Abstract
To optimize culture conditions that enhance production of a highly viscous exopolysaccharide of Lactobacillus fermentum TDS030603, a chemically defined medium was examined. The best yield was found to be 199 ± 23 mg/l when 48-hr cultivation was microaerobically performed at 30°C in the chemically defined medium supplemented with 5% glucose and 1% ammonium citrate without pH control. In response to the optimized exopolysaccharide production, the mRNA expression levels of epsB, epsE, and epsG elevated significantly. Our results indicated that the optimal C/N ratio and/or microaerobic condition can alter the expression levels of several exopolysaccharide biosynthesis-related genes promoting the exopolysaccharide production yield.
Keywords: chemically defined medium, gene expression, highly viscous exopolysaccharide, probiotics
Lactic acid bacteria (LAB) are commonly used in the food industry, especially contributing to the production of fermented dairy products, and are considered highly safe, having been designated as generally recognized as safe. Many food-grade microorganisms including LAB are capable of synthesizing exopolysaccharides (EPSs), which are widely used in the food industry as biothickeners, food emulsifiers, and gelling agents. There are two types of EPS, homo- and hetero-EPS, composed of a single and several species of monosaccharides, respectively (hereafter EPS indicates hetero-EPS unless otherwise noted). Two well-established EPSs that are commercially available are xanthan, produced by Xanthomonas campestris [1], and gellan, produced from Pseudomonas elodea [2]. During the fermentation process, some LAB secrete EPSs that confer a good mouthfeel to fermented products by modifying their texture [3]. Despite beneficial aspects of EPSs produced by LAB, their low yield (40 to 800 mg/l) [4, 5], in contrast to those of xanthan (10 to 25 g/l) [6] and gellan (14 to 17 g/l) [7], prevents commercial use.
It is well known that production yield, structure, and rheological properties such as the viscosity of the microbial EPS vary owing to several factors such as composition of the culture medium, type of strain, and fermentation conditions [8, 9, 10]. De Man-Rogosa-Sharpe (MRS) medium is commonly used for LAB cultivation, but it contains many undefined compounds that interfere with quantification, compositional and structural analyses of EPSs [11]. On the other hand, a chemically defined medium (CDM) is suitable for investigation of influences of medium components on growth and metabolic pathways of a target microorganism. We developed a CDM for Lactobacillus fermentum TDS030603 (hereafter TDS030603), a highly viscous EPS producing LAB [12]. The strain can metabolize several carbohydrates, and the maximum EPS production (69 mg/l) was achieved when glucose was used as the sole carbon source. Another study revealed that EPS-related genes of the strain form a cluster in the genome [13]. Their nucleotide sequences were partially revealed; however, there is so far no information about the relationship between EPS production and EPS-related gene expression levels. In the present work, we explored the optimum conditions for EPS production by TDS030603 using a CDM supplemented with several components under various culture conditions, monitoring the EPS-related gene expression levels by real-time quantitative PCR.
