Abstract
Amongst enzymes which relieve torsional strain and maintain chromosome supercoiling, type IA topoisomerases share a strand-passage mechanism that involves transient nicking and re-joining of a single deoxyribonucleic acid (DNA) strand. In contrast to many bacterial species that possess two type IA topoisomerases (TopA and TopB), Actinobacteria possess only TopA, and unlike its homologues this topoisomerase has a unique C-terminal domain that lacks the Zn-finger motifs characteristic of type IA enzymes. To better understand how this unique C-terminal domain affects the enzyme's activity, we have examined DNA relaxation by actinobacterial TopA from Streptomyces coelicolor (ScTopA) using real-time single-molecule experiments. These studies reveal extremely high processivity of ScTopA not described previously for any other topoisomerase of type I. Moreover, we also demonstrate that enzyme processivity varies in a torque-dependent manner. Based on the analysis of the C-terminally truncated ScTopA mutants, we propose that high processivity of the enzyme is associated with the presence of a stretch of positively charged amino acids in its C-terminal region.
INTRODUCTION
In prokaryotes, and similarly in eukaryotes, deoxyribonucleic acid (DNA) needs to be highly compacted to fit the space limited by the cell size (1–4). Although the chromosome is highly condensed within a cell, it also has to be accessible for dynamic processes such as replication, transcription, recombination and segregation, the proper execution of which is dependent on spatial DNA structure. Local chromosome organization is exerted by nucleoid-associated proteins and histones in prokaryotes and eukaryotes, respectively, whilst global compaction is induced by condensins (SMC) (5–7). However, in most cases negative supercoiling is the first step of DNA compaction which within the cell is maintained by enzymes called topoisomerases.
Topoisomerases act by removing or introducing supercoils into the DNA molecule. The two evolutionarily and functionally distinct types—type I and type II (8,9)—exhibit different reaction mechanisms. Whilst type II topoisomerases (e.g. gyrase) catalyze cleavage and religation of both DNA strands, type I topoisomerases cut a single strand. Formation of an intermediate covalently linked to DNA allows for topological manipulation of the DNA prior to religation. In the case of type I topoisomerases, transfer or rotation of the intact strand occurs due to topological tension, and so the relaxation they perform does not require adenosine triphosphate (bacterial reverse gyrase is a notable exception). Type I topoisomerases are further divided into subtypes IA, IB and IC, according to the origin, the differences in amino acid sequence and/or the relaxation mechanisms. Whilst topoisomerases of type IB are present in eukaryotes, poxviruses and a few bacteria species [their presence in bacteria may be the effect of horizontal gene transfer (10)], topoisomerases of type IA and IC are characteristic of prokaryotes and archaea. Topoisomerases IB and IC form linkage with the 3′-end of DNA and remove several positive or negative supercoils (Lk, linking number; the number by which one DNA helix contacts with the second one and which cannot be altered without strand breakage) per cleavage/religation cycle using a constrained swiveling mechanism, whilst topoisomerases IA (TopA and TopB) form a 5′-phosphotyrosine intermediate and can relax only one negative supercoil per cleavage/religation event using an enzyme-bridged strand passage mechanism (11–13).
The similarity between TopA and TopB proteins manifests itself mostly in the conserved N-terminal region, containing a TOPRIM (topoisomerase-primase) domain and the catalytic tyrosine residue required for the trans-esterification reaction (14,15). Although TopA and TopB share the same overall mechanism of supercoil removal, they differ in details at both the structural and mechanistic levels. TopA homologues contain a long C-terminal domain with zinc-finger motifs that are absolutely required for DNA binding and relaxation, whilst TopB homologues possess a short C-terminal domain lacking those structures. The difference in the structure of TopA and TopB reflects their cellular function and the mechanism by which they relax supercoiled DNA. Whilst Escherichia coli TopA removes negative supercoils efficiently and plays a major role in maintaining the appropriate superhelical density of the chromosome, TopB relaxes negatively supercoiled DNA more slowly and pauses between each relaxation burst (16). However, TopB is more efficient in decatenating single-stranded DNA rings due to the presence of a structural motif named the decatenation loop (17,18). Thus, in vivo TopB may be involved in several types of DNA transactions, but its cellular role remains still unclear.
Although many bacterial species, in particular rapidly growing ones, possess genes encoding both type IA topoisomerases, the linear chromosome of mycelial, multigenomic and sporulating bacteria, Streptomyces coelicolor contains only one gene encoding topoisomerase of type I (ScTopA) (19) that is essential for cell viability (20,21). In contrast to other TopA homologues, the enzyme identified in S. coelicolor (20), as in other Actinobacteria [(22,23) and Supplementary Figure S1], contains a very long C-terminal domain lacking zinc fingers but encompassing the stretch of positively charged amino acids. Thus, it exhibits poor homology to either TopA or TopB families and seemingly belongs to an evolutionarily distinct branch of enzymes (Supplementary Figure S1). The earlier studies (22,24) on topoisomerase I from Mycobacterium smegmatis (MsTopA) suggested its specificity towards recognized sequences and high processivity of DNA relaxation. The unusual features of the actinobacterial TopA proteins sequence prompted us to analyse characteristics of ScTopA-dependent relaxation using single-molecule magnetic tweezer and classical agarose gels. Both strategies gave us direct access to important biological parameters of the enzyme such as velocity and processivity, allowing us to describe in detail the relaxation process. In particular, we show that ScTopA is characterized by an extremely high processivity not reported earlier for any known topoisomerase of type I which is correlated with the presence of a stretch of positively charged amino acids in C-terminus.
MATERIALS AND METHODS
In silico analysis
All similarity analyses were performed using Basic Local Alignment Search Tool. Amino acid sequences of bacterial topoisomerases type IA were retrieved from UniProtKB Protein Knowledgebase (www.uniprot.org). Phylogenetic analysis of bacterial topoisomerases was prepared using MEGA5.1 software and MUSCLE algorithm for multiple sequence alignment. The guide tree was calculated using the neighbour-joining Method. Secondary structures of ScTopA and EcTopA, respectively, were predicted using GOR, HNN and SOPMA algorithms (www.expasy.org/tools).
Protein overexpression and purification
Topoisomerase I (ScTopA) was overproduced using the construct pETtopA and purified as described earlier (21). To overproduce the truncated versions of ScTopA protein, the construct pETtopA was modified by λRed-mediated recombination with the use of the apramycin resistant cassette amplified using oligonucleotides topA_delC_FW and topA_delC_RV (for ScTopAΔC carrying 343-aa C-terminal deletion) and topA881_FW and topA_delC_RV (for ScTopA881 carrying 71-aa C-terminal deletion) yielding pETtopAΔC and pETtopA881, respectively. Both ScTopA-truncated versions were subsequently overexpressed in E. coli BL21 strain, purified and tested for nuclease contamination; 250 ng of the purified protein was incubated 24 h in 37ºC with 200 ng of supercoiled pUC19 plasmid in 25-mM NaH2PO4 pH 8.1, 150-mM NaCl, 5% glycerol, 10-mM MgCl2 and supplemented with 1-mg/ml bovine serum albumin (BSA). After this time DNA was extracted using phenol:chlorophorm:isoamyl alcohol (25:24:1). DNA was analysed by standard gel electrophoresis in 1% agarose in 1x Tris-borate-EDTA (TBE) buffer to confirm no nicking occurred.
