1 and RAP1) termed as shelterin binds to and protects telomeres and regulates their maintenance (21). Two of the most extensively studied shelterin proteins, TRF1 and TRF2, are both telomere-specific, double-stranded DNA binding proteins that play differing roles (22–24); notably TRF2 also possesses structure-specific DNA binding properties (25–27). While TRF1 is thought to be a regulator of telomere length through modulation of telomerase (28), TRF2 is essential for telomere protection. Loss of TRF2 function leads to telomere fusions or activation of ATM/p53-mediated DNA damage response at unprotected telomeres that results in apoptosis or cellular senescence (29–33). The protective nature of TRF2 may derive from its ability to promote formation and stabilization of the T-loop, a structure that embeds the 3′ G-rich telomeric overhang within the homologous duplex region through an invasive mechanism similar to recombination, thereby sequestering telomeric ends and preventing them from triggering DNA damage responses (25,34). Notably, telomeres shortened to a critical length lose this protective mechanism and trigger DNA damage responses with their accompanying consequences (35,36).
Regarding its putative telomeric role, WRN's enzymatic activities have been shown to be influenced by a physical and functional interaction with TRF2 (17,18,37,38), suggesting that these proteins may cooperate during telomere metabolism. Moreover, our lab has previously demonstrated that WRN coordinates its helicase and annealing activities to perform strange exchange, indicating its possible involvement in recombination pathways (7). In agreement, certain telomeric abnormalities associated with WRN deficiency are consistent with a role for WRN in recombinational processes involving telomeric DNA (39–42). Toward a possible role for WRN in telomeric recombination events, in this study we have further investigated the functional interaction between WRN and TRF2, particularly in the context of strand exchange of telomere-related DNA substrates. Here, we report that TRF2 enhances WRN's strand exchange activity preferentially on substrates that possess telomeric sequence as well as structural characteristics that mimic the telomeric terminus. We also find that TRF2 can be displaced from telomeric DNA by WRN, in a manner independent of its ATPase and helicase activities. These results support a potential in vivo function for WRN in coordination with TRF2 during telomeric recombination processes.
MATERIALS AND METHODS
Proteins
Overexpression and purification of His-tagged WRN-E84A and WRN-K577M were performed as previously described (43), except with inclusion of 0.1% Nonidet P40 (NP40) in all chromatography buffers. WRN-E84A contains a point mutation that eliminates its 3′ to 5′ exonuclease activity but does not alter its helicase and annealing activities (7,44); the point mutation in WRN-K577M eliminates ATPase and helicase activities, but retains exonuclease activity (6,45). Titia de Lange (Rockefeller University) provided baculovirus with His-tagged TRF1 and TRF2 constructs. TRF1 and TRF2 were overexpressed and purified as previously reported (46), except a different elution buffer was used (38). His-tagged TRF2ΔB, kindly provided by Jack Griffith and Brian Bower (University of North Carolina at Chapel Hill), was purified as described (26).
DNA substrates
All polyacrylamide gel electrophoresis (PAGE)-purified oligonucleotides (see Supplementary Table S1 for sequences) were from Integrated DNA Technologies. G77telo and G77scr were 5′ radiolabeled using 32P-γ-ATP (Perkin-Elmer) and T4 polynucleotide kinase (New England Biolabs). Unincorporated radionucleotides were removed using Mini Quick Spin Oligo Columns (Roche). These labeled oligomers were annealed to unlabeled complementary oligonucleotides by heating at 90°C followed by step-wise cooling (lowering the temperature in 5°C increments and holding at each temperature for 5–15 min) to promote proper alignment of repeating sequences. Likewise, 5′ biotinylated-G68telo (b-G68telo) was annealed to C50telo, and G24telo to C24telo. Annealed substrates were then purified by native PAGE. Gel slices corresponding to substrate bands were excised and minced, and the substrates eluted by passive diffusion for at least 16 h at 4°C in 10 mM Tris (pH 8.0) and 10 mM NaCl.
Helicase assays
Helicase assays were carried out in 20 μl WRN reaction buffer (40 mM Tris-HCl [pH 8.0], 4 mM MgCl2, 5 mM dithiothreitol [DTT], 100 μg/ml bovine serum albumin [BSA] and 0.1% NP40) supplemented with ATP or ATPγS (1 mM) as indicated and containing *G77telo/C38telo or *G77scr/C38scr (0.05–0.1 nM) and TRF2 (0.52–5.2 nM), WRN-E84A (0.5 nM) and/or UvrD (0.5 nM) as specified. Following pre-incubation of proteins at 4°C for 5 min, DNA substrate was added and the samples were incubated at 37°C for 10 or 15 min. Reactions were stopped by addition of one-sixth volume of helicase stop buffer (30% glycerol, 0.9% SDS, 50 mM EDTA, 0.25% bromophenol blue [BPB] and 0.25% xylene cyanol). DNA species from these reactions were separated by native PAGE (8%). After gel drying, labeled products were detected and quantified using a Storm 860 Phosphoimager and ImageQuant software (GE Healthcare). Unwinding (%) was calculated by determining the amount of labeled single-stranded (*G77telo or *G77scr) product compared to total radioactivity for that lane. Data points here and for strand exchange assays below were compared using paired, two-tailed t-tests.
Strand exchange assays
Strand exchange assays were carried out in 10 μl WRN reaction buffer (pH 7.0, unless otherwise specified) containing radiolabeled substrate (0.05–0.1 nM) plus or minus non-labeled substrate (0.03–1 nM), ATP (1 mM), TRF2 (0.5–7.2 nM), TRF2ΔB (0.6–11.6 nM), TRF1 (0.6–5.8 nM) and/or WRN-E84A (0.5–4.1 nM), with specific DNA and protein components for each experiment described in the Results and/or figure legends. Reactions containing DNA substrates were pre-incubated with TRF2, TRF1 and/or WRN-E84A at 4°C for 5 min followed by incubation at 37°C for 15 min, then stopped as described for helicase assays. Alternatively, some assays were stopped by adding 0.4 mg/ml proteinase K for 10 min at 37°C followed by one-sixth volume of helicase dyes lacking sodium dodecyl sulphate (SDS). DNA species in samples were separated using native PAGE (6–12%) with various acrylamide:bis-acrylamide ratios (19:1, 37.5:1 or 100:1), as specified, to optimize resolution of key DNA products. Labeled products were viewed and quantified as described above. Strand exchange was calculated as the percentage of exchange product in relation to the total radioactivity in each respective lane after subtraction of proper background.
Electrophoretic Mobility Shift Assays
In WRN reaction buffer (pH 7.0 or 8.0) with ATP (1 mM), radiolabeled DNA substrates were incubated at 37°C for 10 or 15 min with TRF1 (0.73–2.9 nM), TRF2 (0.29–2.9 nM) or TRF2ΔB (0.6–5.8 nM) as specified in Results and/or figure legends. In certain experiments, pUC19 plasmid DNA (0–47 pg/μl) or unlabeled (G77scr/C38scr) substrate (0.03–0.2 nM) was added to examine the specificity of TRF2 binding to telomeric duplexes. After addition of one-sixth volume of loading buffer (30% glycerol without or with 50 mM EDTA, 0.25% BPB and 0.25% XC), DNA and DNA–protein complexes were separated using native PAGE (6%, 100:1 acrylamide:bis-acrylamide ratio) gels at 4°C or 25°C as specified in figure legends. Labeled products were viewed as previously described and DNA–protein binding was quantified by subtracting the percentage of unbound DNA substrate signal remaining relative to reactions without protein.
TRF2 displacement assays
WRN-mediated displacement of TRF2 was examined after stably binding TRF2 to a biotin-tagged, telomeric DNA substrate immobilized on streptavidin-coated beads. Briefly, b-G68telo/C50telo DNA substrate (32.5 nM) in 60 μl bead buffer (40 mM Tris-HCl [pH 8.0], 5 mM DTT, 100 μg/ml BSA, 0.1% NP40 and 50 mM NaCl) was incubated (with mixing) with 20 μl pre-equilibrated streptavidin-agarose beads (Life Technologies) at 25°C for 30 min followed by TRF2 (4.3 nM) binding in bead buffer at 25°C for another 30 min. Following each binding step above, beads were collected by centrifugation and washed with bead buffer to remove unbound DNA or TRF2. Subsequently, bead buffer supplemented with 4 mM MgCl2 plus or minus WRN-E84A (60 nM) was added to the beads and incubation continued at 25°C for 5 min. To prevent rebinding of TRF2 to bead-associated telomeric DNA, another DNA substrate, G24telo/C24telo (67 nM), with 24 bp of telomeric sequence was incubated with the reactions for an additional 10 min at 37°C. After centrifugation to pellet beads, supernatants were transferred to new tubes and the beads were washed once and then resuspended in bead buffer. Denaturation of proteins was achieved by addition of equal volume of SDS dyes (4% SDS, 20% glycerol, 0.05% BPB and 2 M 2-mercaptoethanol) to the supernatants and bead suspensions followed by heating at 90°C for 5 min. Supernatant and bead fractions as well as a purified TRF2 marker were analyzed by SDS-PAGE (10%) and Western blotting, using mouse anti-TRF2 primary antibody (Imgenex) overnight followed by HRP-linked anti-mouse secondary antibody (GE Healthcare) for 1 h. TRF2 protein was visualized following chemiluminescent development using ECL2 (GE Healthcare). Densitometry of immunoblots was performed to determine the percentage of TRF2 displaced by WRN-E84A relative to control reactions without WRN, and statistical comparisons were achieved using paired, one-tailed t-tests.
