Abstract
Cathepsin K, a lysosomal cysteine protease critical for bone remodeling by osteoclasts, was recently identified as the deficient enzyme causing pycnodysostosis, an autosomal recessive osteosclerotic skeletal dysplasia. To investigate the nature of molecular lesions causing this disease, mutations in the cathepsin K gene from eight families were determined, identifying seven novel mutations (K52X, G79E, Q190X, Y212C, A277E, A277V, and R312G). Expression of the first pro region missense mutation in a cysteine protease, G79E, in Pichia pastoris resulted in an unstable precursor protein, consistent with misfolding of the proenzyme. Expression of five mature region missense defects revealed that G146R, A277E, A277V, and R312G precursors were unstable, and no mature proteins or protease activity were detected. The Y212C precursor was activated to its mature form in a manner similar to that of the wild-type cathepsin K. The mature Y212C enzyme retained its dipeptide substrate specificity and gelatinolytic activity, but it had markedly decreased activity toward type I collagen and a cathepsin K–specific tripeptide substrate, indicating that it was unable to bind collagen triple helix. These studies demonstrated the molecular heterogeneity of mutations causing pycnodysostosis, indicated that pro region conformation directs proper folding of the proenzyme, and suggested that the cathepsin K active site contains a critical collagen-binding domain.
J. Clin. Invest. 103:731–738 (1999)
Introduction
Pycnodysostosis (Pycno), an autosomal recessive sclerosing skeletal dysplasia, recently was shown by a positional candidacy approach to result from the deficient activity of the lysosomal cysteine protease cathepsin K (EC 3.4.22.38; ref. 1). The disease is characterized by reduced stature, osteosclerosis, acro-osteolysis of the distal phalanges, frequent fractures, clavicular dysplasia, and skull deformities with delayed suture closure (2, 3). The fact that Pycno resulted from deficient cathepsin K activity proved the important physiological role of this lysosomal cysteine protease in bone matrix degradation and implied that inhibition of cathepsin K activity might be therapeutic for bone diseases characterized by excessive bone degradation, such as osteoporosis and certain forms of arthritis.
The cathepsin K gene, which was cloned originally from rabbit osteoclasts (4), and subsequently from several human tissues (5–8), was highly expressed in osteoclasts, the site of the Pycno pathology (9). The predicted 329-residue polypeptide sequence was highly homologous with those of cathepsins S and L (6, 7) and had the typical prepropeptide organization of cysteine proteases of the papain family, including a 15–amino acid signal sequence and a 99-residue pro piece. A putative N-glycosylation site in the pro region, which is conserved among cathepsin K genes from all species sequenced to date (10), presumably facilitates lysosomal targeting of the proenzyme via the mannose 6-phosphate receptor pathway. The catalytic triad within the active site contains active cysteine, histidine, and asparagine residues that are conserved completely among all papain family members (6).
The human cathepsin K gene has been overexpressed in the baculovirus system, and the purified recombinant enzyme's physical and kinetic properties have been determined (11, 12). Cathepsin K was synthesized as a prepropeptide of 37 kDa, and the mature enzyme was a monomeric protein with an apparent molecular mass of about 29 kDa. The enzyme had a broad bell-shaped pH-activity profile with an optimum at 6.1 and had strong collagenolytic, elastase, and gelatinase activities, exceeding those of cathepsins S or L (11). These properties are consistent with the enzyme's ability to degrade organic matrix proteins in the subosteoclastic space.
Previously, three mutations in the mature region of the cathepsin K prepropeptide were reported in four unrelated Pycno families, including a missense (G146R), a nonsense (R241X), and a stop codon (X330W) mutation (1, 13). Transient expression of the X330W allele resulted in no immunologically detectable protein, despite normal message levels (1).
In this report, seven novel cathepsin K mutations are described, comprising four mature region missense mutations, a mature nonsense mutation, and two pro region point mutations. A nonsense mutation in the mature enzyme was found in several unrelated Hispanic Pycno families and suggested a common Iberian descent. Six missense mutants, expressed in Pichia pastoris, lacked detectable residual activity, with the exception of Y212C. This mutant had residual activity in vitro against the fluorescent substrate and gelatin, but the mature Y212C enzyme had little, if any, activity toward type I collagen.
Methods
Specimen collection and DNA sequencing.
Blood samples were obtained with informed consent from eight unrelated families with one or more individuals affected with Pycno. The ethnic background of these families was as follows: northern European (two), Czech, Spanish, Portuguese (two), Indian, and Honduran. Genomic DNA was extracted from blood leukocytes using the Puregene Genomic DNA Isolation kit (Gentra Systems, Minneapolis, Minnesota). Exons 2–8 of the cathepsin K gene (14) were amplified from genomic DNA of Pycno patients by PCR, isolated, and sequenced by cycle sequencing with an ABI 377 Sequencer (Perkin-Elmer Corp., Norwalk, Conneticut, USA).
Confirmation of detected mutations.