TDS030603 was from the culture collection in our laboratory. The stock culture was prepared using MRS (Oxoid, Basingstoke, UK) containing 20% (v/v) glycerol and kept at –80°C until used. A single colony was inoculated into 10 mL MRS broth and incubated statically for 24 hr at 30°C. Cells were harvested by centrifugation at 12,000 × g for 5 min and washed with phosphate buffered saline (PBS) 3 times. The harvested cells were inoculated into 100 mL of CDM to give an absorbance at 600 nm of 0.3 and incubated under various conditions in the absence of pH control as described below. Different concentrations of glucose (1, 5, 10 and 15%), ammonium citrate (0.1, 0.5, 1 and 2%), adenine (0.0005, 0.001, 0.005, 0.01 and 0.05%), orotic acid (0.00001, 0.0001, 0.001, 0.005 and 0.01%) and bile salt mixture (0.001, 0.01 and 0.1%; Sigma-Aldrich, St. Louis, MO, USA) were tested. We also examined microaerobic cultivation using an Anaero Pack system (Mitsubishi Gas Chemical, Tokyo, Japan) that achieves a partial oxygen pressure of the atmosphere of less than 0.1%. To check the effect of agitation, cells were inoculated into CDM in a 500-ml flask and incubated with stirring (180 rpm). Furthermore, cultivation was performed with different initial pH values (4.0, 4.5, 5.0 and 7.0, adjusted by addition of 0.1% lactate or 1% calcium carbonate) and different incubation temperatures (25, 30, 35 and 37°C). After 48-hr cultivation, cell growth was monitored by measurement of optical density at 600 nm or viable cell count. Semi-purified or purified EPS was lyophilized and quantified by weight measurement as described previously [12, 14]. All the experiments, unless otherwise noted, performed in this study were done in triplicate. Values obtained from the experiments were presented as means ± standard deviation (SD). Data from three independent replicates were subjected to one-way analysis of variance (ANOVA) for single-factor analysis using the Microsoft Excel software. Any ANOVA data with p<0.05 were classified as statistically significant.
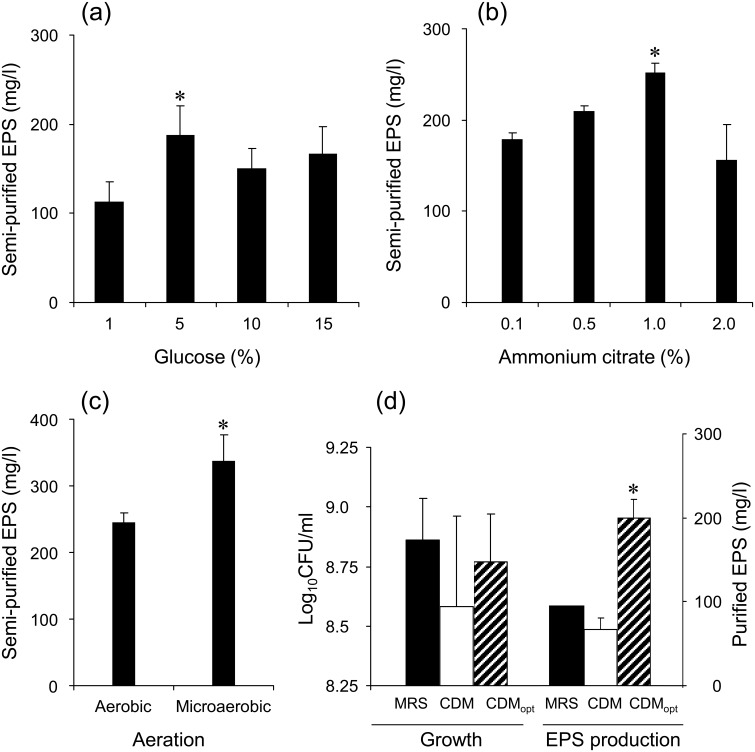
To date, many studies have been performed to improve the EPS yield by several approaches. Modification of the culture conditions including the medium composition, e.g., carbon source [15, 16, 17], C/N molar ratio [18, 19], minor nutritional compounds such as vitamins [20], and toxic compounds such as bile salts [21], and culture environment, e.g., partial oxygen pressure [22], pH control [23, 24] and cultivation temperature [23, 25], was most frequently done due to their accessibility. These factors were generally strain dependent, and they mostly increased EPS production by a couple of folds. In this study, when 5% glucose was added as a supplement, EPS production (183.7 ± 24 mg/l) was significantly higher than 1% (113 ± 22 mg/l) (Fig. 1a). EPS production was enhanced with increased ammonium citrate up to 1.0%, resulting in the highest level of production (245 ± 15 mg/l) (Fig. 1b). From these results, the optimum carbon to nitrogen (C/N) molar ratio was calculated to be 6.3. For further experiments, 5% glucose and 1% ammonium citrate were added to CDM as