Abstract
Expression of histocompatibility leukocyte antigen (HLA) class I molecules on the cell surface depends on the heterodimer of the transporter associated with antigen processing 1 and 2 (TAP1 and TAP2), which transport peptides cleaved by proteasome to the class I molecules. Defects in the TAP2 protein have been reported in two families with HLA class I deficiency, the so-called bare lymphocyte syndrome (BLS) type I. We have, to our knowledge, identified for the first time a splice site mutation in the TAP1 gene of another BLS patient. In addition, class I heavy chains (HCs) did not form the normal complex with tapasin in the endoplasmic reticulum (ER) of the cells of our patient.
J. Clin. Invest. 103:649–652 (1999)
Introduction
Histocompatibility leukocyte antigen (HLA) class I molecules present peptides derived from intracellular proteins to cytotoxic T cells. The peptides, 8–11 amino acids in length, are transported after cleavage by cytosolic proteasomes into the inner lumen of the endoplasmic reticulum (ER) by transporter associated with antigen processing 1 and 2 (TAP1 and TAP2) complexes. Thereafter, the peptides are associated with class I heavy chains (HCs), which form complexes with β2 microglobulin (β2m), tapasin, and calreticulin on the ER lumen (1, 2). After loading of the peptides, the HC–β2m complex, dissociated from TAP, tapasin, and calreticulin, is transported to the cell surface. It has been shown that the expression levels of class I molecules are decreased in several cell lines deficient in TAP, β2m, or tapasin (2–4) and that free HCs are stabilized on the cell surface after the addition of β2m and specific class I molecule–binding peptides in cell culture media (5).
Bare lymphocyte syndrome (BLS) type I is known to be characterized by HLA class I deficiency, sinusitis, and chronic bronchitis and was first reported by Touraine et al. (6). BLS is believed to be caused by deficiency of the TAP2 protein, based on studies of two families (7, 8). In this study, we examined the gene defect of BLS in patient KMW, of the TSU family, reported previously by Maeda et al. (9). KMW was a 46-year-old Japanese female who continues to suffer from sinusitis, chronic bronchitis, and a punched-out ulcer on the left pretibial region. Her parents, HST and UTT, are first cousins (Fig. 1b). In this study, we analyzed the molecules involved in the maturation pathway of class I molecules in KMW-B2 cells, a B-cell line derived from the lymphocytes of KMW, and for the first time found a mutation in the TAP1 gene in a BLS patient.
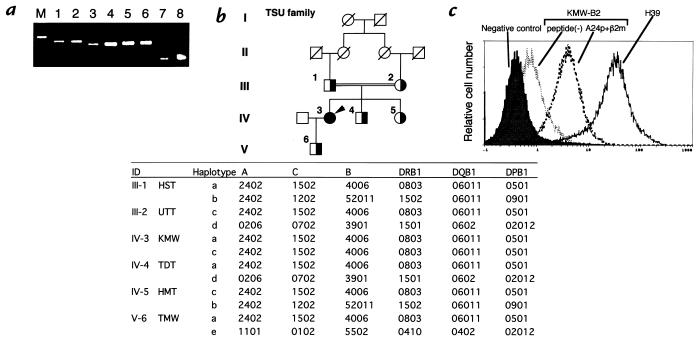
Figure 1.
(a) Expression of the mRNA of HLA-A (lanes 1 and 2), HLA-B (lanes 3 and 4), HLA-C (lanes 5 and 6), and G3PDH (lanes 7 and 8) of the lymphocytes of a healthy donor (lanes 1, 3, 5, and 7) and those of KMW (lanes 2, 4, 6, and 8). M represents a molecular weight marker. HLA-A, HLA-B, and HLA-C fragments were amplified by PCR from the cDNA derived from lymphocytes. The PCR products were sequenced by automated sequencing. (b) Consanguineous pedigree and HLA haplotypes of the TSU family. The filled circle represents an affected woman (IV-3, KMW). Partially filled circles (women) and partially filled squares (men) indicate the family members whose samples were analyzed. HLA class I and class II were typed by the sequencing-based typing system as described in Methods, and HLA haplotypes were estimated. (c) Expression of HLA class I molecules on KMW-B2 cells, a B-cell line of KMW, after incubation in the presence or absence of the A24 consensus peptide (A24p, YYEEQHPEL) and β2m. The KMW-B2 cells were preincubated in the presence or absence of 100 mg/ml A24p and 10 mg/ml human β2m for 18 h. KMW-B2 cells or H39 cells, a normal control B-cell line, were stained by FITC-conjugated mouse IgG (shaded histogram) or FITC-conjugated w6/32 and analyzed using the EPICS XL flow cytometer (Coulter Corp.). HLA, histocompatibility leukocyte antigen.