Putative cathepsin K mutations were confirmed by PCR-based analyses of normal, obligate heterozygous, and/or affected members from each family. In addition, each putative mutation was analyzed in genomic DNAs isolated from 50–55 unrelated normal Caucasian individuals to rule out possible polymorphisms. PCR products containing the mutation were amplified from genomic DNA, digested overnight with the appropriate restriction endonuclease (New England Biolabs Inc., Beverly, Massachusetts, USA), separated by horizontal electrophoresis in 2% agarose gels, and directly viewed with ethidium bromide. The five mutations that created or destroyed restrictions sites were as follows: R241X (AvaI), Y212C (RsaI), A277E (AciI), A277V (AciI), and R312G (TaqI). For the three mutations that neither created nor destroyed convenient restriction sites, either mismatch PCR (NlaIV for G79E; PstI for Q190X) or the amplification refractory mutation system (ARMS) (K25X) was used for mutation detection (15, 16).
Expression and characterization of missense mutations.
The human procathepsin K cDNA that had been cloned in pBluescript SK(+) phagemid and mutated to eliminate the N-glycosylation site in the mature enzyme was used to generate constructs for the six Pycno missense mutations. Each point mutation was introduced individually into the cDNA using the PCR ligation method with Vent Taq polymerase (New England Biolabs Inc.), and each mutant construct was confirmed by sequencing. Unique EcoRI and NotI sites, introduced at the 5′ and 3′ ends of each amplified mutant proenzyme PCR product, were used to ligate each construct into the pPIC9K expression vector (Invitrogen Corp., Carlsbad, California, USA). The expression constructs for the wild-type and the six mutant cathepsin K (gly–) proenzymes were linearized with BglII and then electroporated into P. pastoris GS115 cells (Invitrogen Corp.). After genotype selection and phenotype screening according to the manufacturer's instructions, several clones were obtained for each of the constructs.
Clones were grown up in shaker flasks, and the liquid culture media were concentrated with a spin column (Amicon Inc., Beverly, Massachusetts, USA). The presence of the cathepsin K (gly–) proenzyme was detected by Western blot, using a rabbit polyclonal antibody raised against human cathepsin K (MS2) (provided by D. Brömme). Standard Northern analysis was performed after extraction of total RNA from lyticase-treated cells with RNazol (Tel-Test Inc., Friendswood, Texas, USA). Blots were hybridized with human cathepsin K cDNA and Saccharomyces cervisiae actin (a gift from A. Caplan, Mount Sinai School of Medicine) that had been radiolabeled with [32P]-dCTP (Amersham Pharmacia Biotech, Piscatawy, New Jersey, USA) by the random hexamer method.
To assess cathepsin K enzyme activity, the concentrated medium with the highest yield of proenzyme for each construct was selected. The mature cathepsin K (gly–) proteins were activated from their precursor forms by diluting the medium to 2.5 volumes, to a final solution containing 50 mM sodium acetate, 2.5 mM dithiothreitol, and 2.5 mM EDTA (pH 4.0); adding porcine pepsin (Sigma Chemical Co., St. Louis, Missouri, USA) to a final concentration of 20 μg/ml; and then incubating the mixture at 37°C for 30 min.
The enzymatic activity of the wild-type and mutant cathepsin K (gly–) proteins was assayed using the fluorescent substrate Z-Leu-Arg-MCA at 5 μM in an assay buffer of 100 mM sodium acetate, 2.5 mM EDTA, 2.5 mM dithiothreitol (pH 5.5). Fluorescence was detected by a luminescence spectrometer (Perkin-Elmer Corp.), and enzyme kinetics were obtained by nonlinear regression analysis using the Grafit software package (Erithacus Software Ltd., Staines, United Kingdom). To establish the pH profile for active cathepsin K enzymes, initial rates of substrate hydrolysis with 5 μM Z-Leu-Arg-MCA were determined in 100 mM sodium citrate for pH 2.8–5.6 and in 100 mM sodium phosphate for pH 5.8–9.7. The pH stability of wild-type and active mutant proteins was studied by incubating at 37°C in 100 mM sodium acetate buffer (pH 4.5 and 5.5) or in 100 mM potassium phosphate buffer (pH 6.5), containing 2.5 mM dithiothreitol and 2.5 mM EDTA. At several time points, the residual activity was determined using 5 μM Z-Leu-Arg-MCA as substrate. Substrate specificity of mutant enzymes with residual activity was determined by measuring enzyme kinetics with a panel of synthetic di- and tripeptide substrates. In addition, the mutant enzymes with residual activity against the synthetic substrates were also assayed with biologically relevant substrates. Activated wild-type and mutant enzymes were incubated at 28°C for 12 h with 0.4 mg/ml of soluble calf skin type I collagen (U.S. Biochemical Corp., Cleveland, Ohio) in 100 mM sodium acetate buffer (pH 5.5) containing 2.5 mM dithiotreitol and 2.5 mM EDTA. To measure the gelatinase activity, type I collagen was heated at 68°C for 15 min and then incubated with varying concentrations of the activated wild-type and mutant enzymes for 1 h at 28°C. Samples from the collagenase and gelatinase assays were subjected to SDS-PAGE, fixed with trichloroacetate, and viewed by Coomassie blue staining.
Determination of the processing sites of Y212C and wild-type cathepsin K enzymes.
