Divergence of junction complex genes in rice implicates their importance in cereal growth, development, reproduction, and adaptive evolution.
Abstract
The exon junction complex (EJC) plays important developmental roles in animals; however, its role in plants is not well known. Here, we show various aspects of the divergence of each duplicated MAGO NASHI (MAGO) and Y14 gene pair in rice (Oryza sativa) encoding the putative EJC core subunits that form the obligate MAGO-Y14 heterodimers. OsMAGO1, OsMAGO2, and OsY14a were constitutively expressed in all tissues, while OsY14b was predominantly expressed in embryonic tissues. OsMAGO2 and OsY14b were more sensitive to different stresses than OsMAGO1 and OsY14a, and their encoded protein pair shared 93.8% and 46.9% sequence identity, respectively. Single MAGO down-regulation in rice did not lead to any phenotypic variation; however, double gene knockdowns generated short rice plants with abnormal flowers, and the stamens of these flowers showed inhibited degradation and absorption of both endothecium and tapetum, suggesting that OsMAGO1 and OsMAGO2 were functionally redundant. OsY14a knockdowns phenocopied OsMAGO1OsMAGO2 mutants, while down-regulation of OsY14b failed to induce plantlets, suggesting the functional specialization of OsY14b in embryogenesis. OsMAGO1OsMAGO2OsY14a triple down-regulation enhanced the phenotypes of OsMAGO1OsMAGO2 and OsY14a down-regulated mutants, indicating that they exert developmental roles in the MAGO-Y14 heterodimerization mode. Modified gene expression was noted in the altered developmental pathways in these knockdowns, and the transcript splicing of UNDEVELOPED TAPETUM1 (OsUDT1), a key regulator in stamen development, was uniquely abnormal. Concomitantly, MAGO and Y14 selectively bound to the OsUDT1 premessenger RNA, suggesting that rice EJC subunits regulate splicing. Our work provides novel insights into the function of the EJC locus in growth, development, and reproduction in angiosperms and suggests a role for these genes in the adaptive evolution of cereals.
The exon junction complex (EJC) is one of the fundamental machineries in the postsplicing processes in eukaryotes (Tange et al., 2004). The complex includes over 10 different proteins, such as MAGO NASHI (MAGO), Y14 (Tsunagi or RNA Binding Motif8), Eukaryotic Initiation Factor 4A III, and Barentsz (but also Metastatic Lymph Node51; Kim et al., 2001; Le Hir et al., 2001; Mohr et al., 2001; Palacios et al., 2004; Park et al., 2009; Zhao et al., 2000). MAGO and Y14 are core subunits of the EJC (Kataoka et al., 2001; Le Hir et al., 2001). Both subunits have been confirmed to direct the normal development of animals. In Drosophila spp., a single point mutation in the MAGO locus causes several developmental defects, such as improper development of the posterior lobe of the embryo, nonviable egg sacs in female offspring, and impairment of germplasm cell polarity and germline stem cell differentiation (Boswell et al., 1991; Newmark and Boswell, 1994; Micklem et al., 1997; Newmark et al., 1997; Parma et al., 2007). In Caenorhabditis elegans, the mag-1 gene is involved in hermaphrodite germline sex determination (Li et al., 2000). In mouse, the MAGO gene is related to cell cycle regulation (Inaki et al., 2011). Tsunagi, the Y14 ortholog (Kataoka et al., 2000), is involved in embryogenesis and germline sexual switching (Kawano et al., 2004) and regulating oocyte specification (Parma et al., 2007; Lewandowski et al., 2010). MAGO and Y14 were recently shown to be involved in the eye development of Drosophila species (Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010). MAGO regulates the proliferation and expansion of neural crest-derived melanocytes (Silver et al., 2013), whereas Y14 targets neuronal genes to regulate anxiety behaviors in mice (Alachkar et al., 2013). Therefore, these studies confirm the role of MAGO and Y14 genes in multiple related developmental roles in animal species. However, limited evidence for the developmental role of these genes is found in plants. PFMAGO proteins (MAGO homologs from Physalis floridana) interacting with the MADS domain protein MPF2 (for MADS protein2 from P. floridana) are responsible for male fertility (He et al., 2007). The Arabidopsis (Arabidopsis thaliana) AtMAGO gene is required for pollen grain development, and its knockout is lethal (Johnson et al., 2004; Park et al., 2009). Mutation of MvMAGO disrupts spermatogenesis in Marsilea vestita (van der Weele et al., 2007; Boothby and Wolniak, 2011). Besides, plant MAGO genes also seem to have broad roles in the growth and development of other plant organs. Down-regulating AtMAGO affects the development of root, shoot, floral meristem, and seed (Park et al., 2009). TcMago is preferentially expressed in root hairs in Taiwania cryptomerioides, and overexpressing this gene produces taller transgenic tobacco (Nicotiana tabacum) plants with increased root hairs (Chen et al., 2007). The knockdown of AtY14 yields a lethal phenotype (Park et al., 2009).
MAGO and Y14 proteins form obligate heterodimers (Shi and Xu, 2003; Gong et al., 2014) in eukaryotes. However, only the MAGO-Y14 heterodimer in metazoans binds to the mRNAs 20 to 24 nucleotides upstream of the exon-exon junctions, affecting multiple steps of postsplicing processes, such as mRNA intracellular export, cytoplasmic localization, nonsense-mediated mRNA decay (NMD), and translational enhancement (Hachet and Ephrussi, 2001; Tange et al., 2004; Lee et al., 2009). These subunits were recently demonstrated to be specifically involved in the splicing process of genes with large introns and heterochromatin localization in Drosophila spp. (Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010). The regulation of plant development by EJC is poorly understood. Furthermore, the retention of two copies for each gene family in cereals (Gong et al., 2014) suggests a specific evolutionary role of these genes in cereal evolution. In this study, we characterized OsMAGO1, OsMAGO2, OsY14a, and OsY14b in rice (Oryza sativa) to address the developmental role of EJC subunits in plants. Our study showed the divergence of each paralogous gene pair and that they are essential to both vegetative and reproductive development. The transcript splicing of the rice UNDEVELOPED TAPETUM1 (OsUDT1), a major regulator of early tapetum development (Jung et al., 2005), was found to be specifically affected by the depletion of these EJC subunits in rice. To our knowledge, our data provide the first insight into the essential role of the EJC in growth, development, and reproduction of rice with an implication of adaptive evolution in cereals.
RESULTS
Divergence in the Expression and Sequences of Rice MAGO and Y14 Paralogs
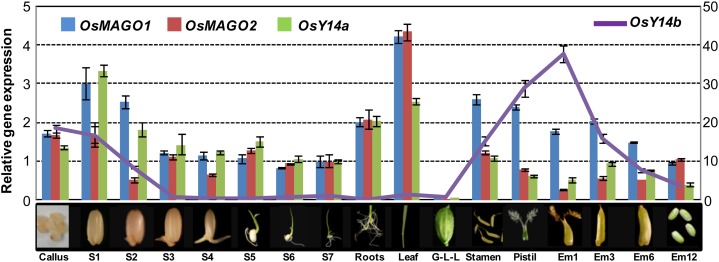
Using quantitative reverse transcription (qRT)-PCR, we showed that, during the normal development of rice, the genes OsMAGO1, OsMAGO2, and OsY14a were constitutively expressed in callus, all tissues of seedlings, floral organs, and seeds (Fig. 1). The difference in expression of these genes was not significant, suggesting coregulation of their expression in rice development. However, OsY14b had a different expression pattern and was expressed in callus, the early developmental stage of seed germination, and floral tissues but not in seedlings (Fig. 1). Thus, OsY14b may have a specialized role in rice development. The expression of OsMAGO1 and OsY14a was studied in response to various stimuli. We observed that OsMAGO2 expression was significantly (P ≤ 0.001) induced by brassinolide (BR), abscisic acid (ABA), GA3, and indole acetic acid (IAA) while OsY14b expression was significantly (P < 0.001) induced by BR, GA3, ABA, and IAA (Supplemental Fig. S1, A–D). OsY14b was also significantly influenced (P < 0.01) by high temperature, salinity stress, and water deficiency (Supplemental Fig. S1, E–H). Therefore, the mRNA expression of OsMAGO2 and OsY14b is more sensitive to various external stimuli as compared with that of OsMAGO1 and OsY14a. Furthermore, OsMAGO1 and OsMAGO2 shared 93.8% sequence identity while OsY14a and OsY14b shared 46.9% identity (Gong et al., 2014). Thus, the relative rapid divergence of rice Y14 might drive the functional divergence of the MAGO-Y14 dimers, and OsY14b might be under a neofunctionalization or subfunctionalization process.
Figure 1.
Expression patterns of rice MAGO and Y14 genes during rice development. Callus, 2-week-old callus on 1/2 MS medium; S1 to S7, 1- to 7-d-old seedling shoots (without roots) on 1/2 MS medium; Roots, 12-d-old seedlings on 1/2 MS medium; Leaf, flag leaves of mature plants at heading stage; G-L-L, mixture of glumes, lemma, and lodicules; Stamen and Pistil, stamen and pistil from mature florets; Em1 to Em12, embryo at 1, 3, 6, and 12 d after fertilization. OsACTIN1 was used as an internal control. Expression was detected with three independent biological samples. The expression for each gene in S7 was set to 1. Average expression and sd are presented.