supplements. Gamar-Nourani et al. [22] demonstrated that a partially anaerobic condition (the partial oxygen pressure 0−10%) was favorable to cell growth and EPS production of Lb. rhamnosus strain C83. This also seems to be the case for our strain, because microaerobic cultivation enhanced cell growth and EPS production (338 ± 39 mg/l) compared with aerobic cultivation (245 ± 15 mg/l) (Fig. 1c). In contrast, minor nutritional compounds such as adenine and orotic acid, incubation temperature, and agitation showed no effect. TDS030603 was damaged and the EPS production was abolished by 2.4 mM bile salts, in contrast to Bifidobacterium animalis [21]. There was no detectable EPS production when the initial medium pH was adjusted to 4.0 and 4.5, whereas pH values of 5.0 and 7.0 resulted in 200 and 314 mg/l EPS, respectively (n = 1). The pH value of CDM supplemented with 5.0% glucose and 1.0% ammonium citrate was approximately 6.5, resulting in 338 ± 39 mg/l EPS under microaerobic conditions, and hence a pH value of 6.5 was optimum. This observation supports the previous findings that high acidity of the medium causes acid stress in cells [9], leading to suppression of bacterial growth and EPS production [26, 27]. Therefore, a neutral pH level controlled by a fermentation bioreactor may promote further EPS production. Consequently, TDS030603 achieved 3.0-fold higher EPS production in the optimized CDM (supplemented with 5.0% glucose and 1.0% ammonium citrate, resulting in an initial pH value of 6.5; static cultivation was performed at 30°C under the microaerobic conditions) than in the non-optimized CDM (Fig. 1d), which is similar to the results in previous reports.
Fig. 1.
Factors affecting on the EPS production of L. fermentum TDS030603.
(a) Glucose concentration versus semi-purified EPS yield; (b) ammonium citrate concentration versus semi-purified EPS yield; (c) aeration condition versus semi-purified EPS yield; (d) cell growth and purified EPS yield in MRS (solid bars), CDM (the non-optimized CDM, hollow bars) and CDMopt (the optimized CDM cultivated under microaerobic conditions, diagonal bars). All experiments were performed in triplicate, except for EPS purification from MRS in panel (d). * p<0.05.
As was previously reported, lactose resulted in higher EPS production than glucose in response to a significant increase in the transcription level of a gene encoding a priming glycosyltransferase [15]. Assuming that increased EPS production of TDS030603 may also be involved in changes in the expression levels of the EPS-related genes, we quantified the transcriptional levels of six EPS-related genes, epsB (encoding a putative polymerization and chain length determination protein), epsC (encoding a putative non-specific protein-tyrosine kinase), epsD (encoding a putative protein tyrosine phosphatase), epsE (encoding a putative glycosyltransferase), epsF (encoding a putative glycosyltransferase) and epsG (encoding a putative glycosyltransferase) [13]. Primers were designed using Primer-3 software version 0.4.0 [28], as shown in Table 1. To evaluate the specific amplification by a set of sense and antisense primers, conventional