RESULTS
WRN has been strongly implicated in telomere metabolism, primarily due to the telomere anomalies that occur in its absence (15,16,19,39,40,42,47). Furthermore, our lab and others have shown that a direct physical interaction with telomeric factor TRF2 influences WRN's exonuclease and helicase activities (17,18,38). These studies, although provocative, do not provide much insight into the specific function of WRN and its cooperation with TRF2 during telomere metabolism. Notably, our lab has also demonstrated that WRN coordinates its helicase and annealing activities to perform strand exchange (7), suggesting a possible function in recombination processes impacting telomere length and structure such as the alternative lengthening of telomeres (ALT) pathway. Such a function would be consistent with the telomeric abnormalities associated with WRN deficiency. To explore a potential role for WRN in telomeric recombination, we examined the effect of TRF2 on not only WRN helicase activity but also, and more importantly, on the ability of WRN to perform strand exchange on DNA substrates, particularly those with telomeric characteristics. Notably, use of 3′-overhang substrates with G-rich sequence composition in helicase and strand exchange assays reflects structures that (i) naturally occur at telomeric ends and (ii) are required for strand invasion in recombination processes. Notably, these overhang substrates also possess the requisite structure to initiate WRN-mediated 3′ to 5′ unwinding—i.e. a single-stranded region 3′ to the duplex to be unwound (48–50). Since these studies were focused on WRN's helicase and strand exchange activities, we almost exclusively used WRN-E84A mutant protein (hereafter referred to as WRN), which lacks the exonuclease function but still retains robust helicase and annealing activities (7,44). Importantly, use of exonuclease-deficient WRN prevents possible degradation of DNA that would alter substrate structure during reactions and significantly complicate interpretation of assay results. In order to optimally determine potential contributions of TRF2, WRN concentrations in both the helicase and strand exchange reactions described below were initially titrated to limiting levels—i.e. concentrations that generated low amounts of relevant reaction products in the absence of TRF2.
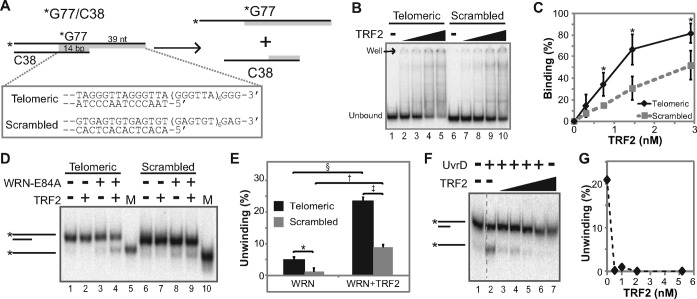
TRF2 enhances WRN-mediated unwinding of telomeric DNA
Initially, the effect of TRF2 on WRN's helicase activity, a crucial component of strand exchange, was examined. All DNA substrates used in this and subsequent assays contained telomeric (or scrambled) repeat sequences adjacent to 24 bp of random sequence included to mediate proper in-frame annealing of repeated sequences during substrate preparation. The partial duplex telomeric DNA substrate (*G77telo/C38telo) used only for helicase assays contained a 39 nt G-strand 3′ overhang adjacent to 38 bp of duplex sequence (Figure 1A). This amount of duplex DNA permitted unwinding by the weak helicase activity of WRN alone and also contained sufficient telomeric duplex sequence (14 bp) to allow TRF2 binding (51). In parallel, assays were performed with a similar substrate (*G77scr/C38scr) with scrambled (GAGTGT) repeat sequence at precisely the same position (Figure 1A) and thus lacking a consensus TRF2 binding sequence. After TRF2 was incubated with each substrate in a manner identical to standard helicase reaction conditions, Electrophoretic Mobility Shift Assays (EMSA) were performed to compare TRF2 binding to telomeric versus scrambled DNA substrate, employing several different electrophoresis conditions found to impact binding. Under the lowest stringency conditions tested (electrophoresis performed at 4°C without BPB and XC in the sample), TRF2 bound to both substrates in a concentration-dependent manner, but binding to telomeric *G77telo/C38telo substrate was consistently higher at each concentration as compared to scrambled *G77scr/C38scr substrate; this differential binding reached statistical significance except at the lowest TRF2 concentration (Figure 1B and C). Under more stringent conditions (using dyes and/or electrophoresis at 25°C), TRF2 binding to both substrates decreased as stringency increased; however, TRF2 binding to the telomeric substrate compared to scrambled substrate was consistently higher under each condition (Supplementary Figure S1A and B). Additional experiments showed that unlabeled G77scr/C38scr substrate could compete away the (non-specific) binding of TRF2 to labeled *G77scr/C38scr but had minimal effects on TRF2 binding to labeled telomeric *G77telo/C38telo substrate (Supplementary Figure S1C and D). Moreover, DNase I footprinting experiments clearly show that TRF2 binds, as expected, to the telomeric duplex region of the *G77telo/C38telo substrate (Supplementary Figure S1E). These results suggest that, although some binding to the scrambled overhang substrate can be observed, TRF2 preferentially binds to the overhang substrate containing telomeric duplex sequences.
Figure 1.
TRF2 stimulates WRN's helicase activity. (A) Diagram of helicase reaction using 3′ overhang (*G77/C38) substrates. Positions of telomeric and scrambled sequences (specified in box) are shaded gray, and radiolabeled strands are indicated by asterisks. (B) To examine DNA binding by TRF2, telomeric (*G77telo/C38telo) or scrambled (*G77scr/C38scr) substrates (0.05 nM) were incubated with TRF2 (0, 0.3, 0.7, 1.5 or 2.9 nM) at 37°C for 10 min in WRN reaction buffer (pH 8.0) and then analyzed by EMSA, performed at 4°C without dyes. DNA and DNA–protein complexes were visualized by phosphorimaging; positions of unbound DNA and the well are indicated at left. (C) Binding (%) of TRF2 to *G77telo/C38telo (black) and *G77scr/C38scr (gray) from four experiments as in B was calculated as described in Materials and Methods based on decreases in unbound substrate. Means and standard deviations were plotted with respect to TRF2 concentration. Significant differences (all P values <0.011) between TRF2 binding to telomeric versus scrambled substrate were determined using unpaired, one-tailed t-tests and are indicated by asterisks. (D) Helicase assays were performed by incubating telomeric (*G77telo/C38telo) or scrambled (*G77scr/C38scr) substrates (0.05 nM) without or with TRF2 (2.9 nM) and WRN-E84A (0.5 nM) at 37°C for 10 min. Products and single-stranded product markers (M) were separated by native PAGE and visualized as above. DNA species are indicated at left. (E) Bar graph of data from helicase assays, as in D with either telomeric (black) or scrambled (gray) overhang substrates and WRN without or with TRF2. Results are from three independent experiments, except for reactions containing WRN plus TRF2 on scrambled substrate (two repeats). Unwinding (%) was calculated as the percentage of helicase product (labeled *G77telo or *G77scr oligomer), compared to the total signal for each respective lane, with means and standard deviations shown. P-values were determined, comparing bracketed data points using paired two-tailed t-tests: *P = 0.0055; §P = 0.0000035; †P = 0.0041; ‡P = 0.00081. (F) Helicase assays using *G77telo/C38telo substrate (0.01 nM) with UvrD (0.5 nM) minus or plus TRF2 (0, 0.5, 1.0, 2.1 or 5.2 nM) were performed at pH 8.0 at 37°C for 10 min. Products were analyzed as above. Dashed vertical lines here (and in other figures) indicate where separated lanes on the same gel were spliced together. Relevant DNA species are indicated at left. (G) Line graph of data from F, depicting the percentage of UvrD-mediated unwinding versus TRF2 concentration.
To investigate the effect of TRF2 on the helicase activity of WRN, limiting concentrations of WRN were pre-incubated with or without TRF2 before addition of *G77telo/C38telo and *G77scr/C38scr substrates with immediate incubation at 37°C, and the products were resolved by native PAGE (Figure 1D); results from multiple experiments are depicted in Figure 1E, which reveals several interesting observations. Using WRN alone, low levels of unwinding were observed for both substrates (Figure 1D, lanes 3 and 8), although slightly better unwinding was observed on the telomeric substrate (Figure 1E). Most prominently, inclusion of TRF2 enhanced WRN-mediated unwinding on both substrates (Figure 1D, lanes 4 and 9, and E). This effect was not due to unwinding catalyzed by TRF2, as TRF2 alone had no detectable unwinding activity on these substrates (Figure 1D, lanes 2 and 7). Notably, TRF2 stimulated WRN unwinding to a higher extent (23.4%) on the telomeric substrate as compared to the scrambled substrate (8.9%). Compared to respective levels of unwinding achieved with WRN alone, TRF2 stimulation of WRN unwinding on the telomeric substrate was much more statistically significant than its effect on unwinding of the scrambled substrate. Importantly, unwinding was catalyzed by the ATPase-dependent helicase activity of WRN, as no unwinding was observed in the presence of ATPγS or when helicase-deficient WRN-K577M mutant protein was used (Supplementary Figure S1F). In another control to determine whether this effect of TRF2 is specific to WRN unwinding activity, TRF2 was tested with another 3′ to 5′ helicase, Escherichia coli UvrD, on the telomeric substrate. In sharp contrast to its stimulatory effect on the helicase activity of WRN, TRF2, at concentrations ranging from 0.5 to 5.2 nM, dramatically inhibited UvrD-mediated unwinding (Figure 1F and G). Taking into account the results of both DNA binding and unwinding assays, our findings suggest a mechanism by which TRF2 recruits WRN to DNA substrate which is then unwound by WRN, with perhaps the moderate preference for TRF2 binding to the telomeric overhang leading to the substantially higher extent of unwinding observed on that substrate. Importantly, the preference of TRF2 for this *G77telo/C38telo substrate containing only 14 bp of telomeric duplex region used in unwinding assays is modest in comparison to the much stronger binding preference of TRF2 for telomeric substrates (G77telo/C50telo, G77telo/C71telo and G77telo/C77telo) containing 26, 47 and 53 bp of telomeric duplex sequence used in the strand exchange assays below.
