Abstract
Isoprenoids make up a remarkably diverse class of more than 25000 biomolecules that include familiar compounds such as cholesterol, chlorophyll, vitamin A, ubiquinone, and natural rubber. The two essential building blocks of all isoprenoids, isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), are ubiquitous in the three domains of life. In most eukaryotes and archaea, IPP and DMAPP are generated through the mevalonate pathway. We have identified two novel enzymes, mevalonate-3-kinase and mevalonate-3-phosphate-5-kinase from Thermoplasma acidophilum, which act sequentially in a putative alternate mevalonate pathway. We propose that a yet unidentified ATP-independent decarboxylase acts upon mevalonate 3,5-bisphosphate, yielding isopentenyl phosphate, which is subsequently phosphorylated by the known isopentenyl phosphate kinase from T. acidophilum to generate the universal isoprenoid precursor, IPP.
Cholesterol, chlorophylls, hemes, ubiquinones, natural rubbers, and archaeal membrane lipids are just a few examples of more than 25000 biomolecules that make up the diverse class of organic molecules called isoprenoids.1−3 Isoprenoids are found in all three domains of life and are involved in essential processes such as electron transport, post-translational modification, regulation of membrane fluidity, and cytoskeleton assembly.4 All isoprenoids are composed of two or more isoprene building blocks (five-atom branched hydrocarbons) derived from isopentenyl pyrophosphate (IPP) and its isomer, dimethylallyl pyrophosphate (DMAPP). Plants and most eubacteria generate IPP and DMAPP from pyruvate and glyceraldehyde 3-phosphate via the deoxyxylulose 5-phosphate (DXP) pathway.5 Eukaryotes and archaea use a separate pathway called the mevalonate pathway, which relies on acetyl-CoA as the sole carbon source.6 The mevalonate pathway in archaea is especially important because IPP and DMAPP are used to make branched lipids that are connected to glycerol through ether linkages to form membrane lipids.7 These branched lipids and ether linkages are thought to promote membrane stability at high temperatures.8,9
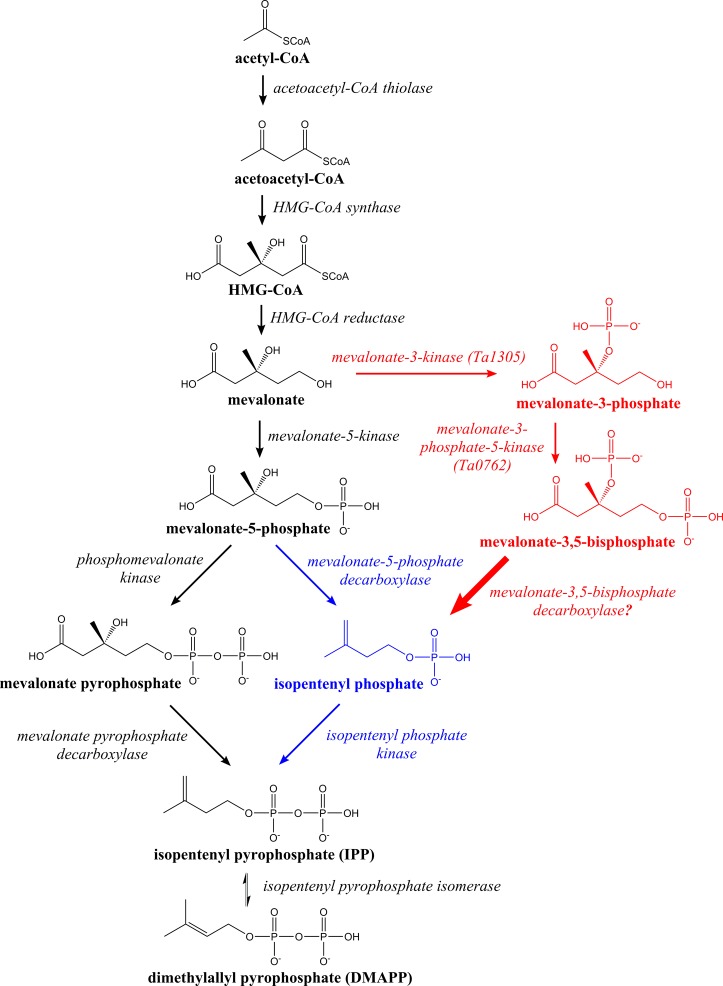
The canonical mevalonate pathway of eukaryotes can be conceptually separated into two parts, which we term the upper and lower stages (Figure 1). In the upper stage, three acetyl-CoAs are condensed and reduced by NADPH to yield mevalonate. In the lower stage, mevalonate is sequentially phosphorylated to make mevalonate pyrophosphate and then decarboxylated to yield IPP, which can be converted to DMAPP by an isomerase.10 Recent phylogenetic analyses of archaeal genomes identified strong homologues for the first three enzymes in the pathway leading up to mevalonate. However, in most cases, no homologues for phosphomevalonate kinase or mevalonate pyrophosphate decarboxylase are found.11 Furthermore, some archaea such as Thermoplasma, Flavobacteria, and Gramella have no detectable mevalonate kinase (MVK) homologues.11
Figure 1.