Methods
Materials.
The compounds used were as follows: w6/32, anti–HLA class I monoclonal antibody (Serotec, Kidlington, United Kingdom); rabbit anti-calreticulin antibody (Affinity BioReagents Inc., Golden, Colorado, USA); rabbit anti-β2m antibody (Cosmo Bio Company, Tokyo, Japan); HLA-A24 consensus peptide (A24p, YYEEQHPEL; Sawady Technology, Tokyo, Japan); and human β2m (Chemicon International Inc., Temecula, California, USA). HC10 (anti–HLA class I monoclonal antibody) was kindly provided by H.L. Ploegh (Harvard Medical School, Boston, Massachusetts, USA). Rabbit anti-peptide antibodies were raised for each of the following regions: RGGPGGRSQKAEGQRWLC of tapasin, CYWAMVQAPADAPE of TAP1 and QEGQDLYSRLVQQRLMD of TAP2 (KMW is homozygous for TAP2*Bky2, the translated protein of which would be expected to react with our anti-TAP2 antiserum). Preparation of rabbit antisera against LMP2, LMP7, and MECL1 have been described previously (10).
Cell preparation.
Peripheral blood mononuclear cells (PBMCs) were isolated from the peripheral blood of KMW and healthy adult volunteers by Ficoll centrifugation (Pharmacia Biotech, Uppsala, Sweden). The B-cell lines KMW-B2 and H39 were generated from Epstein-Barr virus–transformed B cells of KMW and a healthy volunteer, respectively.
PCR amplification and sequencing.
We prepared the mRNA and genomic DNA from PBMCs, using an mRNA direct kit (Dynal AS, Oslo, Norway) and a QIAamp Blood kit (QIAGEN GmbH, Hilden, Germany), respectively. HLA-A, HLA-B, HLA-C, TAP1, and TAP2 fragments were amplified by PCR from lymphocyte cDNA or genomic DNA. The PCR products were sequenced by automated sequencing. HLA class I and class II were typed using an HLA sequencing-based typing system and ABI 310 (Perkin-Elmer Corp., Norwalk, Connecticut, USA).
Flow cytometric analysis.
KMW-B2 cells were incubated in the presence or absence of 10 μg/ml β2m and 100 μg/ml A24p at 37°C for 18 h. The cells were stained by FITC-conjugated mouse IgG or FITC-conjugated w6/32 and analyzed using the EPICS XL flow cytometer (Coulter Corp., Hialeah, Florida, USA).
Sedimentation velocity analysis.
The microsome fractions (precipitates of postmitochondrial fractions by centrifugation at 105,000 g for 1 h) from KMW-B2 and H39 cells were lysed in 1% digitonin lysis buffer. The digitonin-solubilized lysates were loaded onto a linear gradient of 10%–40% (vol/vol) glycerol, as described previously (11). After centrifugation for 30 h at 4°C at 110,000 g, the gradient was separated into 34 fractions of 1 ml each.
Western blot analysis.
Proteins were separated by SDS-PAGE and blotted onto an Immobilon-P membrane (Millipore Corp., Bedford, Massachusetts, USA). The membranes were incubated with antibodies followed by horseradish peroxidase–conjugated second antibodies, and developed using the Renaissance chemiluminescence reagent (NEN Life Science Products, Boston, Massachusetts, USA).
Submission of nucleotide sequence data.
The nucleotide sequence data reported in this article have been submitted to the DNA Data Bank of Japan (DDBJ) nucleotide database and have been assigned the following accession numbers: TAP1*0102N cDNA sequence, AB012644; TAP1*0102N genomic sequence, AB012645. The name TAP1*0102N was officially assigned by the World Health Organization Nomenclature Committee in November 1998.
