Abstract
K-ras is involved in the EGFR pathway that regulates cell survival, motility and proliferation, as well as angiogenesis and metastasis. It is also essential for carcinogenesis. The K-ras mutation status can be used to predict the therapeutic efficacy of targeted drugs such as cetuximab. The aim of this study was to compare K-ras mutation in different types of cancer. Nested and COLD-PCR were used to detect K-ras mutations. The Chi-squared test was used for statistical analysis. In this study, the total K-ras mutation frequency was found to be 9.09, 18.61 and 6.67% in lung, colorectal and gastric cancer, respectively. Similar K-ras mutation frequencies were detected among sample types and genders for lung and gastric cancer, with the exception of colorectal cancer. However, age had no impact on the K-ras mutation rates.
Keywords: K-ras mutation, lung cancer, colorectal cancer, gastric cancer, COLD-PCR
Introduction
Cancer is a global disease with a high incidence of mortality, having caused 7.6 million mortalites in 2008 alone. Over the past 10 years, there has been a decreasing trend in mortality due to cancer, allowing for the prevention of ~1.18 million cancer-related mortalities (1,2).
Lung, colorectal and gastric cancer are the leading cancer types in terms of occurrence and severity: lung cancer is the most common cancer worldwide and the first leading cause of cancer mortality, colorectal cancer is the third most common cancer worldwide and the fourth leading cause of cancer-related mortality, while gastric cancer is the fourth most common cancer worldwide and the third leading cause of cancer-related mortality (1). The incidence of cancer has seen a steady decline in males while it has remained stable in females, resulting in a gradual decrease in the overall mortality rate with regard to cancer (2). Thus NCCN guidelines suggest that the K-ras mutation be detected prior to applying medication such as cetuximab.
Materials and methods
Patients
Clinical samples were obtained from 100 hospitals in China, including 131 tissue samples, 51 plasma samples, and 5 pleural and ascites samples for lung cancer; 445 tissue and 60 plasma samples for colorectal cancer; and 126 tissue and 9 plasma samples for gastric cancer. Approval for this study was obtained from the Shanghai Clinical Research Center Ethics Committee. All patients participating in this study provided written informed consent.
Tissue samples were stored and transported under controlled temperatures, while plasma, and pleural and ascites samples were transported on ice packs. The following materials were purchased: Taq DNA polymerase (Takara Biotechnology Co., Ltd., Dalian, China), dNTP (Shi Ze Biotechnology Co., Ltd., Shanghai, China), the DNA extraction kit (DN10, Aidlab Biotechnologies Co., Ltd., Beijing, China), PCR instrument (EDC-810, Eastwin Biotechnology Co., Ltd., Beijing, China), BioSafe Centrifuge Systems (L420, Xiangyi LXJ Centrifuge Instruments Co., Ltd.).
Methods
Nested and COLD-PCR were used to detect the K-ras mutations. Regular PCR was used to amplify the 465-bp outer product. The primers used were: forward, 5′-GTCGATGGAGG AGTTTGTAAATGAAGT-3′ and reverse 5′-TTCAGATAACTTAACTTTCAGCATAATTATCTTG-3′. This was followed by 10 μl PCR reaction mixture including 0.25 mM dNTP, 0.5 μM primers, 0.5 units Taq DNA polymerase and 10 ng DNA template. The PCR program was conducted under the following conditions: 3 min at 95°C for 1 cycle, 32 amplification cycles for 30 sec at 94°C, 30 sec in 57°C, and 30 sec at 72°C, and maintained for 5 min at 72°C. COLD-PCR was used to amplify the 155-bp inner product. The primers used were: forward, 5′-GTCACATTTT CATTATTTTTATTATAAGG-3′ and reverse 5′-TTTACCTCTATTGTTGGATCATATTC-3′. This was followed by 50 μl PCR reaction mixture including 0.25 mM dNTP, 0.5 μM primers, 0.5 units Taq DNA polymerase and 1 μl outer PCR product. The PCR program was conducted under the following conditions: 3 min at 95°C for 1 cycle, 40 amplification cycles for 30 sec at 80°C, 30 sec in 58°C, 30 sec at 72°C, followed by 15 cycles for 30 sec at 94°C, 30 sec at 58°C, and 30 sec at 72°C, maintained for 5 min at 72°C.