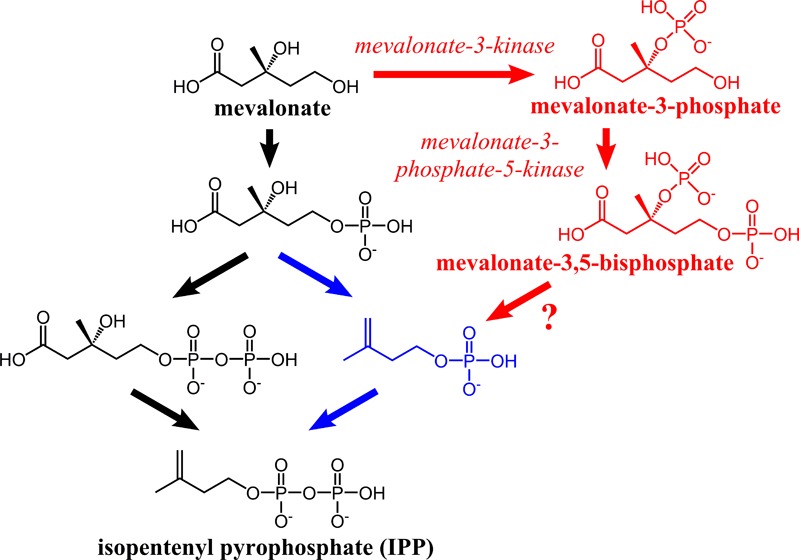
Mevalonate pathways. Eukaryotes use the left side of the fork after mevalonate 5-phosphate (black), and archaea use the right side (blue). The two enzymes identified in this study suggest an alternative pathway branching from mevalonate (red). A predicted decarboxylase (bold arrow) connects mevalonate 3,5-bisphosphate to isopentenyl phosphate.
While it has been hypothesized for more than a decade that archaea possess an alternative mevalonate pathway based on computational genomics,12 a complete pathway was not elucidated until 2013 in the organisms Roseiflexus castenholzii(13) and Haloferax volcanii.14 As shown by the blue arrows in Figure 1, instead of two phosphorylations followed by a decarboxylation as seen in eukaryotes, some archaea phosphorylate once, decarboxylate with mevalonate-5-phosphate decarboxylase, and then phosphorylate again with isopentenyl phosphate kinase. The end product is the same, IPP, but the enzymatic reactions are distinct.
We were interested in obtaining thermophilic enzymes for in vitro reconstruction of the mevalonate pathway.15 As part of this effort, we targeted enzymes from an archeon, Thermoplasma acidophilum, which is naturally found in coal refuse piles in the northeast United States, where it grows optimally at 59 °C.16 One of the target enzymes from T. acidophilum, encoded by the ta1305 gene, was annotated as a mevalonate pyrophosphate decarboxylase in GenBank [genes in italics (e.g., ta1305) and their protein products capitalized in Roman type (e.g., Ta1305)].17 Upon characterization of the expressed enzyme, however, we discovered that it acts on mevalonate to generate mevalonate 3-phosphate, a previously unknown activity (see Figure 1). We were then able to identify a second novel kinase in T. acidophilum encoded by the ta0762 gene, which phosphorylates mevalonate 3-phosphate to form mevalonate 3,5-bisphosphate, another previously unknown metabolite. We propose that these two new enzyme activities are part of a novel mevalonate pathway utilized by T. acidophilum (Figure 1).
Materials and Methods
Materials
Miller LB medium (BD Difco) was used for growth of bacterial strains. Escherichia coli BL21 Gold (DE3) (Agilent) was used as the host strain for both cloning and expression of recombinant proteins. Plasmid pET28a(+) was purchased from Novagen. HotStart Taq Mastermix (Denville) was used for gene amplification. Phusion DNA polymerase (Finnzymes), Taq DNA ligase (MCLab), and T5 exonuclease (Epicenter) were purchased separately and used to make the assembly master mix (AMM) used for cloning. Ni-NTA resin and miniprep reagents were purchased from Qiagen. Primers were synthesized by ValueGene. All other chemicals were purchased from Sigma-Aldrich unless otherwise noted.
Cloning
Genes were amplified by polymerase chain reaction (PCR) from T. acidophilum genomic DNA (ATCC catalog no. 25905D). A modified Gibson method was used to assemble all constructs.18 Primers included 15–20 bp complementary to the ends of the target gene and 15–20 bp complementary to the NdeI and XhoI insertion sites of pET28a(+), which allowed for the addition of an N-terminal six-His tag. Ten nanograms of pET28a(+) digested with NdeI and XhoI was mixed with 30 ng of PCR product and 7 μL of assembly mix [0.1 M Tris-HCl (pH 7.5), 0.2 mM dNTPs, 1 mM NAD+, 5% PEG 8K, 10 mM DTT, 10 mM MgCl2, 0.00375 unit/μL T5 exonuclease, 0.012 μL Phusion DNA polymerase, and 4 units/μL TaqDNA ligase]. After incubation at 50 °C for 2 h, 5 μL was then used to transform BL21 Gold (DE3), and transformants were selected by being plated on LB agar containing 50 μg/mL kanamycin.
Expression and Purification
All E. coli strains were grown at 37 °C in LB medium with 50 μg/mL kanamycin. One liter of LB medium was inoculated with 5 mL of overnight starter culture. Protein expression was induced during the log phase (OD600 of 0.5–0.8) with 0.5 mM IPTG. After 20 h, cells were pelleted, resuspended in 12 mL of buffer A [50 mM Tris-HCl (pH 7.5) and 100 mM NaCl], and lysed by sonication, and cell debris was removed by centrifugation at 30000g for 20 min. The lysate was mixed with 3 mL of a Ni-NTA slurry and incubated at 4 °C while being gently mixed. After 1 h, the lysate mixture was applied to a column and the Ni-NTA beads were washed three times with 25 mL of buffer A containing 10 mM imidazole. Protein was then eluted with 4 mL of buffer A containing 250 mM imidazole. For kinetic characterization, enzymes were further purified on an AKTA FPLC system using a Superdex S200 10/300 GL gel filtration column run with buffer A, at a rate of 0.5 mL/min.