To determine the site(s) of processing of the cathepsin K propeptides, mature Y212C and wild-type cathepsin K proteins were separated on an SDS-PAGE gel and viewed by the colloidal blue staining method (Novex, San Diego, California, USA). The bands of interest were excised and subjected to in-gel digestion with trypsin or endopeptidase Lys-C, and the resulting proteolytic mixtures were extracted as described previously (17, 18). The molecular masses of the peptide mixtures were accurately determined by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry using delayed ion extraction and ion reflection (Voyager-DE STR; Perseptive Biosystems Inc., Framingham, Massachusetts, USA). NH2- and COOH-terminal peptides were identified by comparing the measured masses with those calculated from the known protein sequence and the cleavage rules of the protease, using the software tool PepFound (19, 20). The mass accuracy, determined using internal calibration, was within 0.0001 kDa.
Results
Mutation detection and confirmation.
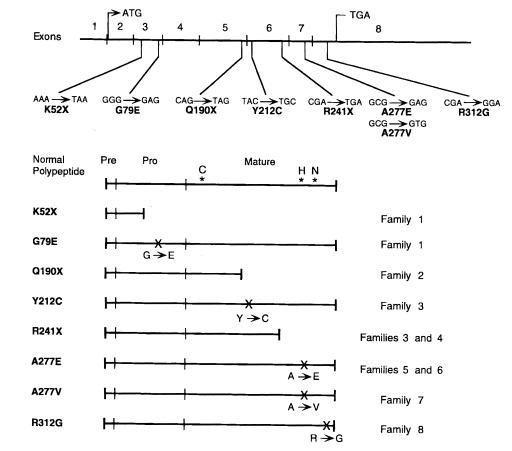
Analysis of the cathepsin K coding region and adjacent intron/exon boundaries from genomic DNAs of patients from eight unrelated Pycno families revealed eight mutations, including seven novel lesions (Fig. 1). These mutations included two pro region changes consisting of a nonsense and a missense lesion, and six mature region changes consisting of two nonsense and four missense mutations. Of note, no readily apparent genotype–phenotype correlation was identified, because all patients appeared to be similarly affected.
Figure 1.
Novel pycnodysostosis mutations in the cathepsin K gene. DNA changes to codons and their predicted effects on the cathepsin K protein are indicated above and positioned on the cathepsin K gene, which is divided into eight exons. The initiation and stop codons are indicated in exons 2 and 8, respectively. The effects of these mutations on the cathepsin K prepropeptide are shown below. The positions of the cysteine, histidine, and asparagine residues that form the active triad in the active cleft are indicated on the normalpolypeptide. The families in which individual mutations were found are indicated on the right.
To rule out the possibility that these cathepsin K base substitutions could be polymorphisms and not disease-causing mutations, alleles from ≥50 normal Caucasian individuals were analyzed for the respective lesions. Using the appropriate PCR-based assays, no K52X, G79E, Q190X, Y212C, R241X, or R312G alleles were detected among greater than 100 control alleles. In one control individual, the restriction assay for the A277E and A277V mutations produced a pattern identical to the A277E and A277V heterozygotes, but sequencing of the PCR product revealed only heteroallelism for a G→A change at nucleotide 936, which did not alter the amino acid sequence, rather than the A277E and A277V mutations, which altered nucleotide 935. Thus, the possibility that these eight coding sequence changes were polymorphisms rather than mutations was considered highly unlikely.
Expression analysis of cathepsin K missense mutations.
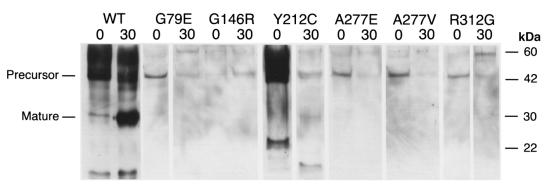
The wild-type and mutant cathepsin K proenzymes were expressed in the P. pastoris system, which has been shown previously (21) to result in high-level production and secretion into the medium (38.4 mg/l of purified active enzyme). In addition, as revealed by the completed S. cervisiae genome project, yeast strains contain no papain-like cysteine proteases, except bleomycin hydrolase, which is an aminopeptidase with no overlap in substrate specificity. Coomassie blue staining of sodium dodecyl sulfate–polyacrylamide electrophoretic gels containing aliquots of media from the G79E, G146R, Y212C, A277E, A277V, and R312G constructs revealed the presence of the expected 43-kDa bands, which were quantitatively less than the band for the wild-type protein (data not shown). Immunologically detectable proteins of the expected mass from each mutant precursor were observed (Fig. 2), but, excepting Y212C, with reduced quantities relative to the wild-type control. In addition, degradative products of smaller mass were noted in the wild-type and Y212C media. Western blot analysis of whole cell lysates revealed a similar discrepancy in cathepsin K production, excluding a failure to secrete the mutant precursors (data not shown). Only supernatant from wild-type had a band of the expected mass (30 kDa) of the mature enzyme, presumably generated by autoactivation. Northern analysis of total RNA extracted from the yeast revealed approximately equal levels of the cathepsin K transcripts in wild-type and mutant clones (data not shown), strongly suggesting instability of these mutant precursor proteins. These results were consistent with instability and/or improper folding of the mutant cathepsin K precursor proteins that were hydrolyzed by pepsin and other proteases.
Figure 2.