Topoisomerase relaxation assay
A 20-μl mixture containing 200 ng of negatively supercoiled plasmid, 10-mM MgSO4, 1-mg/ml BSA, 50-mM NaH2PO4 pH 8.1, 150-mM NaCl, 5% glycerol and increasing concentration of ScTopA protein (0–500 nM) was incubated in 37ºC for the time indicated. To compare the activity of ScTopA and ScTopA881 proteins, we incubated 200 ng of pUC19 plasmid in the presence of 100-nM enzyme for up to 60 min. The reaction was stopped by addition of 1.5 μl of 0.5-M ethylenediaminetetraacetic acid (EDTA). DNA was subsequently extracted by vortexing with 1 volume of phenol:chlorophorm:isoamyl alcohol (25:24:1). DNA was resolved in 1% agarose in Tris-acetate-EDTA (TAE) buffer for 14–16 h at low voltage (2 V/cm). Topoisomers distributions were visualized by staining in ethidium bromide/TAE solution at room temperature for 30 min. Band intensities and relaxation profiles were analysed on Typhoon 8600 Imager using software provided by the producer. To estimate the kinetic parameters (Km and Vmax), 10 ng of ScTopA (5 nM) or 150 ng of ScTopA881 mutant truncated in positively charged C-terminus (75 nM) was incubated with increasing amount of pUC19 plasmid (100–2500 ng) carrying in each DNA molecule 8–10 supercoils that are the substrate for ScTopA. The Vmax parameters were subsequently linearly extrapolated to the same enzyme concentration allowing their comparison. The reaction was stopped at different stages (30–120 s), plasmid was resolved and visualized as described above. To estimate the initial velocity (V0), the intensity of the band corresponding to the supercoiled form was measured using ImageJ2x Software and compared with non-relaxed plasmid −R value (after time 0 s) according to the protocol described by Xu and Leng (25). V0 was calculated from linear decrease of supercoiled form; V0 = [scDNA] × (1 − R) × 1/t, where t is the reaction time. Obtained values were fitted to the Michaelis–Menten model using R Software and drc package.
Single-molecule experiments: sample preparation, magnetic trap calibration and data analysis
Topoisomerase I activity can be assayed in real time on mechanically supercoiled, extended DNA as an increase in DNA extension resulting from topological relaxation of plectonemic supercoils. Controlling the extending force on negatively supercoiled DNA makes it possible to alter the balance between plectonemic and denatured states, making the appearance of denatured states more frequent by increasing the force. The analysis of ScTopA relaxation mechanism was performed for a broad range of DNA substrates (2–51 kb) in Hepes buffer (25-mM Hepes-KOH pH 8.0, 150-mM NaCl, 10-mM MgSO4, 1-mg/ml BSA 0.1% Tween), in which the same activity of ScTopA was observed as in bulk experiment. For single-molecule experiments, DNA fragments to be nanomanipulated were cut with XbaI and SacI, gel purified and subsequently ligated with purified 1-kb DNA labels XbaI-biotin and SacI-digoxigenin, respectively. One-kilobase DNA labels were prepared by polymerase chain reaction-based incorporation of either 2'-deoxyuridine-5'triphosphate-biotin (dUTP-biotin) or dUTP-digoxigenin into 1-kb fragments bearing an appropriate restriction site according to the protocol described previously by Revyakin et al. (26). The 11-kb DNA built in this manner is based on the Charomid 9–11-kb DNA; 2-kb DNA is a fragment of the S. coelicolor chromosome (oriC proximal region). The 17-kb DNA is pFX357 (a kind gift from Dr François-Xavier Barre), and 51-bp molecules are occasional trimers of pFX357 observed in the magnetic trap to be ∼16 µm in length. These longer molecules were prepared by ligating AatII/XhoI-digested pFX357 to 1-kb DNA fragments bearing an AatII or XhoI site and labelled with multiple biotin or digoxigenin groups, respectively, as above.
Subsequently, labelled DNA fragments were attached first to streptavidin-coated magnetic beads (Dynal MyOne, Life Technologies), then to an antidigoxigenin-coated glass flow cell and placed on a home-built magnetic trap running the PicoTwist software suite. Preparation of flow cells for all micromanipulation experiments and measurement of stretching force were performed according to the procedure described in (26). The flow cell was placed on a magnetic trap instrument based on an inverted microscope. Here the magnetic beads were acted upon by a magnetic field gradient generated by a pair of rare earth magnets located above the sample. The gradient generated a vertical stretching force on the bead. The stretching force could be increased by moving the magnets closer to the sample, and rotation of the magnets caused the bead to rotate in lock-step register (Figure 1A). The DNA extension was measured in real time (30 Hz) by analysing the position of the bead above the surface using nm-resolution particle tracking algorithms.
Figure 1.
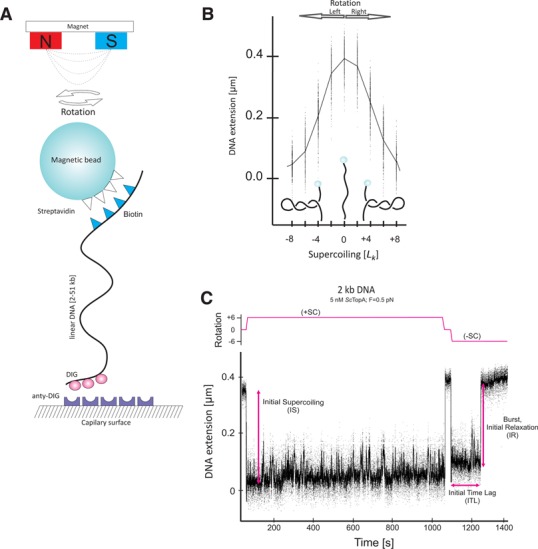
Single-molecule assay for analysis of ScTopA relaxation activity. (A) The scheme of the experiment. (B) Extension versus supercoiling curve for 2-kb dsDNA extended by a force F = 0.3 pN. (C) Time-trace of ScTopA activity acting on 2-kb dsDNA for positive (+SC) and negative (−SC) supercoiling.
To make certain whether the bead was tethered to the surface via a single, intact DNA molecule, the DNA was extended at a low force (F = 0.3 pN) and subsequently supercoils were introduced by rotating the magnet on the tweezers. In these conditions, the extension of a single supercoiled DNA molecule should decrease with the number of magnet rotations at a rate of ∼50–55 nm/turn. Nicked molecules that did not show this characteristic correlation between DNA length and the number of rotations were omitted from subsequent analysis. Similarly, beads attached to more than one DNA molecule could also be omitted from subsequent analysis based on characteristically peaked extension versus supercoiling curves (27).