Enzyme Assays
Enzymes were assayed for the ability to hydrolyze ATP to ADP by coupling the reactions to pyruvate kinase (PK) and lactate dehydrogenase (LDH) from rabbit muscle.19 Assays were conducted in duplicate using a 200 μL volume at 55 °C and contained 1 mM ATP, 10 mM KCl, 5 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM PEP, 0.5 mM NADH, 1.4 μg of enzyme, 1.0 μL of LDH/PK mix (Sigma), and 50 mM bis-tris propane (pH 8.5). The pH of all buffers was adjusted at 25 °C. All reagents were incubated for 10 min at the desired temperature, and then substrate was added to a final concentration of 1 mM. The absorbance at 340 nm was recorded over 20 min on a Spectramax M5 microplate reader. Assays for activity with mevalonate 3-phosphate were conducted in a sequential manner using 0.5 mM mevalonate, 3 mM PEP, and 0.75 mM NADH. The second enzyme was added after complete conversion of mevalonate to mevalonate 3-phosphate as monitored on the microplate reader. All assay conditions included a control with direct addition of 1 mM ADP to ensure PK and LDH activities were not rate-limiting.
Free Phosphate Assays
Phosphate assays were conducted in a 96-well plate using reagents from a glycosyltransferase activity kit (R&D Systems).20 Two microliters of an enzyme reaction mixture was combined with 48 μL of doubly distilled water (ddH2O), followed by 30 μL of malachite green reagent A, 100 μL of ddH2O, and 30 μL of malachite green reagent B. After a 20 min incubation, the absorbance at 620 nm was measured on a Spectramax M5 microplate reader alongside five standards of 0.0–1.0 mM KH2PO4 prepared identically. Mevalonate pyrophosphate decarboxylase from Saccharomyces cerevisiae, prepared as described previously,15 served as a positive control for the release of free phosphate through the decarboxylation of (RS)-mevalonate pyrophosphate.
Gas Chromatography
Isoprenol was extracted from 200 μL of an enzymatic reaction mixture with 600 μL of hexanes. Two microliters of the hexane layer was injected into the GC-FID (HP5890II) instrument equipped with an HP-INNOWAX column (0.320 mm × 30 m, Agilent). The carrier gas was helium with a flow rate of 5 mL/min. The oven temperature was kept at 50 °C for 2 min, increased to 100 °C at a rate of 10 °C/min, increased to 250 °C at a rate of 25 °C/min, and finally held at 250 °C for 2 min. The inlet and detector temperatures were kept at 250 and 330 °C, respectively. The isoprenol concentration was determined by comparison to authentic standards.
Product Identification by NMR
Enzymatic reactions for NMR were conducted in a 250 μL volume using 1.4 μg of enzyme, 20 mM (R)-mevalonate, 20 mM PEP, 1 mM ATP, 1 mM MgCl2, and 10 μL of LDH/PK enzyme mix (Sigma), which was diluted and reconcentrated to remove glycerol. The pH was adjusted to 7.5 at 25 °C using 1.0 M KOH with 20 mM PEP serving as a phosphate buffer in addition to being a substrate for pyruvate kinase. The reaction mixtures were incubated for 6 h at 42 °C. The completed 250 μL reaction mixture was diluted to 500 μL with 99.9% D2O from Cambridge Isotope Laboratories and aliquoted into an NMR tube. All spectra were acquired at ambient temperature on a 500 MHz Bruker AV500 spectrometer equipped with a cryoprobe. Data were processed using Topspin version 3.1. For the spectra of mevalonate 3,5-bisphosphate, the PEP concentration in the enzymatic reaction mixture was doubled to 40 mM to allow for full conversion of 20 mM (R)-mevalonate to mevalonate 3,5-bisphosphate.
Product Identification by ESI Mass Spectrometry
Negative ion electrospray mass spectrometry data were collected with a Waters LCT Premier XE time-of-flight instrument controlled by MassLynx version 4.1. Samples from the NMR tubes were transferred to GC vials and injected into the multimode ionization source with a Waters Acquity UPLC system. The flow injection solvent was a 50/50 MeOH/MeCN mixture (LCMS grade, VWR Scientific), and water blanks were run between all samples. The lock mass standard for accurate mass determination was leucine enkephalin (Sigma catalog no. L9133).
Biochemical Characterization
The optimal pH for the enzymes was determined by kinase assays in 0.5 pH unit increments ranging from pH 6.5 to 9.5 using 50 mM bis-tris propane. The optimal temperature of the enzymatic assay was determined in 5 °C increments ranging from 25 to 60 °C. We were unable to obtain direct kinetic measurements above 60 °C because it is the maximal temperature of the Spectramax M5 microplate reader. To complement this data, the temperature stability was assayed by incubating 30 μL of 0.170 mg/mL Ta1305 or 0.067 mg/mL Ta0762 for 1 h at 30–90 °C in 4–6 °C increments using a thermocycler gradient (Eppendorf ProS PCR machine). Kinetic measurements for Ta1305 were taken at pH 8.5 and 55 °C with 1.4 μg of enzyme over a range of 0.03–4.00 mM (R)-mevalonate. Ta0762 kinetic measurements were performed at pH 8.0 and 60 °C with 0.5 μg of enzyme over the range of 0.03–1.00 mM mevalonate 3-phosphate (produced enzymatically). kcat values were not corrected for temperature because the temperature of each experiment is indicated.