TRF2 stimulates WRN-mediated strand exchange
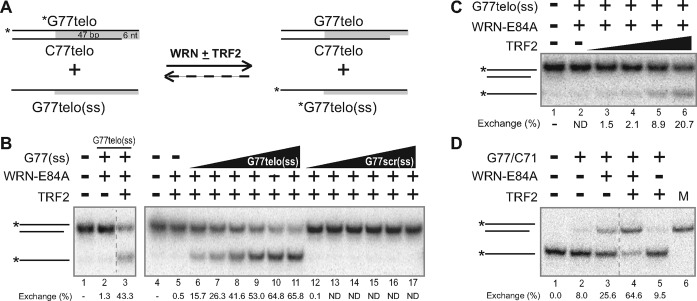
WRN's helicase activity is required for strand exchange, the coordinated invasion of a single-stranded region into a complementary double-stranded substrate, followed by branch migration of the resulting three-way junction (7). Importantly, the process of strand exchange is a key step in many recombination pathways. Since TRF2 stimulated WRN unwinding activity, we wanted to investigate their potential coordination during strand exchange relevant to telomeric recombination processes. To this end, strand exchange was evaluated using assays similar to the helicase assay utilized above, except two DNA substrates were generally used in each reaction, one double-stranded substrate and the other substrate either single-stranded (Figure 2A) or double-stranded (Figure 3A). As before, telomeric or scrambled repeat sequences were present at comparable positions on the respective substrates. However, in these strand exchange reactions, DNA substrates contained longer duplex (telomeric or scrambled) repeat sequences (26, 47 and 53 bp) than in helicase reactions (14 bp), presumably increasing the specificity of TRF2 for telomeric substrates. Importantly, the presence of single-stranded G-rich telomeric sequence specifically at 3′ ends of certain substrates would be the expected arrangement after recombination-related resection of truncated telomeres, and also analogous to invasion of single-stranded telomeric overhangs present on the 3′ ends of linear chromosomes.
Figure 2.
TRF2 enhances WRN-mediated strand exchange between single-stranded DNA and an overhang. (A) Depiction of strand exchange reactions between G77telo/C71telo and G77telo, with positions of telomeric sequences shaded (gray). (B) Strand exchange assays were performed (at pH 8.0) with *G77telo/C71telo (0.05 nM) and unlabeled G77telo (0.5 nM) with TRF2 (3.6 nM) and/or WRN-E84A (0.5 [left panel] or 4.1[right panel] nM) as indicated at 37°C for 15 min. DNA species were separated by native PAGE (8%, 19:1 acrylamide:bis-acrylamide) and visualized by phosphorimaging. Positions of relevant DNA species are indicated at left. (C) In assays performed and analyzed as in B, *G77telo/C71telo (0.05 nM) and G77telo (0.5 nM) were incubated without or with WRN-E84A (2.0 nM) and various concentrations of TRF2 (0, 0.5, 0.9, 1.8 or 3.6 nM) as indicated at 37°C for 15 min. (D) In the reverse reaction scenario, *G77telo (0.1 nM) and G77telo/C71telo (1 nM) were incubated plus or minus TRF2 (5.8 nM) and/or WRN-E84A (1.8 nM) as specified at 37°C for 15 min. DNA species were separated and depicted as in B. For panels B–D, strand exchange levels (below each respective lane) were quantified with respect to control reactions without enzyme; ND = not detectable—i.e. product levels ≤ controls without enzyme.
Figure 3.
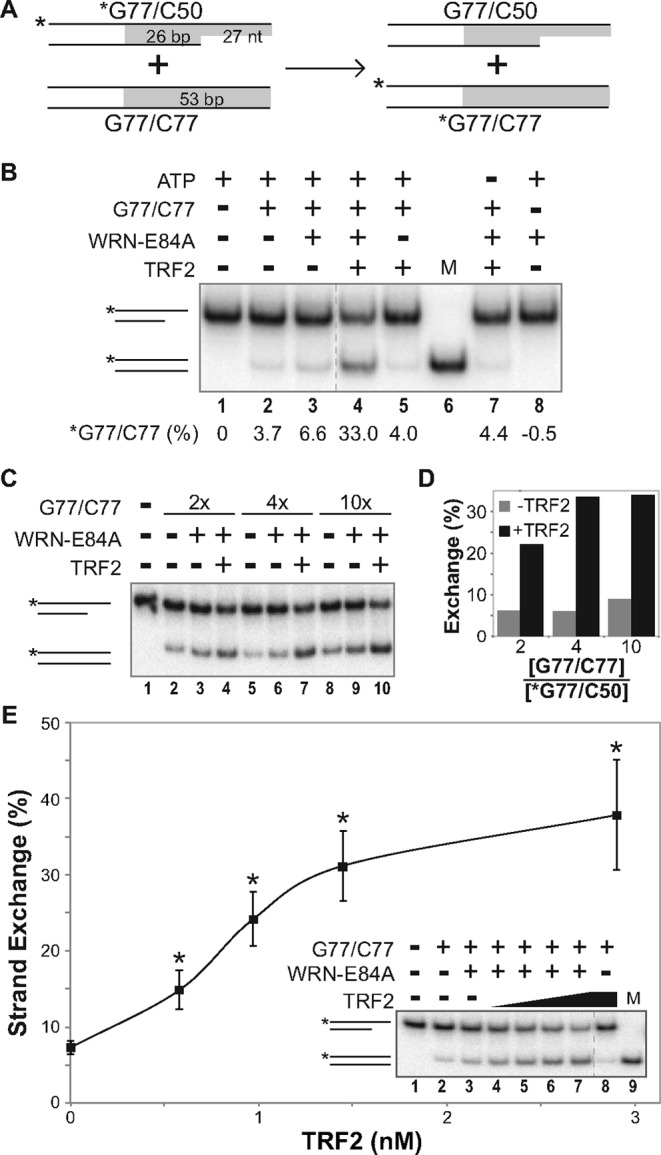
TRF2 enhances WRN-mediated strand exchange between DNA duplex substrates. (A) Depiction of strand exchange reactions between *G77/C50 and G77/C77 substrates, with positions of telomeric (or scrambled) sequences shaded (gray). (B) Strand exchange assays were carried out at pH 7.0 on *G77telo/C50telo (0.1 nM) and G77telo/C77telo (1 nM) using TRF2 (2.9 nM) and/or WRN-E84A (3.4 nM) as indicated at 37°C for 15 min. As controls, ATP (lane 7) or G77telo/C77telo (lane 8) was withheld from individual reactions. Samples and a *G77telo/C77telo DNA marker (M) were separated by native PAGE (12%, 37.5:1); positions of relevant DNA species are indicated at left. Strand exchange (below each lane) was quantified as percentage of G77telo/C77telo product compared to the total signal in each respective lane, after subtracting background levels in *G77telo/C50 telo substrate only reactions (lane 1). (C) Reactions were performed as in B using *G77telo/C50telo (0.1 nM) and G77telo/C77telo (0.2, 0.4 or 1 nM) without or with WRN-E84A (3.8 nM) and TRF2 (2.9 nM) as indicated. DNA species were resolved by native PAGE (12%, 100:1). (D) Strand exchange (%) from assays as in C without (gray bars) or with (black bars) TRF2 was calculated as described above, except amounts of *G77telo/C77telo product present in reactions containing both substrates without enzyme were subtracted from all enzyme-containing reactions, and plotted against the ratio of unlabeled to labeled DNA substrate in the initial reaction. (E) Reactions performed as in B, using *G77telo/C50telo (0.1 nM) and G77telo/C77telo (0.4 nM) with WRN-E84A (3.8 nM) and/or TRF2 (0, 0.6, 1.0, 1.5 or 2.9 nM), were analyzed as described in C, along with a *G77telo/C77telo marker (M). Strand exchange from at least three independent experiments was calculated as described in D and plotted versus TRF2 concentration. Using unpaired, two-tailed t-tests, significant differences (P < 0.0005) in results obtained with WRN and TRF2 reactions versus WRN only controls are denoted by asterisks.