Subcellular Localization and Heterodimerization of Rice MAGO and Y14
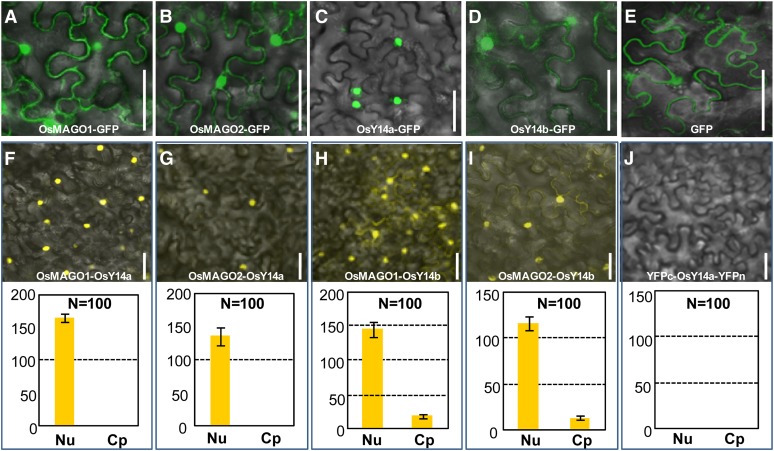
The open reading frames (ORFs) of rice MAGO and Y14 were fused in frame with GFP and were transiently expressed in plant cells to prove that sequence divergence may alter the subcellular localization of a protein. We found that OsMAGO1, OsMAGO2, and OsY14b showed identical distribution in cells and were localized in the nuclei and cytoplasm (Fig. 2, A, B, and D), similar to their homologs in other reported species. Surprisingly, OsY14a seemed to be uniquely localized in the nuclei (Fig. 2C), deviating from the established subcellular localization of these EJC subunits. GFP alone, as the control, was expressed in both nuclei and cytoplasm (Fig. 2E). These, again, reflect a more rapid divergence of the rice Y14 than MAGO pair after duplication.
Figure 2.
Transient expression of the rice MAGO and Y14 proteins. A to E, Subcellular localization. A to D, Expression of the fusion proteins OsMAGO1-GFP (A), OsMAGO2-GFP (B), OsY14a-GFP (C), and OsY14b-GFP (D). E, Expression of GFP as a control. Bars = 50 µm. F to J, Rice MAGO-Y14 heterodimerization revealed in BiFC assays. F to I, Coexpression of OsMAGO1-YFPc and OsY14a-YFPn (F), OsMAGO2-YFPc and OsY14a-YFPn (G), OsMAGO1-YFPc and OsY14b-YFPn (H), and OsMAGO2-YFPc and OsY14b-YFPn (I). The detection of YFP signals indicates an interaction of the combined proteins. J, Coexpression of YFPc and OsY14a-YFPn proteins as a negative control. The quantification of the fluorescence intensity of YFP signals in nucleus (Nu) and cytoplasm (Cp) is presented for each merged image. N, Number of cells quantified. Bars = 50 µm.
Rice MAGO and Y14 formed heterodimers in yeast (Saccharomyces cerevisiae strain AH109) (Supplemental Fig. S2, A and B; Gong et al., 2014). This study used bimolecular fluorescence complementation (BiFC) analyses in plant cells to study this heterodimerization. The yellow fluorescent protein (YFP) was divided into N-terminal (YFPn) and C-terminal (YFPc) halves. The ORFs of these genes were separately fused with either YFPn or YFPc and coexpressed. Fluorescence was observed when two proteins fused to each YFP half interacted with one another. A YFP signal was detected when the fusion proteins OsMAGO1 (or OsMAGO2)-YFPc and OsY14a (or OsY14b)-YFPn were coexpressed (Fig. 2, F–I). In contrast, no YFP signal was observed when the fusion proteins YFPc and OsY14a-YFPn were used as controls (Fig. 2J). In line with the subcellular localization pattern of rice Y14 proteins, the interaction signal of OsMAGO1 (or OsMAGO2) with OsY14a was observed in the nuclei only, whereas their interactions with OsY14b occurred in both nuclei and cytoplasm. Nonetheless, OsMAGO1 preferentially interacted with OsY14a while OsMAGO2 heterodimerized with OsY14b (P < 0.01; Supplemental Fig. S2, C and D), indicating a potential functional separation between OsMAGO1-OsY14a and OsMAGO2-OsY14b.
The above-revealed divergence may cause a functional divergence in each paralogous gene pair in rice. The developmental role of these EJC genes was further characterized using RNA interference (RNAi). Supplemental Tables S1 and S2 detail the independent transgenic lines that were generated and characterized for RNAi.
OsY14a Is Essential for Rice Growth, Development, and Reproduction
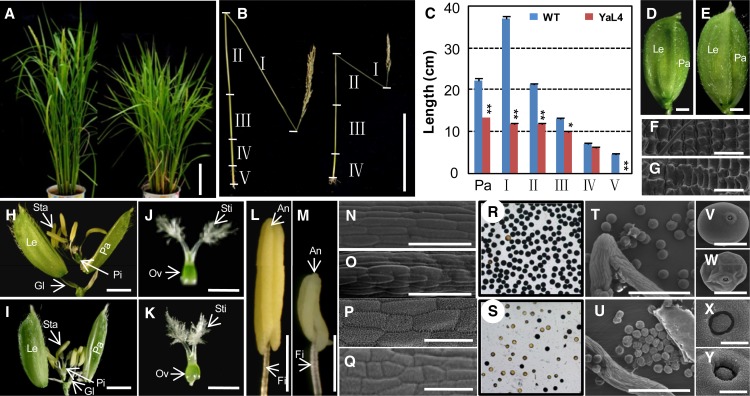
Seven independent Ubiqutin (Ubi):OsY14a-RNAi transgenic lines (YaL1–YaL7) were analyzed, and six of them showed significant down-regulation of OsY14a mRNA, while the mRNAs of OsY14b, OsMAGO1, and OsMAGO2 were not affected (Supplemental Fig. S3A). The six OsY14a knockdowns showed similar phenotypic variations (Supplemental Table S2) but were significantly different from the wild type. The transgenic plants at the heading stage were dwarfed in height as compared with the wild type (Fig. 3A). In mature plants, the wild type had five internodes in total, which were named I to V from the top to the bottom (Fig. 3B). The panicle length (YaL4, 13.23 ± 0.13 cm; wild type, 22.07 ± 0.26 cm; P < 0.001) and the culm length (YaL4, 49.32 ± 0.21 cm; wild type, 82.90 ± 0.56 cm; P < 0.001) of Ubi:OsY14a-RNAi transgenic plants were significantly shorter as compared with the wild type (Fig. 3, B and C). The decreased culm length was mainly due to the reduced length of each internode. In particular, internode V was absent in the OsY14a down-regulated transgenic plants. Furthermore, in comparison with the wild type (Fig. 3, D and H), the florets developed bigger lemma and palea (Fig. 3, E and I; Supplemental Table S2). Furthermore, the epidermal cells on the lemma were significantly longer than those of the wild type (Fig. 3, F and G; Supplemental Fig. S4A). The female organs were fertile, since they could produce normal seeds when crossed with the wild-type pollen (Fig. 3, J and K; Supplemental Table S3). However, the stamen and its filament became pale and were shorter than those in the wild type (Fig. 3, L and M; Supplemental Table S2). Cell size on the filament and stamen became significantly smaller than in the wild type (Fig. 3, N–Q; Supplemental Fig. S4, B and C) and resulted in reduced stamen and filament size. Pollen maturation was inhibited in the transgenic plants as compared with the wild type (Supplemental Table S2). A substantial proportion of pollen grains in the transgenic plants (Fig. 3S) were not stained dark blue using the iodine-potassium iodide (I2-KI) staining method as compared with the wild type (Fig. 3R), further proving that pollen maturation was inhibited in the RNAi lines. The germination pole was normal, but pollen morphology was abnormal (Fig. 3, T–Y). YaL7 without affecting OsY14a expression had no phenotypic defect (Supplemental Fig. S3A; Supplemental Table S2).
Figure 3.
Phenotypic comparisons of wild-type and OsY14a-RNAi transgenic rice plants. A, Rice plants at the heading stage of the wild type (left) and OsY14a-RNAi line 4 (right). Bar = 20 cm. B, Internodes (I–V) and panicle of wild-type and YaL4 seeds after maturation. Bar = 20 cm. C, Size quantification of the internodes (I–V) and panicle (Pa) between the wild type (WT) and YaL4. Significance was evaluated by two-tailed Student’s t test: *P < 0.05, **P < 0.01. D and E, Florets of the wild type and YaL4. Bars = 1 mm. F and G, Epidermal cells on lemma of the wild type and YaL4 by scanning electron microscopy. Bars = 50 µm. H and I, Artificially opened florets of the wild type and YaL4. Bars = 1 mm. J and K, Pistils of the wild type and YaL4. Bars = 1 mm. L and M, Stamens of the wild type and YaL4. Bars = 1 mm. Glume (Gl), lemma (Le), palea (Pa), stamen (Sta), anther (An), filament (Fi), pistil (Pi), stigma (Sti), and ovary (Ov) are indicated in D, E, and H to M. N and O, Cells on filaments of the wild type and YaL4 by scanning electron microscopy. Bars = 50 µm. P and Q, Cells on anthers of the wild type and YaL4 by scanning electron microscopy. Bars = 50 µm. R and S, I2-KI-stained pollen grains of the wild type and YaL4. T to W, Pollen of the wild type and YaL4 by scanning electron microscopy at different magnifications. Bars = 200 µm (T and U) and 20 µm (V and W). X and Y, Germ pore of the wild type and YaL4 by scanning electron microscopy. Bars = 20 µm.