Results
Ta1305 Is Active on Mevalonate
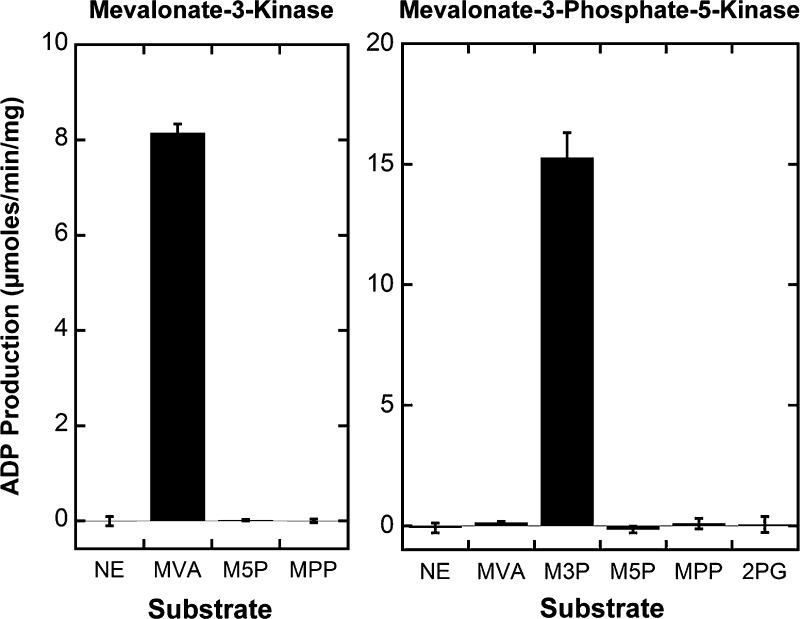
Three genes from T. acidophilum annotated as mevalonate pyrophosphate decarboxylases in GenBank,17ta1305, ta0893, and ta0461, were cloned and expressed in E. coli. Known mevalonate pyrophosophate decarboxylases hydrolyze ATP to ADP as part of their mechanism21 and can therefore be conveniently studied using standard kinase assays.19 No activity was detected in a kinase assay using the expected substrate, 1 mM (RS)-mevalonate pyrophosphate. We therefore retested the three enzymes with mevalonate and mevalonate 5-phosphate. To our surprise, Ta1305 showed significant ATP consumption in the presence of 1 mM (RS)-mevalonate (Figure 2). No activity was detected with any other substrate or enzyme combination.
Figure 2.
Enzyme specificity. The enzymes were incubated with the following substrates each at 1.0 mM: mevalonate (MVA), mevalonate 5-phosphate (M5P), mevalonate 5-pyrophosphate (MPP), mevalonate 3-phosphate (M3P), and 2-phosphoglycerate (2PG). A no enzyme control (NE) was also performed. ATP (1.0 mM) was included, and ADP production was monitored at 55 °C and pH 8.5 for Ta1305 and 60 °C and pH 8.0 for Ta0762.
Ta1305 Is Not a Mevalonate-5-kinase
The only known enzyme to act on mevalonate is mevalonate-5-kinase, which phosphorylates the 5-OH group. We therefore tested if Ta1305 was a mevalonate-5-kinase using a sequential assay. Mevalonate and ATP were incubated with Ta1305, followed by addition of phosphomevalonate kinase (PMVK), which consumes ATP in the presence of mevalonate 5-phosphate. No activity was detected upon addition of PMVK. When we replaced Ta1305 with an authentic mevalonate-5-kinase from S. cerevisiae, however, robust activity was observed. Thus, Ta1305 is not a mevalonate-5-kinase.
Ta1305 Shows No Decarboxylase Activity As Suggested by Homology
The fact that Ta1305 is 18% identical with mevalonate pyrophosphate decarboxylase from S. cerevisiae(22) yet acts on mevalonate suggests that it might directly decarboxylate mevalonate. The expected product of mevalonate decarboxylation would be 3-methylbut-3-en-1-ol (isoprenol) instead of isopentenyl pyrophosphate. To test this hypothesis, we allowed the reaction to go to completion from 1.0 mM mevalonate and analyzed the mixture for any isoprenol production by extracting with hexane followed by gas chromatography. No isoprenol was detected, even after allowing the reaction mixture to incubate for 48 h at 37 °C to promote spontaneous decarboxylation. Positive controls made by spiking the enzymatic reaction mixture with authentic isoprenol indicated that we would have detected isoprenol production as low as 0.01 mM.
Ta1305 Is a Kinase
While the Ta1305 enzyme consumed ATP in the presence of mevalonate, the fate of the phosphate remained unclear. To confirm that this enzyme does not decarboxylate mevalonate and release free phosphate, we assayed the reaction mixture for free phosphate after completion of the reaction using 1 mM ATP and 1 mM (RS)-mevalonate.20 We found no detectable free phosphate, suggesting that the substrate is phosphorylated. The same experiment utilizing authentic mevalonate pyrophosphate decarboxylase from S. cerevisiae as a positive control and its substrate, (RS)-mevalonate pyrophosphate, yielded 0.57 mM free phosphate as expected for the racemic substrate mixture.
Phosphorylation of mevalonate was further verified by ESI mass spectrometry. High-resolution negative ion electrospray ionization mass spectra of the reaction products were recorded. We observed a mass of m/z 227.0313, which is within m/z 0.00008 of the mass expected for a phosphorylated mevalonate [C6H13O7P – H]− (Figure S1 of the Supporting Information). These results suggest that Ta1305 phosphorylates mevalonate in an ATP-dependent manner.
Ta1305 Generates Mevalonate 3-Phosphate
The results so far indicate that Ta1305 is a mevalonate kinase, but there are three potential sites of phosphorylation on mevalonate: the 3-OH group, 5-OH group, and the carboxylate. To identify the site of phosphorylation, we used 13C NMR. While mevalonate alone produces six single peaks in its 13C NMR spectrum, coupling of the carbons to a phosphate will generate doublets for any carbon within three bonds of a 31P atom.23 The three possible phosphorylated products of Ta1305 would each generate a unique set of doublets, which allows for positive identification of the phosphorylated species.