The Chi-squared test was used for statistical analysis. P<0.05 was considered statistically significant.
Results
The K-ras mutation frequency was detected in lung cancer (Table I), colorectal cancer (Table II) and gastric cancer (Table III). COLD-PCR was used to detect the K-ras mutations. Fig. 1 shows the representative results, which showed the G12D (GGT>GAT) mutation.
Table I.
The mutation frequencies of K-ras gene in different sample types, genders and age groups of lung cancer patients detected with COLD-PCR and sequencing.
| Mutation frequency (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Type of samples | Gender | Age (years) | |||||||
|
|
|
|
|||||||
| Amino acid change | K-ras mutation | Plasma N=51 |
Tumor tissue N=131 |
Pleural and ascites N=5 |
Male N=123 |
Female N=64 |
Youth (25–44) N=9 |
Middle age (45–59) N=72 |
Elderly (60–90) N=106 |
| G12S | GGT>AGT | ||||||||
| G12R | GGT>CGT | ||||||||
| G12C | GGT>TGT | 2 (1.53) | 2 (1.63) | 1 (1.39) | 1 (0.94) | ||||
| G12D | GGT>GAT | 5 (3.82) | 1 (20) | 4 (3.25) | 2 (3.13) | 1 (1.39) | 5 (4.72) | ||
| G12A | GGT>GCT | ||||||||
| G12V | GGT>GTT | 4 (3.05) | 2 (1.63) | 2 (3.13) | 1 (1.39) | 3 (2.83) | |||
| G13S | GGC>AGC | ||||||||
| G13R | GGC>CGC | ||||||||
| G13C | GGC>TGC | ||||||||
| G13D | GGC>GAC | 1 (1.96) | 4 (3.05) | 3 (2.44) | 2 (3.13) | 1 (11.11) | 2 (2.78) | 2 (1.89) | |
| G13A | GGC>GCC | ||||||||
| G13V | GGC>GTC | ||||||||
| Q61K | CAA>AAA | ||||||||
| Q61L | CAA>CTA | ||||||||
| Q61H | CAA>CAT | ||||||||
| Total (%) | 1.96 | 11.45 | 20 | 8.94 | 9.38 | 11.11 | 6.94 | 10.38 | |
Table II.
The mutation frequencies of K-ras gene in different sample types, genders, and age groups of colorectal cancer patients detected with COLD-PCR and sequencing.
| Mutation frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Type of samples | Gender | Age (years) | |||||||
|
|
|
|
|||||||
| Amino acid change | K-ras mutation | Plasma N=60 |
Tumor tissue N=445 |
Pleural and ascites N=0 |
Male N=294 |
Female N=211 |
Youth (25–44) N=60 |
Middle age (45–59) N=195 |
Elderly (60–90) N=250 |
| G12S | GGT>AGT | 2 (0.45) | 2 (0.95) | 1 (0.51) | 1 (0.4) | ||||
| G12R | GGT>CGT | 1 (0.22) | 1 (0.34) | 1 (0.4) | |||||
| G12C | GGT>TGT | 5 (1.12) | 3 (1.02) | 2 (0.95) | 1 (0.51) | 4 (1.6) | |||
| G12D | GGT>GAT | 1 (1.67) | 35 (7.87) | 14 (4.76) | 22 (10.43) | 3 (5) | 12 (6.15) | 21 (8.4) | |
| G12A | GGT>GCT | 4 (0.90) | 3 (1.02) | 1 (0.47) | 2 (1.03) | 2 (0.8) | |||
| G12V | GGT>GTT | 1 (1.67) | 14 (3.15) | 9 (3.06) | 6 (2.84) | 8 (4.10) | 7 (2.8) | ||
| G13S | GGC>AGC | 2 (0.45) | 2 (0.95) | 2 (0.8) | |||||
| G13R | GGC>CGC | 1 (0.22) | 1 (0.34) | 1 (0.51) | |||||
| G13C | GGC>TGC | 1 (0.22) | 1 (0.34) | 1 (0.4) | |||||
| G13D | GGC>GAC | 1 (1.67) | 24 (5.39) | 11 (3.74) | 14 (6.64) | 2 (3.33) | 15 (7.69) | 8 (3.2) | |
| G13A | GGC>GCC | ||||||||
| G13V | GGC>GTC | ||||||||
| Q61K | CAA>AAA | ||||||||
| Q61L | CAA>CTA | 2 (0.45) | 1 (0.34) | 1 (0.47) | 1 (0.51) | 1 (0.4) | |||
| Q61H | CAA>CAT | ||||||||
| Total (%) | 5 | 20.45 | 14.97 | 23.70 | 8.33 | 21.03 | 19.2 | ||
Table III.