The analysis of the influence of supercoiling on a DNA molecule attached to a magnetic bead has shown that DNA can be extended by low forces and negatively supercoiled by several percent without provoking extensive DNA denaturation. At low stretching force (F ∼ 0.3 pN in the salt conditions used here) negatively supercoiled DNA adopts a stable, plectonemic form. When such DNA is stretched with higher force (F ∼ 0.5 –1 pN in the salt conditions used here), the equilibrium between plectonemic and denatured states is shifted towards denatured states and locally single-stranded regions appear (28). On the other hand, for equally positively supercoiled DNA and within the force range discussed here, plectonemic supercoiling dominates throughout and denatured states are repressed. Unless stated otherwise, experiments were carried out at F = 0.3 pN.
RESULTS
ScTopA relaxes only negatively supercoiled DNA in a force- and torque-dependent manner
Comparison of ScTopA with its homologues from other bacteria revealed the unusual presence of a long C-terminal domain lacking zinc fingers but terminated with a stretch of positively charged amino acids (Supplementary Figure S1A–C), thus suggesting possible differences in the DNA binding and relaxation activity of ScTopA in comparison to, for instance, the enzyme from E. coli (EcTopA). The sequence analysis and the bulk assays of ScTopA activity, which suggested rapid supercoil removal [(21) and see also Figure 7A] prompted us to analyse the properties of ScTopA in single-molecule experiments using magnetic tweezers. In these experiments, the DNA substrates are tethered between a glass surface and a 1-μm magnetic bead and manipulated as described in the Materials and Methods section (see Figure 1A and B).
Figure 7.
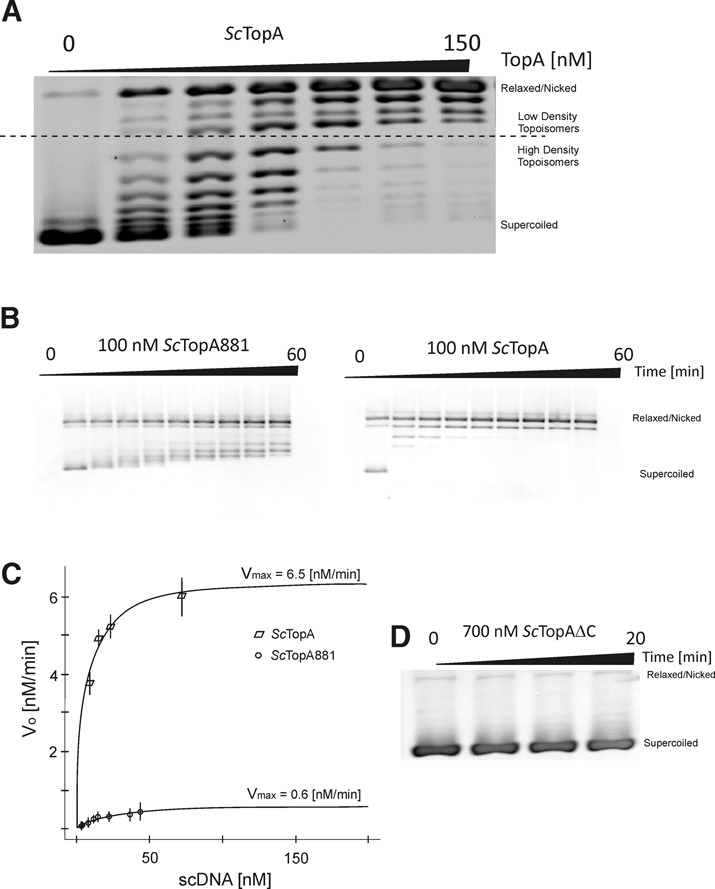
The relaxation of negatively supercoiled pUC19 plasmid by ScTopA is dependent on its C-terminal domain. (A) Agarose gel-based assay demonstrating rapid appearance of low-density topoisomers produced by ScTopA even at low protein concentration (25–50 nM; lanes 1 and 2). (B) Comparison of processive and distributive removal of supercoils during the reaction sustained by wild-type ScTopA (100 nM) and mutant ScTopA881 truncated in C-terminus by 71 amino acids (100 nM), respectively. (C) Estimation of kinetic parameters Vmax and Km by fitting to the Michaelis–Menten equation for wild-type ScTopA and mutant ScTopA881. (D) Deletion of C-terminal domain (343 aa) completely abolishes the activity of the ScTopAΔC mutant (700 nM).
We first analysed the relaxation of positively and negatively supercoiled 2-kb DNA in the presence of ScTopA (5 nM) at two stretching forces (low force F = 0.3 pN, medium force F = 0.5 pN). When positive supercoils were introduced into the DNA molecule, no DNA relaxation by ScTopA was observed either at low or at medium extending forces (Figure 1C and Supplementary Figure S2). On the other hand, when the DNA molecule was negatively supercoiled its relaxation in the presence of ScTopA could be detected (Figure 1C).
Next, we analysed the kinetics of onset of supercoil relaxation using two related parameters: the initial time lag (ITL), defined as the time between mechanical introduction of supercoils into DNA and the first relaxation event caused by ScTopA activity, and the number of molecules for which no relaxation has been observed after 800 s (the maximal recording time). Histograms of individual ITLs show that they are distributed according to single-exponential statistics (Figure 2A). At the medium force (F = 0.5 pN), the ITL was shorter than 100 s for 83% (Figure 2A) of molecules and less than 2% of molecules were not relaxed after 800 s (Figure 2A, grey bar). At the low stretching force (F = 0.3 pN), the number of relaxed DNA molecules decreased dramatically: the ITL was shorter than 100 s for only 26% of observed beads (Figure 2, inset) and almost 50% of observed molecules were not relaxed after 800 s (Figure 2A, inset, grey bar). Thus, we observed that relaxation of negatively supercoiled molecules by ScTopA occurred more frequently at a medium extending force (F = 0.5 pN) than at a low extending force (F = 0.3 pN). These experiments thus show that ScTopA, in a manner similar to EcTopA, exhibits the ability to relax supercoiled DNA in a force- and torque-dependent manner.
Figure 2.
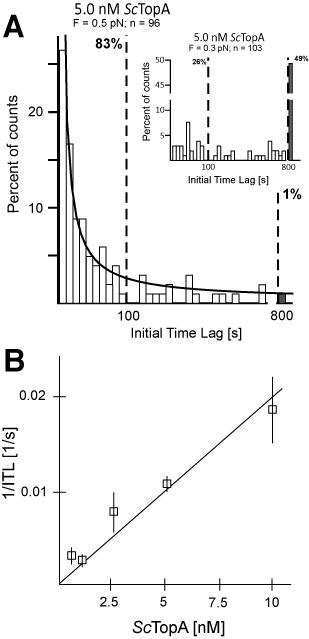
Force and concentration dependence of the rate at which ScTopA initiates DNA relaxation. (A) The initial time lag (ITL) for ScTopA activity displays a single-exponential distribution and decreases with increasing extending force. Histograms show the distribution of ITL values measured for 5-nM ScTopA and DNA extended by a force F = 0.5 pN (main panel) compared to ITL values measured at low extending force (inset, F = 0.3 pN). The lower extending force resulted in a near abolition of DNA relaxation events and a dramatic elevation of the number of unrelaxed molecules (grey bars in both panels). The number of molecules analysed (n) is shown at the top of each panel. The line corresponds to a single-exponential fit to the data obtained at 5 nM and for which unrelaxed molecules are proportionally very infrequent, giving a mean ITL = 29 s. Dashed vertical bars indicate the fraction of molecules relaxed within 100 s and 800 s, respectively. (B) ScTopA activity initiates more rapidly as concentration increases. The rate of initiation 1/ITL is obtained at each concentration by fitting a single-exponential decay to the cumulative probability distribution of individual ITLs. The cumulative probability distribution allows one to include in the analysis the unrelaxed DNA molecules (which become more frequent at lower concentrations), and by fitting at short timescales the characteristic decay time of the distribution is obtained. For 0.5 nM, n = 43 of which 15 are molecules unrelaxed after 800 s; for 1 nM, n = 39 of which 12 are unrelaxed molecules; for 1 nM, n = 59 of which four are unrelaxed molecules; statistics for 5 nM are given in (A); and for 10 nM, n = 42 of which one is an unrelaxed molecule. Error bars represent SEM obtained from the single-exponential fit.