PCR was carried out. The reaction mixture (15 µL of master mix: 2 µL of 10 x Ex Taq buffer, 1.6 µL of 20 mM dNTP mixture, 0.2 µL of 20 μM primers, 0.1 µL of Ex Taq and 10.9 µL of distilled water) was mixed with 1 µL of template cDNA eight times diluted with Easy Dilution (Takara, Otsu, Japan). The cycle conditions were as follows: 94°C for 3 min; 35 cycles of 94°C for 30 sec, 58°C for 30 sec and 72°C for 30 sec; 72°C for 10 min; and then maintenance at 4°C. A single band of each amplicon was confirmed by electrophoresis in 2% agarose gel. The 48-hr cultured cells proliferated in 100 mL of MRS, CDM and the optimized CDM (supplemented with 5% glucose and 1% ammonium citrate; cultivated under the microaerobic conditions) were collected by centrifugation (15,000 × g, 1 hr) and resuspend in 10 mL PBS. A 250-μl cell suspension was used for total RNA extraction using TRIzol reagent (Invitrogen, Scotland, UK), according to the manufacturer’s instructions. To eliminate the contamination of chromosomal DNA, RNA were treated with RQ1 RNase-free DNase (Promega, Madison, WI, USA). The quality of RNA was considered sufficient if the ratio of absorbance at 260 and 280 nm was found to be more than 1.8. Single-stranded cDNA was synthesized from 1 µg of the DNase-treated RNA using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Obtained cDNA was stored at −20°C until used. Real-time quantitative PCR was performed using an Applied Biosystems 7300 HT Sequence Detection System (Applied Biosystems) equipped with a 96-well reaction plate. A partial sequence of 16S rRNA of Lb. fermentum LAB9 (DDBJ accession number: JN039355.1) was used as a reference. PCR products were detected with a Power SYBR green fluorescent dye (Applied Biosystems). A 2-μl aliquot of the diluted template cDNA was added to a mixture of 12.5 µL of the 2 x SYBR green PCR master mix (Applied Biosystems), 0.2 μM primers and water, giving a final volume of 25 µL. The cycle conditions were as follows: 95°C for 10 min; 40 cycles of 95°C for 15 sec, 58°C for 30 sec, and 72°C for 30 sec, followed by a single dissociation step of 95°C for 15 sec, 60°C for 30 sec and 95°C for 15 sec to determine arbitrarily-placed threshold (CT) values of amplicons. The expression level of each EPS gene was normalized against that of the 16S rRNA gene using the 2-ΔΔCt method [29].
Table 1. Primers used for real-time quantitative PCR of the EPS-related genes and 16S rRNA in L. fermentum TDS030603.
| Gene | Primer | Sequence (5’→3’) | Tm (°C) | Product size (bp) |
| epsB | BF2 sense | GAAGGTAAGCACCCAAACCA | 60.0 | 234 |
| BR2 antisense | ATCAAGAACCCAAGCACCAA | 60.5 | ||
| epsC | CF2 sense | ATCCGGACCAACATTCACTT | 59.3 | 214 |
| CR2 antisense | TGGTTAACCCCTTTTGGTTG | 59.7 | ||
| epsD | DF2 sense | TATTCTGGAGGCGTTTTTGG | 60.1 | 205 |
| DR2 antisense | AAACTCACGGGCCATTTTT | 59.4 | ||
| epsE | EF2 sense | CGAGTAGGCCGTAATGGAAA | 60.1 | 209 |
| ER2 antisense | GGTCGTGGACCAATAAAGGA | 59.8 | ||
| epsF | FF2 sense | TCAAGCGGGTATTTGACTTCTT | 60.1 | 238 |
| FR2 antisense | AACATTGGGCCATCAACATC | 60.6 | ||
| epsG | GF2 sense | GTTGGCCTTATGGAAGCAGA | 60.2 | 158 |
| GR2 antisense | CAATTGAATCCCGAGTGAAAG | 59.6 | ||