The mutation frequencies of K-ras gene in different sample types, genders, and age groups of gastric cancer patients detected with COLD-PCR and sequencing.
| Mutation frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Type of samples | Gender | Age (years) | |||||||
|
|
|
|
|||||||
| Amino acid change | K-ras mutation | Plasma N=9 |
Tumor tissue N=126 |
Pleural and ascites N=0 |
Male N=92 |
Female N=43 |
Youth (25–44) N=22 |
Middle age (45–59) N=61 |
Elderly (60–90) N=52 |
| G12S | GGT>AGT | ||||||||
| G12R | GGT>CGT | ||||||||
| G12C | GGT>TGT | ||||||||
| G12D | GGT>GAT | 3 (2.38) | 3 (3.26) | 1 (4.55) | 1 (1.64) | 1 (1.92) | |||
| G12A | GGT>GCT | ||||||||
| G12V | GGT>GTT | ||||||||
| G13S | GGC>AGC | ||||||||
| G13R | GGC>CGC | ||||||||
| G13C | GGC>TGC | ||||||||
| G13D | GGC>GAC | 6 (4.76) | 4 (4.35) | 2(4.65) | 1 (4.55) | 2 (3.28) | 3 (5.77) | ||
| G13A | GGC>GCC | ||||||||
| G13V | GGC>GTC | ||||||||
| Q61K | CAA>AAA | ||||||||
| Q61L | CAA>CTA | ||||||||
| Q61H | CAA>CAT | ||||||||
| Total (%) | 0 | 7.14 | 7.61 | 4.65 | 9.09 | 4.92 | 7.69 | ||
Figure 1.
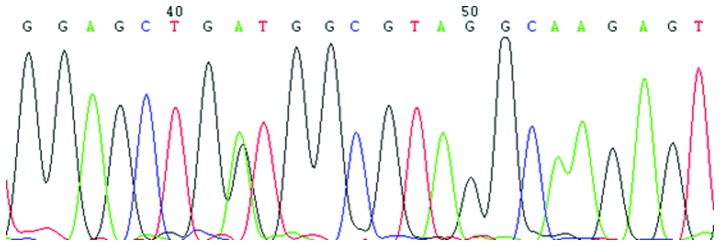
Representative sequencing chromatogram of COLD-PCR product, showing the GGT mutation to GAT at codon 12 of K-ras gene in a colorectal cancer patient.
The total K-ras mutation frequency was 9.09, 18.61 and 6.67% in lung, colorectal and gastric cancer, respectively, as detected in all types of samples which suggested that the K-ras mutations occurred more frequently in colorectal cancer than in the other two types of cancer investigated.
Of 187 lung cancer patients investigated, four mutation types were detected, including G12C (GGT>TGT), G12D (GGT>GAT), G12V (GGT>GTT) and G13D (GGC>GAC). The mutation frequency was 1.96, 11.45 and 20% in plasma, tumor tissue, and pleural and ascites samples, respectively, with no statistical significance being identified (P=0.0935). The ratio of male to female patients was 8.94 and 9.38%, respectively, which did not indicate statistical significance (P=0.9223). Similarly, the ratio for youth, middle age, and elderly patients was 11.11, 6.94 and 10.38%, respectively, which did not indicate statistical significance (P=0.7196).