ScTopA removes several supercoils independently of enzyme concentration
Although in experiments at the single-molecule level ScTopA exhibited substrate preference characteristic of the TopA family, we also observed the atypical occurrence of full relaxation of DNA molecules in a single relaxation burst (i.e. the relaxation event consists in a continuous increase in DNA extension until nearly maximal extension). The relaxation of DNA molecules by removal of several supercoils in a rapid burst may result from the concurrent action of several enzyme molecules, although this is statistically unlikely given the long ITLs observed earlier. To further rule this possibility out, we analysed the relaxation of 2-kb DNA at different ScTopA concentrations and plotted the correlation between initial supercoiling (IS, the length by which the molecule is shortened as a result of magnet rotation) and initial relaxation (IR, the length by which DNA extension increases as a result of ScTopA activity during the first burst of relaxation). Similar values of IS and IR parameters (IS ∼ IR; Figure 3, marked with a dotted line) mean nearly complete DNA relaxation in a single relaxation burst, whilst IR < IS and IR = 0 correspond to relaxation in more than one burst and no relaxation at all, respectively. We noticed that ScTopA at a high concentration (2.5–5.0 nM) relaxed a large fraction of molecules in one burst (63.3% and 67.9%, respectively). Interestingly, decreasing the concentration of ScTopA did not lead to an increase in the fraction of molecules in the IR<IS category, corresponding to relaxation of DNA supercoils in multiple bursts. Instead, using lower ScTopA concentrations (0.5–1.0 nM) resulted in an increase of the fraction of molecules which remained supercoiled, although there was still a significant fraction of molecules that were relaxed in a single burst (41.2% and 44.1%, respectively). Thus, the removal of many supercoils in one relaxation event even at low enzyme concentration is most likely carried out by a single ScTopA molecule or its active multimer, rather than several ScTopA molecules/complexes acting independently.
Figure 3.
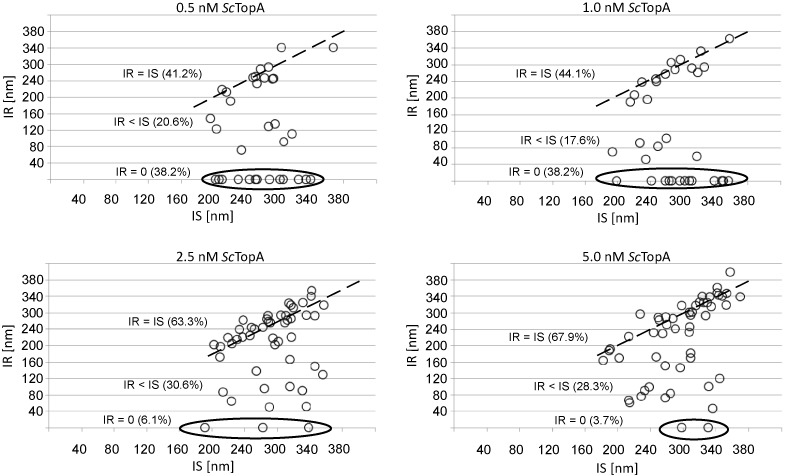
ScTopA concentrations do not influence enzyme processivity. The enzyme processivity as expressed by the ratio between initial supercoiling (IS) and initial relaxation (IR). Each circle represents a single DNA relaxation event; circles accumulated near the dotted line correspond to nearly full relaxation of supercoiled DNA in single relaxation event (IS = IR), circles marked with a ellipse represent the fraction of DNA molecules remaining supercoiled (IR = 0 within the 800-s recording time); circles located in between correspond to molecules relaxed in more than one burst (IS > IR). Percentage of molecules belonging to each population are shown on each diagram in brackets.
We next estimated the rate of onset of supercoil relaxation by considering both the ITL and the number of molecules that have not been relaxed at the end of the experiment. We thus considered an initial population consisting of all of the initially supercoiled DNA molecules and calculated its decay over 800 s to a final state where only the unrelaxed molecules remain. We fit this decay to a single-exponential distribution and obtain the rate as the inverse of the decay time and plot the rate as a function of concentration (Figure 2B and Supplementary Figure S3). The rate of onset of supercoil relaxation increased linearly with topoisomerase I concentration, suggesting that diffusion is limiting in this concentration range and reinforcing the idea that these relaxation events result from the activity of a single topoisomerase I molecule/complex. From these measurements, we estimated an apparent first-order binding rate for the 2-kb DNA of about 2 × 106/M·s, consistent with diffusion-limited activity.
To summarize, the magnetic trap experiments indicate that DNA subjected to ScTopA action remains supercoiled until the enzyme removes nearly all supercoils in a single burst of activity. This explains the apparent bimodal distribution of topoisomers observed in gel-based measurements of ScTopA activity. Although increasing ScTopA concentration shortened the initial time necessary for enzyme activity to be observed, it did not affect enzyme activity itself.
ScTopA preferentially relaxes DNA in a single relaxation burst
Our observation that ScTopA activity can remove nearly all DNA supercoils in a single burst of activity prompted us to check if this was still the case for longer DNA fragments that can accommodate more extensive supercoiling. Thus, we measured the number of relaxation bursts required to remove nearly all supercoils from the DNA substrates in the 2–51-kb range. For the 2-kb DNA substrate, the vast majority of molecules were relaxed in a single burst of ScTopA activity (∼63%). For the remaining third of DNA molecules relaxation occurred in multiple bursts separated by long pauses (Figure 4B, inset); 26% and 11% of all relaxation time-traces showed one and two pauses, respectively, corresponding to two or three relaxation bursts (Figure 4A). Interestingly, when we used longer DNA fragments—11 kb and 17 kb—in the magnetic trapping experiments, the fraction of molecules relaxed in a single burst remained similar to that calculated for 2-kb fragments (51% and 58%, respectively; Figure 4A). Even when the length of the DNA molecule was 51 kb (which allowed us to introduce up to 150 supercoils per molecule) in 49% of relaxation events nearly all supercoils were removed in a single burst, whilst for 51% of molecules we observed one or more pauses separating each burst. The pause duration between two bursts was calculated to be 10-fold shorter than the time necessary for reaction initiation (Figure 4B).
Figure 4.
