Disruption of multiple auxin-input pathways reveals developmental roles for these auxin forms and a feedback loop necessary for maintaining optimal free hormone levels.
Abstract
Auxin is a phytohormone involved in cell elongation and division. Levels of indole-3-acetic acid (IAA), the primary auxin, are tightly regulated through biosynthesis, degradation, sequestration, and transport. IAA is sequestered in reversible processes by adding amino acids, polyol or simple alcohols, or sugars, forming IAA conjugates, or through a two-carbon elongation forming indole-3-butyric acid. These sequestered forms of IAA alter hormone activity. To gain a better understanding of how auxin homeostasis is maintained, we have generated Arabidopsis (Arabidopsis thaliana) mutants that combine disruptions in the pathways, converting IAA conjugates and indole-3-butyric acid to free IAA. These mutants show phenotypes indicative of low auxin levels, including delayed germination, abnormal vein patterning, and decreased apical dominance. Root phenotypes include changes in root length, root branching, and root hair growth. IAA levels are reduced in the cotyledon tissue but not meristems or hypocotyls. In the combination mutants, auxin biosynthetic gene expression is increased, particularly in the YUCCA/Tryptophan Aminotransferase of Arabidopsis1 pathway, providing a feedback mechanism that allows the plant to compensate for changes in IAA input pathways and maintain cellular homeostasis.
The phytohormone auxin regulates cell elongation and division. Indole-3-acetic acid (IAA) is the primary auxin, although related compounds have been identified in plants and several synthetic compounds produce auxin-like responses. IAA signaling affects expression of thousands of genes (Paponov et al., 2008; Simon and Petrášek, 2011), and nontranscriptional activity, including changes in membrane transport, regulates additional physiological responses (Perrot-Rechenmann, 2010; Simon and Petrášek, 2011). Plants require auxin for root development, shoot apical dominance, tropic responses, and flowering as well as many other aspects of growth and development. Auxin application to wild-type Arabidopsis (Arabidopsis thaliana) roots increases lateral root formation and decreases primary root length, although excess auxin leads to plant death. Mutants with low IAA levels have decreased lateral root number and increased primary root length (Evans et al., 1994; Rampey et al., 2004; Strader et al., 2011).
As might be expected with the critical roles of this hormone, IAA levels are tightly regulated through biosynthesis, degradation, and sequestration (Fig. 1A; Korasick et al., 2013). These alternative strategies for IAA metabolism seem to underlie the macroevolutionary differences among land plants in the regulation of developmental processes, including embryo development and meristem structure (Ester Sztein et al., 1999). IAA can be synthesized de novo through pathways that are either Trp dependent or Trp independent (Normanly, 1997; Woodward and Bartel, 2005). In the best characterized pathway, Trp is converted to IAA by Tryptophan Aminotransferase of Arabidopsis1 (TAA1) and the YUCCA (YUC) family of flavin monooxygenases (Mashiguchi et al., 2011). IAA degradation occurs in a nonreversible process, in which IAA is oxidized to oxindole-3-acetic acid directly or by first being converted to IAA-Asp or IAA-Glu (Normanly et al., 1995; Ljung, 2013).
Figure 1.
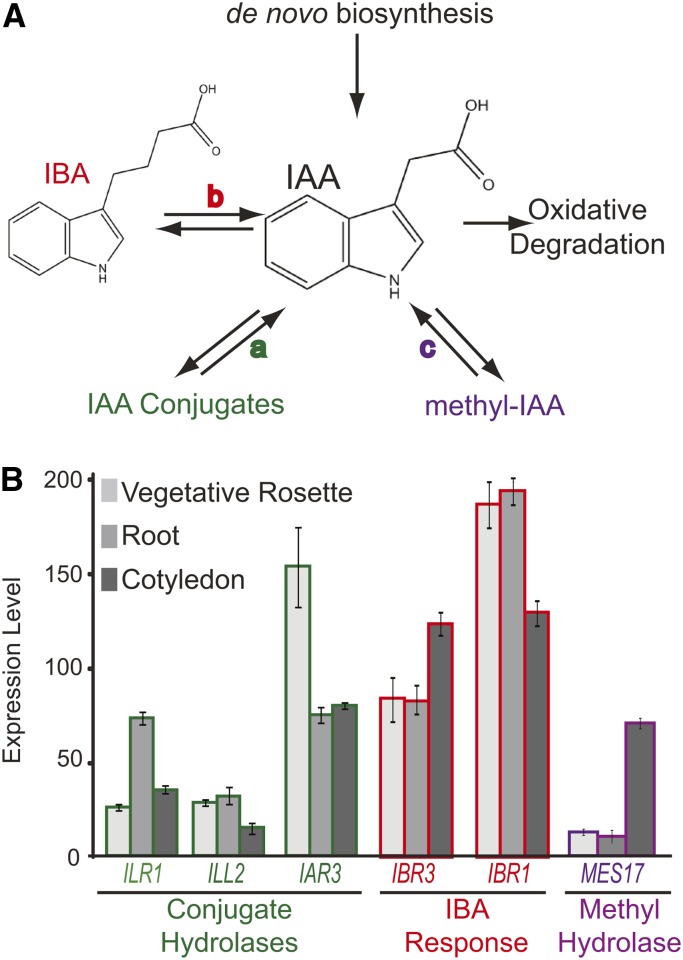
IAA homeostasis pathways. A, Free IAA levels are maintained in plant cells by several mechanisms, including de novo biosynthesis and oxidative degradation. IAA-conjugate hydrolases (a) hydrolyze IAA conjugates to free IAA. IBA-response genes (b) are necessary to metabolize IBA to free IAA. MeIAA is converted to free IAA by MES17 (c). B, Genes involved in IAA input pathways are differentially expressed throughout development, which was revealed by the Arabidopsis eFP browser (Winter et al., 2007). Expression levels at three stages are shown here as an example.
Another mechanism to reduce the level of active auxin in a cell is sequestration. The cellular auxin pool includes both free IAA (the active signaling molecule) as well as inactive precursors or conjugated forms (Korasick et al., 2013). IAA can be stored in an inactive state to allow for quick release or vascular transport or prevent oxidative degradation (Cohen and Bandurski, 1982). Sequestration occurs by reversible processes, including addition of amino acids, peptides, or sugars. Additionally, indole-3-butyric acid (IBA), a two-carbon elongation of IAA, has been described as an IAA input. Gas chromatography-mass spectrometry analysis of many plants, including Arabidopsis, tobacco (Nicotiana tabacum), soybean (Glycine max), bean (Phaseolus vulgaris), rice (Oryza sativa), and maize (Zea mays), has identified IBA and conjugated forms of IAA (Cohen and Bandurski, 1982; Slovin et al., 1999; Tam et al., 2000; Walz et al., 2002; Normanly et al., 2010; Spiess and Zolman, 2013). IBA is found at lower levels in plants than IAA, leading to questions regarding its assignment as a storage form (Strader et al., 2011; Hu et al., 2012), and another study has reported that IBA was not detected in Arabidopsis, aspen (Populus spp.), or wheat (Triticum aestivum; Novák et al., 2012). IAA conjugates are found at high levels, especially in plants grown in the presence of IAA (Bajguz and Piotrowska, 2009).