Of 505 colorectal cancer patients investigated, 11 mutation types were detected, including G12S (GGT>AGT), G12R (GGT>CGT), G12C (GGT>TGT), G12D (GGT>GAT), G12A (GGT>GCT), G12V (GGT>GTT), G13S (GGC>AGC), G13R (GGC>CGC), G13C (GGC>TGC), G13D (GGC>GAC) and Q61L (CAA>CTA). The mutation frequency for plasma, and tumor tissue samples was 5 and 20.45%, respectively, indicating statistical significance (P=0.0039). The ratio for male to female patients was 14.97 and 23.70%, respectively, indicating statistical significance (P=0.0129). Similarly, the ratio for youth, middle age, and elderly patients was 8.33, 21.03 and 19.2%, respectively, which did not indicate statistical significance (P=0.0824).
Of 135 gastric cancer patients investigated, the mutation types G12D (GGT>GAT), and G13D (GGC>GAC) were detected. The mutation frequency for plasma, and tumor tissue samples was 0 and 7.14%, respectively, which did not indicate statistical significance (P=1.0000). The ratio for male to female patients was 7.61 and 4.65%, respectively, which indicated no statistical significance (P=0.7860). Similarly, the ratio for youth, middle age, to elderly patients was 9.09, 4.92 and 7.69%, respectively, which indicated no statistical significance (P=0.7425).
Of all the K-ras mutation types in lung cancer, G12C accounted for 1.07%, G12D for 3.21%, G12V for 2.14%, G13D for 2.67% of the total mutation frequency. Analysis of colorectal cancer mutations showed that, G12D accounted for 7.13%, G12V for 2.97%, G13D for 4.95%, while the remaining mutation types collectively accounted for 3.56% of the total mutation frequency (Table II). In gastric cancer patients, only two K-ras mutation types were identified, with G12D accounting for 2.22%, whereas G13D accounted for 4.44% of the total mutation frequency. Thus, G12D and G13D are the two most frequently occurring mutation types in the three types of cancer investigated.
Discussion
The mammalian ras gene family comprises H-ras, K-ras, N-ras, encoding H-ras, K-ras, N-ras proteins, respectively, with a similar structure and function. The Ras protein is located in the inner region of the cell membrane, tranforms signals from EGFR to mitogen-activated protein kinases (MAPKs), to control cell growth, proliferation, and motility, as well as metastasis and angiogenesis (3). The K-ras gene usually contains point mutations at codons 12, 13 and 61 (Tables I–III), and these mutations often activate the K-ras oncogene (4,5). The K-ras mutation status is associated with the therapeutic efficacy of EGFR-targeting monoclonal antibodies, rendering patients with K-ras mutation as not suitable for Erbitux treatment (6).
Various methods have been developed to improve detection sensitivity, such as denaturing high-performance liquid chromatography (DHPLC) (7), nested Allele-Specific Blocker (ASB-)PCR (8), PCR single-strand conformation polymorphism (SSCP) (9), restriction fragment length polymorphism (RFLP) (10), and the amplification refractory mutation system (ARMS) (11). Due to the need for simple equipment, high sensitivity, COLD-PCR (co-amplification at lower denaturation temperature-PCR) (12–15) has been widely used, it can enrich variant DNA sequences and improve detection sensitivity.
In the present study, the results showed that the mutation frequency of K-ras was different in the three types of cancer, indicating statistical significance (P=0.0001). The ratio for the variables compared was highest in colorectal cancer. Thus, detection of K-ras mutation status is more important for colorectal cancer patients when personalized medicine is involved.
The mutation frequency was not statistically significant for the different sample types for lung and gastric cancer. Therefore, plasma samples may be substituted by tissue samples when the latter are not readily available, particularly for lung cancer patients, from whom pleural and ascites samples are also feasible. However, other types of samples cannot be substituted for colorectal cancer tissues for K-ras mutation detection, considering that the detection frequency of K-ras mutations in tumor tissues is 4-fold that of plasma samples, with the difference between sample types being statistically significant for colorectal cancer patients.
For lung cancer and gastric cancer patients, the mutation frequency indicated no statistical significance for gender, although a difference was identified for colorectal cancer. The frequency for male to female was 14.97 and 23.70% (P=0.0129), respectively, suggesting the likelihood of mutation in female colorectal cancer patients as compared with their male counterparts.