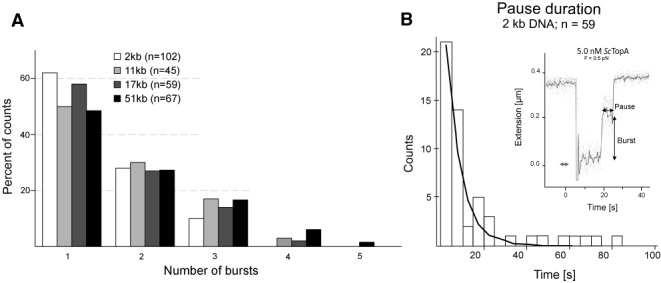
Relaxation bursts and pausing by ScTopA. (A) The number of relaxation bursts required to remove negative supercoils from DNA is not dependent on the length of DNA substrates. Histograms show the fractions of 2–51-kb DNA molecules relaxed in the specified number of bursts (1–5). (B) Example of a pause between two successive relaxation bursts and distribution of pause lifetimes.
Independently of the length of the DNA substrate ScTopA removes all negative supercoils from it in the first relaxation burst. However, we observed that occasionally the enzyme pauses during a relaxation burst and requires some time to start the next relaxation burst. Moreover, comparison of the ITLs and the pause duration suggests that during relaxation cycle ScTopA does not dissociate form DNA, since the pause time is remarkably shorter than ITLs calculated for the same enzyme concentration.
ScTopA exhibits higher processivity and velocity than other TopA homologues
Originally all type IA (29,30) topoisomerases were demonstrated to use a strand-passage mechanism to decrease gradually the superhelical density (ΔLk = −1) of negatively supercoiled DNA resulting in a distributive pattern of relaxation products. However, differences in the mechanism of relaxation of negatively supercoiled DNA between TopA and TopB (both belonging to the subtype IA) have recently been described (16–18). The ability of ScTopA to remove a large number of supercoils in a single burst independently of enzyme concentration and substrate length prompted us to further explore its properties, in particular its reaction rate and processivity, using a long DNA fragment as substrate.
Reaction rates, given in μm/s, correspond to the speed at which the DNA extension increases as supercoils are removed by ScTopA (Figure 5A), and the distribution of reaction rates is given in Figure 5B. The velocity distribution appears to be Gaussian, with a mean reaction rate of 0.45 μm/s. The reaction rate in units of supercoils (Lk) removed per second is obtained by dividing the reaction rate given in μm/s by the size of a single supercoil (∼60 nm/supercoil in the experimental conditions used here), giving a mean on the order of 8–10 Lk/s (Figure 5B).
Figure 5.
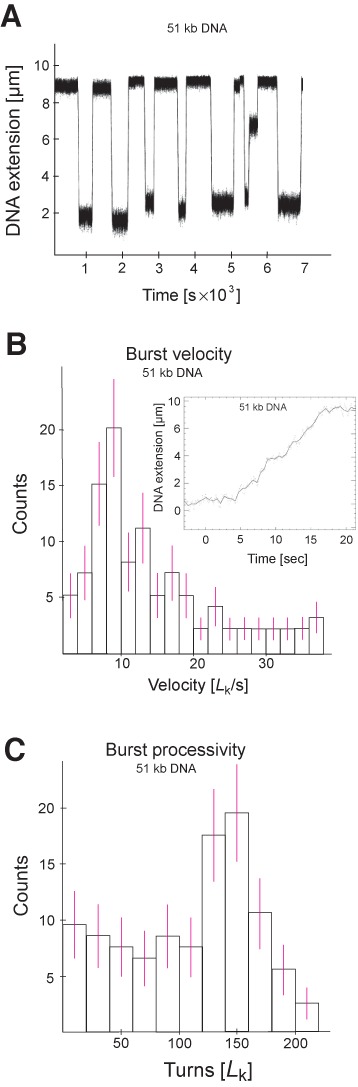
ScTopA relaxation exhibits high velocity and processivity of supercoils removal (Lk/s) in a single burst. (A) Typical time-trace for supercoils relaxation as observed on 51-kb DNA substrate supercoiled by −150 turns. (B) Burst velocity distribution and (inset) expanded view of a relaxation burst. (C) Burst processivity distribution.
To estimate the processivity of the enzyme, we determined the number of supercoils (Lk) removed only in the first burst of ScTopA activity; a plot of the resulting probability distribution is shown in Figure 5C. The distribution is characterized by a weak decay at small values as well as a ‘brick wall’ at 7.0 μm corresponding to relaxation events that are substrate-limited. Clearly the very high processivity of ScTopA makes it difficult to quantify as even the longest single-molecule DNA substrate is too short for the processivity of most events to be determined. Although a significant number of events were observed for which the change in DNA extension was less than 7.5 μm and frequency of shorter bursts was constant (5–7% per 1 μm) suggesting the stochastic pause of relaxation process (Figure 5C), the most likely events corresponded to those for which nearly all supercoils were removed in the first burst of ScTopA activity. We note that some measurements show more than 150 turns removed in one burst, which is presumably due to the fact that individual molecules can occasionally be only partially relaxed during a prior relaxation run. When supercoiled by an additional 150 negative turns, the population of molecules obtained thus includes some individuals with more negative supercoiling than the 150 added turns yet which can still be removed in a single burst of catalysis.
Three parameters (ITL, processivity and velocity) influence the overall reaction velocity. Based on the collected data, we calculated the average total relaxation rate (TRR) for two DNA substrates: 2-kb DNA and pUC19 (∼2.7 kb) in single molecule and bulk experiments, respectively (Supplementary Figure S4A and B). TRR is defined by the time lapse between the mechanically introduced negative supercoils on DNA molecule to the point at which the bead returns to the starting position (as the result of enzyme activity). The TRR parameter calculated for 2 kb was similar (the values for most of them were not higher than 0.3 Lk/s) to the TRRbulk calculated for pUC19 plasmid based on the Michaelis–Menten equation (0.2 Lk/s) (Supplementary Figure S4C). The comparison of relaxation patterns of two negatively supercoiled plasmids differing in length and linking number (Lk) shows that the ScTopA relaxes both substrates with the same high processivity if the molar ratio of enzyme:plasmid is retained (Supplementary Figure S7). Thus, our results suggest that the overall relaxation velocity is the intrinsic property of the enzyme and is not dependent on the substrate type.
In summary, ScTopA differs from its TopA homologue from E. coli in terms of both burst velocity of supercoil removal and processivity. Comparing with E. coli TopA, for which average velocity and processivity were estimated as 3.3 Lk/s and 20 Lk/run, respectively (16), ScTopA is about 3-fold faster (10 Lk/s) and almost 10-fold more processive (150 Lk/burst). However, the waiting time preceding the supercoils removal by ScTopA makes the overall enzyme less effective than if calculated only based on the burst velocity and processivity.
ScTopA switches between processive and distributive DNA relaxation in a torque-dependent manner
To determine how efficient ScTopA was at completely removing all supercoils, we measured the number of residual supercoils left unremoved by ScTopA activity. This was done by initially supercoiling the DNA by a fixed number of turns, denoted by n, and determining the resulting extension of the molecule, denoted by l1. After ScTopA has been allowed to act on the DNA for a fixed amount of time, it is supercoiled anew by the same number of turns, n, and the resulting extension l2 is recorded. This extension l2 is systematically smaller than l1 (Figure 6), indicating that there remained non-zero residual supercoiling after ScTopA action. The difference in extensions l2–l1 is thus related to the number of residual, unrelaxed supercoils, nres, according to nres = (l2–l1)/δ, where δ is the change in DNA extension associated with one supercoil.