Immunoblot of mutant and wild-type cathepsin K proteins. Aliquots of Pichia pastoris culture supernatants, before and after 30 min of pepsin treatment (indicated as 0 and 30, respectively), were electrophoresed in SDS-PAGE and transferred to blots that were hybridized with polyclonal rabbit anti–human cathepsin K antibodies. The wild-type cathepsin K (WT) proenzyme (lane 1) was detected at a higher than expected molecular mass, which resolved after pepsin digestion (lane 2). Molecular mass standards (measured in kDa) are indicated in the right margin. There was no immunologically detectable cathepsin K protein in supernatants from pPIC9K vector–only control (data not shown). The G146R precursor protein, which was less abundant before pepsin treatment in this Western blot, was more abundant at 0 min than at 30 min in all subsequent repetitions
Incubation of the precursor proteins with pepsin resulted in reduction of the wild-type and Y212C peptides to the expected mass of the mature enzyme (Fig. 2). The amount of Y212C enzyme generated was substantially less than the quantity of initial precursor, suggesting degradation by pepsin and other proteases. For the other mutant precursors, however, incubation with pepsin resulted in loss of the precursor peptide but no emergence of a 30-kDa protein.
As anticipated from the Western analyses, the fluorescent enzyme assays revealed no detectable protease activity of the G79E, G146R, A277E, A277V, or R312G mutant proteins toward the synthetic substrate. In contrast, low levels of activity were detected for Y212C mutant protein compared with the wild-type cathepsin K, which showed activity levels and kinetics comparable to those published previously (11). To further characterize the residual activity of the Y212C mutant protein, conditioned medium from a large-scale preparation was concentrated. The percentage of residual cathepsin K activity in the Y212C mutant protein was quantitated using a stoichiometric assay of the enzyme with the irreversible cysteine cathepsin inhibitor E-64, which binds to these proteases in a 1:1 molar ratio. After normalizing the activities of the Y212C and wild-type enzymes to molarity, the Y212C protein had a hydrolysis rate with the dipeptide substrate Z-Leu-Arg-MCA that was 41% of that with the wild-type cathepsin K (kcat 0.64 ± 0.07/s vs. 1.56 ± 0.53/s). The Michaelis-Menten constants for the two enzymes were similar (Km 6.81 ± 1.66 μM vs. 6.57 ± 1.46 μM, respectively), so the catalytic efficiency (kcat/Km) of the Y212C protein was only 40% of that for the wild-type enzyme (94,000 vs. 237,400 1/M·s, respectively).
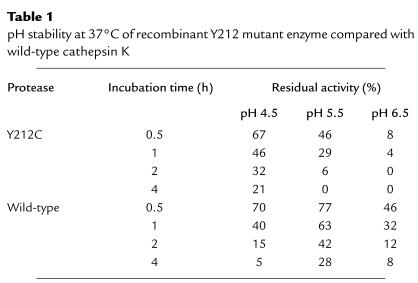
The pH-activity profiles of the Y212C and wild-type enzymes were comparable with pH optima at 6.1 and similarly broad profiles (Y212C pK1 = 4.2 and pK2 = 8.4; wild-type CTSK pK1 = 3.9 and pK2 = 8.5). Compared with wild-type enzyme, however, the Y212C activity was unstable at pH 5.5 and 6.5, although it was more stable than wild-type activity at the pH 4.5 (Table 1).
Table 1.
pH stability at 37°C of recombinant Y212 mutant enzyme compared with wild-type cathepsin K
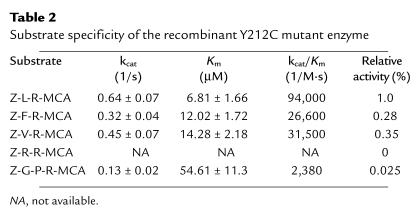
The substrate specificity of Y212C was determined using synthetic di- and tripeptide substrates (Table 2). In a pattern very similar to wild-type (11), the order of catalytic efficiency (kcat/Km) for the Y212C enzyme against four dipeptide substrates was Z-LR-MCA > Z-VR-MCA ≈ Z-FR-MCA >> Z-RR-MCA. The Y212C enzyme had minimal catalytic activity toward the tripeptide substrate Z-GPR-MCA, which has been shown to be efficiently hydrolyzed by recombinant rabbit cathepsin K (22).
Table 2.
Substrate specificity of the recombinant Y212C mutant enzyme
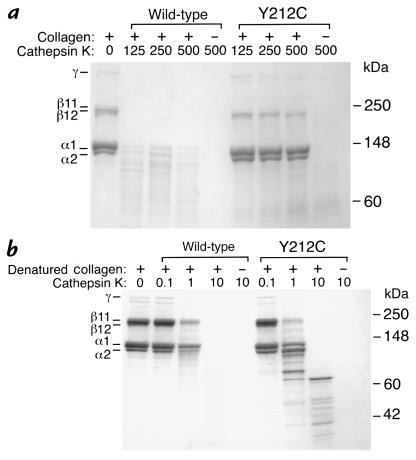
The ability of the Y212C and wild-type enzymes to degrade type I collagen was determined in vitro at pH 5.5 and 28°C. As shown in Fig. 3, there was pronounced collagen degradation by the wild-type enzyme after 12 hours, but only minimal collagenolysis by the Y212C enzyme. In contrast, the Y212C enzyme retained significant proteolytic activity against gelatin, although it did not hydrolyze the substrate as efficiently as did the wild-type activity (Fig. 3).