OsY14b Is Required in Rice Embryonic Organogenesis and Development
Surprisingly, transgenic plants could not be regenerated from Ubi:OsY14b-RNAi lines with extra effort (Supplemental Table S1). The callus formation was normal (Supplemental Fig. S5A), and tiny shoot-like structures were sometimes regenerated (Supplemental Fig. S5B). However, they were not able to develop into plantlets and finally withered and died (Supplemental Fig. S5C). These observations suggest that OsY14b is essential in shoot formation from the callus and in early shoot development, as evident from gene expression analyses (Fig. 1). Five independent calli with tiny green shoots were examined and were found to have extremely low levels of OsY14b expression, whereas the expression levels of OsY14a, OsMAGO1, and OsMAGO2 were not affected (Supplemental Fig. S5D). The failure in plant regeneration of OsY14b-RNAi suggested that down-regulation of this gene was lethal to the growth and development of rice plants.
Two MAGO Genes Play a Redundant Role in Rice Growth and Development
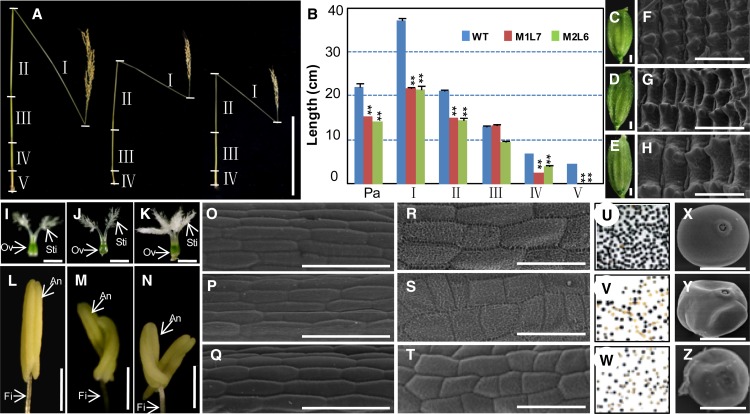
The 3′ untranslated region (UTR) of OsMAGO1 was first used to silence the OsMAGO1 gene in rice (Supplemental Table S1). Five of six Ubi:OsMAGO1-3′UTR-RNAi transgenic plants (M1L1-M1L6) were associated with severe and specific down-regulation of OsMAGO1 (Supplemental Fig. S3B). However, they did not produce any phenotypic variations in comparison with the wild type (Supplemental Fig. S6; Supplemental Table S2), indicating a potential functional redundancy between OsMAGO1 and OsMAGO2. Therefore, we tried to use their coding regions (CDS) to silence the two genes simultaneously (Supplemental Table S1). Altogether, we characterized 13 independent transgenic rice lines for Ubi:OsMAGO1-CDS-RNAi (five lines) and Ubi:OsMAGO2-CDS-RNAi (eight lines). Three lines (M2L1–M2L3) with severely down-regulated OsMAGO2 were identified in Ubi:OsMAGO2-CDS-RNAi transgenic plants (Supplemental Fig. S3D). The phenotypes of these three plants were not different from those of the wild type (Supplemental Table S2). However, six lines with severe and specific down-regulation of both OsMAGO1 and OsMAGO2 were further obtained from Ubi:OsMAGO1-CDS-RNAi (M1L7–M1L9) and Ubi:OsMAGO2-CDS-RNAi (M2L4–M2L6; Supplemental Fig. S3, C and D). These transgenic plants showed dramatic phenotypic variations (Supplemental Table S2). These transgenic plants at the heading stage were smaller than the wild type. At the maturation stage, the panicle length (M1L7, 15.30 ± 0.14 cm; M2L6, 14.07 ± 0.20 cm; wild type, 22.07 ± 0.26 cm; P < 0.001) and culm (M1L7, 54.67 ± 0.32 cm; M2L6, 48.38 ± 0.24 cm; wild type, 82.90 ± 0.56 cm; P < 0.001) of these transgenic plants were significantly shorter in comparison with the wild type (Fig. 4, A and B). The decreased culm length can be attributed to the reduced length of each internode, especially the I internode. Furthermore, these transgenic plants lacked the V internode (Fig. 4, A and B). Compared with the wild type (Fig. 4C), the florets were bigger in the RNAi lines (Fig. 4, D and E). The increased lemma resulted from an increase in cell expansion (Fig. 4, F–H; Supplemental Fig. S4A). The female organs looked normal and were fertile (Fig. 4, I–K; Supplemental Table S3), while the stamen and its filament became significantly shorter than in the wild type. Unlike the wild type (Fig. 4L), one filament bearing two stamens often developed in these transgenic plants (Fig. 4, M and N; Supplemental Table S2). Cells on the stamen and filament became smaller as compared with the wild type (Fig. 4, O–T; Supplemental Fig. S4, B and C). A substantial proportion of pollen grains were not only immature as revealed by I2-KI staining (P < 0.001; Fig. 4, U–W; Supplemental Table S2) but were also deformed (Fig. 4, X–Z). The two rice MAGO genes, therefore, are functionally redundant.
Figure 4.
Phenotypic variations in OsMAGO1OsMAGO2 down-regulated transgenic rice plants. A, Internodes (I–V) and panicle comparison of the wild type (left), M1L7 (Ubi:OsMAGO1-CDS-RNAi; middle), and M2L6 (Ubi:OsMAGO2-CDS-RNAi; right) after seed maturation. Bar = 20 cm. B, Size quantification of the internodes (I–V) and panicle (Pa) between the wild type (WT), M1L7, and M2L6. Significance was evaluated by two-tailed Student’s t test: **P < 0.01. C to E, Florets of the wild type, M1L7, and M2L6. Bars = 1 mm. F to H, Cells on lemma of the wild type, M1L7, and M2L6 by scanning electron microscopy. Bars = 50 µm. I to K, Pistils of the wild type, M1L7, and M2L6. Bars = 1 mm. L to N, Stamens of the wild type, M1L7, and M2L6. Bars = 1 mm. Anther (An), filament (Fi), stigma (Sti), and ovary (Ov) are indicated in I to N. O to Q, Cells on filaments of the wild type, M1L7, and M2L6 by scanning electron microscopy. Bars = 50 µm. R to T, Cells on anthers of the wild type, M1L7, and M2L6 by scanning electron microscopy. Bars = 50 µm. U to W, I2-KI-stained pollen grains of the wild type, M1L7, and M2L6. X to Z, Pollen of the wild type, M1L7, and M2L6 by scanning electron microscopy. Bars = 20 µm.
OsMAGO1OsMAGO2 double knockdowns showed similar phenotypic variations to those observed in Ubi:OsY14a-RNAi plants, suggesting that these genes may exert their roles through the MAGO-Y14 heterodimers.
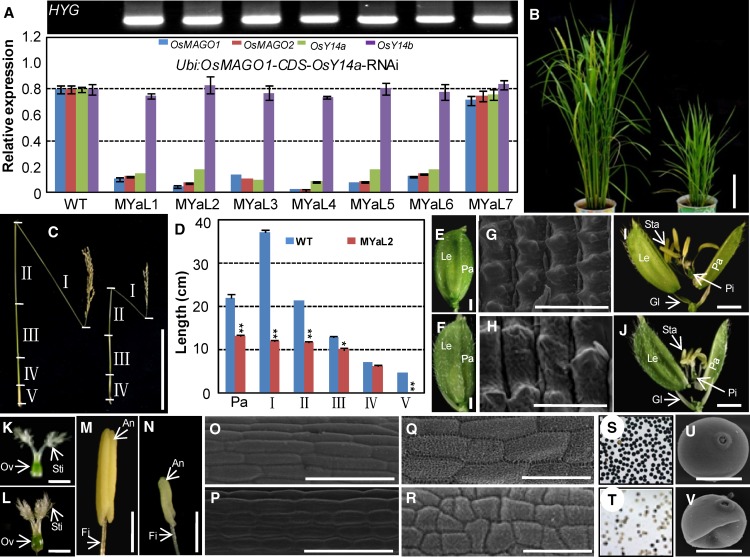
Rice MAGO and Y14 Exert Their Roles in an Obligate Heterodimerization Mode
In order to evaluate the developmental roles of the MAGO-Y14 heterodimers, we generated OsMAGO1OsMAGO2OsY14a knockdowns (OsY14b was excluded due to the lethality of single knockdowns) using the Ubi:OsMAGO1-CDS-OsY14a construct. Seven rice RNAi lines (MYaL1–MYaL7) were analyzed, and six of them showed a strong down-regulation of OsMAGO1, OsMAGO2, and OsY14a (Fig. 5A). These triple gene knockdowns featured similar phenotypic variation to that observed in the knockdowns of either double OsMAGO1OsMAGO2 or single OsY14a. The transgenic plants were smaller than the wild-type plants (Fig. 5B). Panicle lengths (MYaL2, 11.23 ± 0.11 cm; wild type, 22.07 ± 0.26 cm; P < 0.001) and culm lengths (MYaL2, 43.90 ± 0.23 cm; wild type, 82.90 ± 0.56 cm; P < 0.001) of these transgenic plants were significantly shorter than in the wild type (Fig. 5, C and D). The decreased culm length can again be attributed to the reduced length of each internode. The V internode was also absent in these transgenic plants (Fig. 5, C and D). Compared with the wild type, the floret developed bigger lemma and palea (Fig. 5, E and F; Supplemental Table S2) with bigger cells (Fig. 5, G and H; Supplemental Fig. S4A). The female organs were normal (Fig. 5, K and L; Supplemental Table S3), whereas the stamen and its filament became pale and shorter as compared with those in the wild type (Fig. 5, M and N; Supplemental Table S2) with smaller cells (Fig. 5, O–R; Supplemental Fig. S4, B and C), suggesting a functional abnormality. A large portion of pollen grains were immature (P < 0.001; Fig. 5, S–T; Supplemental Table S2) and deformed (Fig. 5, U and V). These findings verify that both rice MAGO and OsY14a have important roles through heterodimerization in the same developmental pathways. The essential role in male fertility was particularly investigated.