Figure 6.
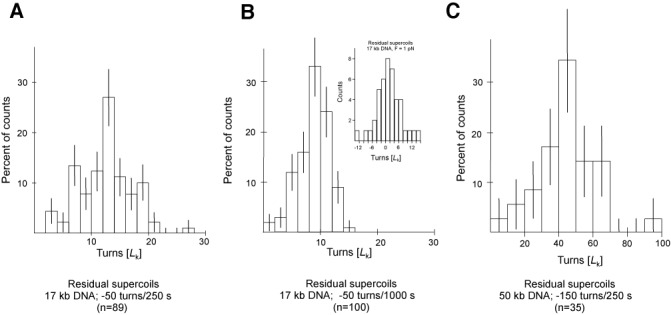
Residual supercoiling left unremoved by ScTopA relaxation bursts indicates ScTopA switches between high and low processivity as a function of torque acting on the DNA. Seventeen-kilobase DNA supercoiled by −50 turns and left to react with TopA shows (A) relaxation of ∼38 supercoils in the first 250 s of incubation but only (B) relaxation of ∼3 additional supercoils in the following 750 s of the reaction, for an extending force of about 0.3 pN. (Inset) When the extending force is increased to 1 pN, the average residual supercoiling is zero after a 250-s reaction. (C) Fifty-one-kilobase DNA supercoiled by −150 turns and left to react with TopA for 250 s shows an increase in residual supercoils roughly three times greater than what is observed on 17-kb DNA.
We first determined the distribution of nres for 17-kb substrate DNA supercoiled by −50 turns and allowed to react with ScTopA for 250 s at a low extending force (F = 0.3 pN). In this timeframe, all molecules are nearly completely relaxed by one or more bursts of ScTopA activity, however, as shown in Figure 6A, there remains on average 15 supercoils in the DNA. We next determined the distribution of nres for the same DNA and the same IS, but allowing ScTopA to react for 1000 s (Figure 6B). The latter distribution shows that allowing the enzyme to act for longer leads to a slightly smaller number of residual supercoils but comparing the two distributions indicates that the enzyme has lost nearly all of its processivity and is only able to remove a small number of supercoils at a time. We next repeated this experiment using 51-kb substrate DNA supercoiled by −150 turns and allowed to react for 250 s; the distribution of residual supercoiling is shown in Figure 6C. Comparing the data obtained with the shorter and longer DNA molecules shows that nres is proportional to the length of the substrate DNA, as would be expected for a topoisomerase with torque-dependent processivity. In agreement with this, we found that when the first experiment in this series was repeated but for an increased extending force (F = 1 pN), the distribution of nres became centred about nres = 0 (Figure 6B, inset).
We also calculated the superhelical densities of 17-kb and 51-kb DNA at the point at which the ScTopA processivity dropped as σ17 = −0.008 ± 0.001 and σ51 = 0.009 ± 0.001, respectively (Figure 6B and C). These results suggest that decreasing the absolute density below ∼0.01 inhibits enzyme processivity. From extension versus supercoiling curves (Supplementary Figure S7) we note that this supercoiling density corresponds to the buckling transition for DNA extended at this low force (F = 0.3 pN), and at this force the buckling transition occurs at a torque on the order of 4 pN·nm (or ∼1 kBT) (31). This value is close to the denaturation torque for DNA, suggesting that the existence of denatured regions may be crucial to maintaining a high degree of processivity for ScTopA.
To conclude, these data indicate that ScTopA removes supercoils in a highly processive fashion as long as the torque acting on the DNA is high enough, but at low torques displays distributive activity.
Processivity of ScTopA protein is provided by the positively charged C-terminus
ScTopA processivity was also analysed by resolving the reaction intermediates in agarose gel electrophoresis. In this assay, a broad range of enzyme concentrations (0–150 nM) was used in reactions with negatively supercoiled pUC19 plasmid. ScTopA-mediated plasmid relaxation led to appearance of low-density topoisomers even when supercoiled plasmid or its partially relaxed topoisomers were still noticeable (Figure 7A) confirming the processive behaviour of ScTopA and underscoring the difference in relaxation mechanism between ScTop protein and its homologue from E. coli. The presence of positively charged amino acids at the C-terminal end of ScTopA prompted us to analyse the DNA relaxation properties of mutants lacking either the whole C-terminal domain (343 aa; ScTopAΔC) or the stretch of basic amino acids at the C-terminus (71 aa; ScTopA881). Interestingly, removal of the whole C-terminal domain almost completely abolished enzyme activity (Figure 7D), whilst the pattern of ScTopA881 relaxation products was distributive (Figure 7B, left panel) instead of processive as observed for the wild-type protein (Figure 7B, right panel). By titrating the pUC19 reaction substrate from 10 to 70 nM, we measured the reaction velocities for ScTopA and ScTopA881 within their linear range and fitted the reaction kinetics to the Michaelis–Menten model (see the Materials and Methods section). Based on differential equation of the model, the maximal enzymatic rates Vmax were calculated to be 6.5 nM/min for 5-nM ScTopA and Michaelis–Menten constant Km 5.1 nM and 0.62 nM/min and 20.7 nM, respectively, for the mutant ScTopA881 (Figure 7C).
In summary, these bulk experiments confirm the high processivity of ScTopA and demonstrate that complete removal of the C-terminal domain abolishes enzyme activity whilst truncation of its positively charged C-terminus affects affinity for substrate and processivity.
DISCUSSION
Many bacteria, especially rapidly growing species (E. coli, Bacillus subtilis), possess two topoisomerases of type IA—topoisomerase I (TopA) and topoisomerase III (TopB). In contrast, Actinobacteria possess only one gene encoding a type IA topoisomerase, TopA. Although in E. coli both proteins TopA and TopB are closely related and share the same catalytic mechanism, they relax DNA with different efficiencies and processivities and play key roles in different cellular processes (16,32–34). Sequence and structure alignments suggest that S. coelicolor TopA (ScTopA), similar to homologous M. smegmatis TopA (MsTopA), possesses a unique C-terminal domain that terminates with a stretch of positively charged amino acids but which, unlike E. coli TopA, lacks Zn fingers (22,23). In contrast to previous observations made by Bao and Cohen (20), who reported a slightly decreased relaxation activity of ScTopA protein lacking the C-terminal domain, our data show that removal of the C-terminal domain of ScTopA completely abolished enzyme activity. Although our ScTopAΔC mutant (lacking 343 aa) and the mutant constructed by Bao and Cohen (20) (lacking 353 aa) differ somewhat in length, it is difficult to explain the discrepancy in the observed enzyme activity. Interestingly, Jain and Nagaraja (35) demonstrated that the activity of MsTopA lacking the C-terminal 34 kDa domain was also abolished; however, it could be restored if supplemented with recombinant C-terminal domain. Our studies showed that removal of the short positively charged C-terminus (71 aa) from ScTopA greatly affected enzyme activity, reducing its processivity and affinity for the substrate. It has been proposed that basic amino acids may replace Zn fingers in efficient DNA binding and that their elimination decreases enzyme affinity for DNA substrates (36). Recently, Ahmed et al. (37) have shown that the C-terminal domain of MsTopA protein may play a role in strand-passage and suggested that the presence of a positively charged C-terminus may provide an additional binding site, ensuring efficient DNA relaxation. Our calculation of Km for the wild-type and truncated ScTopA protein (5.1 and 20.7 nM, respectively) corroborates with this notion. It was earlier reported that M. smegmatis MsTopA binds both single- and double-stranded DNA oligomers with similar affinity, and Mg2+ ions are not necessary for DNA binding and cleavage (24), however their concentration affected processivity of the enzyme (23). We also found that, although ScTopA requires the presence of single-stranded regions in a substrate for relaxation, it effectively binds to double-stranded DNA (dsDNA) (Supplementary Figure S5).