To prepare samples for natural abundance NMR analysis, we wanted to keep the ATP concentrations low to simplify the NMR spectra. We therefore regenerated ATP in situ from PEP using pyruvate kinase. In this manner, ATP is kept at a very low concentration and recycled, leaving only contributions from the much simpler spectrum of PEP. A spectrum of the no enzyme control sample [i.e., 10 mM (R)-mevalonate] showed six singlet peaks as expected (Figure 3A), and the carbons were assigned with the guidance of computationally predicted spectra from the Human Metabolome Database24 and ChemNMR Pro version 13.0.25 Another control sample employing yeast mevalonate-5-kinase showed splitting at carbons 4 and 5 as expected for mevalonate 5-phosphate (Figure 3B). The product of Ta1305 activity, however, showed splitting at carbons 2–4 and 6, which is consistent with phosphorylation at the 3-OH position (Figure 3C). Thus, we can annotate Ta1305 as mevolonate-3-kinase (full 13C NMR spectra are available as Figures S2–S4 of the Supporting Information).
Figure 3.
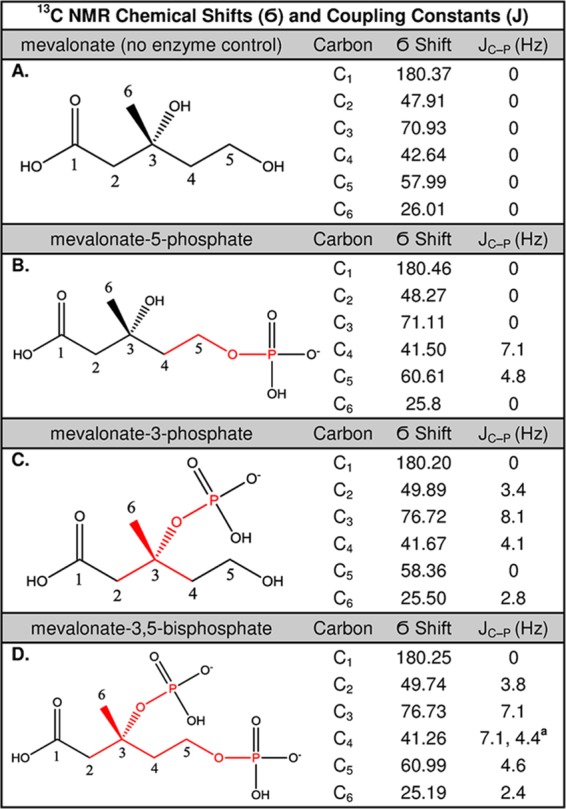
13C NMR of completed enzymatic reactions (10 mM products) in 50% D2O. The chemical shifts of the six mevalonate carbons are listed along with the J couplings from 31P (red): (A) no enzyme, (B) yeast mevalonate-5-kinase, (C) Ta1305 activity, and (D) Ta1305 and Ta0762 activity. Reactions were monitored to >96% completion before spectra were acquired. Full spectra are shown in Figures S2–S4 and S6 of the Supporting Information.
Identification of an Enzyme That Acts on Mevalonate 3-Phosphate
To identify the next step in a possible new mevalonate pathway, we screened enzymes that might further phosphorylate the newly identified metabolite, mevalonate 3-phosphate. Seven genes annotated in GenBank as kinases from T. acidophilum were cloned, and the proteins were expressed and purified. We chose two putative kinase genes found near ta1305 in the genome (ta1304 and ta1307), three genes that were homologous to those of the mevalonate kinase family (ta0344, ta0436, and ta0546), and two additional genes that were annotated as small molecule kinases (ta0762 and ta0364) but, to the best of our knowledge, had not been characterized.
We screened the putative kinases for their ability to phosphorylate mevalonate 3-phosphate using a sequential kinase assay. We first used Ta1305 to generate mevalonate 3-phosphate from mevalonate in situ (monitored by ADP production). After the reaction had reached completion, we added the second test enzyme. Of the seven enzymes tested, only Ta0762 showed activity with mevalonate 3-phosphate (Figure 2). No activity was detected with mevalonate, mevalonate 5-phosphate, or mevalonate pyrophosphate. Ta0762 had been computationally annotated as 2-phosphoglycerate kinase, though no activity was detected with 2-phosphoglycerate (Figure 2). This result suggests Ta0762 is misannotated and acts in a pathway directly after mevalonate-3-kinase. To determine if Ta0762 is a kinase, we conducted a free phosphate assay with 2.0 mM ATP and 1.0 mM mevalonate in a sequential enzymatic reaction. After monitoring the reaction until it reached completion on the microplate reader, we detected only 0.02 mM free phosphate, suggesting mevalonate 3-phosphate is also phosphorylated. To confirm the production of a doubly phosphorylated product, we again employed ESI mass spectrometry. The reaction product had a mass of m/z 307.0016, which is within m/z 0.0031 of the mass expected for mevalonate diphosphate [C6H14O10P2 – H]− (Figure S5 of the Supporting Information).
Ta0762 Generates Mevalonate 3,5-Bisphosphate
13C NMR was employed to determine the position of phosphorylation by Ta0762 using the same procedure used to identify mevalonate 3-phosphate. Splitting was observed at carbons 2–6, which is consistent with phosphates attached to the 3-OH and 5-OH positions simultaneously (Figure 3D, full spectrum in Figure S6 of the Supporting Information). In addition, carbon 4 showed a doublet of doublets pattern consistent with contributions from 31P atoms bonded in both the 3-OH and 5-OH positions simultaneously. The observed spectrum is consistent with the structure of mevalonate 3,5-bisphosphate.