Figure 3.
SDS-PAGE of type I collagen (soluble calf skin collagen) after digestion with recombinant wild-type and Y212C cathepsin K enzymes. (a) Collagenase activity. Digestion of soluble calf skin collagen at 28°C and pH 5.5 for 12 h by wild-type and Y212C cathepsin K enzymes is shown. The presence (+) or absence (–) of collagen and the concentration of the cathepsin K in nM are indicated. (b) Gelatinase activity. Digestion of denatured soluble calf skin collagen by wild-type and Y212C cathepsin K enzymes is shown. The presence (+) or absence (–) of gelatin and the concentration of the cathepsin K in nM are indicated. Molecular mass standards (measured in kDa) are indicated in the right margin, and the collagen proteins are labeled in the left margin.
To ensure that the altered substrate specificity of the Y212C enzyme was not an artifact of abnormal processing of the propeptide, the NH2-terminal residues of this mutant, as well as the wild-type cathepsin K proteins, were compared. MALDI-TOF mass spectrometry of endopeptidase Lys-C digested mature proteins indicated that both the Y212C and wild-type enzymes were constituted by a mixture of enzymes with W111EGRAPD, G113RAPD, and A115PD NH2-termini. In addition, the Y212C enzymes contained peaks corresponding to NH2-termini of Y107IPEWEGRAPD and I108PEWEGRAPD, suggesting less-complete maturation of some species. Finally, MALDI-TOF analysis of a tryptic digest of the mutant and wild-type enzymes demonstrated identical traces, except for peaks of 2.664 and 2.653 kDa in the former and latter, respectively, which corresponded to the predicted 24-residue tryptic fragments containing the C212 and Y212 residues. The ragged NH2-termini were not detectable after the tryptic digest because trypsin cleaved at R112, producing very short NH2-terminal fragments that were lost during the elution and mass spectrometry.
Discussion
The seven novel cathepsin K mutations identified in eight unrelated Pycno families further document that defects in the gene cause this skeletal dysplasia and emphasize the degree of molecular genetic heterogeneity underlying this disease. Each point mutation was confirmed by PCR-based allele-specific analysis of genomic DNA from the affected individuals and heterozygous parents, and none was found among more than 100 chromosomes from unrelated normal individuals. Notably, among the 12 families studied to date (1, 13), all of the disease-causing mutations were missense and nonsense mutations that altered exonic coding sequences. In addition, two of the lesions identified here involved a missense and a nonsense mutation in the pro region of the polypeptide. A single, rare polymorphism was found at nucleotide 936 (G→A change) that did not alter the amino acid sequence.
Among the detected mutations, R241X was found in family 3 from Spain, family 4 from Portugal, a previously described Mexican-American family (1), and an extended Mexican kindred (13). The finding of the R241X mutation in four unrelated families of Iberian ancestry, and its absence in other Pycno families, suggests that this lesion may be relatively common in that geographic region. Support for this concept would require population studies to determine the frequency of the R241X allele in the Iberian Peninsula and in other European and North African populations. If the mutation is of Iberian origin, then this mutation may be relatively ancient, because its transfer to one or both Mexican families probably occurred more than 300 years ago. Alternatively, but less likely, the R241X mutations could have arisen independently on several occasions, because it occurred at a CpG dinucleotide, a known hot spot for mutations (23). Of note, the A277E mutation was detected in two families that share no apparent ethnic background (Indian and Portuguese), although Vasco de Gama first traveled around the western coast of India in 1498 (24). This mutation also occurred at a CpG dinucleotide and could have arisen independently by unrelated mutational events. Future development of intragenic cathepsin K polymorphic markers would permit further analysis of the ancestral background of the R241X and A277E mutations.
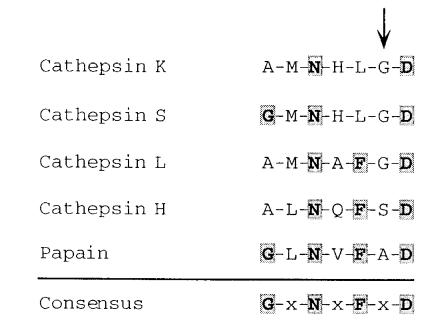
The K52X and G79E mutations are the first identified in the 99-residue pro piece region of the cathepsin K prepropolypeptide. The K52X nonsense mutation is predicted to terminate polypeptide synthesis in the pro region, thereby eliminating the entire mature enzyme. In contrast, the potential effect of the G79E missense mutation may be subtler. In general, lysosomal cysteine proteases are synthesized as prepropolypeptides. The prepeptide is the signal sequence, and the pro piece subserves several functions, including promotion of protein folding, protection of the nascent enzyme from neutral pH, trafficking of the polypeptide to the lysosome, and inhibition of protease function until the proenzyme reaches the lysosome, where intramolecular or intermolecular proteolysis will remove the propeptide and activate the enzyme. The cathepsin K pro region contains the only completely conserved potential N-glycosylation site (Asn103) for lysosomal trafficking via the mannose 6-phosphate receptor pathway. Based on homology with the closely related procathepsin L polypeptide, for which the three-dimensional structure was recently solved (25), a portion of the cathepsin K pro piece sits in the active cleft, so that the enzyme remains nonfunctional until the pro piece is clipped in the lysosome. The G79E missense mutation altered the sixth residue of a seven–amino acid conserved propeptide motif, G-x-N-x-F-x-D (26), that is found among the members of the papain family of cysteine proteases (Fig. 4). This motif does exhibit some variability, and the amino acid residues found in position 6 among papain family members are alanine, serine, glycine, and threonine. Site-directed mutagenesis of residues 1, 3, 5, and 7 in the G-L-N-V-F-A-D motif in papain demonstrated that some substitutions resulted in complete loss of enzymatic function secondary to improper protein folding (note that residues 2, 4, and 6 were not mutagenized; ref. 26).