IAA-amino acid conjugates are formed by GRETCHEN HAGEN3 (GH3) enzymes (Staswick et al., 2005). These genes were identified as auxin inducible and maintain auxin homeostasis at high IAA levels by conjugating, hence inactivating, IAA (Hagen and Guilfoyle, 1985; Staswick et al., 2005). Dominant gh3 mutants have low auxin phenotypes, including rolled leaves, smaller leaves, and decreased lateral roots (Park et al., 2007). Conjugates include linkages to Leu, Ala (Kowalczyk and Sandberg, 2001), Val (Ludwig-Müller et al., 2009), Gly, Phe (Pencík et al., 2009), and Trp (Staswick, 2009). Other conjugates, including IAA-Asp and IAA-Glu, seem to be degradation precursors, because application to Arabidopsis does not promote auxin responses (Ljung et al., 2002; Woodward and Bartel, 2005). Amidohydrolases convert storage forms back to free IAA. In vitro, Indole-3-Acetic Acid Alanine-Resistant3 (IAR3) and Indole-3-Acetic Acid Leu-Resistant1-Like2 (ILL2) hydrolyze IAA-Ala to free IAA (LeClere et al., 2002). Indole-3-Acetic Acid Leu-Resistant1 (ILR1) and ILL2 hydrolyze IAA-Phe and IAA-Leu to free IAA (Davies et al., 1999; LeClere et al., 2002). ill2 ilr1 iar3 triple mutants show decreased lateral root formation and increased primary root length (Rampey et al., 2004). Free IAA levels are reduced (Rampey et al., 2004), suggesting the importance of this input pathway for IAA homeostasis.
Methyl indole-3-acetic acid (MeIAA) is made by esterification of a carboxyl group by indole-3-acetic acid carboxyl methyltransferase1 (IAMT1); Methyl Esterase17 (MES17) hydrolyzes MeIAA to free IAA, which is shown in feeding experiments (Qin et al., 2005; Yang et al., 2008). IAMT1 was identified by its biochemical activity in vitro (Zubieta et al., 2003). An iamt1-D dominant mutant causes curled leaves, and IAMT overexpression disrupts gravitropic responses and root elongation mediated by changes in auxin-induced gene expression (Qin et al., 2005). The mes17 mutant has increased hypocotyl length (Qin et al., 2005; Yang et al., 2008). These results suggest a role for IAMT1 and MES17 in auxin metabolism. However, MeIAA has not been identified in plants (Qin et al., 2005), and the importance of MeIAA as an independent metabolite or a precursor for longer ester conjugates has not been established.
In Arabidopsis seedlings, IBA has been reported to account for 25% to 30% of the total auxin pool (Ludwig-Müller, 2000; Simon and Petrášek, 2011). The mechanism for IBA synthesis is unknown; IBA may be made similarly to IAA de novo biosynthesis or in a process similar to fatty acid biosynthesis (Ludwig-Müller, 2000). Whereas IAA conjugates and MeIAA are converted to IAA in single-step processes, IBA is converted to free IAA in a multistep process similar to fatty acid β-oxidation (Zolman et al., 2008; Strader et al., 2011). Four enzymes have been proposed to act in this pathway: Indole-3-Butyric Acid Response1 (IBR1), IBR3, IBR10, and Enoyl-CoA Hydratase2 (ECH2). Feeding experiments with labeled IBA showed that loss of these enzymes directly disrupted IBA conversion to IAA (Zolman et al., 2008; Strader et al., 2011). Mutants in these four enzymes have defects in plant development, including lateral rooting, vasculature patterning, and root hair expansion, mediated by changes in auxin-induced gene expression (Zolman et al., 2000; Strader et al., 2011).
Auxin input pathways are differentially expressed in individual tissues and developmental stages (Fig. 1B; Winter et al., 2007). For example, ILR1, which hydrolyzes IAA-Leu and IAA-Phe to free IAA, is expressed primarily in 2-d-old cotyledons, whereas IAR3 is primarily expressed in vascular tissue (Rampey et al., 2004). IBA response genes are most highly expressed in early seedling development but are expressed throughout the lifecycle (Zolman et al., 2008). MES17 expression is low, with the highest levels in cotyledons and stems (Yang et al., 2008). In addition, expression and activity of these pathways are connected to environmental stress conditions in many plant species (Titarenko et al., 1997; Tognetti et al., 2010; Kinoshita et al., 2012; Woldemariam et al., 2012; Salopek-Sondi et al., 2013).
Although details about the individual pathways have been elucidated, we do not know how plants use each pathway in combination to maintain auxin homeostasis. When one auxin input pathway is lost, specific auxin responses change (Table I). In this article, we examine the overlapping roles of multiple pathways producing IAA. Our data indicate that input pathways work together to regulate IAA levels. Mutations in two pathways lead to reductions in auxin levels with increasing disruptions to physiological responses. To compensate for low auxin levels, the plant then activates a specific set of auxin-biosynthetic genes, including those involved in de novo IAA biosynthesis.
Table I. Combination mutants used in this study were made by crossing previously characterized single and double mutants.
| Mutants | Mutation(s) | Description |
|---|---|---|
| ibr1a | Arg → Cys | Short-chain dehydrogenase; disruption leads to IBA response phenotypes in root elongation, lateral root initiation |
| ibr3b | Glu → Lys, splice site mutation | Acyl-CoA dehydrogenase; disruption leads to IBA response phenotypes in root elongation, lateral root initiation |
| mes17c | Transfer DNA (T-DNA) insertion | Methyl esterase; disruption leads to increased hypocotyl length and resistance to MeIAA |
| ill2 iar3d | T-DNA in ILL2 | IAA-conjugate hydrolases; disruption leads to resistance to IAA-Ala |
| T-DNA in IAR3 | ||
| ill2 ilr1d | T-DNA in ILL2 | IAA-conjugate hydrolases; disruption leads to resistance to IAA-Leu and IAA-Phe |
| Deletion in ILR1 | ||
| ill2 iar3 ibr3 | Triple mutant combining disruptions in IBA response and IAA-conjugate hydrolysis | |
| ill2 ilr1 iar3 ibr1 | Quadruple mutant combining disruptions in IBA response and IAA-conjugate hydrolysis | |
| mes17 ill2 iar3 | Triple mutant combining disruptions in MeIAA hydrolysis and IAA-conjugate hydrolysis | |
| mes17 ibr3 | Double mutant combining disruptions in MeIAA hydrolysis and IBA response |
RESULTS
To better understand the pathways influencing IAA levels, we made higher order mutants that disrupted these pathways in combination (Table I). We combined defects in IBA metabolism and IAA-conjugate hydrolases to study the effects of losing these sequestration forms. These mutants are ill2 iar3 ibr3 and ill2 ilr1 iar3 ibr1. We also made mutants with defects in mes17 and IBA metabolism, mes17 ibr3, as well as mes17 and IAA-conjugate hydrolases, mes17 ill2 iar3, to determine the importance of MES17-mediated metabolism. Together, these mutants ablate three potential IAA input pathways but leave de novo biosynthesis functional (Fig. 1A).
Each mutation was confirmed by PCR-based genotyping (Supplemental Table S1) and analysis of root length on exogenous auxins (Supplemental Fig. S1). For instance, the ill2 iar3 ibr3 and ill2 iar3 ilr1 ibr1 mutants showed resistance to root inhibition on IAA-Ala and IBA, which was expected based on the parent mutant phenotypes. mes17 containing mutants were confirmed by expression analysis (Supplemental Fig. S1; data not shown).