| 16S rRNA | 16SF1 sense | GCGTTATCCGGATTTATTGG | 59.3 | 240 |
| 16SR1 antisense | CTGTTCGCTACCCATGCTTT | 60.3 | ||
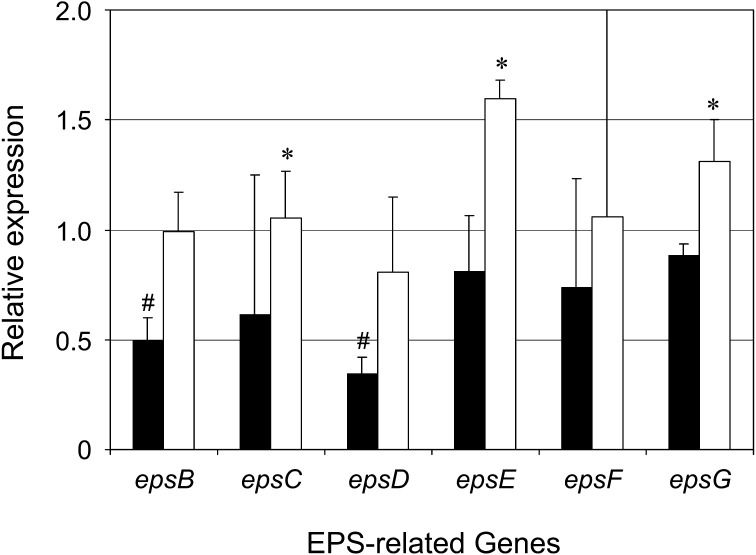
There are several proteins involved in EPS production, which is categorized into two distinct processes: (i) uptake of sugars and formation of sugar nucleotides and (ii) synthesis of EPS and its extracellular export. Proteins involved in the former process are encoded by housekeeping genes. Meanwhile, genes encoding proteins related to the latter form a cluster in the genome (in thermophilic LAB) or in the plasmid (in mesophilic LAB). The transcriptional levels of several genes involved in both processes have been revealed to alter the EPS production yield. For example, a 3-fold increase in the eps expression level, which was guided by a 9-fold higher EPS plasmid copy number, resulted in an almost 4-fold increase in the EPS production level of Lactococcus lactis NIZO B40 [30]. On the other hand, homologous overexpression of the UDP-glucose pyrophosphorylase gene in combination with the phosphoglucomutase gene led the EPS yield of Streptococcus thermophilus LY03 to increase from 0.17 to 0.31 g/mol of carbon from lactose [31]. Audy et al. [15] reported that Bifidobacterium longum subsp. longum CRC 002 promoted EPS production with significantly high transcription levels of genes encoding a priming glycosyltransferase and other enzymes involved in the Leloir pathway during exponential growth. In this study, we performed real-time quantitative PCR to elucidate the relationship between enhanced EPS production and changes in the expression level of six EPS-related genes (Fig. 2). Among them, epsB, epsE and epsG showed 2.0-, 2.0- and 1.5-fold higher mRNA expression levels, respectively, under the optimized conditions than under the non-optimized conditions, indicating that these three genes are upregulated in response to either the optimal C/N molar ratio or microaerobic environment. Interestingly, the transcriptional levels of epsB and epsD were downregulated in the non-optimized CDM compared with MRS. These results are not completely in agreement with the previous observation that EPS production per viable cell counts was 39-fold higher in non-optimized CDM than MRS [12]. Therefore, we speculate that the most influential gene or genes with respect to the EPS production of our strain are present in other genomic regions. Whole genome sequencing and DNA microarray analysis are currently being performed for genome-wide analysis.
Fig. 2.
Relative expression of six EPS-related genes in CDM (solid bars) and CDMopt (hollow bars).
The amounts of mRNA expression of six EPS-related genes (epsB, epsC, epsD, epsE, epsF and epsG) in the cells obtained by cultivation in MRS, CDM and CDMopt were estimated by real-time quantitative PCR and normalized against the 16S rRNA gene. The normalized values of the six genes in CDM and CDMopt are shown relative to those in MRS. Analysis was performed for three independent experiments with duplicate wells, and the average values were indicated (means ± SD). * p<0.05 (CDM versus CDMopt). # p<0.05 (CDM versus MRS).