Figure 4.
Alignment of conserved motif from the pro region of several members of the papain family of cysteine proteases. The seven-residue motif from four human cysteine cathepsins (GenBank/EMBL accession numbers: cathepsin K, U13665; cathepsin L, X12451; cathepsin S, M90696; cathepsin H, X16832) and papain (M15203) are aligned, and the consensus sequence described by Vernet et al. (26) is shown below. The site of the cathepsin K G79E mutation is indicated by the arrow.
To model the changes produced by the G79E mutation, the method of conformational memories (27) was used to explore the conformational accessibility of the backbone and side-chain torsional angles in procathepsin K, using peptides consisting of nine amino acid residues with the wild-type or mutated residue occupying the central position; the electrostatic potentials (ESPs) of these constructs were calculated and displayed on the solvent-accessible surface with the program GRASP (28). The conformational search with the nine-residue peptide containing the E79 mutation demonstrated a shift in the population of accessible torsion angles of Ψ79 compared with that observed in the crystal structure of cathepsin L (25) and readily available to the wild-type cathepsin K nonapeptide. This result implied a decreased flexibility for this conserved motif in the mutant pro piece, because decreased stability might result if Ψ79 of the E79 mutant were to assume a value near that observed in cathepsin L. Comparison of the ESPs showed that the mutation had far-reaching consequences for the electrostatic properties of this portion of the propeptide. Whereas the negative ESP was confined to a relatively small region in the wild-type sequence near D80, it was extended to a surface covering about four adjacent amino acid residues in the mutant. The results also showed the shift of the ESP over the entire nonapeptide to significantly more negative values that could impact the reactivity and recognition properties of this region of the protein. These predicted conformational and electrostatic effects on the pro piece suggest that the G79E mutation might cause improper folding of the cathepsin K proenzyme, resulting in a loss of protease activity, and were consistent with the results of the expression studies with the recombinant G79E mutant. Despite the production of an immunologically detectable proenzyme protein that included a mature region with the normal cathepsin K primary structure, no mature protein or enzymatic activity was detected. This finding strongly implicated a perturbation in the secondary structure of the precursor protein caused by the G79E change in the propeptide that resulted in complete degradation, rather than activation, by pepsin.
Expression studies with the five missense mutants affecting the mature region of cathepsin K showed that four (G146R, A277V, A277E, and R312G) had no detectable mature protein or enzyme activity. The three-dimensional structure of cathepsin K shows that G146 lies deep within the active-site cleft, so the substitution of the large, charged Arg residue was expected to have significant adverse effects upon this protein. The A277E and A277V mutations altered another residue in the active site, A277, which is immediately next to the invariably conserved H276 residue that forms the ion pair with the active cysteine (C139) to effect protein catalysis. Moreover, the A277 residue is highly conserved among the papain family members, existing as either an alanine or a glycine residue in nearly all instances (29). It was anticipated that the replacement of A277 by a charged residue in the A277E mutation would obliterate the protease activity, but these expression studies revealed that even the A277V change, which appeared to introduce a less drastic alteration, resulted in a precursor protein that was unable to autoactivate and was unstable in the presence of pepsin. Thus, the obliterative effects of A277V suggest that there is little tolerance for larger amino acid side groups at this critical position in the active cleft .
The effects of the fourth missense mutation with no residual activity, R312G, are less obvious. The R312 residue does not lie within the active site, but rather resides on the surface. Based on the three-dimensional structure of human procathepsin L (25), R312 would not be expected to interact with the pro piece, so the R312G lesion presumably affects the precursor protein structure directly. The R312 residue is conserved among approximately two thirds of the papain family members, never existing as a glycine residue (7), and the R312G mutation eliminates a positive charge, all consistent with the conclusion that this mutation significantly disrupts the surface structure of the cathepsin K proenzyme in this region.