Our results show that ScTopA is able to remove up to 150 supercoils per relaxation burst and that the enzyme is characterized by a torque-dependent processivity which leads to more distributive and/or lower activity at low torque when most of the DNA supercoiling has been relaxed. The apparent processivity of ScTopA could be the result of either simultaneous action of many protein molecules on DNA or the ability of a single ScTopA molecule or multimeric complex to remove several supercoils each time it binds to DNA. Analysis of the concentration dependence of the reaction rules out the possibility that apparent processivity is due to multiple active complexes acting independently as processive activity was observed at all concentrations tested. The formation of a multimeric complex composed of many ScTopA molecules acting and removing supercoils cooperatively could explain the long ITL observed for low ScTopA concentration (conditions in which a long ITL could be due to shifting the complex-assembly equilibrium towards an inactive monomeric state). It has been reported earlier that vaccinia virus topoisomerase IB is able to form a filament spreading along dsDNA forming synapses between distinct DNA fragments in a highly cooperative fashion (38). Detailed analysis of enzyme activity supports the idea that a single ScTopA molecule is able to act processively, although the hypothesis of multimer assembly cannot be excluded. The burst rate of supercoil removal by ScTopA was determined to be 8 Lk/s, comparable to E. coli TopA (3.3 Lk/s) but significantly lower than that of E. coli TopB (129 Lk/s) or human type IB topoisomerase (6.7 μm/s; which corresponds to 134 Lk/s) (12,16). Nevertheless, the overall relaxation velocity results from the combination of the burst rate and the processivity related to affinity to substrate. Our single-molecule studies show that efficient DNA relaxation by ScTopA is the result of extremely high processivity (150 Lk/burst), much higher than reported for E. coli TopB (28 Lk/burst) and human TopIB (50 Lk/burst) estimated at similar extending force (12,16). E. coli TopB acts more processively than E. coli TopA presumably thanks to the creation of a stable enzyme:DNA complex (17,39) (Table 1). The high processivity of EcTopB was linked to the presence of the positively charged region known as the decatenation loop in its N-terminal domain (17). Disruption of the decatenation loop resulted in inhibition of the enzyme via an apparent reduction in processivity. ScTopA does not possess a bona fide decatenation loop, although some homologous amino acids are present in its N-terminal domain (Supplementary Figure S1D). Thus, increased enzyme processivity is presumably linked to the C-terminal stretch of basic amino acids as indicated by Ahmed et al. (37). Predictions of secondary structure suggest that the C-terminus of ScTopA forms an α-helix with basic amino acids precisely oriented in a helical fashion on the surface of the α-helix and which may constitute the domain interacting with the highly compacted supercoiled DNA, providing higher affinity to substrate. This could confer the ability of a single ScTopA molecule to remove several supercoils each time it binds to DNA and leading to processive removal of supercoils. Thus, our most favoured hypothesis explaining rapid removal of several supercoils is the formation of a stable and processive ScTopA:DNA complex, perhaps thanks to the presence of denatured regions in the DNA.
Table 1.
Characterization of DNA relaxation by type I topoisomerases: E. coli (EcTopA and EcTopB), human type IB topoisomerase and S. coelicolor (ScTopA)
| Parameter | EcTopA | EcTopB | Human type IB topo | ScTopA |
|---|---|---|---|---|
| Relaxation rate per burst (Lk/s) | 3.3 | 129 | 134 | 8 |
| Initial time lag (ITL)a (s) | 1.6 | 129 | nd | 29 |
| Turns removed in a single burst (Lk) (low torque) | 20 | 28 | 50 | 150b |
aITL was estimated for 2-nM ScTopA and was compared with 2-nM EcTopA and 2-nM EcTopB (16).
bThe number of turns removed by ScTopA in a single relaxation burst may be higher than 150 Lk as the experimental estimation is limited by the 51-kb DNA substrate.
In summary, we suggest that in Streptomyces, which lacks a copy of the topB gene encoding for topoisomerase III, ScTopA exhibits some of the properties of TopA and TopB proteins. Although the portfolio of topology-modifying enzymes in S. coelicolor is simplified in comparison with other species, the topological problems it encounters during replication and segregation of a large number of long, linear chromosomes are presumably non-trivial. This could lead to the requirement for a highly processive topoisomerase, and indeed such features have been observed in vitro using bulk and single molecule techniques. The unusually high processivity of ScTopA combined with its previously demonstrated telomerase activity (20) and its ability to resolve topological problems during chromosome segregation (21) suggests specific adaptation of the only type I topoisomerase in S. coelicolor to efficiently fulfil multiple functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors acknowledge financial support from the Parent Bridge Programme of Foundation for Polish Science, co-financed by European Union, Regional Development Fund, and from EURYI.
Authors Contributions: M.J.S. contributed substantially to conception and design, acquisition of data, analysis and interpretation of data and drafting the article. T.S. contributed substantially to conception and design, acquisition of data, or analysis and interpretation of data and revising the manuscript critically for important intellectual content. A.S. contributed to acquisition of data. J.Z.C. contributed to revising the manuscript critically for important intellectual content. D.J. contributed substantially to conception and design, drafting the article and final approval of the version to be published.
FUNDING
Parent Bridge Programme of Foundation for Polish Science; co-financed by European Union, Regional Development Fund; EURYI [to T.S.]. Funding for open access charge: Parent Bridge Programme of Foundation for Polish Science (POMOST/2010-2/5).
Conflict of interest statement. None declared.