Figure 5.
Phenotypic variations of OsMAGO1OsMAGO2OsY14a down-regulated transgenic rice plants. A, Gene expression in Ubi:OsMAGO1-CDS-OsY14a-RNAi transgenic lines. Hygromycin gene expression (HYG) was detected by RT-PCR. OsACTIN1 was used as an internal control. Three independent biological samples were used to detect expression. The expression for each gene in the wild type (WT) was set to 1. Average expression and sd are presented. B, Rice plants at the heading stage of the wild type (left) and MYaL2 (OsMAGO1OsMAGO2OsY14a knockdown; right). Bar = 20 cm. C, Internodes (I–V) and panicle of the wild type (left) and MYaL2 (right) after seed maturation. Bars = 20 cm. D, Size quantification of the internodes (I–V) and panicle (Pa) between the wild type and MYaL2. Significance was evaluated by two-tailed Student’s t test: *P < 0.05, **P < 0.01. E and F, Florets of the wild type and MYaL2. Bars = 1 mm. G and H, Cells on lemma of the wild type and MYaL2 by scanning electron microscopy. Bars = 50 µm. I and J, Artificially opened florets of the wild type and MYaL2. Bars = 1 mm. K and L, Pistils of the wild type and MYaL2. Bars = 1 mm. M and N, Stamens of the wild type and MYaL2. Bars = 1 mm. Glume (Gl), lemma (Le), palea (Pa), stamen (Sta), anther (An), filament (Fi), pistil (Pi), stigma (Sti), and ovary (Ov) are indicated in E to N. O and P, Cells on filaments of the wild type and MYaL2 by scanning electron microscopy. Bars = 50 µm. Q and R, Cells on anthers of the wild type and MYaL2 by scanning electron microscopy. Bars = 50 µm. S and T, I2-KI-stained pollen grains of the wild type and MYaL2. U and V, Pollen of the wild type and MYaL2 by scanning electron microscopy. Bars = 20 µm.
Down-Regulating MAGO or Y14 Causes Abnormal Pollen Development in Rice
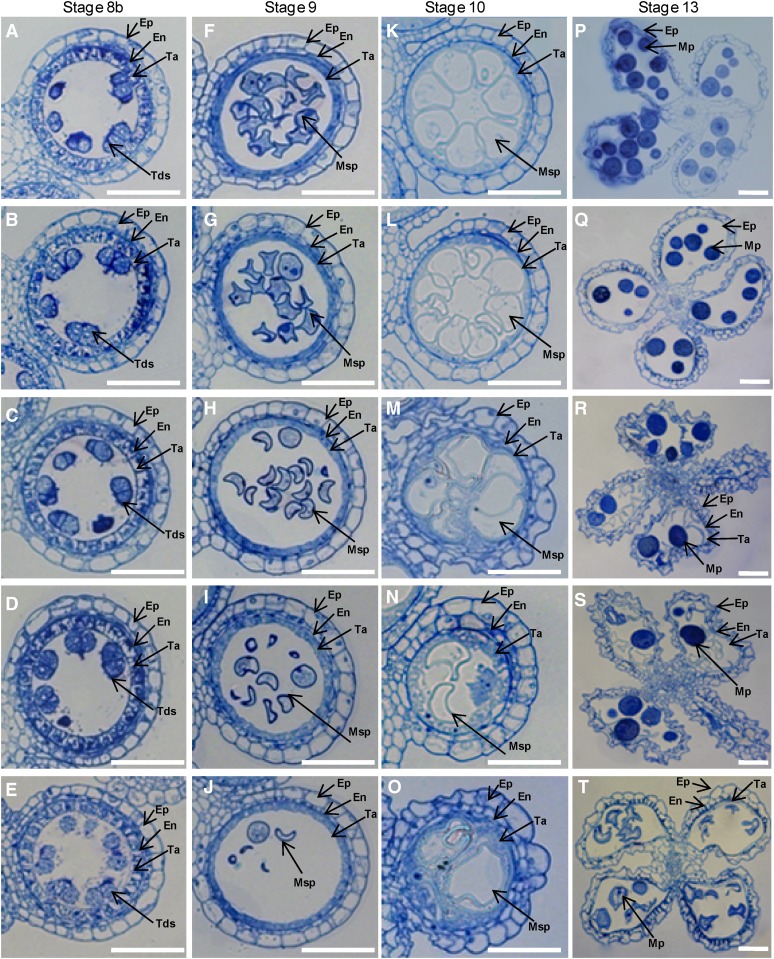
RNAi transgenic plants showed a decreased seed-setting rate once MAGO and Y14 genes were down-regulated (Supplemental Table S2). However, their seed-setting rate was similar to that of artificially pollinated wild-type plants when crossed with wild-type pollen (Supplemental Table S3), suggesting that the female functionality was not altered. Pollen maturation and morphology were severely influenced in transgenic plants; therefore, the abnormality in male fertility was established as a major cause for low seed-setting rate. A histological analysis of stamen development was performed to further understand the role of male sterility in seed set. According to Zhang and Wilson (2009), rice anther development is divided into 14 stages. The developmental defects were not detected in anthers unless the genes were down-regulated until stage 8b. At this stage, the tetrads with four haploid microspores were formed, the middle cell layer degenerated, and the endothecium became narrower in all plants (Fig. 6, A–E). At stage 9, the young microspores were released from the tetrads. The microspores in the wild type and the Ubi:OsMAGO1-RNAi transgenic plants (M1L1) had large nuclei, and homogenous cytoplasmic constituents were densely stained (Fig. 6, F–G). On the other hand, the RNAi knockdown of OsMAGO1OsMAGO2 double (M2L6), OsY14a single (YaL4), and OsMAGO1OsMAGO2OsY14a triple (MYaL2) genes showed more vacuolated microspores that stained less as compared with the wild type (Fig. 6, H–J). Specifically, typical falcate pollen grains were formed. At stage 10, the wild-type and M1L1 tapetum appeared to undergo degradation. The microspores were larger and more vacuolated as compared with the wild type (Fig. 6, K and L). In the M2L6, YaL4, and MYaL2 knockdowns, the vacuolated microspores were irregular and shrunken and the tapetum became swollen and broken irregularly (Fig. 6, M–O). Thus, the anther lobes were filled with the debris of abolished microspores and cytosolic constituents of the tapetum. At stage 13, the wild-type and M1L1 tapetal layer and the endothecium were completely degraded, and round and densely stained mature pollen grains were formed (Fig. 6, P and Q). This showed that the normal microspores contain starch, lipids, and other storage metabolites, and they are important for pollen viability and function. However, anther wall layers persisted at this stage in M2L6, YaL4, and MYaL2 knockdowns. The entire endothecium and the partial tapetum were maintained, and most of the pollen grains disintegrated into debris (Fig. 6, R–T), thus leading to defects in pollen maturation and morphology. Therefore, OsMAGO1 and OsMAGO2, OsY14a, and their heterodimers OsMAGO1 (or OsMAGO2)-OsY14a play an important role in anther development.
Figure 6.
Transverse sections of anthers in the wild type and down-regulated mutants. Four stages of anther development in wild-type and transgenic plants were compared. A, F, K, and P show the wild type; B, G, L, and Q show the OsMAGO1 down-regulated plant M1L1; C, H, M, and R show the OsMAGO1/OsMAGO2 down-regulated plant M2L6; D, I, N, and S show the OsY14a down-regulated plant YaL4; and E, J, Q, and T show the OsMAGO1OsMAGO2OsY14a down-regulated plant MYaL2. A to E, Stage 8b. F to J, Stage 9. K to O, Stage 10. P to T, Stage 13. En, Endothecium; Ep, epidermis; Mp, mature pollen; Msp, microspore; Ta, tapetum; Tds, tetrads. Bars = 50 µm.
Rice MAGO and Y14 Genes Affect Transcript Levels of Putative Target Genes
To determine the molecular basis of phenotypic variation in the knockdowns of rice MAGO and Y14 genes, we investigated the effects of EJC disruption on the transcript levels of randomly selected genes that were involved in vegetative and flower development and embryogenesis. For this purpose, we designed gene-specific primers flanking the full coding region or covering all exons for each gene (Supplemental Table S4). In reverse transcription (RT)-PCR, the alteration of mRNA abundance of the investigated genes without any defects in splicing was predominant in Ubi:OsMAGO2-CDS-RNAi (M2L6), Ubi:OsY14a-RNAi (YaL4), and Ubi:OsMAGO1-CDS-OsY14a-RNAi (MYaL2), with down-regulation of OsMAGO1OsMAGO2 and OsY14a genes in comparison with wild-type and transgenic controls (M1L1 with a severe down-regulation of OsMAGO1 and YaL7 without any down-regulation of OsY14a; Supplemental Fig. S7). The genes showing the down-regulation of mRNA expression were OsGA20ox2 (GA 20-OXIDASE2; Qiao and Zhao, 2011), OsGAMYB (an R2R3 MYB transcription factor as a positive GA-signaling component; Kaneko et al., 2004), OsEUI (for elongated uppermost internode; Zhu et al., 2006), OsCIPK23 (CALCINEURIN B-LIKE INTERACTING PROTEIN KINASE23; Yang et al., 2008), OsC6 (encoding a lipid transfer protein; Zhang et al., 2010a), OsTDR (for tapetum degeneration retardation; Li et al., 2006), OsCSA (for carbon-starved anther; Zhang et al., 2010b), and OsMST8 (MONOSACCHARIDE TRANSPORTER8; Mamun et al., 2006). However, the PERSISTENT TAPETAL CELL1 transcripts (Li et al., 2011) increased. Furthermore, the mature mRNA expression of some genes was not altered in these knockdowns. These were OsRBP (a double-stranded RNA-binding protein; Tang et al., 2002), OsRab5a (Wang et al., 2010), OsGSR1 (for GA-stimulated gene; Wang et al., 2009), OsSIN (for short internodes; Han et al., 2005), OsMSP1 (MULTIPLE SPOROCYTE1; Li et al., 2011), OsINV4 (a rice anther-specific cell wall invertase gene; Oliver et al., 2005), OsLEAFY (NM_001060277), OsMADS18 (Fornara et al., 2004), OsMADS8 (Kang et al., 1997), and OsACTIN1 (NM_001057621). Interestingly, multiple splicing products of the OsUDT1 (Jung et al., 2005) gene were visible in the down-regulation mutants of OsMAGO1OsMAGO2 and OsY14a genes (Supplemental Fig. S7).