Combination Mutants Disrupt Germination and Seedling Development
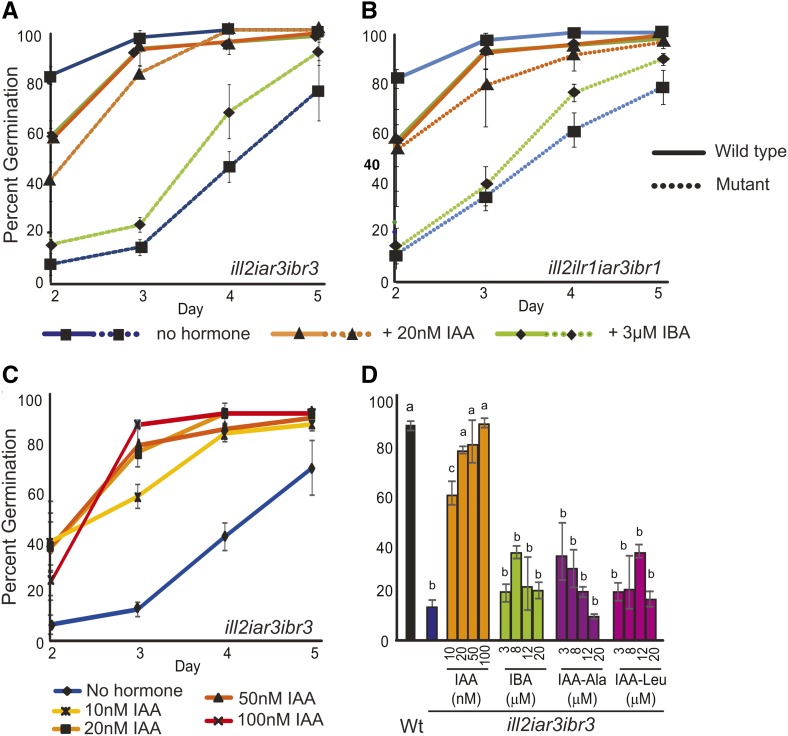
Germination defects were observed in plants combining IBA metabolism and IAA-conjugate hydrolase mutations (Fig. 2). Germination defects are not observed in any of the parental lines (data not shown), indicating that the combination of mutations in the input pathway is responsible for the phenotype. Conversely, the mes17 ill2 iar3 and mes17 ibr3 mutants showed weaker germination defects (Supplemental Fig. S2), suggesting that IBA and IAA conjugates play a larger role in germination than MES17 activity. Notably, the mutant lines germinate to approximately wild-type levels overall but show a delay in germination of 2 to 3 d.
Figure 2.
Combination mutants with defects in IBA metabolism and IAA-conjugate hydrolysis have germination delays. A and B, Wild-type and ill2 iar3 ibr3 (A) or ill2 ilr1 iar3 ibr1 (B) germination was measured on media with no hormone, 3 µm IBA, or 20 nm IAA. Data points are the averages from three germination experiments. Values on d 2 to 4 for the mutants on no hormone or IBA are significantly different than corresponding lines grown on 20 nm IAA or the wild type (P < 0.05). C, IAA rescues the ill2 iar3 ibr3 germination defect in a concentration-dependent manner. D, The percentage of plants germinated by d 3 for the wild type and the ill2 iar3 ibr3 mutant on no hormone or increasing concentrations of IAA, IBA, IAA-Ala, and IAA-Leu. n = 3; error bars represent the se, and a to c indicate significant differences using one-way ANOVA (P < 0.05). Wt, Wild type.
This result suggests that either decreases in free IAA or accumulation of IBA or IAA conjugates to toxic levels are delaying germination in mutant seeds. The germination delays observed in the IBA metabolism/IAA-conjugate hydrolase mutants were rescued with low amounts of exogenous IAA (Fig. 2, A and B) in a concentration-dependent manner (Fig. 2C). Control experiments using a low concentration of IBA or auxin conjugates (Fig. 2; Supplemental Fig. S2) did not rescue the germination delays. To determine if a buildup of IAA conjugates or IBA was contributing to germination defects, germination was tested on wild-type and mutant seeds using increasing concentrations of IAA-Ala, IAA-Leu, or IBA. Wild-type seed germination was only minimally disrupted by addition of these compounds and not in a concentration-dependent manner (Supplemental Fig. S2B). Mutant germination rates were specifically rescued by IAA but not significantly affected by addition of other compounds (Fig. 2D; Supplemental Fig. S2, C–E). These results suggest that the germination defects are primarily caused by decreases in free IAA levels.
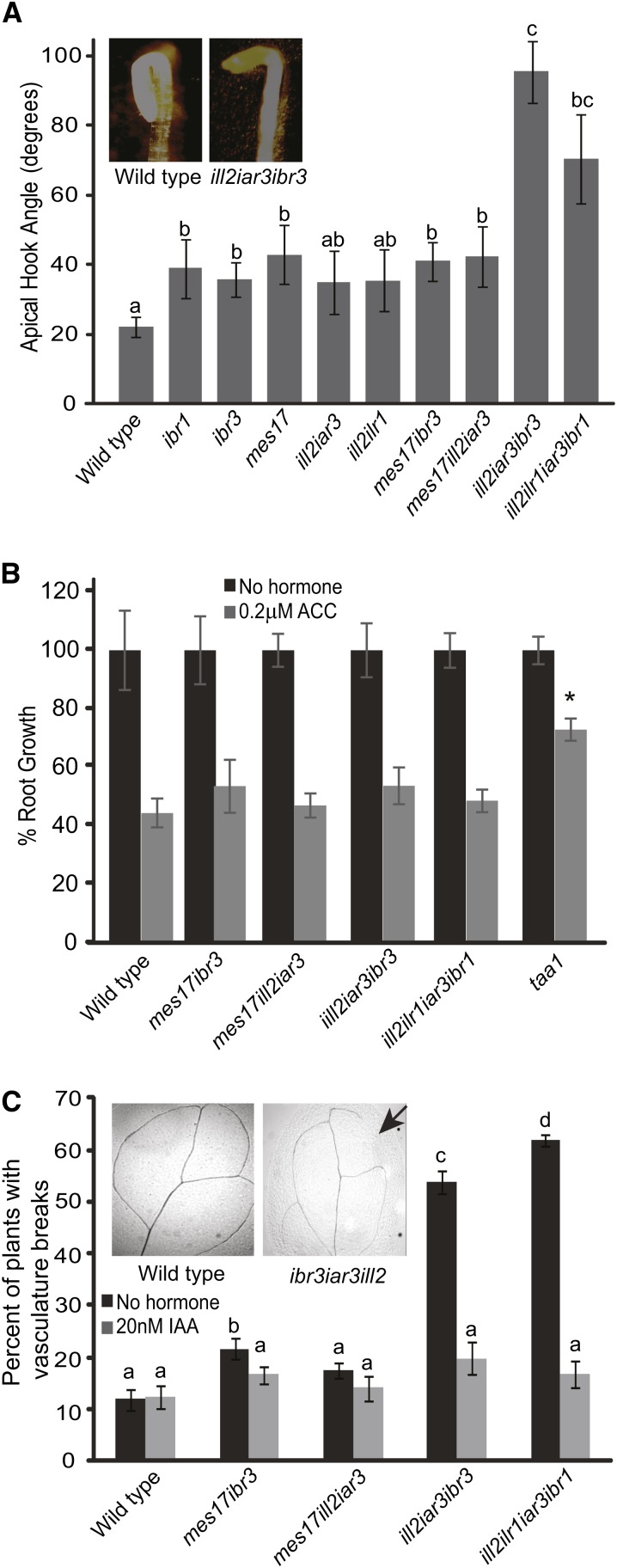
Auxin is required for apical hook initiation and maintenance in etiolated seedlings. Apical hook initiation and maintenance are lost in mutants with lower IAA levels, including the auxin biosynthesis mutant taa1 tryptophan aminotransferase2 (tar2; Stepanova et al., 2008; Vandenbussche et al., 2010; Strader et al., 2011). Apical hooks were examined in all mutants. The mutants defective in both IBA metabolism and IAA-conjugate hydrolases, ill2 iar3 ibr3 and ill2 ilr1 iar3 ibr1, had decreased apical hook curvature (Fig. 3A). These mutants had open apical hooks compared with the parental lines, indicating that both IAA conjugates and IBA play a role in apical hook development. Conversely, the mes17 ill2 iar3 and mes17 ibr3 mutants showed apical hook curvature, suggesting that IAA derived by MES17 is less important for this aspect of seedling development.