Results
As shown in Fig. 1a, the mRNAs of the HLA-A, HLA-B, and HLA-C genes were detected in PBMCs of KMW by reverse transcriptase (RT)-PCR. cDNA sequence analysis revealed no mutation in the HLA-A, HLA-B, or HLA-C genes (data not shown). The HLA haplotypes of the members of the TSU family were determined by sequencing-based typing. KMW was homozygous for the haplotype A*2402-Cw*1502-B*4006-DRB1*0803-DQB1*06011-DPB1*0501 (Fig. 1b). In flow cytometric analysis using the anti–class I monoclonal antibody w6/32, the class I molecule expression level of the lymphocytes of KMW was determined to be less than 5% of that of the lymphocytes of healthy donors, as reported previously (9), whereas the lymphocytes of the family members (Fig. 1b, partially filled symbols) showed normal levels of expression of class I molecules (data not shown). The expression level of class I molecules on the surface of KMW-B2 cells was also lower than that on a B-cell line derived from the lymphocytes of a healthy volunteer (H39 cells) (Fig. 1c). The class I molecule expression level on the surface of KMW-B2 cells did not change even after 42 hours of incubation with 1,000 U/ml interferon-γ (data not shown). However, the expression level of class I molecule on KMW-B2 cells increased after incubation for 18 hours with β2m and the A24-specific peptide (A24p) (Fig. 1c). These observations suggest that the cause of the HLA class I deficiency in KMW is a defect in the supply of the peptides bound to class I molecules.
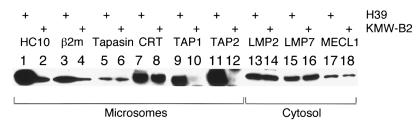
Therefore, we examined the expression levels of molecules involved in the processing/presentation of intracellular antigens by Western blot analysis. As shown in Figure 2, HC, β2m, calreticulin, tapasin, and subunits of immunoproteasomes (LMP2, LMP7, and MECL1) (12) were detected in KMW-B2 cells at a level equivalent to that in H39 cells, although the expression levels of HC and β2m were considerably lower. However, both TAP1 and TAP2 proteins were undetectable in KMW-B2 cells, in marked contrast to their high expression levels in H39 cells. RT-PCR analysis was used to examine the expression of the mRNAs of TAP1 and TAP2 in the PBMCs of KMW and revealed that similar amounts of PCR products were amplified from cDNA of the PBMCs of KMW and healthy controls (data not shown). Then the sequences of the TAP1 and TAP2 genes of KMW were identified.
Figure 2.
Western blot analysis of H39 and KMW-B2 cells. The microsomes and cytosol were prepared from H39 and KMW-B2 cells, purified by centrifugation and lysed in 1% digitonin lysis buffer. The lysates were subjected to Western blot analysis. Proteins of microsomes (lanes 1–12) or cytosol (lanes 13–18), derived from the control H39 cells (lanes 1, 3, 5, 7, 9, 11, 13, 15, 17) or KMW-B2 cells (lanes 2, 4, 6, 8, 10, 12, 14, 16, 18) were analyzed by Western blot analysis using various antibodies.
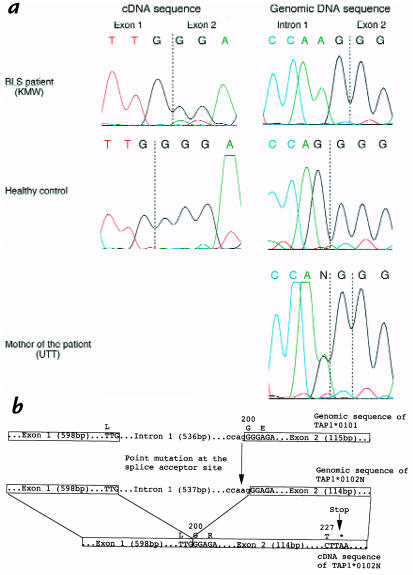
The results revealed a G deletion in codon 200 in the cDNA sequence of the TAP1 gene of KMW (Fig. 3a). This deletion causes a frameshift, which results in the premature appearance of a stop codon at 228 and the synthesis of a truncated TAP1 protein that cannot be detected by the presently used antibodies against the COOH-terminal peptide. We then performed genomic sequencing of the gene, which revealed that KMW was homozygous for TAP1*0102N, the allele with a point mutation of G→A at the splice acceptor site of the 3′ end of intron 1 (Fig. 3a). This point mutation may lead to the use of a shifted splice site with a consequent one-base deletion in the processed mRNA, because the mutated splice acceptor site of TAP1*0102N did not match with the conventional splice consensus sequences (Fig. 3b; ref. 13). Many instances in which a single-base substitution at a splice junction resulted in abnormal splicing have been reported (3, 14). Members of the TSU family (Fig. 1b, partially filled symbols) were revealed to be heterozygous for TAP1*0102N (only the genomic sequence of the mother, UTT, is shown in Fig. 3a). Although the HLA class I deficiency has been reported previously as being due to a defect in TAP2 (7), we identified a splice site mutation in the TAP1 gene in an HLA class I–deficient patient.