The expression of these genes was also investigated (red box in Supplemental Fig. S7) in Ubi:OsY14b-RNAi callus (YbL1), which showed increased expression of OsRBP in comparison with the wild type. On the other hand, the mature transcripts of OsEUI, OsCIPK23, OsGAMYB, and OsCSA genes were severely down-regulated, and OsRab5a, OsGSR1, OsSIN, OsMSP1, OsMST8, and OsACTIN1 expression seemed unchanged. The expression of other genes was not detectable in the callus.
Rice MAGO and Y14 Regulate the Splicing of the OsUDT1 Transcripts
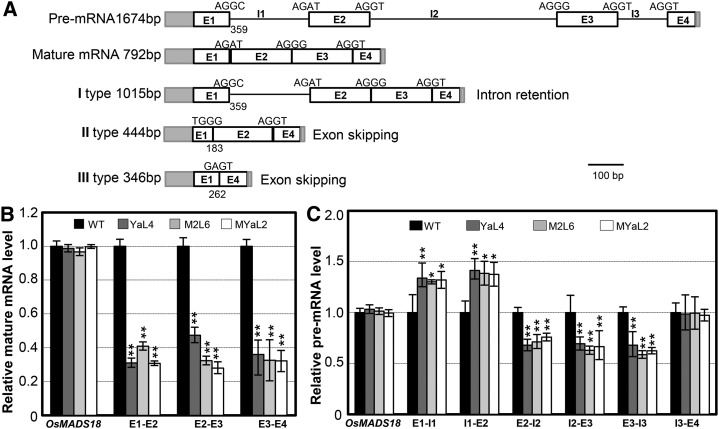
The occurrence of multiple splicing products of the OsUDT1 transcripts in the knockdowns of the rice EJC genes (Supplemental Fig. S7) indicated that the splicing of this gene was affected in these RNAi lines. In order to ascertain the nature of these transcript species, we cloned and sequenced the RT-PCR products from both the wild-type rice and Ubi:Y14a-RNAi plants. As compared with the expected mature transcripts, three abnormal transcript species (types I, II, and III) of the OsUDT1 gene were produced (Fig. 7A). Intron retention was observed in the type I species, and exon skipping events occurred in the type II and III transcript species. Interestingly, the first exon was misspliced, resulting in a different size of the first intron observed in the type II and III transcript species. These results suggested that these EJC subunits play a critical role in faithful splicing of the OsUDT1 premRNA.
Figure 7.
Rice MAGO and Y14 affect the OsUDT1 transcripts. A, OsUDT1 transcript species from the knockdown of EJC genes. The premRNA and mature mRNA are given as controls. Gray rectangles, UTRs; white rectangles, exons (E); lines, introns (I). The numbers represent the lengths of the 5′UTR and E1. Three abnormal transcript species were obtained: I type; the first intron was retained because of the GC-AG boundary; II type, lacking partial E1 and entire E3; III type, lacking partial E1 and the whole E2 and E3. B, The OsUDT1 mature mRNA level is reduced in knockdowns of rice EJC genes. Primers spanning each exon-exon junction were used in qRT-PCR. C, PremRNA variations of the OsUDT1 gene in knockdowns of rice EJC genes. Total RNAs from the inflorescence of the wild type (WT) and three RNAi lines, M2L6, YaL4, and MYaL2, as indicated were used for qRT-PCR using primers spanning each exon-intron junctions. In B and C, the MADS box gene OsMAGO18 was used as a control. Three independent biological samples were used, and error bars represent sd. Significance was evaluated by two-tailed Student’s t test: *P < 0.05, **P < 0.01.
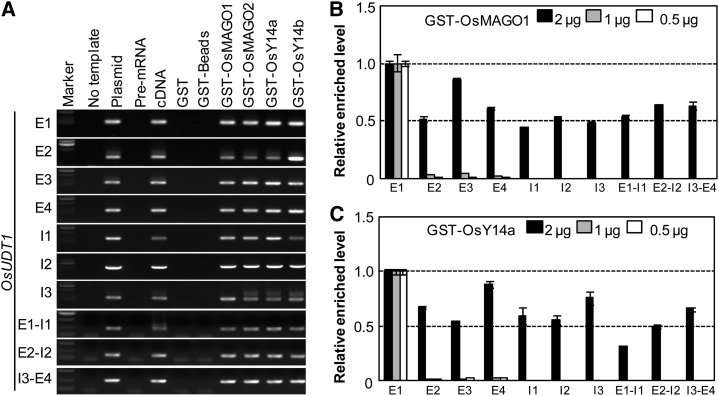
To further evaluate this, we examined the expression level of the premRNA and mRNA of the OsUDT1 gene by qRT-PCR using primers to amplify both intron-exon and exon-exon junctions (Fig. 7, B and C). A reduction in the amount of the mature OsUDT1 transcripts in transgenic plants M2L6, YaL4, and MYaL2 was apparent as compared with the wild type (Fig. 7B). The variability of premRNA levels over the length of the OsUDT1 gene (Fig. 7C) further suggested an abnormal splicing of the OsUDT1 transcripts in knockdowns of rice EJC genes. We next synthesized the fusion proteins of rice MAGO and Y14 with glutathione S-transferase (GST) and incubated them with premRNA of 14 genes (see “Materials and Methods”). All of these fusion proteins could specifically bind to the OsUDT1 premRNA after elution, as demonstrated by saturated RT-PCR amplification (Supplemental Fig. S8). Surprisingly, we found that the rice MAGO and Y14 proteins could bind to all regions of the OsUDT1 premRNA when a high dosage of each premRNA segment was incubated with each GST fusion protein (Fig. 8A). To clarify this, we did competitive binding assays through reducing the concentrations of the EJC subunits. Once a low concentration of GST-OsMAGO1 (Fig. 8B) or GST-OsY14a (Fig. 8C) was input, these proteins competitively bound to the first exon region of the OsUDT1 transcripts. Similar findings were observed for GST-OsMAGO2 and OsY14b (Supplemental Fig. S9, A and B). The capability of binding to premRNA, particularly the preferential binding to the first exon region, might form a key basis for regulating the splicing of the OsUDT1 transcripts by these EJC subunits.
Figure 8.
Rice MAGO and Y14 bind to the OsUDT1 premRNA. A, Preselection of the binding region. Saturated RT-PCR was performed using each exon (E) and intron (I) premRNA from the GST fusion protein beads indicated. The no-template and premRNA samples were used as negative controls, while the plasmids and cDNAs were used as positive controls. For GST, GST-Beads, GST-MAGO, and GST-Y14 samples, 1 μg was incubated with 250 ng of the premRNAs as indicated. B and C, PremRNA competitive binding assays. The mixture of each exon and intron premRNA fragment (125 ng each) was incubated with the indicated concentration of GST-OsMAGO1 (B) or GST-Y14a (C). The relative enriched level of the bound premRNA is shown by qRT-PCR assays. The bound E1 premRNA level was set to 1. Each experiment was performed with three independent samples, and error bars represent sd.
DISCUSSION
MAGO and Y14 are the core components of EJC and play essential roles in the posttranscriptional processes and organism development in animals. However, these processes are poorly understood in plants. In this study, we show the divergence of rice MAGO and Y14 duplicated gene pairs and establish that they are all essential to the growth and development of multiple organs. Furthermore, divergent expression in response to stresses implies a clear divergence of these genes for adaptation. Moreover, OsUDT1, a key regulatory gene in stamen development, was found to be a posttranscriptional splicing target of the EJC subunits in rice.
Functional Divergence of MAGO and Y14 Duplicated Gene Pairs in Rice
We found that OsMAGO1 and OsMAGO2 were highly conserved (more than 93%) and that their genes had a similar and constitutive expression pattern during rice development. Moreover, single gene knockdown of either OsMAGO1 or OsMAGO2 did not show any phenotypic variation, while obvious phenotypic deviations from the wild type were observed in the double OsMAGO1OsMAGO2 knockdowns. Therefore, both OsMAGO1 and OsMAGO2 are redundant in rice development (Fig. 9). The OsMAGO1OsMAGO2 knockdowns affected floral growth and showed retarded vegetative growth. The lemma and palea in these knockdowns became larger, while the stamen and its filament became shorter. Furthermore, the organ size appeared to correlate to the cell size in these organs, indicating that rice MAGO could affect cell expansion. These knockdowns further showed inhibited stamen development, altered pollen morphology, and inhibited pollen maturation processes. The cytohistological assays we conducted revealed that normal degradation and absorption of both endothecium and tapetum were inhibited, thus leading to defects in pollen maturation and morphology. Defects in pollen development were also observed in hapless1 (atmago) and AtMAGO-RNAi plants in Arabidopsis (Johnson et al., 2004; Park et al., 2009) and in the knockdowns of MvMAGO in M. vestita (van der Weele et al., 2007; Boothby and Wolniak, 2011). However, the transgenic rice plants in our study did not show any defects in floral meristem and seed development, as were observed in Arabidopsis (Park et al., 2009). Thus, MAGO genes probably went through functional conservation and diversification in plants.