Figure 3.
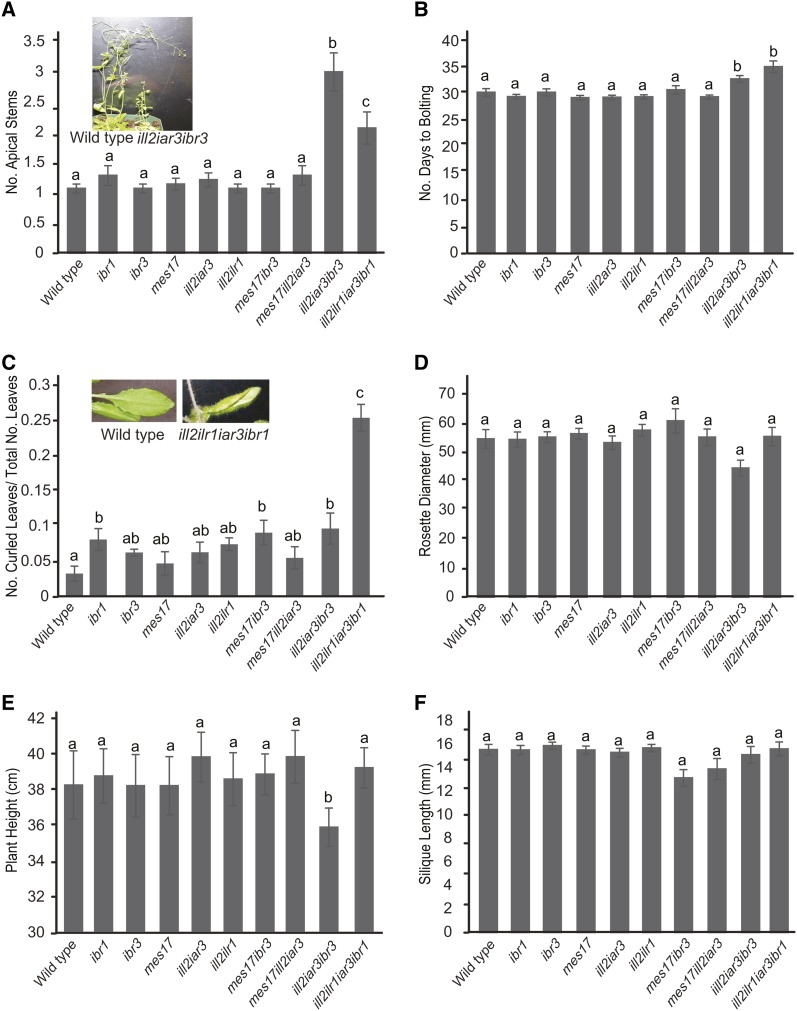
Combination mutants have phenotypes indicative of low auxin levels. A, Combination mutants have defects in apical hook maintenance compared with the wild type. Apical hooks were measured on seedlings grown 1 d in the light and 4 d in the dark. ImageJ software was used to measure the apical hook angle. Inset, One representative wild-type plant and one representative iar3 ill2 ibr3 plant. n ≥ 15; error bars represent se, and a to c indicate significant differences using one-way ANOVA (P < 0.05). B, Combination mutants show wild-type sensitivity to ethylene. Roots were measured from 5-d-old seedlings grown on no hormone or 0.2 µm ACC. Data are shown as percent root growth compared with no hormone controls. A taa1 single mutant is included as an ACC-resistant control. n ≥ 10; error bars represent se. *, Statistically significant result by Student’s t test compared with the wild type under the same conditions (P < 0.05). C, Combination mutants have breaks in vasculature tissue that could be rescued by IAA application; 7-d-old cotyledons were cleared and imaged after growth on media with no hormone or supplemented with 20 nm IAA. The graph shows the percentage of plants with cotyledon breaks. Inset, One representative wild-type leaf and one ill2 iar3 ibr3 leaf. The arrow indicates a break in vein pattern. n ≥ 35; a to d indicate significant differences using one-way ANOVA (P < 0.05).
Because of the positive feedback loop between auxin and ethylene, we tested ethylene sensitivity for all mutants. taa1 was used as a control with elongated roots in the presence of 1-aminocyclopropane-1-carboxylic acid (ACC) compared with the wild type (Stepanova et al., 2008). Percent root length was unchanged in the mutant lines compared with the wild type (Fig. 3B), consistent with the idea that ethylene specifically affects the de novo auxin biosynthesis pathway (He et al., 2011; Zheng et al., 2013).
Combination Mutants Alter Root Morphology
IAA influences both primary and lateral root growth. Under high levels of auxin by either endogenous production or application, roots show decreased primary root length but increased lateral rooting. These phenotypes are observed in the yuc1D mutant, which overexpresses YUC1, causing the plant to have elevated free IAA levels (Zhao et al., 2001). Conversely, mutants with low free IAA levels, such as the ill2 ilr1 iar3 conjugate hydrolase triple mutant, have an increased primary root length and decreased number of lateral roots (Rampey et al., 2004).
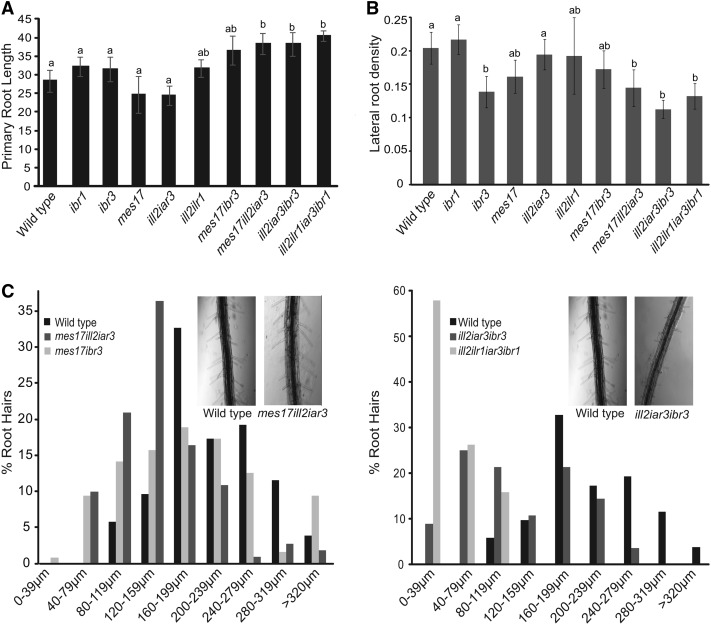
To determine if the combination mutants had root defects, primary root length and lateral root formation were evaluated. The ill2 ilr1 iar3 ibr1, ill2 iar3 ibr3, mes17 ill2 iar3, and mes17 ibr3 mutants all showed an increase in primary root length (Fig. 4A) that was rescued by application of low levels of auxin (Supplemental Fig. S3A). Additionally, the mutants had decreased lateral root density (Fig. 4B; Supplemental Fig. S3B), although changes in lateral root number were rescued by the synthetic auxin 1-napthaleneacetic acid (Supplemental Fig. S3C). These results further suggest that the mutant plants have decreased auxin levels. Because both the ill2 iar3 ibr3 and ill2 ilr1 iar3 ibr1 mutants showed phenotypes similar to the ibr3 single mutant, IBA metabolism seems to play the largest role in lateral root initiation.
Figure 4.