Biochemical Characterization of Mevalonate-3-kinase and Mevalonate-3-phosphate-5-kinase
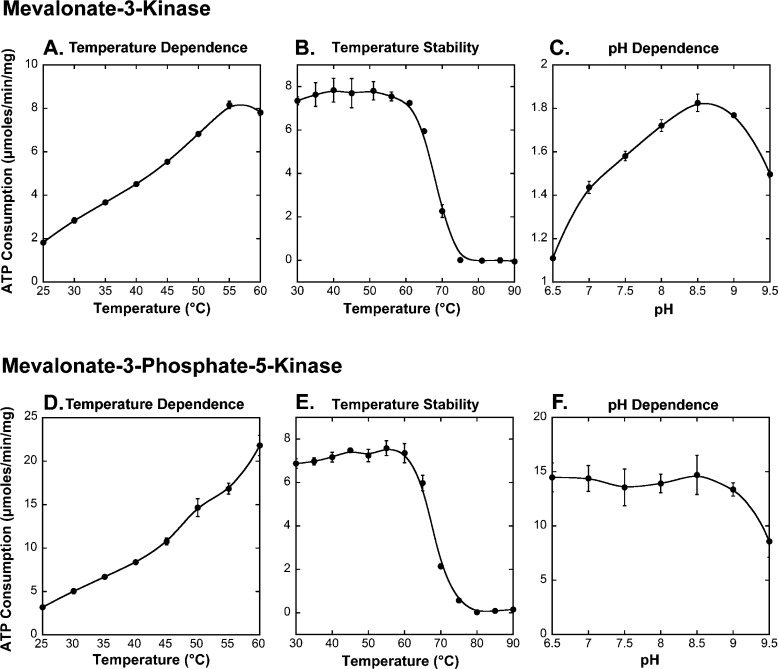
To characterize the kinetic parameters of the new enzymes, we first determined the optimal pH and temperature for each enzyme. As shown in Figure 4, mevalonate-3-kinase performs optimally at pH 8.5 and 55 °C. Mevalonate-3-phosphate-5-kinase performs optimally at 60 °C but does not demonstrate significant pH dependence in the pH range of 6.5–9.0. Both enzymes are quite stable, retaining more than 95% of their activity after incubation at 60 °C for 1 h. Under optimal conditions with (R)-mevalonate as a substrate, mevalonate-3-kinase was found to have a Km of 97 ± 6 μM and a kcat of 5.0 ± 0.1 s–1 (Figure S7 of the Supporting Information), which are comparable to those of mevalonate-5-kinase from the archeon Methanosarcina mazei (Km of 68 ± 4 μM and kcat of 4.3 ± 0.2 s–1).26 Mevalonate-3-phosphate-5-kinase is significantly faster with a kcat of approximately 9.0 s–1. We were unable to accurately determine the Km for mevalonate-3-phosphate-5-kinase, however, because the substrate is not commercially available and the detection limit of our assay is 30 μM substrate. Nevertheless, the enzyme was still at Vmax when it was assayed with 60 μM mevalonate 3-phosphate, indicating that the Km is well below 60 μM. For both enzymes, ATP was confirmed to be at saturation because for both enzymes, increasing the ATP concentration from 1 to 5 mM saw no change in the observed rates.
Figure 4.
Biochemical characterization of mevalonate-3-kinase (A–C) and mevalonate-3-phosphate-5-kinase (D–F). (A) Activity at pH 8.5 over a temperature range of 25–60 °C in 5 °C increments. (B) Residual activity after the sample had been incubated at the indicated temperature for 1 h and then assayed at 55 °C and pH 8.5. (C) Activity at 25 °C over a pH range of 6.5–9.5 in pH increments of 0.5. (D) Activity at pH 8.0 over a temperature range of 25–60 °C in 5 °C increments. (E) Residual activity after the sample had been incubated at the indicated temperature for 1 h and then assayed at 60 °C and pH 8.0. (F) Activity at 60 °C over a pH range of 6.5–9.5 in pH increments of 0.5. Curve fits in panels A–F were generated by a Stineman function34 and included simply to highlight data trends.
Discussion
We propose the two enzymes described here constitute part of a novel mevalonate pathway in T. acidophilum that splits at mevalonate and rejoins the known archaeal mevalonate pathway at isopentenyl phosphate (Figure 1, red arrows). In T. acidophilum, there is strong evidence of the presence of acetoacetyl-CoA thiolase, HMG-CoA synthase, HMG-CoA reductase, isopentenyl phosphate kinase, and isopentenyl pyrophosphate isomerase based on homology10 and experiment.27,28 However, no homologues have been identified for mevalonate-5-kinase or phosphomevalonate kinase in this organism. Most surprising is the absence of mevalonate-5-kinase because it is the only enzyme previously known to act on mevalonate. We propose that T. acidophilum contains a novel pathway in which mevalonate-3-kinase phosphorylates mevalonate at the 3-OH position, followed by mevalonate-3-phosphate-5-kinase, which phosphorylates the 5-OH position, and finally an unidentified decarboxylase converts mevalonate 3,5-bisphosphate to isopentenyl phosphate (Figure 1, red arrows).
The proposed decarboxylase enzyme would conduct the same chemical transformation as mevalonate pyrophosphate decarboxylase but in an ATP-independent manner because the substrate is already phosphorylated in the correct position for decarboxylation. We tested apparent mevalonate pyrophosphate decarboxylase homologues Ta0461 and Ta0893 for activity on mevalonate 3,5-bisphosphate, but none was detected in our hands. Both enzymes expressed to inclusion bodies, and after many attempts, we were unable to refold these proteins in high yield; therefore, it remains possible that one of them is the missing decarboxylase. We also considered the possibility of an alternative pathway in which IP kinase acts directly on mevalonate 3,5-bisphosphate. When we incubated mevalonate 3,5-bisphosphate with IP kinase, however, we saw no release of phosphate (decarboxylation) or ATP consumption (kinase activity).