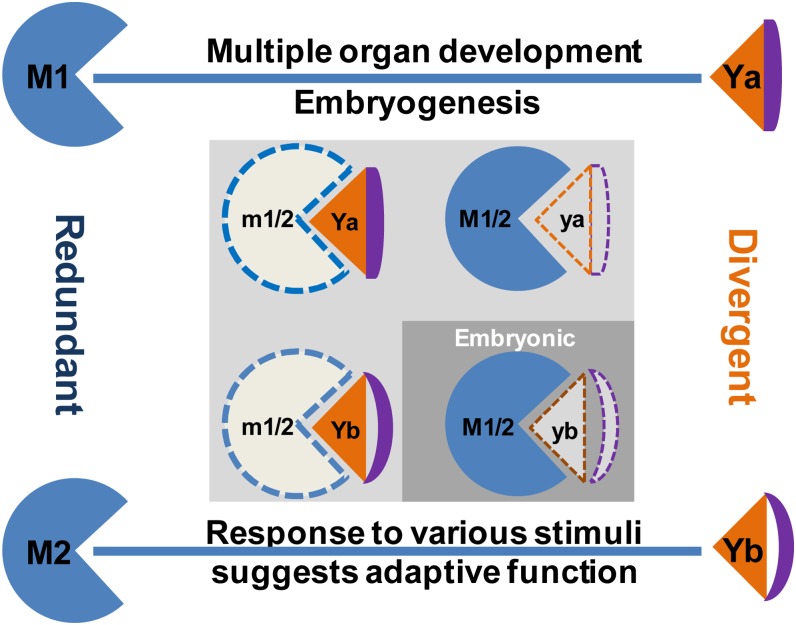
Figure 9.
Functional divergence of OsMAGO1/OsMAGO2 and OsY14a/OsY14b in rice. M1/2 and Ya/b represent OsMAGO1/OsMAGO2 and OsY14a/OsY14b, respectively. Solid patterns indicate wild-type M1/2 or Ya/b, and dotted patterns indicate m1/2 or ya/b mutants (null or knockdown). Blue lines represent heterodimerization. Only the preferential heterodimerizations are shown.
Unlike the paralogous OsMAGO1/OsMAGO2 gene pair, the OsY14a/OsY14b pair showed distinct divergence that was supported by dramatic sequence variations (less than 47%) and distinct differential gene expression patterns during rice development. OsY14a had a broad expression domain similar to that of the OsMAGO1/OsMAGO2 pair; therefore, severe down-regulation of this gene led to similar phenotypic variation to that observed in OsMAGO1OsMAGO2 double knockdowns. In contrast, OsY14b was only expressed in embryonic callus and earlier vegetative and floral development. Moreover, OsY14b protein was under positive selection, and the two paralogs became extensively diverged in their sequences (Gong et al., 2014). These facts together support a specialization of function for this gene. Similar to the mutation in AtY14 in Arabidopsis (Park et al., 2009), we also observed a lethal phenotype associated with failure in shoot induction from the embryonic callus in down-regulating OsY14b. Thus, we concluded that OsY14b plays a key role in embryo development and early embryonic organogenesis (Fig. 9). This function could not be replaced by OsY14a, suggesting that the two paralogous genes underwent dramatic functional divergence after duplication.
Similar mutational phenotypic variations were observed in the knockdowns of OsMAGO1OsMAGO2 or OsY14a, suggesting that they might exert their roles by forming heterodimers. The role of the MAGO-Y14 heterodimer has not been demonstrated, possibly due to its lethality in a species having a single copy. Due to the essential role of OsY14b, it is hard to reveal the developmental role of OsY14b and its heterodimers with OsMAGO1 and OsMAGO2 in rice. Nonetheless, using RNAi technology, we generated the triple gene (OsMAGO1OsMAGO2OsY14a) down-regulated mutants. These transgenic rice plants showed enhanced phenotypic variations that occurred in Ubi:OsMAGO1OsMAGO2-RNAi and Ubi:OsY14a-RNAi plants. As demonstrated in animals (Mohr et al., 2001; Kawano et al., 2004; Parma et al., 2007; Lewandowski et al., 2010), rice MAGO and Y14 form a complex to exert their role in the same or related developmental pathways that control growth, development, and reproduction.
Rice MAGO and Y14 Selectively Regulate the Splicing of the OsUDT1 Transcript
Abnormal growth and development in transgenic rice plants can be attributed to the abnormal expression of related genes. This was consistent with the previous observation (Qiao and Zhao, 2011) that down-regulation of OsGA20ox2 reduced plant height in rice. The multiple defects in pollen development and male sterility are thought to be associated with abnormal expression of various genes, such as OsCSA encoding an MYB domain protein that regulates sugar partitioning (Zhang et al., 2010b), OsGAMYB (Kaneko et al., 2004), OsUDT1 (Jung et al., 2005), OsTDR (Li et al., 2006), and OsC6 (Zhang et al., 2010a). Another rice gene, OsINV4, is involved in Suc accumulation and pollen sterility in rice (Oliver et al., 2005), while OsMSP1 controls early sporogenic development (Li et al., 2011). However, the expression of neither of these genes was affected by down-regulating the rice EJC genes. Our observations also support that multiple regulatory pathways are involved in pollen development in rice (Wang et al., 2013). The interconnections of EJC subunits are important for correct gene expression in Arabidopsis (Mufarrege et al., 2011). This is corroborated by our data, which show that rice EJC subunits may fulfill their essential roles by regulating a substantial set of gene expression.
The altered expression levels of these genes are unknown; however, the altered splicing of the OsUDT1 transcripts was clearly supported by the occurrence of multiple splicing products of this gene in our rice EJC knockdowns. Additional sequencing suggested that the regulation of OsUDT1 transcript splicing by rice MAGO proteins and OsY14a was fulfilled in a combined manner consisting of intron retention, exon skipping, and recognition of alternative splice sites in the first exon. Furthermore, we found that OsMAGO1/OsMAGO2, OsY14a, and their heterodimers could bind to the OsUDT1 premRNA, particularly at the first exon. In line with our observations, the EJC subunits also control the transcript splicing of the MITOGEN-ACTIVATED PROTEIN KINASE gene in Drosophila spp. (Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010). In eukaryotic cells, NMD, as a surveillance mechanism, eliminates mRNAs that contain nonsense mutations or acquire premature termination codons because of aberrant splicing (Chang et al., 2007). Therefore, NMD is an effective safeguard for eliminating aberrant gene expression (Chang et al., 2007; Kalyna et al., 2012; Chuang et al., 2013; Nyikó et al., 2013). However, the aberrant OsUDT1 transcripts were specifically maintained despite the deletions of the rice MAGO and Y14 genes. In contrast, the expression levels of most altered genes were affected either by down- or up-regulation, suggesting a role for these EJC subunits in transcriptional regulation. Considering the nature of these EJC subunits, the ancillary subunits might be essential for the transcriptional regulatory role. This was shown by the interaction of PFMAGO with MPF2, a MADS domain regulatory factor in P. floridana (He et al., 2007). Alternatively, the observed transcriptional regulation might be a consequence of the abnormal splicing of OsUDT1, an upstream regulator in the network of male fertility in rice (Wang et al., 2013). Nevertheless, further investigations in rice are required to support these notions.
The expression of genes from various developmental pathways was affected by down-regulating rice MAGO and Y14 genes, suggesting no specificity of the EJC targets in rice. However, not all monitored gene expression was altered on disruption of either MAGO or Y14 genes in rice, suggesting that gene expression in general does not require the EJC, hence arguing for the specificity and priority of the EJC targets in rice. However, in animals, the EJC is proposed to bind to intron-containing genes (Le Hir et al., 2000), and the intron length determines the EJC’s effect on splicing in Drosophila spp. (Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010). The presence of the alternative splicing sites within the large introns containing repetitive sequences could interfere with the splicing of the introns they occupy (Ponicsan et al., 2010). However, a bias for long introns in human target units was not detected (Michelle et al., 2012) or found in rice (Supplemental Table S4). The target specificity of the rice EJC subunits is not clear yet, but OsUDT1, the upstream key regulator for male fertility (Jung et al., 2005), is apparently the direct target for the rice EJC subunits. The discrepancy and diversity of mechanisms for EJC target selection between different species suggest that they act in a species-specific evolutionary mode.
Therefore, our study suggests that MAGO and Y14 genes were separately duplicated in cereals and their sequence divergence shaped their functional evolution. OsMAGO1 and OsY14a partitioned their fundamental roles in growth, development, and reproduction through specific regulations of gene expression in rice, while OsY14b specialized in embryo organogenesis (Fig. 9). Moreover, both OsMAGO2 and OsY14b evolved to be sensitive to external changes. These findings in rice indicate the importance of these EJC genes in cereal development and adaptive evolution.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa ‘Zhonghua 10’) and its transgenic plants generated in this work were cultivated in a growth chamber under short-day conditions (10-h-light/14-h-dark cycle at a temperature of 28°C and 25°C, respectively) with a relative humidity of 50%. Nicotiana benthamiana plants were grown in an incubator (RXZ-380C; Ningbo) under long-day conditions (16-h-light/8-h-dark cycle) at a constant temperature of 22°C.