Age did not affect the mutation frequency in the three types of cancer investigated, suggesting that K-ras mutation does not play a role in patient age Previously, an anticorrelation pattern of K-ras mutation status with the therapeutic effect, progression-free survival and overall survival following patient treatment with Erbitux was demonstrated (4,6). By contrast, results of other studies have shown that many patients cannot improve efficacy end-points after receiving Erbitux (16,17) without K-ras mutation detection. That finidng suggests that other key signal transduction molecules also play an important role in the downstream of Erbitux against EGFR, for example, B-raf, PIK3CA (17). Therefore, the mutation status of genes such as B-raf, and PIK3CA should be detected at the same time as the K-ras mutation status.
Acknowledgements
This study was supported by Science and Technology Development Foundation in Baoshan District, Shanghai, China (11-B-9), and Development Center Foundation for Medical Science and Technology, Ministry of Health, China (W2012FZ139).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Kiaris H, Spandidos DA. Mutations of ras genes in human tumours. International Journal of Oncology. 1995;7:413–429. [PubMed] [Google Scholar]
- 4.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 5.Bos JL, Fearon ER, Hamilton SR, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 6.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 7.Lilleberg SL, Durocher J, Sanders C, Walters K, Culver K. High sensitivity scanning of colorectal tumors and matched plasma DNA for mutations in APC, TP53, K-RAS, and BRAF genes with a novel DHPLC fluorescence detection platform. Ann NY Acad Sci. 2004;1022:250–256. doi: 10.1196/annals.1318.039. [DOI] [PubMed] [Google Scholar]
- 8.Mostert B, Jiang Y, Sieuwerts AM, et al. KRAS and BRAF mutation status in circulating colorectal tumor cells and their correlation with primary and metastatic tumor tissue. Int J Cancer. 2013;133:130–141. doi: 10.1002/ijc.27987. [DOI] [PubMed] [Google Scholar]
- 9.Abdul Murad NA, Othman Z, Khalid M, et al. Missense mutations in MLH1, MSH2, KRAS, and APC genes in colorectal cancer patients in Malaysia. Dig Dis Sci. 2012;57:2863–2872. doi: 10.1007/s10620-012-2240-2. [DOI] [PubMed] [Google Scholar]
- 10.Sinha R, Hussain S, Mehrotra R, et al. Kras gene mutation and RASSF1A, FHIT and MGMT gene promoter hypermethylation: indicators of tumor staging and metastasis in adenocarcinomatous sporadic colorectal cancer in Indian population. PLoS One. 2013;8:e60142. doi: 10.1371/journal.pone.0060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bando H, Yoshino T, Tsuchihara K, et al. KRAS mutations detected by the amplification refractory mutation system-Scorpion assays strongly correlate with therapeutic effect of cetuximab. Br J Cancer. 2011;105:403–406. doi: 10.1038/bjc.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Makrigiorgos GM. COLD-PCR: a new platform for highly improved mutation detection in cancer and genetic testing. Biochem Soc Trans. 2009;37:427–432. doi: 10.1042/BST0370427. [DOI] [PubMed] [Google Scholar]
- 13.Zuo Z, Chen SS, Chandra PK, et al. Application of COLD-PCR for improved detection of KRAS mutations in clinical samples. Mod Pathol. 2009;22:1023–1031. doi: 10.1038/modpathol.2009.59. [DOI] [PubMed] [Google Scholar]
- 14.Carotenuto P, Roma C, Cozzolino S, et al. Detection of KRAS mutations in colorectal cancer with Fast COLD-PCR. Int J Oncol. 2012;40:378–384. doi: 10.3892/ijo.2011.1221. [DOI] [PubMed] [Google Scholar]
- 15.Pennycuick A, Simpson T, Crawley D, et al. Routine EGFR and KRAS mutation analysis using COLD-PCR in non-small cell lung cancer. Int J Clin Pract. 2012;66:748–752. doi: 10.1111/j.1742-1241.2012.02961.x. [DOI] [PubMed] [Google Scholar]
- 16.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 17.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]