Initially, reactions were set up to examine strand exchange between G77telo (ss), a single-stranded 77-mer with 53 nt of G-telomeric sequence at its 3′ end and G77telo/C71telo, a substrate containing the same G77telo oligomer annealed to a complementary 71-mer (C71telo), resulting in a 71 bp double-stranded region containing 47 bp of duplex telomeric sequence adjacent to a 6 nt 3′ overhang of the G-rich strand (Figure 2A). In these assays, strand exchange is measured by swapping of unlabeled and labeled G77telo strands with the complementary C71telo strand (Figure 2A). Since we have shown that WRN alone catalyzes strand exchange (7), limiting concentrations of WRN were used for these reactions. Under these conditions, WRN alone mediated minimal strand exchange (Figure 2B, lane 2). Importantly, the combination of TRF2 and WRN resulted in increased exchange, as indicated by generation of faster-migrating, single-stranded product (Figure 2B, lane 3). In this reaction scenario, single-stranded products might result from simple unwinding, as observed previously (Figure 1D). However, this is clearly not the case, as generation of single-stranded product does not occur in the absence of the complementary G77telo oligomer (Figure 2B, lane 5); moreover, WRN alone could not detectably unwind the *G77telo/C71telo or *G77telo/C50telo substrates in the absence of homologous DNAs (data not shown and Figure 3B, lane 8). We attribute the lack of WRN-mediated (and TRF2-assisted) unwinding in these reactions to the much longer duplex region (71 bp) of the G77telo/C71 substrate as compared to the 38 bp region of the G77telo/C38 substrate used for helicase assays. Generation of single-stranded, labeled product indicative of strand exchange absolutely depended upon the presence of completely complementary G77telo, as no detectable products were observed when another 77-mer (G77scr) containing scrambled GAGTGT repeats in place of telomeric repeats was used (Figure 2B, lanes 6–17). Furthermore, increasing amounts of complementary G77telo mediated increasing levels of strand exchange, reaching 65.8% at the highest G77telo concentration (Figure 2B, lanes 6–11). Importantly, under these conditions WRN-mediated strand exchange was also dependent upon the presence of TRF2 and its concentration in the reaction (Figure 2C, lanes 2–6). While low concentrations of TRF2 showed little or no enhancement of strand exchange compared to reactions containing WRN alone, higher TRF2 concentrations substantially increased the levels of strand exchange (Figure 2C, lanes 3–6). To further confirm that we were observing strand exchange and determine its requirements, we performed the reverse reaction—i.e. using labeled *G77telo and unlabeled G77telo/C71telo and measured strand exchange by generation of *G77telo/C71telo product. Although WRN alone mediated some strand exchange in this scenario, much higher levels of the *G77telo/C71telo product were generated in the presence of both WRN and TRF2 (Figure 2D, lanes 3–4). As expected, TRF2 alone could not mediate strand exchange (Figure 2D, compare lanes 2 and 5). These results indicate that TRF2 enhances strand exchange performed by WRN in reactions containing both a substrate with duplex telomeric sequence and a single-stranded substrate containing G-rich telomeric sequence.
Although the results above suggested that TRF2 stimulated WRN's strand exchange activity on telomeric DNA, we wanted to design assays that more closely model telomeric recombination events that might be mediated by cooperation between WRN and TRF2. In contrast to the partial duplex used above (Figure 2A), we constructed a duplex (G77telo/C77telo) substrate (Figure 3A) that possesses no single-stranded character, making it more closely related to the in vivo telomeric duplex region as well as a low-affinity substrate for WRN (52). To better mimic both a normal telomeric end and a strand invasion precursor, in the same reactions we utilized a labeled partial duplex (*G77telo/C50telo) substrate containing a 27 nt G-rich telomeric 3′ overhang adjacent to 26 bp of telomeric duplex sequence (Figure 3A). Thus, in contrast to the previous assay, both labeled and unlabeled substrates contained sufficient duplex telomeric sequence to bind TRF2. Strand exchange in this particular assay was measured by swapping of strands between the unlabeled and labeled duplexes, resulting in conversion of *G77telo/C50telo to *G77telo/C77telo (Figure 3A). Notably, the *G77telo/C77telo product migrates faster than the *G77/C50 substrate on the gel system (native 12% PAGE, 37.5:1 or 100:1 acrylamide:bis-acrylamide ratio) used especially to separate these DNA products. Initially, the capability of WRN and TRF2 to perform strand exchange between *G77telo/C50telo and G77telo/C77telo was examined (Figure 3B). At limiting concentrations used here, WRN alone mediated a very modest increase in the *G77telo/C77telo strand exchange product over background levels observed without protein and in the presence of only TRF2 (Figure 3B, compare lanes 2, 3 and 5); we attribute this background to low levels of single-stranded C77telo oligomer in unlabeled substrate preparations, as it is not detected when G77telo/C77telo is absent (Figure 3B, lane 8). Importantly, with both WRN and TRF2, a substantial increase in the faster migrating *G77telo/C77telo product occurred (Figure 3B, lane 4). Importantly, this activity was dependent on ATP (Figure 3B, lane 7), indicating the involvement of WRN's ATPase-dependent helicase activity. This strand exchange activity was further examined by varying the concentration of unlabeled homologous duplex G77telo/C77telo (2–10× molar excess over labeled *G77/C50) in the reaction (Figure 3C and D). Results from these experiments clearly show that, together, WRN and TRF2 generate substantially higher levels (3–5-fold) of strand exchange product than observed with WRN alone (Figure 3C and D). Furthermore, 4- and 10-fold excesses of the unlabeled telomeric duplex promoted higher levels of strand exchange in the presence of both WRN and TRF2 than a 2-fold excess (Figure 3C, lanes 4, 7 and 10, and D), demonstrating that this process is somewhat influenced by substrate concentration. The effect of TRF2 concentration on this strand exchange reaction was also evaluated in multiple experiments (Figure 3E). Consistent with previous results, low levels of WRN-mediated strand exchange (7.3%) occur in the absence of TRF2 (Figure 3E, lane 3). However, increasing TRF2 concentrations resulted in increasing amounts of exchange, with the most efficient exchange (37.9%) occurring at the highest (2.9 nM) TRF2 concentration (Figure 3E, lane 7). Similar to the previous reaction scenario, these results also clearly demonstrate that TRF2 enhances WRN-mediated strand exchange activity.
TRF2 specifically stimulates WRN-mediated strand exchange between telomeric duplexes
The findings above indicate that WRN and TRF2 act together to mediate strand exchange, but they did not examine whether this cooperation is specific to telomeric DNA. To determine the specific influence of telomeric DNA on these strand exchange reactions, we utilized pairs of DNA substrates as depicted in Figure 3A. While one pair of substrates (*G77telo/C50telo and G77telo/C77telo) contained telomeric repeat (TTAGGG/AATCCC) sequences, the other pair (*G77scr/C50scr and G77scr/C77scr) had the same structure but replaced telomeric regions with scrambled (GAGTGT/CTCACA) repeat sequences with identical nucleotide content. Again, strand exchange was measured by transfer of labeled (*G77telo or *G77scr) strand from the overhang substrate to produce a labeled, fully duplex substrate (Figure 3A). We compared the levels of strand exchange using equivalent and fixed amounts of telomeric (*G77telo/C50telo and G77telo/C77telo) or scrambled (*G77scr/C50scr and G77scr/C77scr) substrate pairs, maintaining a fixed WRN concentration while varying TRF2 (Figure 4A and B). As above, limiting amounts of WRN were used in these reactions so that low levels of strand exchange were observed on telomeric and scrambled substrate pairs without TRF2 (Figure 4A, lanes 3 and 12). As expected, TRF2 alone could not mediate strand exchange using either telomeric or scrambled substrate pairs (Figure 4A, lanes 8 and 17). On substrates possessing scrambled sequences in the presence of WRN, increasing TRF2 concentrations resulted in only minor increases in strand exchange (Figure 4A, lanes 13–16). However, TRF2 more substantially stimulated WRN-mediated exchange when substrate pairs contained telomeric sequences (Figure 4A, lanes 3–7). In fact, at every TRF2 concentration evaluated, WRN-catalyzed strand exchange was significantly higher with telomeric than with scrambled DNA substrates; at the highest TRF2 concentration tested, strand exchange reached 37.9% using telomeric substrates compared to only 13.4% using scrambled substrates (Figure 4B). Interestingly, a modest preference for WRN-mediated exchange between the telomeric substrates was also observed even in the absence of TRF2 (Figure 4B), although the levels of exchange were relatively low in these comparisons. These results indicate that TRF2 stimulated WRN-mediated strand exchange much more readily on substrates containing telomeric repeats, most likely because of TRF2's binding specificity for duplex telomeric DNA (23,24). To corroborate this, we used EMSA to examine TRF2 binding to telomeric and scrambled DNA substrates with or without a plasmid DNA competitor (pUC19) lacking telomeric sequences. Both *G77telo/C77telo and G77telo/C50telo were bound by TRF2, as indicated by almost complete loss of the unbound substrate and appearance of a new product at the top of the gel; smearing of the signal in reactions containing G77telo/C50telo likely is due to less stable TRF2 binding to this substrate that contains a shorter (26 bp) duplex telomeric region (Figure 4C). Binding by TRF2 is highly specific, since the presence of plasmid DNA (pUC19) only slightly increases the amount of unbound product (Figure 4C, lanes 2–6 and 14–18). In contrast, *G77scr/C77scr and *G77scr/C50scr (Figure 4C, lanes 7–12 and 19–24) remain largely unbound by TRF2 whether or not pUC19 is present. Taken together, these studies suggest that TRF2 binding specifically to telomeric sequences stimulates WRN-mediated strand exchange between substrates containing duplex telomeric DNA.
Figure 4.