The only mutant cathepsin K with residual protease activity was Y212C. Biochemical characterization performed revealed that the Y212C mutant had only modestly diminished enzymatic activity with dipeptide substrate specificity and pH profiles that were quite similar to those of the recombinant wild-type cathepsin K. The Y212C activity, however, was unstable at pH conditions around its pH optimum. Most critically, the mutant enzyme retained almost no collagenolytic activity, despite retaining good gelatinolytic activity, and showed significantly reduced activity toward Z-GPR-MCA, a tripeptide found previously to be an excellent and relatively specific synthetic substrate for cathepsin K (22). Mass spectrometric analysis demonstrated that the activation of the Y212 propeptide resulted in enzymes with NH2-termini that were identical to those of the wild-type cathepsin K, although additional species with longer NH2-terminal extensions were also noted. The presence of cathepsin K enzymes with variable NH2-termini was also documented in a prior study of the activation of cathepsin K (30), as well as by similar work with cathepsins B and S (31, 32). Inspection of the three-dimensional structure of the mature enzyme (33) revealed that the NH2-terminus is relatively far from the active site, residing on the opposite side of the protein, so that the extension of the NH2-terminus by a small number of residues from the predicted cleavage site at A115 would not be expected to interfere with enzymatic function or substrate specificity. Because the Y212C enzymes are still capable of degrading denatured proteins as well as the synthetic dipeptides favored by wild-type cathepsin K, it suggests that the structural deformation caused by the Y212C mutation prevents engagement of the type I collagen substrate in the active cleft. It remains to be determined whether, like matrix metalloproteinases, cathepsin K will have a second, noncatalytic domain that is also required for collagen binding (34).
The Y212 residue does not lie in the active site, however, but rather on the surface of the mature protein. This residue is completely conserved among the six known human cysteine cathepsins, except for cathepsin H, in which it exists as a phenylalanine residue (7), and is never a cysteine residue among any papain family member (29). Because the Y212C mutation introduces a thiol group that might form a novel disulfide bridge or disrupt existing ones, it suggests that a structural deformation on the surface perturbs the conformation of the active site, preventing proper engagement of type I collagen. Future three-dimensional structural studies with Y212C protein may provide more precise information about the nature of these conformational changes.
In summary, seven new mutations in the cathepsin K gene have been identified, providing further evidence that the deficient activity of this enzyme causes Pycno and demonstrating the molecular genetic heterogeneity underlying this skeletal dysplasia. Among these lesions, two were in the region encoding the propeptide that is presumably responsible for protein folding, lysosomal trafficking, and inactivation of the enzyme until it is cleaved in the lysosome. The lack of activity by the pro region missense mutant G79E suggests that the previously identified seven-residue pro region motif that it alters is critical for protein folding. Several missense and nonsense mutations in the mature polypeptide were detected, R241X being frequent in the Hispanic patients studied. Expression of five mature enzyme missense mutations revealed that four mutant enzymes had no detectable activity and that one, Y212C, had good protease activity and preserved dipeptide substrate specificity but retained minimal activity toward type I collagen and a synthetic tripeptide. This structure/function information suggests that the cathepsin K active site contains a critical collagen-binding domain.
Acknowledgments
This work was supported in part by National Institutes of Health research grants (1 R29 AR-44231 to B.D. Gelb, 5 R37 DK-34045 to R.J. Desnick, 5 PO1 HD-22657 to D.L. Rimoin, and DA00060-18 to H. Weinstein), a grant from the National Center for Research Resources for the Mount Sinai General Clinical Research Center (5 M01 RR00071), a grant for the Mount Sinai Child Health Research Center from the National Institutes of Health (5 P30 HD-28822), and a Basic Research Award from the March of Dimes Birth Defects Foundation (to B.D. Gelb).
References
- 1.Gelb BD, Shi G-P, Chapman HA, Desnick RJ. Pycnodysostosis, a lysosomal disease due to cathepsin K deficiency. Science. 1996;273:1137–1139. doi: 10.1126/science.273.5279.1236. [DOI] [PubMed] [Google Scholar]
- 2.Maroteaux P, Lamy M. La pycnodysostose. Presse Med. 1962;70:999–1002. [PubMed] [Google Scholar]
- 3.Andrén L, Dymling JF, Hogeman KE, Wendeberg B. Osteopetrosis acro-osteolytica: a syndrome of osteopetrosis, acro-osteolysis and open sutures of the skull. Acta Chir Scand. 1962;124:496–507. [PubMed] [Google Scholar]
- 4.Tezuka K, et al. Molecular cloning of a possible cysteine proteinase predominantly expressed in osteoclasts. J Biol Chem. 1994;269:1106–1109. [PubMed] [Google Scholar]
- 5.Inaoka T, et al. Molecular cloning of human cDNA for cathepsin K: novel cysteine proteinase predominantly expressed in bone. Biochem Biophys Res Commun. 1995;206:89–96. doi: 10.1006/bbrc.1995.1013. [DOI] [PubMed] [Google Scholar]