The observation of stable tertiary phosphorylated mevalonate is striking because the mechanism of mevalonate pyrophosphate decarboxylase is thought to proceed through a transient tertiary phosphorylated intermediate.30,31 In a previous mechanistic study of mevalonate pyrophosphate decarboxylase, a heavy oxygen atom was incorporated into mevalonate pyrophosphate at the 3-OH position, and then after ATP-dependent conversion to IPP, the heavy oxygen was detected in free phosphate.30 This suggests a simple mechanism in which the γ-phosphate from ATP is transferred to the 3-OH position of mevalonate pyrophosphate, activating the substrate for decarboxylation. The current literature predicts this tertiary phosphorylated molecule is inherently unstable and falls apart with concomitant decarboxylation.32,33 Our observation of mevalonate 3-phosphate and mevalonate 3,5-bisphosphate as stable metabolites at 55 °C suggests that the decarboxylation step requires enzyme catalysis, however.
To shed light on the reason why mevalonate-3-kinase does not function as a decarboxylase, the active site residues of five bacterial mevalonate pyrophosphate decarboxylases (MDCs) were aligned with mevalonate-3-kinase from T. acidophilum. We focused on nine active site residues shown to interact with mevalonate pyrophosphate in a MDC crystal structure by Barta et al.33 Only four of nine highly conserved residues are preserved in mevalonate-3-kinase (Figure S8 of the Supporting Information). It is likely that these five nonconserved residues (L19, L20, I23, D190, and T276) contribute to the loss of decarboxylase activity.
Because of the apparently low reliability of predicting function through sequence homology, we cannot tell which organisms have a true copy of mevalonate-3-kinase and which have the classical mevalonate pyrophosphate decarboxylase. Sequence homology obtained through a protein BLAST search35 revealed five homologues of mevalonate-3-kinase that were >30% identical. All five are annotated as “mevalonate pyrophosphate decarboxylase” or “hypothetical protein” and belong to the order Thermoplasmatates: Thermoplasma volcanium (67%), Thermoplasmatales archeon (54%), Ferroplasma sp. Type II (45%), Ferroplasma acidarmanus (42%), and Picrophilus torridus (39%). For comparison, the level of sequence identity between mevalonate-3-kinase and classical yeast MDC is 19%.
Taken together, we have identified two novel enzymes, mevalonate-3-kinase and mevalonate-3-phosphate-5-kinase, which act sequentially in a putative alternate mevalonate pathway in T. acidophilum. Full confirmation of this pathway requires identification of a missing mevalonate 3,5-bisphosphate decarboxylase. Our findings raise important questions about the mechanism of mevalonate pyrophosphate decarboxylases because tertiary phosphorylated mevalonate species clearly exist as stable metabolites.
Acknowledgments
We thank members of the Bowie lab for critical reading of the manuscript and Dr. Greg Khitrov (UCLA Molecular Instrumentation Center, Mass Spectrometry Facility, Department of Chemistry) for expertise in the acquisition and analysis of GC–MS data.
Glossary
Abbreviations
- IPP
isopentenyl pyrophosphate
- DMAPP
dimethylallyl pyrophosphate
- PMVK
phosphomevalonate kinase
- MDC
mevalonate pyrophosphate decarboxylase
- PEP
phosphoenolpyruvate
- LDH
lactate dehydrogenase
- PK
pyruvate kinase
- Ni-NTA
nickel-nitrilotriacetic acid
- GC-FID
gas chromatography with flame ionization detection
- UPLC
ultra performance liquid chromatography
- ESI
electrospray ionization
- FPLC
fast protein liquid chromatography
- NMR
nuclear magnetic resonance
- PCR
polymerase chain reaction
- LB
Luria broth
- IPTG
isopropyl β-d-1-thiogalactopyranoside.
Supporting Information Available
Full 13C NMR spectra, ESI mass spectroscopy data, and a Michaelis–Menten curve for mevalonate-3-kinase (Figures S1–S7). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
The work was supported by U.S. Department of Energy Grant DE-FC02-02ER63421 to J.U.B., and J.M.V. received support from the National Institutes of Health Chemistry Biology Interface Training Program (National Institute of General Medical Sciences Grant 5T32GM008496). Use of the Waters LCT Premier XE time-of-flight instrument was supported by Grant S10-RR025631 from the National Center for Research Resources. NMR experiments were supported by the National Science Foundation via Equipment Grant CHE-1048804.
Funding Statement
National Institutes of Health, United States
Footnotes
A doublet of doublets was seen at a JC4–P1 of 7.1 and a JC4–P2 of 4.4. All other splittings are simple doublets.