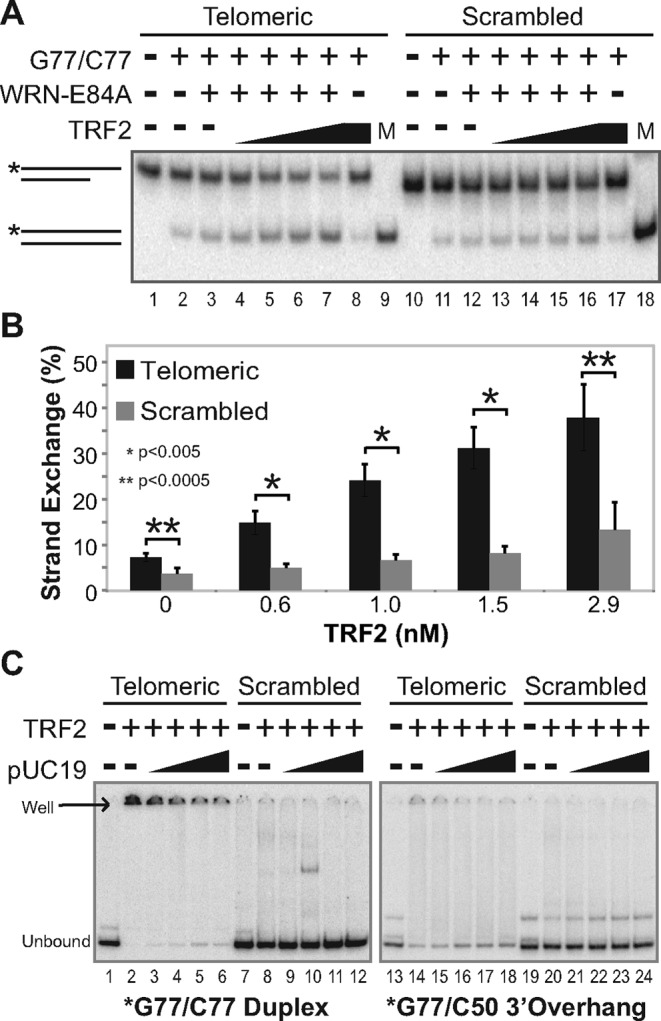
TRF2 specifically stimulates telomeric strand exchange. (A) To examine the sequence specificity of strand exchange, telomeric (*G77telo/C50telo [0.1 nM] and G77telo/C77telo [0.4 nM]) or scrambled (*G77scr/C50scr [0.1 nM] and G77scr/C77scr [0.4 nM]) substrate pairs were incubated at pH 7.0 with WRN-E84A (3.8 nM) and/or TRF2 (0, 0.6, 1.0, 1.5 or 2.9 nM) as indicated at 37°C for 15 min. DNA species in these reactions and *G77telo/C77telo and *G77scr/C77scr markers (M) were separated by native PAGE (12%, 100:1), with relevant DNA species depicted at left. (B) Bar graph of data from at least three independent experiments, performed as in A. Strand exchange (%) was calculated as described in Figure 3D, with means and standard deviations plotted for telomeric (black) compared to scrambled (gray) substrate pairs at individual TRF2 concentrations. Significant differences in strand exchange between bracketed telomeric and scrambled substrate pairs are indicated with asterisks; *P <0.005 and **P <0.0005. (C) TRF2 DNA binding specificity was evaluated by EMSA (performed at 25°C including dyes) after incubating TRF2 (2.9 nM) at pH 7.0 at 37°C for 15 min with (left panel) *G77telo/C77telo or *G77scr/C77scr (0.1 nM each) or (right panel) *G77telo/C50telo or *G77scr/C50scr (0.1 nM each) minus or plus pUC19 (4.7, 9.4, 18.8 or 47 pg/μl = 1-, 2-, 4- or 10-fold molar excess over labeled substrate, respectively). Positions of unbound and TRF2-bound (well) DNA species are depicted at left.
The ability of TRF2 to bind duplex telomeric DNA apparently is involved in its stimulation of the strand exchange activity of WRN on these substrates. Since TRF1 also binds telomeric duplex DNA, we next examined the possibility that it may act in a similar manner. Initially, strand exchange reactions were performed in the presence or absence of WRN with our telomeric substrate pair (*G77telo/C50telo and G77telo/C77telo) with equimolar amounts of TRF1 or TRF2, at a TRF2 concentration that produced efficient stimulation of WRN-mediated strand exchange. Neither TRF2 nor TRF1 alone catalyzed strand exchange (Figure 5A, lanes 4 and 6). As before, TRF2 clearly stimulated WRN-mediated strand exchange compared to reactions containing WRN alone (Figure 5A, lanes 3 and 5). In contrast, the effect of TRF1 on WRN-mediated strand exchange was much less pronounced than with TRF2 (Figure 5A, lanes 3, 5 and 7). To examine this more closely, strand exchange assays were performed in the presence of WRN and over a range of concentrations of TRF1 or TRF2 (Figure 5B). As before, using limiting amounts of WRN resulted in only minimal (5.8%) strand exchange (Figure 5B, lane 3). Although TRF1 could modestly enhance WRN-mediated strand exchange, TRF2 was much better at stimulating this strand exchange reaction at each respective concentration (Figure 5B and C). Notably, the amount of exchange observed at 6 nM TRF1 was only 17.4%, while approximately twice the level of strand exchange (35.1%) was achieved with half the concentration (3 nM) of TRF2 (Figure 5B, lanes 7 and 13, and C). These observed differences were not due to lower TRF1 binding, since TRF1 and TRF2 bound *G77telo/C77telo with similar efficiencies in EMSA assays (Figure 5D). Therefore, these results indicate that TRF2 preferentially stimulates WRN-mediated strand exchange activity specifically on telomeric substrates. Although the specificity of TRF2 for telomeric duplex sequences is necessary for this effect, it is not alone sufficient for optimal stimulation, as TRF1 has similar DNA binding specificity but its effect on WRN-mediated strand exchange is much weaker. Collectively, these experiments indicate TRF2 specifically stimulates WRN-mediated strand exchange between a partial duplex substrate with a G-rich 3′ overhang and a fully duplex substrate, both containing duplex telomeric sequences sufficient for TRF2 binding. Importantly, this scenario reflects recombination processes that might occur between the normal 3′ telomeric overhang (or a resected truncated telomere) and a duplex telomeric region of another or possibly the same duplex DNA molecule.
Figure 5.
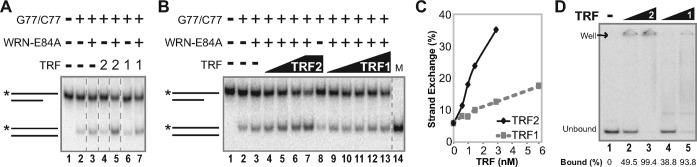
Comparison of effects of TRF1 and TRF2 on WRN-mediated strand exchange. (A) To examine the effects of TRF1 or TRF2 on WRN-mediated strand exchange, *G77telo/C50telo (0.1 nM) and G77telo/C77telo (0.4 nM) were incubated (pH 7.0) with WRN-E84A (3.8 nM), TRF1 (2.9 nM) and/or TRF2 (2.9 nM) as specified at 37°C for 15 min. Products were analyzed as in Figure 4A. (B) Experiments were performed as in A, except varying TRF1 (0.6, 1.0, 1.5, 2.9 or 5.8 nM) or TRF2 (0.6, 1.0, 1.5 or 2.9 nM) concentrations; *G77telo/C77telo marker (M) was included on the gel. (C) Strand exchange from B was calculated as in Figure 3D and plotted versus TRF2 (black diamonds) and TRF1 (gray squares) concentration. (D) Binding of TRF1 or TRF2 (0, 0.7 or 2.9 nM) to *G77telo/C77telo (0.1 nM) was performed at pH 7.0 at 37°C for 15 min and analyzed by EMSA (performed at 25°C with dyes). Unbound and bound (in well) TRF species are indicated. DNA binding (% depicted below each lane) was determined as described in Materials and Methods, based on the percentage of unbound substrate remaining.
TRF2's basic domain is required to stimulate WRN-mediated strand exchange
Subsequently, we explored the dependence of strand exchange upon the direct interaction between WRN and TRF2. In this regard, the basic (B) domain near the N-terminus of TRF2 has been shown to mediate its interaction with WRN (41). Therefore, we examined the ability of a mutant TRF2 protein lacking this domain (TRF2ΔB) to stimulate WRN-catalyzed strand exchange, in comparison to wild-type TRF2. In strand exchange reactions between duplex telomeric DNA substrates *G77telo/C50telo and G77telo/C77telo, addition of TRF2ΔB along with WRN had little or no effect on the level of strand exchange product over that observed with WRN alone, in contrast to the marked stimulatory effect of wild-type TRF2 (Figure 6A and B). Similarly, TRF2ΔB could not detectably stimulate WRN-catalyzed strand exchange between double- and single-stranded telomeric substrates (Supplementary Figure S2). This was not caused by loss of telomeric DNA binding by TRF2ΔB, as it bound to G77telo/C77telo and G77telo/C50telo telomeric duplex substrates with a similar affinity as wild-type TRF2 (Figure 6C and D). These binding results are consistent with the telomere-specific DNA binding function of the Myb domain present in both TRF2 and TRF2ΔB, and earlier comparisons of these proteins on telomeric duplex substrates (24,29,53). Our findings indicate that the basic domain of TRF2 is necessary for optimal stimulation of WRN-mediated strand exchange between telomeric substrates, and strongly suggest that the interaction between WRN and TRF2 mediated by this domain plays a crucial role in this process.
Figure 6.