Abiotic Stresses and Hormonal Treatments
For abiotic stresses, 12-d-old rice seedlings on one-half-strength Murashige and Skoog (1/2 MS) medium were irrigated with 20% (w/v) mannitol or 200 mm NaCl solution. Hot and cold stresses were provided by placing the seedlings in an incubator maintained at 4°C and 42°C, respectively. Seedlings were sampled at 0, 1, 4, 8, 12, 24, 36, and 48 h after treatments. For hormonal treatments, seedlings were treated with 100 μm ABA, 1 μm BR, 10 μm GA3, and 10 μm IAA and then sampled at 0, 1, 3, 8, and 24 h. ABA (A1049), BR (E1641), GA3 (G7645), and IAA (I2886) were purchased from Sigma-Aldrich.
RT-PCR Analyses
Total RNA from the tissues indicated was isolated using the Trizol kit (Invitrogen) and then treated with RNase-free DNase (Promega) to remove genomic DNA contamination. First-strand complementary DNA (cDNA) was synthesized using the MLV RT-PCR kit (Invitrogen) in a 20-µL reaction with the oligo(dT)17 primer. qRT-PCR was performed using the SYBR Premix Ex Taq (Tli RNaseH plus) kit (Takara) in an Mx3000P real-time PCR instrument (Stratagene) in a final 25-μL volume. The reactions were performed at 95°C for 30 s, 40 cycles of 95°C for 5 s, 60°C for 40 s, and then 60 s at 95°C, 30 s at 60°C, and 30 s at 95°C, and one cycle for melting curve analysis. Experiments were performed using three independent biological samples. Means and sd are presented. Semiquantitative RT-PCR was performed using the ExTaq system (Takara) in a final 25-μL volume. The reactions were performed at 95°C for 3 min, 24, 28, or 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min, and then 72°C for 10 min. The PCR products were separated on a 1.2% (w/v) agarose gel. Typical gels from three experiments on independent biological samples are presented. The OsACTIN1 gene was used as an internal control.
Plasmid Construction
The cDNAs containing ORFs were cloned into the pGEM-T EASY vector (Promega) and sequenced (Gong et al., 2014). To generate constructs for RNAi, each coding region of OsMAGO1 and OsMAGO2, the 3′UTR of OsMAGO1, and the gene-specific fragments of OsY14a and OsY14b was inserted into the vector pTCK303 driven by the ubiquitin promoter (Wang et al., 2004) to make the pTCK303-OsMAGO1CDS, pTCK303-OsMAGO2CDS, pTCK303-OsMAGO1-3′UTR, pTCK303-OsY14a, and pTCK303-OsY14b plasmids, respectively. pTCK303-OsMAGO1CDS-OsY14a was also made for triple gene knockdowns. For subcellular localization, the ORFs of rice MAGO and Y14 were cloned into the expression vector pCAMBIA1302 to generate pCAMBIA1302-OsMAGO1, pCAMBIA1302-OsY14a, and pCAMBIA1302-OsY14b using primers with restriction digestion sites NcoI and SpeI. The ORF of OsMAGO2 was cloned into expression vector pSuper1300 to generate pSuper1300-OsMAGO2 using gene-specific primers containing XbaI and BamHI cutting sites. For the BiFC experiments, the ORFs from rice MAGO and Y14 were cloned into the pSPY35SNE and pSPY35SCE vectors (Walter et al., 2004) to generate pSPYNE/CE-OsMAGO1, pSPYNE/CE-OsMAGO2, pSPYNE/CE-OsY14a, and pSPYNE/CE-OsY14b. GST fusion proteins were produced by inserting the ORFs of rice MAGO and Y14 into the pGEX-4T vector to generate pGEX-4T-OsMAGO1, pGEX-4T-OsMAGO2, pGEX-4T-OsY14a, and pGEX-4T-OsY14b.
Sequencing Analyses and Primer Synthesis
All constructs (made) and products of the RT-PCR assays were commercially sequenced. The primers used in this work (Supplemental Table S5) were commercially synthesized by the Beijing Genomic Institute and by Taihe Biotechnology.
Rice Transgenic Analyses
Rice embryonic calli were induced from germinated seeds on N6 nutrient stock with 2 mg/L of 2,4-Dichlorophenyloxy acetic acid solid medium and inoculated with Agrobacterium tumefaciens EHA105 as described previously (Xu et al., 2005). The plantlet regeneration rate is summarized in Supplemental Table S1.
Yeast Two-Hybrid Assay
The indicated combination of bait and prey plasmids in Supplemental Figure S2 was cotransformed into the Saccharomyces cerevisiae strain AH109. The nonlethal β-galactosidase activity was performed on synthetic dropout medium/-Trp-Leu-His-Ade plates. o-Nitrophenyl-β-d-galactoside was used as a substrate to quantify the interacting affinity. The procedures described in the Yeast Protocols Handbook (Clontech) were followed.
Transient Protein Expression Analyses
For subcellular localization, the derived pCAMBIA1302 constructs were first transformed into the A. tumefaciens EHA105 cells and then introduced into the epidermal cells of N. benthamiana by agroinfiltration. The empty GFP vector (pCAMBIA1302) was infiltrated as a control. For the BiFC assay, the resulting pSPY35SNE and pSPY35SCE plasmids were transformed into A. tumefaciens EHA105, and then the YFPn- and YFPc-fused proteins were coexpressed in epidermal cells of N. benthamiana by agroinfiltration (Walter et al., 2004). The GFP or YFP signal was observed with a confocal laser scanning microscope (FV1000 MPE; Olympus). The YFP signal intensity was quantified using ImageJ (http://rsb.info.nih.gov/nih-image; Abramoff et al., 2004).
Anther Histochemical and Microscopic Analysis
For the anther cytohistological assay, the spikelets of various anther developmental stages were fixed with 70% (v/v) ethanol, 5% (v/v) glacial acetic acids, and 3.7% (v/v) formaldehyde for 24 h at room temperature. Fixed anthers were dehydrated through an ethanol series, embedded into Spurr’s resin (Sigma-Aldrich), and polymerized at 60°C for 24 h. The sample blocks were sectioned into 1-μm-thick slices with a microtome (Leica Ultracut R) and stained with 0.1% (w/v) Toluidine Blue O (Merck). Pollen from mature anthers was dissected into a drop of I2-KI solution. The images were captured with a Zeiss Imager A1 Digital Camera System. For scanning electron microscopic analyses, lemma, anthers, and filaments from mature florets were fixed using 70% (v/v) ethanol, 5% (v/v) glacial acetic acids, and 3.7% (v/v) formaldehyde. Then they were dehydrated through an ethanol series followed by replacing them with an isoamyl acetate series and drying with liquid carbon dioxide. Images were captured with a scanning electron microscope (Hitachi S-800) at 10 kV. Florets and organs were photographed using a Nikon SMI 1000 microscopic digital camera. Organ and cell sizes were measured by using the software Carl Zeiss Vision Axiovision release 4.7 (http://microscopy.zeiss.com).
GST Fusion Protein Induction and Abstraction
The GST fusion protein plasmids and the empty pGEX-4T-1 vector were transformed into Escherichia coli Rosetta (DE3; TransGen), and protein expression was induced by a certain concentration (1.0 mM or 0.5 mM) of isopropyl β-d-1-thiogalactopyranoside. The soluble GST fusion proteins were extracted and immobilized onto glutathione Sepharose beads (Amersham Biosciences).
PremRNA Binding with GST Fusion Proteins
The genomic sequence of each gene was amplified using PCR amplification and constructed into T-plasmid (pEASY-T1 or pEASY Blunt Zero vector; TransGen). Sense premRNA was transcribed from the plasmid after linearization by T7 promoter (In Vitro Transcription T7 Kit; Takara). The premRNA binding with GST fusion proteins was performed as described previously (Ramasamy et al., 2006). To search the premRNA target of rice MAGO and Y14, 250 ng each premRNA was incubated with 1 μg of each GST fusion protein. For competitive binding assays, 125 ng of each intron and exon premRNA was mixed and incubated with different concentrations (2, 1, and 0.5 μg) of each GST fusion protein as indicated. Reverse transcription of purified premRNA was performed by random primer (hexadeoxyribonucleotide mixture, pd(N)6; Takara) and used for RT-PCR to detect each interaction of premRNA with a GST fusion protein.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene expression of rice MAGO and Y14 under different types of stress.
Supplemental Figure S2. Interaction of rice MAGO and Y14 proteins in yeast.
Supplemental Figure S3. Genotyping analyses of the RNAi transgenic rice plants.
Supplemental Figure S4. Variation in cell size in wild-type and transgenic rice plants.
Supplemental Figure S5. Ubi:OsY14b-RNAi transgenic rice plants.
Supplemental Figure S6. Phenotype of OsMAGO1-3′UTR-RNAi transgenic rice plants.
Supplemental Figure S7. Gene expression profiles in the transgenic rice plants.
Supplemental Figure S8. Rice MAGO and Y14 specifically bind to the OsUDT1 premRNA.
Supplemental Figure S9. OsMAGO2 and OsY14b proteins bind to the OsUDT1 premRNA.
Supplemental Table S1. Transformation of rice RNAi lines.
Supplemental Table S2. Phenotypic variations in RNAi transgenic rice plants.
Supplemental Table S3. Ovary fertility alteration in transgenic rice plants.
Supplemental Table S4. Gene information used in expression studies.