Acknowledgments
We are grateful to Mr. Shota Katayama for his practical advice on real-time quantitative PCR experiments. This study was supported financially by Yotsuba Milk Product Co., Ltd., and by the Global Center of Excellence Frontier Program for Animal Global Health and Hygiene of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- 1.Becker A, Katzen F, Pühler A, Ielpi L. 1998. Xanthan gum biosynthesis and application: a biochemical/genetic perspective. Appl Microbiol Biotechnol 50: 145–152 [DOI] [PubMed] [Google Scholar]
- 2.Banik RM, Kanari B, Upadhyay SN. 2000. Exopolysaccharide of the gellan family: prospects and potential. World J Microbiol Biotechnol 16: 407–414 [Google Scholar]
- 3.Doleyres Y, Schaub L, Lacroix C. 2005. Comparison of the functionality of exopolysaccharides produced in situ or added as bioingredients on yogurt properties. J Dairy Sci 88: 4146–4156 [DOI] [PubMed] [Google Scholar]
- 4.Ruas-Madiedo P, Hugenholtz J, Zoon P. 2002. An overview of the functionality of exopolysaccharides produced by lactic acid bacteria. Int Dairy J 12: 163–171 [Google Scholar]
- 5.Patel AK, Michaud P, Singhania RR, Soccol CR, Pandey A. 2010. Polysaccharides from probiotics: new developments as food additives. Food Technol Biotechnol 48: 451–463 [Google Scholar]
- 6.García-Ochoa F, Santos VE, Casas JA, Gómez E. 2000. Xanthan gum: production, recovery, and properties. Biotechnol Adv 18: 549–579 [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Xu P, Yuan Y, Liu C, Zhang D, Yang Z, Yang C, Ma C. 2006. Modeling for gellan gum production by Sphingomonas paucimobilis ATCC 31461 in a simplified medium. Appl Environ Microbiol 72: 3367–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petry S, Furlan S, Crepeau MJ, Cerning J, Desmazeaud M. 2000. Factors affecting exocellular polysaccharide production by Lactobacillus delbrueckii subsp. bulgaricus grown in a chemically defined medium. Appl Environ Microbiol 66: 3427–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaningelgem F, Zamfir M, Adriany T, De Vuyst L. 2004. Fermentation conditions affecting the bacterial growth and exopolysaccharide production by Streptococcus thermophilus ST 111 in milk-based medium. J Appl Microbiol 97: 1257–1273 [DOI] [PubMed] [Google Scholar]
- 10.Wang YX, Lu ZX. 2005. Optimization of processing parameters for the mycelial growth and extracellular polysaccharide production by Boletus spp. ACCC 50328. Process Biochem 40: 1043–1051 [Google Scholar]
- 11.Kimmel SA, Roberts RF. 1998. Development of a growth medium suitable for exopolysaccharide production by Lactobacillus delbrueckii ssp. bulgaricus RR. Int J Food Microbiol 40: 87–92 [DOI] [PubMed] [Google Scholar]
- 12.Fukuda K, Shi T, Nagami K, Leo F, Nakamura T, Yasuda K, Senda A, Motoshima H, Urashima T. 2010. Effects of carbohydrate source on physicochemical properties of the exopolysaccharide produced by Lactobacillus fermentum TDS030603 in a chemically defined medium. Carbohydr Polym 79: 1040–1045 [Google Scholar]
- 13.Dan T, Fukuda K, Sugai-Bannai M, Takakuwa N, Motoshima H, Urashima T. 2009. Characterization and expression analysis of the exopolysaccharide gene cluster in Lactobacillus fermentum TDS030603. Biosci Biotechnol Biochem 73: 2656–2664 [DOI] [PubMed] [Google Scholar]
- 14.Leo F, Hashida S, Kumagai D, Uchida K, Motoshima H, Arai I, Asakuma S, Fukuda K, Urashima T. 2007. Studies on a neutral exopolysaccharide of Lactobacillus fermentum TDS030603. J Appl Glycosci 54: 223–229 [Google Scholar]
- 15.Audy J, Labrie S, Roy D, Lapointe G. 2010. Sugar source modulates exopolysaccharide biosynthesis in Bifidobacterium longum subsp. longum CRC 002. Microbiology 156: 653–664 [DOI] [PubMed] [Google Scholar]