Figure 3.
(a) Sequences of the TAP1 gene of KMW, the mother (UTT), and a healthy control. cDNA sequences of the TAP1 gene revealed KMW to be homozygous for TAP1*0102N, an allele with a G deletion in codon 200. Genomic DNA sequencing of the TAP1 gene also revealed a point mutation of G→A on 536 bp in intron 1 of the TAP1*0102N of KMW and the family members (partially filled symbols in Fig. 1b are heterozygous for TAP1*0102N, and only the sequence of the mother is shown). PCR products of the TAP1 gene were sequenced by automated sequencing as described in Methods. (b) Sequences of the TAP1*0101 and TAP1*0102N. G was substituted to A on 536 bp in intron 1 of the TAP1*0102N, which resulted in an altered splice acceptor site and deletion of G in codon 200 of mRNA. This deletion caused a frameshift and resulted in the premature appearance of a stop codon at 228 and the synthesis of a truncated TAP1 protein. (GenBank accession numbers: TAP1*0102N cDNA sequence, AB012644; TAP1*0102N genomic sequence, AB012645.) TAP, transporter associated with antigen processing.
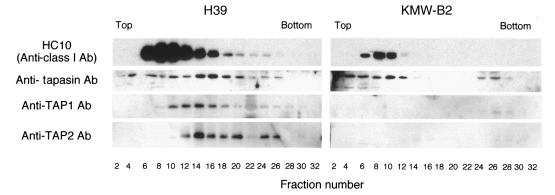
It is noteworthy that the tapasin molecule is critical to the linkage between HC and TAP (1). Therefore, we investigated the relationship between HC and tapasin in KMW-B2 cells. To this end, detergent-solubilized microsomal materials were analyzed by glycerol density gradient centrifugation. As shown in Fig. 4, HC, tapasin, TAP1, and TAP2 were cosedimented in fractions 12–20 in H39 cells, although considerable amounts of HC and tapasin were recovered in the slow-sedimenting fractions 6–12. In contrast, no appreciable quantity of TAP1 or TAP2 was detected, and the amount of HC was extremely low in the microsomes of KMW-B2 cells, as shown in Fig. 2. Interestingly, tapasin shifted to lighter fractions in KMW-B2 cells compared with H39 cells, implying that a lack of TAP proteins disrupts the functional interaction between HC and tapasin. This assumption was further supported by the finding that anti-calreticulin antibodies immunoprecipitated tapasin in H39 cells, but not in KMW-B2 cells, whereas they immunoprecipitated HC in both types of cells (data not shown). These results suggest that the complex containing HC becomes smaller without TAP1 and TAP2 and that most of the HC is probably degraded in KMW-B2 cells.
Figure 4.
Sedimentation velocity analysis of HC, tapasin, and TAP in the microsomes of H39 and KMW-B2 cells. The digitonin-solubilized microsomes were analyzed by glycerol density gradient centrifugation as described in Methods. After centrifugation, the gradient was separated into 34 fractions of 1 ml each. Samples of the fractions were subjected to Western blot analysis. HC, heavy chain; Ab, antibody.
Discussion
It is interesting that TAP2 was undetectable in KMW-B2 cells; nevertheless, no abnormality was found in the sequence analysis of TAP2 cDNA (data not shown). Perhaps, TAP2 is degraded rapidly because of the deficiency of its partner TAP1, as other molecules without normal conformation were in the ER (15). However, TAP1 protein has been reported to be detected in the cells of TAP2-deficient patients (7, 8). This difference may be explained by assuming that TAP1 is capable of forming a complex with a chaperone(s) such as tapasin in the absence of TAP2, thus preventing its destabilization.
HC has been shown to be associated with tapasin in a TAP-deficient cell line, 721.174 (1). To the contrary, our data indicate that HC did not form a complex with tapasin in KMW-B2, TAP-deficient patient cell line. This could be explained by the genetic differences between the cell lines, such as polymorphisms of HLA class I or tapasin, which may cause a difference of affinity between the two molecules (16–19).
In a TAP1-deficient mouse, reduction of cell-surface expression of class I molecules was observed as seen in KMW but TAP2 protein expression has not been analyzed (20). A small number of CD8+ T cells exists in the peripheral blood of KMW (21) as well as in that of the TAP1-deficient mouse (20).
Acknowledgments
The authors thank Hidde L. Ploegh for providing HC10; Mie Nieda and Motoko Nishimura for the B-cell lines; Kohichiro Nakamura for providing clinical information about KMW; Masaru Takamizawa and Naoyuki Tsuchiya for constructive discussions; and Ritsuko Miyamoto for technical assistance.