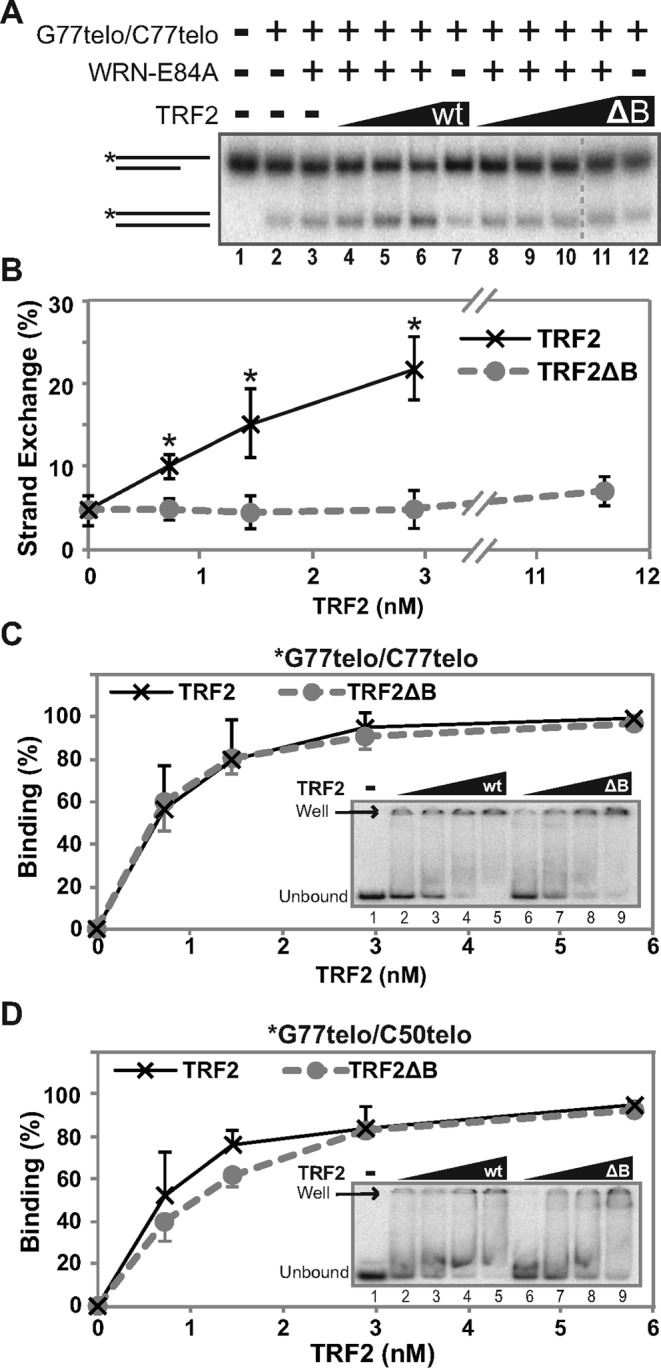
TRF2ΔB does not stimulate WRN-mediated strand exchange. (A) To determine the involvement of TRF2's basic domain in stimulating WRN-mediated strand exchange, *G77telo/C50telo (0.1 nM) and G77telo/C77telo (0.4 nM) were incubated (pH 7.0) with WRN-E84A (3.6 nM) minus or plus wild-type (wt) TRF2 (0.6, 1.5 or 2.9 nM) or TRF2ΔB (ΔB) (0.6, 1.5, 2.9 or 11.6 nM) as indicated at 37°C for 15 min and DNA products analyzed as in Figure 4A. (B) Line graph of data from at least three independent experiments, performed as in A. Strand exchange (%) was calculated as described in Figure 3D, and plotted versus wild-type TRF2 (black) and TRF2ΔB (gray) concentration. At each TRF2 concentration, significant differences (all P values <0.02, indicated by asterisks) between wild-type TRF2 and TRF2ΔB as well as between wild-type TRF2 and WRN only control were calculated using unpaired, two-tailed t-tests. (C–D) Binding of TRF2 or TRF2ΔB (0, 0.6, 1.5, 2.9 or 5.8 nM) to (C) *G77telo/C77telo (0.1 nM) or (D) *G77telo/G50telo (0.1 nM) was performed at pH 7.0 at 37°C for 15 min, followed by EMSA performed at 4°C without dyes (insets). DNA binding (%; means and standard deviations) by wild-type TRF2 (black) and TRF2ΔB (gray) from at least three independent experiments was calculated as described in Materials and Methods based on the percentage of unbound substrate remaining and plotted against TRF2/TRF2ΔB concentration.
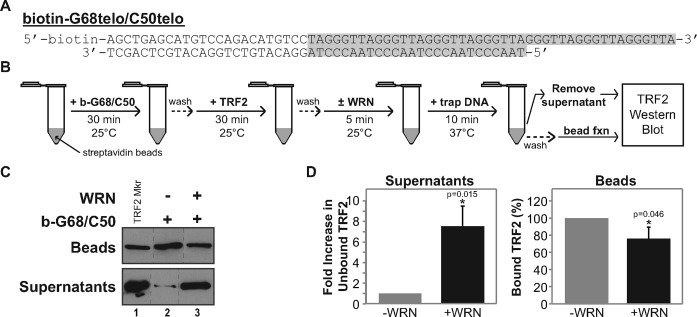
WRN displaces TRF2 bound to telomeric structures
TRF2's ability to enhance WRN's helicase and strand exchange activities on telomeric DNA substrates also suggests that, during these processes, WRN may displace TRF2 bound to telomeric duplex DNA. To assess this possibility, we developed an assay (Figure 7B) to measure TRF2 displacement, utilizing a biotin-tagged telomeric substrate, b-G68telo/C50telo, that can be immobilized on streptavidin beads. This substrate contains a duplex region (24 bp of random sequence and 26 bp of telomeric repeat sequence) identical to the 3′ overhang substrate (G77telo/C50telo) used above, but possesses a slightly shorter (18 nt) G-rich 3′ overhang (Figure 7A). Thus, this substrate should bind TRF2 in a manner essentially the same as observed with G77telo/C50telo (Figure 4C). Following immobilization of b-G68telo/C50telo on streptavidin beads and washing away any unbound DNA substrate, TRF2 was allowed to bind to the immobilized telomeric substrate. After washing away any unbound TRF2, WRN was added to this reaction followed by addition of a short 24-bp telomeric duplex to reduce potential re-association of displaced TRF2 with the immobilized substrate. At this point, displaced TRF2 (in supernatants) and bound TRF2 (on beads) were detected by Western blotting. In the absence of WRN, TRF2 was almost exclusively associated with the bead fraction (Figure 7C, lane 2), which we attribute to stable TRF2 binding to the telomeric duplex region of the immobilized substrate. Importantly, addition of WRN resulted in substantial levels of TRF2 in the supernatant (Figure 7C, lane 3). When TRF2 levels were quantified from multiple experiments (Figure 7D), after WRN was added significantly lower amounts of TRF2 were reproducibly found in the bead fraction while TRF2 present in the supernatant fraction was markedly increased (by ∼7-fold). Importantly, these reactions were performed without ATP thus eliminating WRN helicase activity; therefore, TRF2 displacement could not have been caused by unwinding of the immobilized substrate. These results suggest that WRN can directly displace TRF2 from telomeric duplex DNA in a manner independent of its unwinding activity, probably through direct WRN–TRF2 interactions observed previously (17,38). Taken together, our results suggest that direct interactions between TRF2 and WRN and between TRF2 and telomeric DNA mediate coordinated and preferential strand exchange between telomeric sequences.
Figure 7.
WRN displaces TRF2 from telomeric DNA. (A) Sequence and structure of b-G68/C50telo substrate. Telomeric sequence is shaded (gray); the position of biotin is noted (elsewhere abbreviated as ‘b’). (B) Flow diagram of the TRF2 displacement assay. (C) TRF2 displacement assays were performed as in B with specific details described in Materials and Methods. TRF2 associated with streptavidin beads or in supernatants was detected using Western blotting showing two separate blots from a single experiment, along with purified TRF2 markers (lane 1). (D) Bar graph of bead-bound and displaced TRF2 from the final step of three independent experiments as in C. Bound TRF2 (beads, right panel) was calculated as a percentage of the minus WRN control. Fold change of released TRF2 (supernatant, left panel) was calculated with respect to the minus WRN control, which was normalized to 1. Statistical comparisons were performed between minus and plus WRN samples with P-values (*) calculated using paired, one-tailed t-tests.
DISCUSSION
A possible role for WRN in telomere metabolism was initially promoted by the finding that the premature senescence of WS cells can be rescued by telomerase expression (14). A telomeric role for WRN was more strongly supported by phenotypes observed in mice lacking WRN and telomerase (15,16) and by telomeric abnormalities in cells having altered WRN status (19,39–42,47). Additionally, shelterin component and telomeric DNA binding factor TRF2 was shown to interact with WRN and influence its enzymatic activities (17,18,37,38), although these studies did not provide much clarity as to the specific function of WRN at telomeres. Notably, WRN may play a role in promoting proper recombination between telomeric regions, as some studies have reported that WRN deficiency or dysfunction is associated with aberrant telomeric recombination (39,40). In the present study, we examined the molecular coordination between WRN and TRF2 in more detail with emphasis on possible cooperation in telomeric recombination processes. Since we had previously demonstrated that WRN catalyzes strand exchange (7), we were particularly interested in TRF2's effect on WRN-mediated recombinational actions on DNA substrates that possess telomeric characteristics. Our findings show that TRF2 stimulates WRN's helicase and strand exchange activities preferentially on telomeric DNA, and that WRN displaces TRF2 from telomeric DNA during this process. This coordination between TRF2 and WRN during strand exchange of telomeric sequences may also play a key role in telomeric recombination processes and/or T-loop dynamics at telomeres.