- 16.Cerning J, Renard CM, Thibault JF, Bouillanne C, Landon M, Desmazeaud M, Topisirovic L. 1994. Carbon source requirements for exopolysaccharide production by Lactobacillus casei CG11 and partial structure analysis of the polymer. Appl Environ Microbiol 60: 3914–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Looijesteijn PJ, Boels IC, Kleerebezem M, Hugenholtz J. 1999. Regulation of exopolysaccharide production by Lactococcus lactis subsp. cremoris By the sugar source. Appl Environ Microbiol 65: 5003–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SS, Lee JS, Cho JY, Kim YE, Hong EK. 2010. Effects of C/N ratio and trace elements on mycelial growth and exo-polysaccharide production of Tricholoma matsutake. Biotechnol Bioproc Eng 15: 293–298 [Google Scholar]
- 19.Miqueleto AP, Dolosic CC, Pozzi E, Foresti E, Zaiat M. 2010. Influence of carbon sources and C/N ratio on EPS production in anaerobic sequencing batch biofilm reactors for wastewater treatment. Bioresour Technol 101: 1324–1330 [DOI] [PubMed] [Google Scholar]
- 20.Grobben GJ, Sikkema J, Smith MR, de Bont JAM. 1995. Production of extracellular polysaccharides by Lactobacillus delbrueckii ssp. bulgaricus NCFB 2772 grown in a chemically defined medium. J Appl Bacteriol 79: 103–107 [Google Scholar]
- 21.Ruas-Madiedo P, Gueimonde M, Arigoni F, de los Reyes-Gavilán CG, Margolles A. 2009. Bile affects the synthesis of exopolysaccharides by Bifidobacterium animalis. Appl Environ Microbiol 75: 1204–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamar-Nourani L, Blondeau K, Simonet JM. 1998. Influence of culture conditions on exopolysaccharide production by Lactobacillus rhamnosus strain C83. J Appl Microbiol 85: 664–672 [Google Scholar]
- 23.Degeest B, Mozzi F, De Vuyst L. 2002. Effect of medium composition and temperature and pH changes on exopolysaccharide yields and stability during Streptococcus thermophilus LY03 fermentations. Int J Food Microbiol 79: 161–174 [DOI] [PubMed] [Google Scholar]
- 24.Kimmel SA, Roberts RF, Ziegler GR. 1998. Optimization of exopolysaccharide production by Lactobacillus delbrueckii subsp. bulgaricus RR grown in a semidefined medium. Appl Environ Microbiol 64: 659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols CM, Bowman JP, Guezennec J. 2005. Effects of incubation temperature on growth and production of exopolysaccharides by an antarctic sea ice bacterium grown in batch culture. Appl Environ Microbiol 71: 3519–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Berg D, Robijn GW, Janssen AC, Giuseppin M, Vreeker R, Kamerling JP, Vliegenthart J, Ledeboer AM, Verrips CT. 1995. Production of a novel extracellular polysaccharide by Lactobacillus sake 0-1 and characterization of the polysaccharide. Appl Environ Microbiol 61: 2840–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheirsilp B, Shimizu H, Shioya S. 2001. Modelling and optimization of environmental conditions for kefiran production by Lactobacillus kefiranofaciens. Appl Microbiol Biotechnol 57: 639–646 [DOI] [PubMed] [Google Scholar]
- 28.Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols: Methods in Molecular Biology. ed.Krawetz S. and Misener S. pp.365–386. Totowa: Humana Press. [DOI] [PubMed] [Google Scholar]
- 29.Yuan JS, Reed A, Chen F, Stewart CN., Jr2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7: 85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boels IC, Van Kranenburg R, Kanning MW, Chong BF, De Vos WM, Kleerebezem M. 2003. Increased exopolysaccharide production in Lactococcus lactis due to increased levels of expression of the NIZO B40 eps gene cluster. Appl Environ Microbiol 69: 5029–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levander F, Svensson M, Rådström P. 2002. Enhanced exopolysaccharide production by metabolic engineering of Streptococcus thermophilus. Appl Environ Microbiol 68: 784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]