- 6.Shi G-P, et al. Molecular cloning of human cathepsin O, a novel endoproteinase and homologue of rabbit OC2. FEBS Lett. 1995;357:129–134. doi: 10.1016/0014-5793(94)01349-6. [DOI] [PubMed] [Google Scholar]
- 7.Brömme D, Okamoto K. Human cathepsin O2, a novel cysteine protease highly expressed in osteoclastomas and ovary: molecular cloning, sequencing and tissue distribution. Biol Chem Hoppe Seyler. 1995;376:379–384. doi: 10.1515/bchm3.1995.376.6.379. [DOI] [PubMed] [Google Scholar]
- 8.Li Y-P, et al. Cloning and complete coding sequence of a novel human cathepsin expressed in giant cells of osteoclastomas. J Bone Miner Res. 1995;10:1197–1202. doi: 10.1002/jbmr.5650100809. [DOI] [PubMed] [Google Scholar]
- 9.Everts V, Aronson DC, Beertsen W. Phagocytosis of bone collagen by osteoclasts in two cases of pycnodysostosis. Calcif Tissue Int. 1985;37:25–31. doi: 10.1007/BF02557674. [DOI] [PubMed] [Google Scholar]
- 10.Gelb BD, et al. Cathepsin K: isolation and characterization of the murine cDNA and genomic sequence, the homolog of the human pycnodysostosis gene. Biochem Mol Med. 1996;59:200–206. doi: 10.1006/bmme.1996.0088. [DOI] [PubMed] [Google Scholar]
- 11.Brömme D, Okamoto K, Wang BB, Biroc S. Human cathepsin O2, a matrix protein-degrading cysteine protease expressed in osteoclasts. J Biol Chem. 1996;271:2126–2132. doi: 10.1074/jbc.271.4.2126. [DOI] [PubMed] [Google Scholar]
- 12.Bossard MJ, et al. Proteolytic activity of human osteoclast cathepsin K. J Biol Chem. 1996;271:12517–12524. doi: 10.1074/jbc.271.21.12517. [DOI] [PubMed] [Google Scholar]
- 13.Johnson MR, et al. A nonsense mutation in the cathepsin K gene observed in a family with pycnodysostosis. Genome Res. 1996;6:1050–1055. doi: 10.1101/gr.6.11.1050. [DOI] [PubMed] [Google Scholar]
- 14.Gelb BD, et al. Structure and chromosomal assignment of the human cathepsin K gene. Genomics. 1997;41:258–262. doi: 10.1006/geno.1997.4631. [DOI] [PubMed] [Google Scholar]
- 15.Haliassos A, et al. Modification of enzymatically amplified DNA for the detection of point mutations. Nucleic Acids Res. 1989;17:3606. doi: 10.1093/nar/17.9.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton CR, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin J, et al. A strategy for rapid, high-confidence protein identification. Anal Chem. 1997;69:3995–4001. doi: 10.1021/ac970488v. [DOI] [PubMed] [Google Scholar]
- 18.Ogryzko VV, et al. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 19.Fenyo D, Zhang W, Chait BT, Beavis RC. Internet-based analytical chemistry resource: a model project. Anal Chem. 1996;68:721A–726A. [Google Scholar]
- 20.Chait BT, Kent SB. Weighing naked proteins: practical, high-accuracy mass measurement of peptides and proteins. Science. 1992;257:1885–1894. doi: 10.1126/science.1411504. [DOI] [PubMed] [Google Scholar]
- 21.Linnevers CJ, et al. Expression of human cathepsin K in Pichia pastoris and preliminary crystallographic studies of an inhibitor complex. Protein Sci. 1997;6:919–921. doi: 10.1002/pro.5560060421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aibe K, et al. Substrate specificity of recombinant osteoclast-specific cathepsin K from rabbits. Biol Pharm Bull. 1996;19:1026–1031. doi: 10.1248/bpb.19.1026. [DOI] [PubMed] [Google Scholar]
- 23.Cooper DN, Krawczak M. The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum Genet. 1990;85:55–74. doi: 10.1007/BF00276326. [DOI] [PubMed] [Google Scholar]
- 24.Ryan, A.N. 1992. Vasco da Gama. In Encyclopedia Americana. Grolier Inc. Danbury, CT. 259–260.
- 25.Colombe R, et al. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15:5492–5503. [PMC free article] [PubMed] [Google Scholar]
- 26.Vernet T, et al. Processing of the papain precursor. J Biol Chem. 1995;270:10838–10846. doi: 10.1074/jbc.270.18.10838. [DOI] [PubMed] [Google Scholar]
- 27.Guarnieri F, Weinstein H. Conformational memories and the exploration of biologically relevant peptide conformations: an illustration for the gonadotropin-releasing hormones. J Am Chem Soc. 1996;118:5580–5589. [Google Scholar]
- 28.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 29.Berti PJ, Storer AC. Alignment/phylogeny of the papain superfamily of cysteine proteases. J Mol Biol. 1995;246:273–283. doi: 10.1006/jmbi.1994.0083. [DOI] [PubMed] [Google Scholar]
- 30.McQueney MS, et al. Autocatalytic activation of human cathepsin K. J Biol Chem. 1997;272:13955–13960. doi: 10.1074/jbc.272.21.13955. [DOI] [PubMed] [Google Scholar]
- 31.Rowan AD, Mason P, Mach L, Mort JS. Rat procathepsin B. Proteolytic processing to the mature form in vitro. J Biol Chem. 1992;267:15993–15999. [PubMed] [Google Scholar]
- 32.Bromme D, et al. Functional expression of human cathepsin S in Saccharomyces cerevisiae. Purification and characterization of the recombinant enzyme. J Biol Chem. 1993;268:4832–4838. [PubMed] [Google Scholar]
- 33.McGrath ME, Klaus JL, Barnes MG, Bromme D. Crystal structure of human cathepsin K complexed with a potent inhibitor. Nat Struct Biol. 1997;4:105–109. doi: 10.1038/nsb0297-105. [DOI] [PubMed] [Google Scholar]
- 34.Shingleton WD, Hodges DJ, Brick P, Cawston TE. Collagenase: a key enzyme in collagen turnover. Biochem Cell Biol. 1996;74:759–775. doi: 10.1139/o96-083. [DOI] [PubMed] [Google Scholar]