REFERENCES
- 1.Everid A.C., Small J.V., Davies H.G. Electron-microscope observations on the structure of condensed chromatin: evidence for orderly arrays of unit threads on the surface of chicken erythrocyte nuclei. J. Cell Sci. 1970;7:35–48. doi: 10.1242/jcs.7.1.35. [DOI] [PubMed] [Google Scholar]
- 2.Richmond T.J., Davey C.A. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman S.B. Cooperative transitions of isolated Escherichia coli nucleoids: implications for the nucleoid as a cellular phase. J. Struct. Biol. 2006;153:160–175. doi: 10.1016/j.jsb.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman S.B. Shape and compaction of Escherichia coli nucleoids. J. Struct. Biol. 2006;156:255–261. doi: 10.1016/j.jsb.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 5.Nasmyth K., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 6.Carter S.D., Sjögren C. The SMC complexes, DNA and chromosome topology: right or knot. Crit. Rev. Biochem. Mol. Biol. 2012;47:1–16. doi: 10.3109/10409238.2011.614593. [DOI] [PubMed] [Google Scholar]
- 7.Thadani R., Uhlmann F., Heeger S. Condensin, chromatin crossbarring and chromosome condensation. Curr. Biol. 2012;22:R1012–R1021. doi: 10.1016/j.cub.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 9.Forterre P., Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forterre P., Gribaldo S., Gadelle D., Serre M.C. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Belova G.I., Prasad R., Nazimov I.V., Wilson S.H., Slesarev A.I. The domain organization and properties of individual domains of DNA topoisomerase V, a type 1B topoisomerase with DNA repair activities. J. Biol. Chem. 2002;277:4959–4965. doi: 10.1074/jbc.M110131200. [DOI] [PubMed] [Google Scholar]
- 12.Koster D.A., Croquette V., Dekker C., Shuman S., Dekker N.H. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 13.Seol Y., Zhang H., Pommier Y., Neuman K.C. A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16125–16130. doi: 10.1073/pnas.1206480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravind L., Leipe D.D., Koonin E.V. Toprim–a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng B., Sorokin E.P., Tse-Dinh Y.C. Mutation adjacent to the active site tyrosine can enhance DNA cleavage and cell killing by the TOPRIM Gly to Ser mutant of bacterial topoisomerase I. Nucleic Acids Res. 2008;36:1017–1025. doi: 10.1093/nar/gkm1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terekhova K., Gunn K.H., Marko J.F., Mondragón A. Bacterial topoisomerase I and topoisomerase III relax supercoiled DNA via distinct pathways. Nucleic Acids Res. 2012;40:10432–10440. doi: 10.1093/nar/gks780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z., Mondragón A., Hiasa H., Marians K.J., DiGate R.J. Identification of a unique domain essential for Escherichia coli DNA topoisomerase III-catalysed decatenation of replication intermediates. Mol. Microbiol. 2000;35:888–895. doi: 10.1046/j.1365-2958.2000.01763.x. [DOI] [PubMed] [Google Scholar]
- 18.Nurse P., Levine C., Hassing H., Marians K.J. Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J. Biol. Chem. 2003;278:8653–8660. doi: 10.1074/jbc.M211211200. [DOI] [PubMed] [Google Scholar]
- 19.Bentley S.D., Chater K.F., Cerdeño-Tárraga A.M., Challis G.L., Thomson N.R., James K.D., Harris D.E., Quail M.A., Kieser H., Harper D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 20.Bao K., Cohen S.N. Reverse transcriptase activity innate to DNA polymerase I and DNA topoisomerase I proteins of Streptomyces telomere complex. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14361–14366. doi: 10.1073/pnas.0404386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szafran M., Skut P., Ditkowski B., Ginda K., Chandra G., Zakrzewska-Czerwińska J., Jakimowicz D. Topoisomerase I (TopA) is recruited to ParB complexes and is required for proper chromosome organization during Streptomyces coelicolor sporulation. J. Bacteriol. 2013;195:4445–4455. doi: 10.1128/JB.00798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhaduri T., Nagaraja V. DNA topoisomerase I from Mycobacterium smegmatis. Indian J. Biochem. Biophys. 1994;31:339–343. [PubMed] [Google Scholar]
- 23.Bhaduri T., Bagui T.K., Sikder D., Nagaraja V. DNA topoisomerase I from Mycobacterium smegmatis. An enzyme with distinct features. J. Biol. Chem. 1998;273:13925–13932. doi: 10.1074/jbc.273.22.13925. [DOI] [PubMed] [Google Scholar]
- 24.Bhaduri T., Sikder D., Nagaraja V. Sequence specific interaction of Mycobacterium smegmatis topoisomerase I with duplex DNA. Nucleic Acidc Res. 1998;26:1668–1674. doi: 10.1093/nar/26.7.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X., Leng F. A rapid procedure to purify Escherichia coli DNA topoisomerase I. Protein Expr. Purif. 2011;77:214–219. doi: 10.1016/j.pep.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revyakin A., Ebright R.H., Strick T.R. Single-molecule DNA nanomanipulation: improved resolution through use of shorter DNA fragments. Nat. Methods. 2005;2:127–138. doi: 10.1038/nmeth0205-127. [DOI] [PubMed] [Google Scholar]
- 27.Charvin G., Vologodskii A., Bensimon D., Croquette V. Braiding DNA: experiments, simulations, and models. Biophys. J. 2005;88:4124–4136. doi: 10.1529/biophysj.104.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strick T.R., Allemand J.F., Bensimon D., Bensimon A., Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 29.Dean F.B., Cozzarelli N.R. Mechnism of strand passage by Escherichia coli topoisomerase I. The role of the required nick in catenation and knotting of duplex DNA. J. Biol. Chem. 1985;260:4984–4994. [PubMed] [Google Scholar]
- 30.Dekker N.H., Rybenkov V.V., Duguet M., Crisona N.J, Cozzarelli N.R., Bensimon D., Croquette V. The mechanism of type IA topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12126–12131. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosconi F., Allemand J.F., Bensimon D., Croquette V. Measurement of the torque on a single stretched and twisted DNA using magnetic tweezers. Phys. Rev. Lett. 2009;102:078301. doi: 10.1103/PhysRevLett.102.078301. [DOI] [PubMed] [Google Scholar]
- 32.Ahumada A., Tse-Dinh Y.C. The role of the Zn(II) binding domain in the mechanism of E. coli DNA topoisomerase I. BMC Biochem. 2002;3:1–13. doi: 10.1186/1471-2091-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Cheeks B.A., Lee C., Hayama R., Marians K.J. A role for topoisomerase III in Escherichia coli chromosome segregation. Mol. Microbiol. 2012;86:1007–1022. doi: 10.1111/mmi.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain P., Nagaraja V. Indispensable, functionally complementing N and C-terminal domains constitute site-specific topoisomerase I. J. Mol. Biol. 2006;357:1409–1421. doi: 10.1016/j.jmb.2006.01.079. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z., Cheng B., Tse-Dinh Y.C. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6939–6944. doi: 10.1073/pnas.1100300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed W., Bhat A.G., Leelaram M.N., Menon S., Nagaraja V. Carboxyl terminal domain basic amino acids of mycobacterial topoisomerase I bind DNA to promote strand passage. Nucleic Acids Res. 2013;41:7462–7471. doi: 10.1093/nar/gkt506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno-Herrero F., Holtzer L., Koster D.A., Shuman S., Dekker C., Dekker N.H. Atomic force microscopy shows that vaccinia topoisomerase IB generates filaments on DNA in a cooperative fashion. Nucleic Acids Res. 2005;33:5945–5953. doi: 10.1093/nar/gki906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H.L., DiGate R.J. The carboxyl-terminal residues of Escherichia coli DNA topoisomerase III are involved in substrate binding. J. Biol. Chem. 1994;269:9052–9059. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