References
- 1.Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 2.Ortmann B, et al. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997;277:1306–1309. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- 3.Hicklin DJ, et al. Beta2-microglobulin mutations, HLA class I antigen loss, and tumor progression in melanoma. J Clin Invest. 1998;101:2720–2729. doi: 10.1172/JCI498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly A, et al. Assembly and function of the two ABC transported proteins encoded in the human major histocompatibility complex. Nature. 1992;355:641–644. doi: 10.1038/355641a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen HL, et al. A functionally defective allele of TAP results in loss of MHC class I antigen presentation in a human lung cancer. Nat Genet. 1996;13:210–213. doi: 10.1038/ng0696-210. [DOI] [PubMed] [Google Scholar]
- 6.Touraine JL. The bare-lymphocyte syndrome: report on the registry. Lancet. 1981;1:319–321. doi: 10.1016/s0140-6736(81)91922-x. [DOI] [PubMed] [Google Scholar]
- 7.de la Salle H, et al. Homozygous human TAP peptide transported mutation in HLA class I deficiency. Science. 1994;265:237–241. doi: 10.1126/science.7517574. [DOI] [PubMed] [Google Scholar]
- 8.Teisserenc H, et al. A case of primary immunodeficiency due to a defect of the major histocompatibility gene complex class I processing and presentation pathway. Immunol Lett. 1997;57:183–187. doi: 10.1016/s0165-2478(97)00072-2. [DOI] [PubMed] [Google Scholar]
- 9.Maeda H, et al. Defective expression of HLA class I antigens: a case of the bare lymphocyte without immunodeficiency. Immunogenetics. 1985;21:549–558. doi: 10.1007/BF00395879. [DOI] [PubMed] [Google Scholar]
- 10.Hisamatsu H, et al. Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J Exp Med. 1996;183:1807–1816. doi: 10.1084/jem.183.4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimbara N, et al. Double-cleavage production of the CTL epitope by proteasomes and PA28: role of the flanking region. Genes Cells. 1997;2:785–800. doi: 10.1046/j.1365-2443.1997.1610359.x. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka K, Kasahara M. The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunol Rev. 1998;163:161–176. doi: 10.1111/j.1600-065x.1998.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 13.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gantla S, et al. Splice-site mutations: a novel genetic mechanism of Crigler-Najjar syndrome type 1. Am J Hum Genet. 1998;62:585–592. doi: 10.1086/301756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cresswell P, Hughes EA. Protein degradation: the ins and outs of the matter. Curr Biol. 1997;7:R552–R555. doi: 10.1016/s0960-9822(06)00279-x. [DOI] [PubMed] [Google Scholar]
- 16.Peh CA, et al. HLA-B27-restricted antigen presentation in the absence of tapasin reveals polymorphism in mechanisms of HLA class I peptide loading. Immunity. 1998;8:531–542. doi: 10.1016/s1074-7613(00)80558-0. [DOI] [PubMed] [Google Scholar]
- 17.Lewis JW, Sewell A, Price D, Elliott T. HLA-A*0201 presents TAP-dependent peptide epitopes to cytotoxic T lymphocytes in the absence of tapasin. Eur J Immunol. 1998;28:3214–3220. doi: 10.1002/(SICI)1521-4141(199810)28:10<3214::AID-IMMU3214>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa H, et al. Polymorphism of TAPASIN and its linkage disequilibria with HLA class II genes in the Japanese population. Tissue Antigens. 1998;52:279–281. doi: 10.1111/j.1399-0039.1998.tb03044.x. [DOI] [PubMed] [Google Scholar]
- 19.Copeman J, Bangia N, Cross JC, Cresswell P. Elucidation of the genetic basis of the antigen presentation defects in the mutant cell line .220 reveals polymorphism and alternative splicing of the tapasin gene. Eur J Immunol. 1998;28:3783–3791. doi: 10.1002/(SICI)1521-4141(199811)28:11<3783::AID-IMMU3783>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP mutant mice are deficient in antigen presentation, surface class I molecules, and CD4-8+ T cells. Cell. 1992;71:1205–1214. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa H, et al. Tolerance of NK and LAK activity for HLA class I–deficient targets in a TAP-deficient patient (bare lymphocyte syndrome type I) Hum Immunol. 1999;60:32–40. doi: 10.1016/s0198-8859(98)00097-4. [DOI] [PubMed] [Google Scholar]