Initially, we examined TRF2's effect on WRN's helicase activity, using 3′ overhang substrates with single- and double-stranded telomeric (GGGTTA) or scrambled (GAGTGT) repeats (Figure 1A); the former possessed 14 bp of duplex telomeric sequence (specifically TAGGGTTAGGGTTA), sufficient for TRF2 binding (51,54). Our EMSA studies on these substrates consistently showed preferential binding to the telomeric overhang substrate, although binding to the scrambled substrate can be observed particularly under lower stringency conditions (Figure 1B and C; Supplementary Figure S1A–D). Our results demonstrated that TRF2 stimulated the helicase activity of WRN on both telomeric and scrambled overhang substrates but to a greater extent on the telomeric overhang to which TRF2 binds preferentially (Figure 1B–E). Our data is highly consistent with a mechanism by which the DNA binding properties of TRF2 and its association with WRN are important in directing WRN helicase activity to both substrates, but with modest preference toward the telomeric overhang substrate to which it has higher affinity. This interpretation differs somewhat from a previous report showing TRF2 stimulation of WRN's helicase activity on DNA substrates without and with two telomeric (TTAGGG) repeats, neither of which could stably bind TRF2 under their EMSA conditions (17). Opresko et al. interpreted their results as the direct interaction between TRF2 and WRN specifically altering the latter's helicase activity. Although we certainly agree that TRF2 and WRN directly interact and do not exclude the possibility that this interaction can somewhat enhance WRN helicase function, our results indicate that TRF2's DNA binding affinity helps recruit WRN to enhance its helicase activity on TRF2-bound substrate. Importantly, the specificity of TRF2 for telomeric DNA increases as the amount of telomeric duplex sequence increases, as evidenced by DNA binding studies performed with substrates used in strand exchange assays (Figure 4C); this highly preferential binding by TRF2 results in its strikingly specific stimulation of WRN-mediated strand exchange between telomeric substrates compared to substrates containing scrambled sequences (Figure 4A and B). This is also consistent with our earlier observation of the stimulatory effect of TRF2 on WRN exonuclease activity on telomeric substrates (38). Our interpretation would seem to be more physiologically relevant, as TRF2 clearly binds telomeric DNA in vivo and this recruitment mechanism would best explain WRN's observed localization at telomeres that is affected by TRF2 status (18,19,41) and its apparent function in telomere maintenance. It might be expected that helicase function would be hindered by another protein bound to the DNA duplex region, as we observed with the pronounced inhibitory effect on UvrD-mediated helicase activity at TRF2 concentrations that stimulated WRN-catalyzed unwinding (Figure 1F and G). This striking contrast between TRF2's effects on WRN versus UvrD strongly supports a coordinated, functional interaction between TRF2 and WRN relevant to telomere maintenance. Such a coordinated function is further supported by WRN's ability, even in the absence of its ATPase and helicase activities, to directly displace TRF2 from telomeric DNA (Figure 7C and D). Theoretically, the ability of WRN, after its recruitment to telomeric DNA, to displace TRF2 (and potentially larger shelterin complexes) would be crucial in remodeling DNA structures during telomeric metabolic processes. Notably, this type of mechanism has been observed in other scenarios, including the relationship between RPA and RAD51 during the filament formation step of recombinational repair (55,56).
Because of WRN's ability to perform strand exchange (7) and the aberrant telomeric recombination phenotypes of WRN-deficient cells (39–41), it is likely that WRN might somehow be involved in telomeric recombination. Therefore, we examined whether TRF2 could also stimulate WRN's strand exchange activity as well as the specificity of this reaction for telomeric DNA. Using two independent reaction scenarios, WRN-mediated strand exchange was preferentially enhanced by TRF2 in situations where the paired homologous DNA substrates contained telomeric sequence. In one scenario, TRF2 stimulated WRN-mediated strand exchange between a predominantly duplex substrate containing 47 bp of telomeric duplex and 6 nt of single-stranded G-rich telomeric DNA and a homologous single-stranded substrate; this effect was abolished when a non-homologous, single-stranded substrate with scrambled telomeric sequence was employed (Figure 2). A second, more realistic scenario utilized paired homologous duplexes, with both test substrates containing sufficient telomeric duplex sequence to bind TRF2 (Figure 3A). While one substrate was fully duplex and contained 53 bp of telomeric repeat sequence, the other contained both a telomeric duplex region (26 bp) and a 3′ overhang with only G-rich telomeric repeat sequence, thus resembling the recombinational structure present at physiological telomeric ends or formed during resection of the ‘centromere-proximal end’ of telomeric double-strand breaks. In these reactions, WRN-mediated strand exchange was markedly stimulated (by as much as nearly 6-fold) by TRF2 in a manner dependent upon TRF2 concentration and ATP (Figures 3–6). In contrast, TRF2 had little or no stimulatory effect on WRN-mediated strand exchange in parallel control reactions with structurally identical substrates but with scrambled repeat sequences replacing telomeric regions (Figure 4A and B). Compared to TRF2, TRF1 showed only a very modest enhancement of WRN-mediated activity in strand exchange reactions with paired telomeric substrates, even though TRF1 bound these substrates similarly as TRF2 (Figure 5). These results convincingly demonstrate that TRF2 markedly and specifically stimulates WRN-mediated strand exchange between substrates containing telomeric sequences. It is noteworthy that, in both types of strand exchange reactions here, there were no exposed, complementary single-stranded regions on the paired substrates to initiate spontaneous or WRN-mediated annealing, in contrast to the original observation of WRN-mediated strand exchange (7). Instead, it would appear that coordinated unwinding and strand invasion by WRN is required for these particular reactions and that WRN-mediated, telomeric strand invasion is substantially enhanced by TRF2, which alone does not perform strand exchange (Figures 3–5). TRF2's stimulatory effect on WRN's helicase and strand exchange activities is probably mediated through its ability to directly bind both WRN and telomeric DNA (17,23,24,38). We examined the involvement of the direct TRF2–WRN interaction in this stimulatory effect using a TRF2ΔB mutant lacking the basic domain previously shown to mediate its interaction with WRN (41). While TRF2ΔB maintains its ability to bind telomeric DNA substrates with affinity similar to wild-type TRF2 (Figure 6C and D), it cannot stimulate WRN-mediated strand exchange (Figure 6A and B), suggesting that the interaction between WRN and TRF2 is crucial for this process. However, because this basic domain has been shown to also influence DNA structure-related binding by TRF2 (26), we cannot rule out the possibility that this property may also contribute to the stimulatory effect of TRF2 on WRN-mediated strand exchange between telomeric substrates. Yet another possible contributing factor is suggested by TRF2's pronounced stimulation on WRN-mediated strand exchange between two substrates containing telomeric duplex regions. Since TRF2 forms homodimers with two DNA binding sites (24), we speculate that telomeric binding sites on two substrate molecules might be bridged and brought into close proximity by TRF2 homodimers, facilitating more efficient strand exchange by WRN. Such a function might also be consistent with TRF2's proposed role in T-loop formation and stability (25), in which it might fold back and align telomeric ends with internal telomeric duplex regions. Regardless of the precise mechanism, our results demonstrate a specific functional relationship between WRN and TRF2 and suggest their coordinated action in telomeric recombination processes. Consistent with these results, WRN can be found localized at telomeres and co-immunoprecipitates with TRF2 in vivo (17–19,41).
In considering specific roles for WRN in conjunction with TRF2 during telomere maintenance, it is certainly crucial that telomeres, because of their repeating sequence composition and presence on the ends of all chromosomes, would seem to be much more susceptible to recombination than the rest of the genome (57). Furthermore, 3′ overhang structures present on normal telomeric ends are recombinogenic structures themselves, although they are normally bound by telomeric factors that suppress illegitimate recombination events. Specifically, evidence indicates that these 3′ overhangs may be folded back and inserted into internal telomeric duplex regions on the same chromosome (34), creating T-loop structures that prevent telomeric ends from activating checkpoint pathways triggering apoptosis or cellular senescence. Importantly, TRF2 plays an essential role in telomere protection and may itself facilitate T-loop formation (25,29,30,36). Telomere deprotection, by critical shortening or loss of TRF2 function, can lead to telomere fusions and uncontrolled homologous recombination, both of which may shorten telomeres stochastically (57). It is noteworthy that alteration or loss of WRN function (as in WS) results in telomere-related phenotypes, including stochastic telomere loss, elevated sister telomere exchange, enhanced rescue of telomere-related crisis via telomeric recombination and premature cellular senescence or apoptosis in primary cells (19,39,42,47). Our findings suggest that WRN and TRF2 function coordinately to promote strand exchange between telomeric sequences. Such an activity might be important in several pathways to maintain telomere stability. First, WRN and TRF2 may act together during T-loop formation or resolution. Invasion of telomeric overhangs into telomeric duplex regions to form T-loops are strand exchange events that are strikingly similar to our reactions catalyzed by WRN that are markedly stimulated by TRF2. Defects in T-loop formation would cause problems with telomere end protection and promote checkpoint activation and senescence/apoptosis or uncontrolled telomeric recombination, consistent with some phenotypes associated with WRN deficiency. Alternatively, TRF2 and WRN may cooperate during replication to catalyze T-loop resolution, which is essentially another strand exchange event; compromised resolution of T-loop structures might result in stochastic telomere loss associated with WRN deficiency. This would be consistent with proposed role of WRN and other RecQ helicases in responding to replication blocking events (58–63). Analogously, WRN might use its (TRF2-stimulated) strand exchange activity to resolve telomeric recombination intermediates in such a way to suppress crossing over, thereby suppressing sister telomeric exchange. An analogous, genome-wide function for its RecQ homolog BLM has been proposed (64).