Combination mutants have root phenotypes consistent with defects in auxin responses. A, Combination mutants were grown on media without hormone, and root length was measured. B, Mutants were grown without hormone and have fewer lateral roots. Root density is the number of lateral roots per millimeter of primary root length. For A and B, n ≥ 15; error bars represent se, and a and b indicate significant differences using one-way ANOVA (P < 0.05). C, Roots were imaged from 7-d-old seedlings, and root hair length was measured using ImageJ software. Results shown are representative of four experiments. Insets, One representative wild-type plant and one representative mutant plant.
To further evaluate the role of these input pathways on root development, root hair length was measured on 5-d-old seedlings. Both ill2 ilr1 iar3 ibr1 and ill2 iar3 ibr3 have shorter root hairs (Fig. 4C). Although the mes17 ill2 iar3 and mes17 ibr3 mutants show small changes in root hair length, the phenotype is less severe than the ill2 ilr1 iar3 ibr1 and ill2 iar3 ibr3 mutants, suggesting that the IAA-amino acid conjugate and IBA sources of IAA play a role in root hair elongation.
Combination Mutant Phenotypes Continue into Adult Development
Mutants combining IAA-conjugate hydrolase and IBA metabolism defects had small breaks in leaf vein patterning (Fig. 3C). This phenotype was seen in slightly more than one-half of the cotyledons examined, significantly more than in wild-type plants or parental lines. Similar vein patterning defects are common in auxin mutants, particularly those disrupting de novo auxin biosynthesis and auxin transport (Cheng et al., 2007; Scarpella et al., 2010; Péret et al., 2012). Potential changes in IAA levels based on reduced IAA input pathways in the ill2 ilr1 iar3 ibr1 and ill2 iar3 ibr3 mutants may be causing decreases in IAA levels that result in discontinuous veination. This phenotype was rescued by growth of seedlings on 20 nm IAA (Fig. 3C), further suggesting the vasculature pattern defects are caused by low endogenous IAA concentrations.
The two combination mutants disrupting IBA metabolism and IAA-conjugate hydrolases have adult phenotypes, including decreased apical dominance and a slight delay in flowering time (Fig. 5, A and B). These phenotypes also are indicative of low free IAA levels in the plant. Many of the leaves in the ill2 ilr1 iar3 ibr1 mutant are rolled (Fig. 5C). Although all the mutants and the wild type displayed a few curled leaves, the ill2 ilr1 iar3 ibr1 mutant had a significantly higher ratio of curled leaves to total number of leaves. Rolled leaves are seen in other mutants with altered auxin levels, including iamt1-D and auxin resistant3 (Qin et al., 2005; Pérez-Pérez et al., 2010), suggesting that this phenotype is a result of changes in auxin levels in the plant. However, whether this phenotype is directly caused by altered IAA levels or buildup of IAA conjugates or IBA in the leaf is unknown. Notably, many aspects of development remain generally unchanged in the mutants, including fertility, rosette diameter, and plant height (Fig. 5, D–F). Fertility was examined by evaluating silique length, number of seeds, and number of aborted embryos (Fig. 5F; data not shown). The mes17 ibr3 and mes17 ill2 iar3 mutants do not have any adult phenotypes compared with their parental lines.
Figure 5.
Combination mutants have altered adult development. Phenotypic responses were examined for the wild type and combination mutants after being grown under continuous light at 22°C, including the number of apical stems (A), the number of days until bolting (B), the ratio of curled leaves to normal rosette leaves (C), rosette diameter (D), and plant height (E). F, Silique length was examined as one indicator of fertility by measuring the length of siliques 5 to 8 at maturity. Insets, One representative wild-type plant and one representative mutant plant. A to E, n ≥ 15; F, n ≥ 30. a to c, Significant differences using one-way ANOVA (P < 0.05).
IAA Levels Are Decreased in Combination Mutants
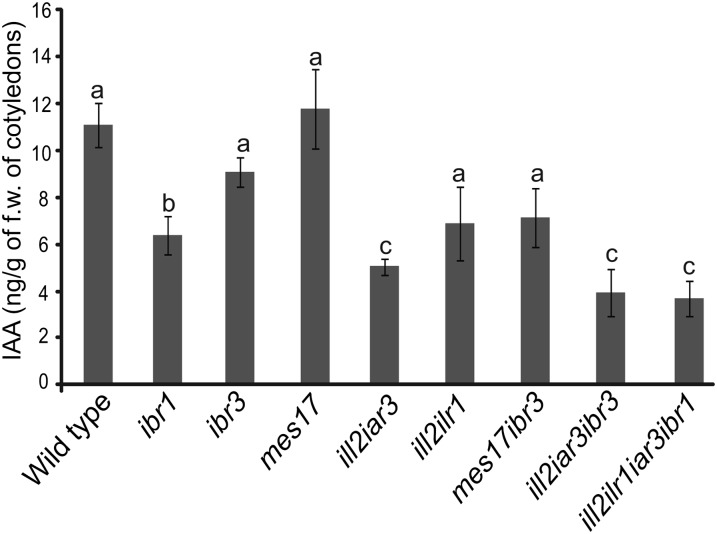
The phenotypes associated with these mutants suggested that the IAA-conjugate hydrolase/IBA-response combination mutant plants had reduced IAA levels. We measured free IAA levels in seedlings, separating storage tissues (cotyledons) and growing tissue (meristems plus hypocotyls). Free IAA levels were reduced in cotyledons in mutants combining IAA-conjugate hydrolases and IBA metabolism disruptions (Fig. 6). However, the mes17 ill2 iar3 and mes17 ibr3 did not show a decrease in free IAA levels. The IAA measurements from the growing tissue were not statistically different between the wild type and mutants (Supplemental Fig. S4). This finding may be because de novo biosynthesis is more important for IAA maintenance in the growing tissue, whereas cotyledons act more as storage organs. Because cotyledons act as storage organs, the storage forms of IAA may be more important for maintaining IAA levels in cotyledons.
Figure 6.
Reduced levels of free IAA are seen in combination mutants defective in IBA metabolism and IAA-conjugate hydrolysis; 5-d-old cotyledon tissue was collected, and free IAA levels were measured using gas chromatography-mass spectrometry. n ≥ 6; error bars represent se. a to c, Significant differences using one-way ANOVA (P < 0.05). f.w., Fresh weight.
Auxin Biosynthesis Gene Expression Is Activated in Response to Altered Auxin Homeostasis
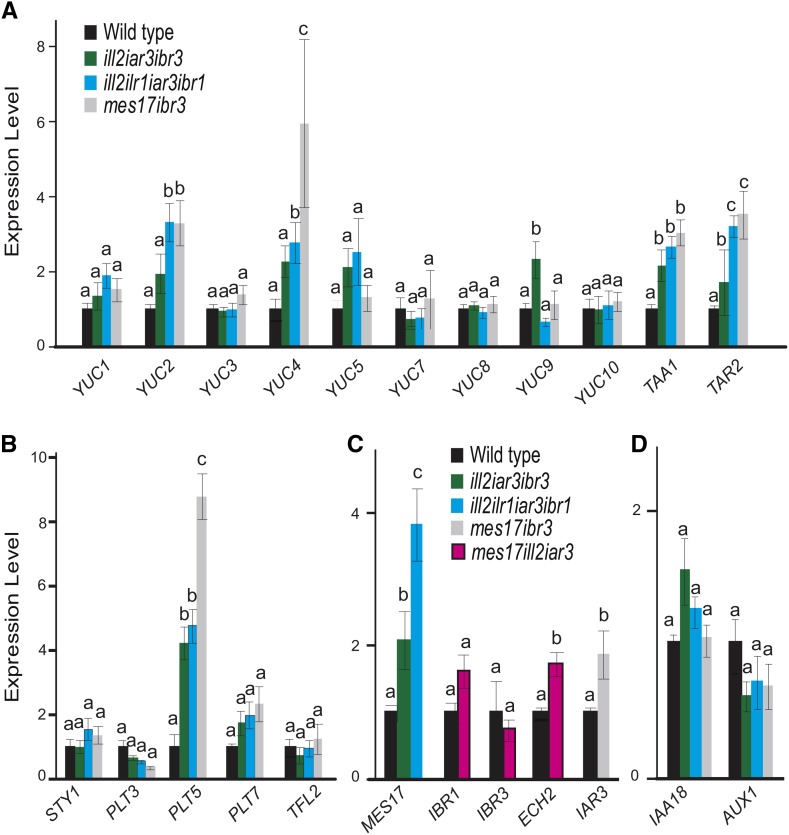
We hypothesized that other IAA input pathways are up-regulated to compensate for the loss of IAA storage metabolism. Quantitative PCR was performed to analyze gene expression of different IAA biosynthetic pathways. In lines combining IBA metabolism and IAA-conjugate hydrolase mutations, expression of YUC and TAA genes was higher, indicating an increase in this main biosynthetic pathway (Fig. 7). TAA1, TAR2, YUC2, and YUC4 are significantly up-regulated (Fig. 7A). Interestingly, YUC3, YUC7, YUC8, and YUC10 were not up-regulated in the mutants, suggesting specificity in how gene expression changes in response to low auxin levels.