Supplementary Material
References
- Reiling K. K.; Yoshikuni Y.; Martin V. J. J.; Newman J.; Bohlmann J.; Keasling J. D. (2004) Mono and diterpene production in Escherichia coli. Biotechnol. Bioeng. 87, 200–212. [DOI] [PubMed] [Google Scholar]
- Holstein S. A.; Hohl R. J. (2004) Isoprenoids: Remarkable Diversity of Form and Function. Lipids 39, 293–309. [DOI] [PubMed] [Google Scholar]
- Summons R. E.; Jahnke L. L.; Hope J. M.; Logan G. A. (1999) 2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature 400, 554–557. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L.; Brown S. B. (1990) Regulation of the mevalonate pathway. Nature 343, 425–430. [DOI] [PubMed] [Google Scholar]
- Dewick P. M. (1997) The biosynthesis of C5–C25 terpenoid compounds. Nat. Prod. Rep. 19, 111–144. [DOI] [PubMed] [Google Scholar]
- Katsuki H.; Bloch K. (1967) Studies on the biosynthesis of ergosterol in yeast. Formation of methylated intermediates. J. Biol. Chem. 242, 222–227. [PubMed] [Google Scholar]
- Boucher Y. (2007) Lipids: Biosynthesis, function, and evolution, in Archaea. Molecular and Celluar Biology (Cavicchioli R., Ed.) pp 341–353, ASM Press, Washington, DC. [Google Scholar]
- Koga Y.; Morii H. (2005) Recent advances in structural research on ether lipids from archaea including comparative and physiological aspects. Biosci., Biotechnol., Biochem. 69, 2019–2034. [DOI] [PubMed] [Google Scholar]
- Faisal K.; Hospital S. (1990) Physicochemical characterization of tetraether lipids from Thermoplasma acidophilum. V. Evidence for the existence of a metastable state in lipids with acyclic hydrocarbon chains. Biochim. Biophys. Acta 1024, 54–60. [DOI] [PubMed] [Google Scholar]
- Boucher Y.; Kamekura M.; Doolittle W. F. (2004) Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol. Microbiol. 52, 515–527. [DOI] [PubMed] [Google Scholar]
- Lombard J.; Moreira D. (2011) Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol. Biol. Evol. 28, 87–99. [DOI] [PubMed] [Google Scholar]
- Smit A.; Mushegian A. (2000) Biosynthesis of isoprenoids via mevalonate in archaea: The lost pathway. Genome Res. 10, 1468–1484. [DOI] [PubMed] [Google Scholar]
- Dellas N.; Thomas S. T.; Manning G.; Noel J. P. (2013) Discovery of a metabolic alternative to the classical mevalonate pathway. eLife 2, e00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannice J. C.; Skaff D. A.; Keightley A.; Addo J.; Wyckoff G. J.; Miziorko H. M. (2013) Identification in Haloferax volcanii of Phosphomevalonate Decarboxylase and Isopentenyl Phosphate Kinase as Catalysts of the Terminal Enzymatic Reactions in an Archaeal Alternate Mevalonate Pathway. J. Bacteriol. 196, 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korman T. P.; Sahachartsiri B.; Li D.; Vinokur J. M.; Eisenberg D.; Bowie J. U. (2014) A Synthetic Biochemistry System for the in vitro Production of Isoprene from Glycolysis Intermediates. Protein Sci. 23, 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A.; Graml W.; Santos-Martinez M. L.; Koretke K. K.; Volker C.; Mewes H. W.; Frishman D.; et al. (2000) The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407, 508–513. [DOI] [PubMed] [Google Scholar]
- Benson D. A.; Clark K.; Karsch-Mizrachi I.; Lipman D. J.; Ostell J.; Sayers E. W. (2014) GenBank. Nucleic Acids Res. 42(Database Issue), D32–D37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G.; Young L.; Chuang R.-Y.; Venter J. C.; Hutchison C. A.; Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Technikova-Dobrova Z.; Sardanelli A. M.; Papa S. (1991) Spectrophotometric determination of functional characteristics of protein kinases with coupled enzymatic assay. FEBS Lett. 292, 69–72. [DOI] [PubMed] [Google Scholar]
- Wu Z. L.; Ethen C. M.; Prather B.; Machacek M.; Jiang W. (2010) Universal phosphatase coupled glycosyltransferase assay. Glycobiology 21, 727–733. [DOI] [PubMed] [Google Scholar]
- Jabalquinto A. M.; Alvear M.; Emil E. C. (1988) Physiological aspects and Mechanism of Action of Mevalonate 5-Diphosphate Decarboxylase. Comp. Biochem. Physiol. 90, 671–677. [DOI] [PubMed] [Google Scholar]
- Krepkiy D.; Miziorko H. M. (2004) Protein Sci. 13, 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson V. J.; Woodside A. B.; Neal T. R.; Stremler K. E.; Muehlbacher M.; Poulter C. D. (1986) Phosphorylation of Isoprenoid Alcohols. J. Org. Chem. 51, 4768–4779. [Google Scholar]
- Wishart D. S.; Knox C.; Guo A. C.; Eisner R.; Young N.; Gautam B.; Hau D. D.; et al. (2009) HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 37(Database Issue), D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultra, ChenDraw 11.0, Chemical Structure Drawing Standard, (2012) Cambridge Soft Corp.: Cambridge, MA. [Google Scholar]
- Primak Y. A.; Du M.; Miller M. C.; Wells D. H.; Nielsen A. T.; Weyler W.; Beck Z. Q. (2011) Characterization of a feedback-resistant mevalonate kinase from the archaeon Methanosarcina mazei. Appl. Environ. Microbiol. 77, 7772–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochowski L. L.; Xu H. M.; White R. H. (2006) Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. J. Bacteriol. 188, 3192–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Poulter C. D. (2010) Characterization of thermophilic archaeal isopentenyl phosphate kinases. Biochemistry 49, 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg M.; Yuan C.; de Waard A.; Bloch K. (1962) On the Formation of Isopentenyl Pyrophosphate. Biochemistry 1, 182–188. [DOI] [PubMed] [Google Scholar]
- Voynova N. E.; Fu Z.; Battaile K. P.; Herdendorf T. J.; Kim J.-J. P.; Miziorko H. M. (2008) Human mevalonate diphosphate decarboxylase: Characterization, investigation of the mevalonate diphosphate binding site, and crystal structure. Arch. Biochem. Biophys. 480, 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byres E.; Alphey M. S.; Smith T. K.; Hunter W. N. (2007) Crystal structures of Trypanosoma brucei and Staphylococcus aureus mevalonate diphosphate decarboxylase inform on the determinants of specificity and reactivity. J. Mol. Biol. 371, 540–553. [DOI] [PubMed] [Google Scholar]
- Barta M. L.; Skaff D. A.; McWhorter W. J.; Herdendorf T. J.; Miziorko H. M.; Geisbrecht B. V. (2011) Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis. J. Biol. Chem. 286, 23900–23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stineman R. W. (1980) A Consistently Well Behaved Method of Interpolation. Creative Computing 6, 54–57. [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.