One intriguing possibility is that, in coordination with TRF2, WRN may act within telomeric regions to facilitate break-induced replication (BIR), the pathway now believed to mediate restarting of collapsed replication forks (65,66). Importantly, the BIR mechanism involves invasion of the detached arm of the replication fork into the homologous duplex, quite similar to the process catalyzed by WRN in our experiments (Figures 3–5). If the replication fork collapses in telomeric regions, WRN may work together with TRF2 bound to telomeric DNA to promote telomeric BIR, by a recombinational process similar to ALT. In this case, a deficiency in WRN and loss of telomeric BIR might result in truncation of a telomere at the point of collapse, due to the lack of distal origins of replication in telomeres. This would be consistent with stochastic telomere loss and downstream phenotypes (such as accelerated cellular senescence) observed in WRN-deficient cells (14–16,19,47,67). Because of possible enhanced damage formation in the G-rich strand (due to the acute reactivity of guanine), the lagging arm may be much more prone to collapse, possibly also explaining the observation that stochastic telomere loss in WRN-deficient cells is a replication-mediated event that preferentially occurs on the sister chromatid derived from lagging strand replication (19). Moreover and also consistent with WRN deficiency, loss of its function in telomeric BIR may result in more aberrant telomeric recombination events, perhaps initiated as a backup mechanism to restore telomere length and stability and rescue cell survival. Notably, such a role for WRN in humans would also be in agreement with the proposed role for its budding yeast homolog Sgs1 in one pathway for telomeric BIR or telomeric recombination allowing type II survivors (65,68). Thus, a putative coordinated function for WRN and TRF2 specifically in telomeric BIR seems an attractive possibility, although additional investigation is needed to support such a role.
In summary, our results have demonstrated that WRN and TRF2 function coordinately to achieve strand exchange of telomeric sequences. These findings substantially increase our understanding of the WRN–TRF2 functional interaction and, more importantly, point to possible specific roles in telomeric recombination processes that are in agreement with telomere-related abnormalities observed when WRN function is lost.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors would like to thank Titia de Lange for providing baculovirus constructs for overproduction and purification of TRF1 and TRF2, and Jack Griffith and Brian Bower for providing purified TRF2ΔB.
FUNDING
National Institutes of Health [R01AG027258 to D.K.O., D.N.E. received support from T32ES007266].
Conflict of interest statement. None declared.
REFERENCES
- 1.Epstein C.J., Martin G.M., Schultz A.L., Motulsky A.G. Werner's syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine. 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Goto M., Miller R.W., Ishikawa Y., Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol. Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 3.Goto M. Hierarchical deterioration of body systems in Werner's syndrome: implications for normal ageing. Mech. Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- 4.Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 5.Matsumoto T., Shimamoto A., Goto M., Furuichi Y. Impaired nuclear localization of defective DNA helicases in Werner's syndrome. Nat. Genet. 1997;16:335–336. doi: 10.1038/ng0897-335. [DOI] [PubMed] [Google Scholar]
- 6.Gray M.D., Shen J.C., Kamath-Loeb A.S., Blank A., Sopher B.L., Martin G.M., Oshima J., Loeb L.A. The Werner syndrome protein is a DNA helicase. Nat. Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 7.Machwe A., Xiao L., Groden J., Matson S.W., Orren D.K. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 8.Shen J.C., Gray M.D., Oshima J., Kamath-Loeb A.S., Fry M., Loeb L.A. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J. Biol. Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 9.Huang S., Li B., Gray M.D., Oshima J., Mian I.S., Campisi J. The premature ageing syndrome protein, WRN, is a 3′–>5′ exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 11.Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 12.Salk D., Au K., Hoehn H., Martin G.M. Cytogenetics of Werner's syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet. Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- 13.Fukuchi K., Martin G.M., Monnat R.J., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc. Natl Acad. Sci. U.S.A. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyllie F.S., Jones C.J., Skinner J.W., Haughton M.F., Wallis C., Wynford-Thomas D., Faragher R.G., Kipling D. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat. Genet. 2000;24:16–17. doi: 10.1038/71630. [DOI] [PubMed] [Google Scholar]
- 15.Chang S., Multani A.S., Cabrera N.G., Naylor M.L., Laud P., Lombard D., Pathak S., Guarente L., DePinho R.A. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 16.Du X., Shen J., Kugan N., Furth E.E., Lombard D.B., Cheung C., Pak S., Luo G., Pignolo R.J., DePinho R.A., et al. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol. Cell. Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 18.Opresko P.L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kolvraa S., May A., Seidman M.M., Bohr V.A. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 19.Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 20.Jog S.P., Reddy S., Comai L. Cell cycle-regulated association between the Werner syndrome protein and its molecular partners. Cell Cycle. 2011;10:2038–2040. doi: 10.4161/cc.10.12.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palm W., de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 22.Zhong Z., Shiue L., Kaplan S., de Lange T. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell. Biol. 1992;12:4834–4843. doi: 10.1128/mcb.12.11.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bilaud T., Brun C., Ancelin K., Koering C.E., Laroche T., Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 24.Broccoli D., Smogorzewska A., Chong L., de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 25.Stansel R.M., de Lange T., Griffith J.D. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fouche N., Cesare A.J., Willcox S., Ozgur S., Compton S.A., Griffith J.D. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J. Biol. Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- 27.Poulet A., Buisson R., Faivre-Moskalenko C., Koelblen M., Amiard S., Montel F., Cuesta-Lopez S., Bornet O., Guerlesquin F., Godet T., et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Steensel B., de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 29.van Steensel B., Smogorzewska A., de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 30.Karlseder J., Broccoli D., Dai Y., Hardy S., de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 31.Takai H., Smogorzewska A., de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 32.Dimitrova N., de Lange T. Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol. Cell. Biol. 2009;29:5552–5563. doi: 10.1128/MCB.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart J.A., Chaiken M.F., Wang F., Price C.M. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 2012;730:12–19. doi: 10.1016/j.mrfmmm.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 35.Chin L., Artandi S.E., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 36.Denchi E.L., de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 37.Nora G.J., Buncher N.A., Opresko P.L. Telomeric protein TRF2 protects Holliday junctions with telomeric arms from displacement by the Werner syndrome helicase. Nucleic Acids Res. 2010;38:3984–3998. doi: 10.1093/nar/gkq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Machwe A., Xiao L., Orren D.K. TRF2 recruits the Werner syndrome (WRN) exonuclease for processing of telomeric DNA. Oncogene. 2004;23:149–156. doi: 10.1038/sj.onc.1206906. [DOI] [PubMed] [Google Scholar]
- 39.Laud P.R., Multani A.S., Bailey S.M., Wu L., Ma J., Kingsley C., Lebel M., Pathak S., DePinho R.A., Chang S. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagelstrom R.T., Blagoev K.B., Niedernhofer L.J., Goodwin E.H., Bailey S.M. Hyper telomere recombination accelerates replicative senescence and may promote premature aging. Proc. Natl Acad. Sci. U.S.A. 2010;107:15768–15773. doi: 10.1073/pnas.1006338107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B., Jog S.P., Reddy S., Comai L. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2DeltaB-mediated telomere shortening. Mol. Cell. Biol. 2008;28:1892–1904. doi: 10.1128/MCB.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crabbe L., Jauch A., Naeger C.M., Holtgreve-Grez H., Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc. Natl Acad. Sci. U.S.A. 2007;104:2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orren D.K., Brosh R.M., Jr, Nehlin J.O., Machwe A., Gray M.D., Bohr V.A. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machwe A., Lozada E.M., Xiao L., Orren D.K. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol. Biol. 2006;7:1. doi: 10.1186/1471-2199-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machwe A., Xiao L., Theodore S., Orren D.K. DNase I footprinting and enhanced exonuclease function of the bipartite Werner syndrome protein (WRN) bound to partially melted duplex DNA. J. Biol. Chem. 2002;277:4492–4504. doi: 10.1074/jbc.M108880200. [DOI] [PubMed] [Google Scholar]
- 46.Bianchi A., Smith S., Chong L., Elias P., de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai Y., Murnane J.P. Telomere instability in a human tumor cell line expressing a dominant-negative WRN protein. Human Genet. 2003;113:337–347. doi: 10.1007/s00439-003-0972-y. [DOI] [PubMed] [Google Scholar]
- 48.Shen J.C., Gray M.D., Oshima J., Loeb L.A. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998;26:2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohaghegh P., Karow J.K., Brosh R.M., Jr, Bohr V.A., Hickson I.D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brosh R.M., Jr, Waheed J., Sommers J.A. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- 51.Bianchi A., Stansel R.M., Fairall L., Griffith J.D., Rhodes D., de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Kobbe C., Thoma N.H., Czyzewski B.K., Pavletich N.P., Bohr V.A. Werner syndrome protein contains three structure-specific DNA binding domains. J. Biol. Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 53.Wang R.C., Smogorzewska A., de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 54.Fairall L., Chapman L., Moss H., de Lange T., Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- 55.Sugiyama T., Kowalczykowski S.C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 56.Wang X., Haber J.E. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004;2:E21. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Opresko P.L. Telomere ResQue and preservation–roles for the Werner syndrome protein and other RecQ helicases. Mech. Ageing Dev. 2008;129:79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 58.Machwe A., Xiao L., Groden J., Orren D.K. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 59.Machwe A., Xiao L., Lloyd R.G., Bolt E., Orren D.K. Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lebel M., Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc. Natl Acad. Sci. U.S.A. 1998;95:13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pichierri P., Franchitto A., Mosesso P., Palitti F. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poot M., Yom J.S., Whang S.H., Kato J.T., Gollahon K.A., Rabinovitch P.S. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–1226. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- 63.Pirzio L.M., Pichierri P., Bignami M., Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu L., Hickson I.D. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 65.McEachern M.J., Haber J.E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 66.Llorente B., Smith C.E., Symington L.S. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- 67.Martin G.M., Sprague C.A., Epstein C.J. Replicative life-span of cultivated human cells. Effects of donor's age, tissue, and genotype. Lab. Invest. 1970;23:86–92. [PubMed] [Google Scholar]
- 68.Johnson F.B., Marciniak R.A., McVey M., Stewart S.A., Hahn W.C., Guarente L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20:905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.