Figure 7.
Biosynthesis genes are up-regulated in combination mutants. Expression analysis was performed using quantitative PCR of IAA de novo biosynthesis genes (A), YUC regulators (B), other IAA input pathways (C), and one gene involved in auxin signaling and one gene involved in transport (D). Expression is shown relative to the wild type, and transcript levels of target genes were normalized to expression of Ubiquitin 10 (At4G05320). n ≥ 3; error bars represent se. a to c, Significant differences using one-way ANOVA (P < 0.05).
The up-regulation of genes involved in de novo biosynthesis implies that a feedback loop is maintaining auxin levels under changing conditions. To better understand how regulation of these auxin biosynthesis genes occurs, we tested expression of several known regulators of auxin biosynthesis genes. Interestingly, PLETHORA5 (PLT5), a member of the PLT family of transcription factors, showed increased expression, although PLT3 and PLT7 family members did not (Fig. 7B). As recently shown by Pinon et al. (2013), PLT transcription factors regulate YUC gene expression, including YUC4. We also examined expression of STYLISH1 (STY1), a known controller of YUC4 expression (Eklund et al., 2010), and TERMINAL FLOWER2 (TFL2), which regulates expression of several YUC genes (Rizzardi et al., 2011), but found that neither STY1 nor TFL2 expression was significantly affected (Fig. 7B).
In addition to biosynthesis pathways, we also checked gene expression in the other input pathways. For instance, in mutants disrupting IBA-response genes and IAA-conjugate hydrolases, auxin biosynthesis pathway activation would increase IAA levels, but an MeIAA-based conversion to IAA also could be increased (Fig. 1). Indeed, MES17 expression was increased in the mutants, providing an additional pathway to maintain IAA levels (Fig. 7C). Similarly, in the mes17 ill2 iar3 mutant, ECH2, one of four known IBA-response genes, showed somewhat increased expression, although IBR1 and IBR3 levels were unchanged (Fig. 7C). In the mes17 ibr3 mutant, levels of the conjugate hydrolase IAR3 also were increased (Fig. 7C).
DISCUSSION
Auxin responses are regulated at three interdependent levels: homeostasis, directional transport, and signaling. The general term homeostasis includes biosynthesis, oxidative degradation, and auxin modifications. Combined with transport, these factors maintain the precise levels of auxin desirable in a cell based on cell type, developmental stage, and environmental conditions. Specific transcriptional activation and repression pathways occur based on the level of free IAA in an individual cell, leading to physiological changes in plant growth and development. In this article, we focused on the potential IAA input pathways apart from biosynthesis, allowing us to determine the specific and overlapping roles of input pathways on auxin levels and physiology.
Modified forms have auxin-like activity in assays and act primarily by their conversion to free IAA. Some exceptions have been suggested: IBA has increased efficacy over IAA for some processes, including adventitious rooting (Ludwig-Müller et al., 2005), and some lateral root initiation seems to be independent of or in addition to IBA conversion to IAA (Chhun et al., 2003; Strader et al., 2011). Furthermore, changing free IBA levels independent of IAA changes in plants overexpressing the URIDINE DIPHOSPHATE GLYCOSYLTRANSFERASE74E2 IBA-conjugating enzyme connects shoot morphogenesis to IBA activity (Tognetti et al., 2010). However, the primary activity of IBA and IAA conjugates seems to be based on conversion to IAA, because conversion mutants lose responses to the storage compound. These modified structures cannot directly facilitate IAA signaling. Transport of these molecules also is distinct from IAA transport streams (Strader and Bartel, 2011; Korasick et al., 2013). The regulated conversion of storage compounds to free IAA may allow for an extended or controlled auxin response distinct from the rapid changes associated with activation of transport or signaling pathways.
Individual pathways contribute to specific physiological responses in unique ways. For example, the ech2 ibr3 ibr1 ibr10 mutant disrupting IBA responses has defects in cotyledon expansion not seen in the ill2 ilr1 iar3 mutant disrupting three conjugate hydrolases (Rampey et al., 2004; Strader et al., 2011). Similarly, mes17 has a moderate hypocotyl elongation defect (Yang et al., 2008) not seen in lines disrupting other input pathways. The genes encoding proteins promoting auxin production are expressed throughout growth and development, with no obvious connection to specific physiological outputs (Fig. 1B). However, proteins could be regulated in a developmental-specific or environmental-specific manner, leading to activation and auxin increases in individual cells. Organelle location also separates auxin pools, and changing import or metabolism may contribute to specific responses (Simon and Petrášek, 2011). The fact that each pathway does play a unique role would suggest one reason why multiple IAA input pathways are required. Each pathway has become somewhat specialized, perhaps in conjunction with other developmental or metabolic pathways or in a plant-specific manner, to aid the plant in maintaining the appropriate cellular auxin levels. Another reason is redundancy, because having multiple auxin input pathways guarantees that a plant will be able to regulate the critical hormone, even if one pathway is disrupted.
Examining higher order mutants disrupting input pathways has allowed a better understanding of how homeostasis is maintained. Mutant plants had developmental phenotypes indicative of changes to the IAA pool. ill2 iar3 ibr3 and ill2 ilr1 iar3 ibr1 mutants had germination defects that were alleviated by low concentrations of IAA (Fig. 2), suggesting that IAA derived from these modified forms is contributing to seed germination. Combination mutants had altered root length and lateral root initiation defects that, again, were rescued by auxin application (Fig. 4; Supplemental Fig. S3). In addition, we saw changes in root hair number and length (Fig. 4). These results support a role for both input pathways in controlling root architecture. Combining IBA response and IAA-conjugate hydrolase defects also led to changes in adult growth, including flowering time, vascular development, and leaf shape (Figs. 3 and 5). As suggested by experiments in which phenotypic responses were alleviated by auxin application, mutant disruptions resulted from reduced auxin levels (Fig. 6). Although there is some specificity based on the pathways that are feeding into the pool, the overall auxin level is critical, and the IAA-conjugate hydrolase and IBA response pathways are interacting to affect auxin levels.