Supplemental Table S5. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Kang Chong for generously providing the vector plasmid to prepare constructs in transgenic rice, Yinghou Xiao for scanning electron microscopic analysis, Dr. Li Wang for rice transgenic analyses, and Fengqin Dong for transverse sections.
Glossary
- EJC
exon junction complex
- NMD
nonsense-mediated mRNA decay
- qRT
quantitative reverse transcription
- BR
brassinolide
- ABA
abscisic acid
- IAA
indole acetic acid
- ORF
open reading frame
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- RNAi
RNA interference
- I2-KI
iodine-potassium iodide
- UTR
untranslated region
- RT
reverse transcription
- 1/2 MS
one-half-strength Murashige and Skoog
- cDNA
complementary DNA
Footnotes
This work was supported by the Chinese Ministry of Agriculture Transgenic Major Project (grant no. 2009ZX08009–011B) and by the National Natural Science Foundation of China (grant no. 31370259).
The online version of this article contains Web-only data.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Alachkar A, Jiang D, Harrison M, Zhou Y, Chen G, Mao Y. (2013) An EJC factor RBM8a regulates anxiety behaviors. Curr Mol Med 13: 887–899 [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrançois M, Chagnon P, Gendron P, Caron-Lizotte O, Bonneil E, Thibault P, et al. (2010) The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143: 251–262 [DOI] [PubMed] [Google Scholar]
- Boothby TC, Wolniak SM. (2011) Masked mRNA is stored with aggregated nuclear speckles and its asymmetric redistribution requires a homolog of Mago nashi. BMC Cell Biol 12: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell RE, Prout ME, Steichen JC. (1991) Mutations in a newly identified Drosophila melanogaster gene, mago nashi, disrupt germ cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development 113: 373–384 [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. (2007) The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chen YR, Shaw JF, Chung MC, Chu FH. (2007) Molecular identification and characterization of Tcmago and TcY14 in Taiwania (Taiwania cryptomerioides). Tree Physiol 27: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Chuang TW, Chang WL, Lee KM, Tarn WY. (2013) The RNA-binding protein Y14 inhibits mRNA decapping and modulates processing body formation. Mol Biol Cell 24: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Parenicová L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM. (2004) Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol 135: 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P, Zhao M, He C. (2014) Slow co-evolution of the MAGO and Y14 protein families is required for the maintenance of their obligate heterodimerization mode. PLoS ONE 9: e84842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. (2001) Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr Biol 11: 1666–1674 [DOI] [PubMed] [Google Scholar]
- Han Y, Jiang JF, Liu HL, Ma QB, Xu WZ, Xu YY, Xu ZH, Chong K. (2005) Overexpression of OsSIN, encoding a novel small protein, causes short internodes in Oryza sativa. Plant Sci 169: 487–495 [Google Scholar]
- He C, Sommer H, Grosardt B, Huijser P, Saedler H. (2007) PFMAGO, a MAGO NASHI-like factor, interacts with the MADS-domain protein MPF2 from Physalis floridana. Mol Biol Evol 24: 1229–1241 [DOI] [PubMed] [Google Scholar]
- Inaki M, Kato D, Utsugi T, Onoda F, Hanaoka F, Murakami Y. (2011) Genetic analyses using a mouse cell cycle mutant identifies magoh as a novel gene involved in Cdk regulation. Genes Cells 16: 166–178 [DOI] [PubMed] [Google Scholar]
- Johnson MA, von Besser K, Zhou Q, Smith E, Aux G, Patton D, Levin JZ, Preuss D. (2004) Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G. (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. (2012) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 40: 2454–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, et al. (2004) Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Jang S, Chung JE, Cho YG, An G. (1997) Characterization of two rice MADS box genes that control flowering time. Mol Cells 7: 559–566 [PubMed] [Google Scholar]
- Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. (2001) Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J 20: 6424–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell 6: 673–682 [DOI] [PubMed] [Google Scholar]
- Kawano T, Kataoka N, Dreyfuss G, Sakamoto H. (2004) Ce-Y14 and MAG-1, components of the exon-exon junction complex, are required for embryogenesis and germline sexual switching in Caenorhabditis elegans. Mech Dev 121: 27–35 [DOI] [PubMed] [Google Scholar]
- Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. (2001) The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J 20: 2062–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Choe J, Chi SG, Kim YK. (2009) Exon junction complex enhances translation of spliced mRNAs at multiple steps. Biochem Biophys Res Commun 384: 334–340 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. (2001) The protein Mago provides a link between splicing and mRNA localization. EMBO Rep 2: 1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. (2000) The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski JP, Sheehan KB, Bennett PE, Jr, Boswell RE. (2010) Mago Nashi, Tsunagi/Y14, and Ranshi form a complex that influences oocyte differentiation in Drosophila melanogaster. Dev Biol 339: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson ZA, Zhang D. (2011) PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol 156: 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Boswell R, Wood WB. (2000) mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev Biol 218: 172–182 [DOI] [PubMed] [Google Scholar]
- Mamun EA, Alfred S, Cantrill LC, Overall RL, Sutton BG. (2006) Effects of chilling on male gametophyte development in rice. Cell Biol Int 30: 583–591 [DOI] [PubMed] [Google Scholar]
- Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture S, et al. (2012) Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol Cell Biol 32: 954–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, González-Reyes A, St Johnston D. (1997) The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol 7: 468–478 [DOI] [PubMed] [Google Scholar]
- Mohr SE, Dillon ST, Boswell RE. (2001) The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev 15: 2886–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufarrege EF, Gonzalez DH, Curi GC. (2011) Functional interconnections of Arabidopsis exon junction complex proteins and genes at multiple steps of gene expression. J Exp Bot 62: 5025–5036 [DOI] [PubMed] [Google Scholar]
- Newmark PA, Boswell RE. (1994) The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development 120: 1303–1313 [DOI] [PubMed] [Google Scholar]
- Newmark PA, Mohr SE, Gong L, Boswell RE. (1997) mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila. Development 124: 3197–3207 [DOI] [PubMed] [Google Scholar]
- Nyikó T, Kerényi F, Szabadkai L, Benkovics AH, Major P, Sonkoly B, Mérai Z, Barta E, Niemiec E, Kufel J, et al. (2013) Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res 41: 6715–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Van Dongen JT, Alfred SC, Mamun EA, Zhao XC, Saini HS, Fernandes SF, Blanchard CL, Sutton BG, Geigenberger P, et al. (2005) Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ 28: 1534–1551 [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E. (2004) An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757 [DOI] [PubMed] [Google Scholar]
- Park NI, Yeung EC, Muench DG. (2009) Mago Nashi is involved in meristem organization, pollen formation, and seed development in Arabidopsis. Plant Sci 176: 461–469 [DOI] [PubMed] [Google Scholar]
- Parma DH, Bennett PE, Jr, Boswell RE. (2007) Mago Nashi and Tsunagi/Y14, respectively, regulate Drosophila germline stem cell differentiation and oocyte specification. Dev Biol 308: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponicsan SL, Kugel JF, Goodrich JA. (2010) Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev 20: 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Zhao KJ. (2011) The influence of RNAi targeting of OsGA20ox2 gene on plant height in rice. Plant Mol Biol Rep 29: 952–960 [Google Scholar]
- Ramasamy S, Wang H, Quach HN, Sampath K. (2006) Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells. Dev Biol 292: 393–406 [DOI] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. (2010) Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143: 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xu RM. (2003) Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev 17: 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Leeds KE, Hwang HW, Miller EE, Pavan WJ. (2013) The EJC component Magoh regulates proliferation and expansion of neural crest-derived melanocytes. Dev Biol 375: 172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XR, Wu H, Jia M, Yu XH, He YK. (2002) Isolation and expressional analysis of cDNA encoding a dsRNA binding protein homologue OsRBP of rice. J Plant Physiol Mol Biol 28: 41–45 [Google Scholar]
- Tange TO, Nott A, Moore MJ. (2004) The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol 16: 279–284 [DOI] [PubMed] [Google Scholar]
- van der Weele CM, Tsai CW, Wolniak SM. (2007) Mago nashi is essential for spermatogenesis in Marsilea. Mol Biol Cell 18: 3711–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang K, Peng X, Ji Y, Yang P, Zhu Y, Li S. (2013) Gene, protein, and network of male sterility in rice. Front Plant Sci 4: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, Xu Z, Wang Z, Chong K. (2009) OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J 57: 498–510 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ren Y, Liu X, Jiang L, Chen L, Han X, Jin M, Liu S, Liu F, Lv J, et al. (2010) OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J 64: 812–824 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen CB, Xu YY, Jiang RX, Han Y, Xu ZH, Chong K. (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22: 409–417 [Google Scholar]
- Xu ML, Jiang JF, Ge L, Xu YY, Chen H, Zhao Y, Bi YR, Wen JQ, Chong K. (2005) FPF1 transgene leads to altered flowering time and root development in rice. Plant Cell Rep 24: 79–85 [DOI] [PubMed] [Google Scholar]
- Yang W, Kong Z, Omo-Ikerodah E, Xu W, Li Q, Xue Y. (2008) Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). J Genet Genomics 35: 531–543, S1–S2 [DOI] [PubMed] [Google Scholar]
- Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D. (2010a) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 154: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DB, Wilson ZA. (2009) Stamen specification and anther development in rice. Chin Sci Bull 54: 2342–2353 [Google Scholar]
- Zhang H, Liang W, Yang X, Luo X, Jiang N, Ma H, Zhang D. (2010b) Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22: 672–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XF, Nowak NJ, Shows TB, Aplan PD. (2000) MAGOH interacts with a novel RNA-binding protein. Genomics 63: 145–148 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, et al. (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.