Combining these defects led to changes in plant growth and response not seen with mutations in either pathway alone, even in lines with stronger disruptions, suggesting that these pathways are together contributing to the auxin pool. It is important to note that the mutant lines that we chose may retain some activity. Both ibr3-1 (Zolman et al., 2007) and ibr1-2 (Zolman et al., 2008) alleles have phenotypes similar to insertion alleles, suggesting a null response. However, ibr3 and ibr1 have weaker phenotypes than other mutants disrupted in the IBA-to-IAA conversion pathway (Spiess and Zolman, 2013). This response may explain partial effects in germination assays, where IBA could slightly rescue the mutant phenotypes (Fig. 2). In addition, unrelated mutations in the mutant backgrounds could be combining to contribute to the triple and quadruple mutant phenotypes. However, use of these single mutants was a conscious choice, designed to remove potential negative consequences of stronger phenotypic disruptions in the individual pathways that could complicate analysis of the additive responses (Strader et al., 2011), and other crosses using these lines have not revealed hidden mutations. Furthermore, two different combinations disrupting IBA-response genes and IAA-conjugate hydrolases show similar responses. Because the phenotypes seen in these combination mutants were distinct from mutants with additional disruption of the individual pathways, we can begin examining the effects of a combined disruption. Alternatively, analysis of MES17 is complicated by the presence of other esterases that could be acting on similar targets, including MES1, MES2, MES7, and MES18 (Yang et al., 2008). Although MES17 has reported activity on MeIAA (Yang et al., 2008), additional higher order mutants may be required for full understanding of this pathway.
Although many aspects of auxin metabolism are affected by these changes in auxin input pathways, other pathways are not affected. The plants are generally healthy (Fig. 5). Gravitropic responses were normal in all mutants (Supplemental Fig. S5). Variation in auxin levels, similar to those seen between cotyledons and hypocotyls (Fig. 6; Supplemental Fig. S4), may allow signaling pathways to generate normal physiological responses in specific root cells, including gravitropic responses, whereas other processes are disrupted (Fig. 4). Such variation could result from alterations in auxin homeostasis or transport for instance. In addition, although a quadruple mutant disrupting IBA-response genes showed cell expansion defects (Strader et al., 2011), these mutants showed no changes in pavement cell growth (data not shown). In this case, the strong disruption of IBA-to-IAA conversion in specific cells may be more relevant than the general changes in IAA levels in combination mutants. Alternatively, in some cases, the phenotypic defects seem contradictory to previous studies. The ill2 ilr1 iar3 ibr1 mutant has rolled leaves (Fig. 5). Because this phenotype is more commonly associated with mutants having increased IAA levels (Pérez-Pérez et al., 2010), its presence here may be because of accumulation of the IAA conjugates or IBA.
The effects of these mutations are moderated by changes to de novo biosynthesis pathways (Fig. 7). By examining which enzymes the plant up-regulates when IAA sources are limited, we can determine the enzymes most important and/or rate limiting for maintaining auxin levels. For example, in the ill2 ilr1 iar3 ibr1 mutant, we saw that YUC2 and YUC4 are up-regulated but YUC7, YUC8, and YUC9 are not, indicating that, under these conditions, YUC2 and YUC4 are playing a larger role in regulating IAA biosynthesis. This may be because YUC7, YUC8, and YUC9 are less active in auxin biosynthesis than YUC2 and YUC4 or because these genes are more responsive to low auxin levels in these cells. These expression changes are increasing the primary biosynthetic pathway combining YUC and TAA (Mashiguchi et al., 2011) and would indicate that the plant is able to sense and respond to the low IAA levels caused by these mutations, providing a feedback mechanism to reduce disruptions from too little auxin in a cell. The regulation of specific YUC/TAA genes reveals a degree of specificity in this response. Gene expression of IAA18, acting in auxin signaling, and AUXIN RESISTANT1 (AUX1), an auxin transport component, were not affected (Fig. 7D).
We also investigated potential mechanisms for how plants regulate these specific biosynthetic enzymes. PLT5, one member of the PLT family of transcriptional regulators, was significantly up-regulated (Fig. 7). Changes in PLT regulators could be directly contributing to increased YUC expression. YUC1 and YUC4 expression levels were reduced in plt3 plt5 plt7 triple mutants (Pinon et al., 2013). Furthermore, YUC4 transcript levels increased quickly after activation of an inducible PLT5 construct (Pinon et al., 2013). Details on the potential connections between PLT5 and other YUC genes have not been explored. Additional analysis will be necessary to determine how these biosynthetic pathways are up-regulated. Analysis of known transcription factor binding sites (Chang et al., 2008) did not reveal obvious inducers.
In addition to IAA biosynthesis pathways, MES17 expression was up-regulated in the mutants combining disruptions of IAA-conjugate hydrolases and IBA-response enzymes, and IAR3 is up-regulated in the mes17 ibr3 mutant (Fig. 7D). These results indicate that, in addition to biosynthesis pathways, input pathways can also respond to changing IAA levels and be used to help a plant respond to and maintain the appropriate IAA levels. In some cases, mutants that disrupt one input pathway seem to be hypersensitive to other sources of auxin (Supplemental Fig. S1), most notably ill2 iar3 on media containing IBA. One hypothesis for this response is that these sources now are contributing more highly to the auxin pool in these mutants; examination of the levels of each form of auxin in these mutant lines will be revealing.
Mutations in input pathways led to increased MES17 expression, consistent with the idea that MES17 is converting an auxin storage form to IAA. Altering MES17 expression causes phenotypic responses consistent with changes in auxin metabolism (Qin et al., 2005; Yang et al., 2008). However, the specifics of this reaction are not known. Loss of MES17 in conjunction with IBA-response pathways or IAA-conjugate hydrolase pathways produced a less dramatic phenotype than that in mutants disrupting other auxin input pathways, which may indicate that this pathway is less active or more specific, perhaps at a developmental or environmental level, than the others in its contributions to overall auxin levels. Alternatively, as described above, other esterases (Yang et al., 2008) may be able to compensate for loss of MES17. Finally, changes in gene expression were apparent in the mes17 combination mutants (Fig. 7), which may normalize auxin levels in these plants (Fig. 6), resulting in fewer physiological changes.
Although these compounds have been recognized as contributing to auxin pools, largely by genetics and metabolite analysis, other compounds may be more relevant to plant physiology. Notably, MeIAA has not been detected in plants, which may be attributed to low abundance or rapid turnover (Qin et al., 2005) or because MeIAA is an intermediate in an esterification pathway. Similarly, IBA has been found in many plants but often at amounts similar to or less than free IAA, which is unexpected for a storage molecule; IBA may not be the relevant substrate but may be an intermediate in metabolism or biosynthesis (Spiess and Zolman, 2013). Alternatively, as described above, IBA may be acting independently of IAA conversion in some responses. Finally, IAA peptides similarly may play into these pathways either in addition to or as indicated by IAA amino acids. Additional studies are necessary to define the specific metabolic pathways, but our results provide support that the input pathways and these enzymes are feeding into IAA homeostasis.
The overall context of these three inputs must now be addressed in the context of other pathways, including transport. Auxin levels also are coordinated by the amount of auxin degradation as well as additional sequestration pathways, including IAA-ester conjugates such as IAA-Glc (Woodward and Bartel, 2005). In particular, understanding the connections to Glc conjugates requires the identification of enzymes that convert glycosylation forms back to free, active IAA; such enzymes have been characterized in other species, including Z. mays (Hall and Bandurski, 1986; Kowalczyk and Bandurski, 1990; Jakubowska et al., 1993; Kowalczyk et al., 2003; Jakubowska and Kowalczyk, 2005). Another complication is the further modifications of these forms, including IBA amino acid (Campanella et al., 2004) and sugar (Korasick et al., 2013) conjugates as well as MeIAA-Glc (Narasimhan et al., 2003). Finally, although sequestration pathways are important in auxin homeostasis, auxin biosynthesis also plays a clear role in auxin homeostasis and must be incorporated. Future studies will be necessary for full understanding of the pathways that influence IAA levels and how they interact under normal growth and development as well as during changing environmental conditions.
MATERIALS AND METHODS
Mutant Analysis
The single mutants iar3-6, ill2-2, ilr1-5, ibr1-1, ibr3-1, and mes17 (Salk_092550) and double mutants ill2-2 iar3-6 and ill2-2 ilr1-5 were previously characterized (Table I). ill2-2 iar3-6 was crossed with ibr3-1 to make the ill2 iar3 ibr3 mutant. ill2-2 iar3-6 was crossed with ill2-2 ilr1-5 and then ibr1-1 to make the ill2 ilr1 iar3 ibr1 mutant. The mes17 single mutant was crossed with ill2-2 iar3-6 to make the mes17 ill2 iar3 mutant. The mes17 and ibr3-1 single mutants were crossed to yield mes17 ibr3. All mutants are in the Columbia background. For simplicity, mutant names are shown without allele designation. Mutant genotypes were confirmed using PCR analysis of base pair changes or the presence of T-DNA insertions (Supplemental Table S1). taa1 (SALK_022743C) lines were used as a control for ACC experiments.
Phenotypic Analysis
Seeds were surface sterilized and suspended in agar for plating. Seedlings were grown on plant nutrient media with 0.5% (w/v) Suc (PNS; Haughn and Somerville, 1986). Micropore surgical tape was used to wrap plates before incubation at 22°C in continuous light. All plates containing hormones were incubated under yellow-filtered light. Except for germination studies, sterilized seeds were imbibed in 0.1% (w/v) agar at 4°C for 3 d to promote germination.
To measure apical hooks, seedlings were grown for 1 d in the light followed by 3 d in the dark. Seedlings were imaged using a dissecting scope, and hook angles were measured using ImageJ software.
Root length was measured after growth on PNS or indicated concentrations of IAA, IBA, or IAA-Ala for 10 d. For lateral root measurements, plants were grown on PNS for 10 d. The number of lateral roots was counted using a Leica Zoom 2000 Microscope. Data are shown as lateral root density (number of lateral roots per millimeter of root length). For lateral root induction experiments, plants were grown on media without hormone for 4 d before transfer to 1-napthaleneacetic acid-containing plates. Root hair length was measured similar to method by Strader et al. (2011). Plants were grown for 7 d and imaged. Root hair length was measured and counted using ImageJ software.
For vein patterning, seedlings were grown for 7 d on media with no added hormone or media supplemented with 20 nm IAA. Cotyledons were removed from the seedling and cleared according to the work by Sieburth (1999).
To measure ethylene responses, root length was measured after plants were grown vertically for 4 d in the dark on media containing no hormone or supplemented with 0.2 µm ACC. Percent root growth was calculated relative to no hormone.
For adult phenotypic assays, plants were grown on PNS for 10 d and then rescued to soil under continuous light at 22°C. Apical stems were counted 10 d after bolting for each plant. Rosette diameter was measured on the day of bolting, and rolled leaves were determined by dividing the number of rolled leaves by the total number of leaves in the rosette. Plant height is indicated as the height from the rosette to the top of plant and was measured 15 d after bolting. Silique length was determined by measuring the lengths of siliques 5 to 8 for each plant.
RNA Extraction and Expression Analysis
Fifteen 5-d-old seedlings were placed in a 1.5-mL tube and frozen in liquid nitrogen. RNA was extracted from the tissue using the IBI Mini Total RNA Kit (Plant; MidSci). Levels of desired transcripts were measured with the BioRad CFX96 Real Time PCR System using SsoAdvanced SYBR Green Supermix. No reverse transcriptase and no template were used as negative controls for all reactions, and all samples were normalized to expression of Ubiquitin 10 (At4G05320). Primers for each gene were designed using QuantPrime (Supplemental Tables S1 and S2). All samples were done as biological triplicates, and each biological replicate was performed in technical triplicate.
IAA Quantification
Meristem plus hypocotyl tissue or cotyledons were removed from the plant, weighed for a mass greater than 2 mg, frozen in liquid nitrogen, and stored at −80°C; 2 to 5 mg of frozen tissue was then homogenized in 20 μL of homogenization buffer (35% [v/v] 0.2 m imidazole, 65% [v/v] isopropanol, pH 7) containing 0.2 ng of [13C6]IAA, and purification and quantification were done according to the works by Liu et al. (2011, 2012). Briefly, the plant extract was purified by weak anion exchange and affinity solid-phase extraction. Final eluent was methylated and analyzed by gas chromatography-selected reaction-monitoring mass spectrometry. Quantification was achieved by comparing the signal from endogenous IAA with that of an internal standard.
Sequence data from this article can be found in The Arabidopsis Information Resource and GenBank (National Center for Biotechnology Information) databases. Accession numbers for the genes discussed are listed in Supplemental Tables S1 and S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Confirmation of mutant lines.
Supplemental Figure S2. Examination of germination in mutant lines.
Supplemental Figure S3. Characterization of rooting responses with auxin.
Supplemental Figure S4. Measurement of IAA levels in meristems and hypocotyls.
Supplemental Figure S5. Analysis of gravitropic responses in mutant lines.
Supplemental Table S1. Genotyping primers.
Supplemental Table S2. qPCR primers.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center at The Ohio State University for seed stocks; the Salk Institute Genomic Analysis Laboratory for generating the sequence-indexed Arabidopsis T-DNA insertion mutants; Bonnie Bartel for contributions during the initiation of this project; Harding University Molecular Genetics Laboratory students and Shelly Boyer for help in generating the higher order mutants; and Shelly Boyer, Elizabeth Haswell, Vanessica Jawahir, and Ying Li for critical comments on the article.
Glossary
- ACC
1-aminocyclopropane-1-carboxylic acid
- IAA
indole-3-acetic acid
- IBA
indole-3-butyric acid
- MeIAA
methyl indole-3-acetic acid
- PNS
plant nutrient media with 0.5% Suc
Footnotes
This work was supported by the National Science Foundation (Plant Genome Research Program grant nos. IOS–0923960 and IOS–1238812 to J.D.C., MCB–1203438 to J.D.C., and IOS–0845507 to B.K.Z.), the Minnesota Agricultural Experiment Station (to J.D.C.), the Gordon and Margret Bailey Endowment for Environmental Horticulture (to J.D.C.), and the University of Missouri Research Board (to B.K.Z.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Bajguz A, Piotrowska A. (2009) Conjugates of auxin and cytokinin. Phytochemistry 70: 957–969 [DOI] [PubMed] [Google Scholar]
- Campanella JJ, Olajide AF, Magnus V, Ludwig-Müller J. (2004) A novel auxin conjugate hydrolase from wheat with substrate specificity for longer side-chain auxin amide conjugates. Plant Physiol 135: 2230–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WC, Lee TY, Huang HD, Huang HY, Pan RL. (2008) PlantPAN: plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics 9: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhun T, Taketa S, Tsurumi S, Ichii M. (2003) The effects of auxin on lateral root initiation and root gravitropism in a lateral rootless mutant Lrt1 of rice (Oryza sativa L.). Plant Growth Regul 39: 161–170 [Google Scholar]
- Cohen JD, Bandurski RS. (1982) Chemistry and physiology of the bound auxins. Annu Rev Plant Physiol 33: 403–430 [Google Scholar]
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B. (1999) IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Ståldal V, Valsecchi I, Cierlik I, Eriksson C, Hiratsu K, Ohme-Takagi M, Sundström JF, Thelander M, Ezcurra I, et al. (2010) The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 22: 349–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester Sztein A, Cohen JD, de la Fuente IG, Cooke TJ. (1999) Auxin metabolism in mosses and liverworts. Am J Bot 86: 1544–1555 [PubMed] [Google Scholar]
- Evans ML, Ishikawa H, Estelle MA. (1994) Responses of Arabidopsis roots to auxin studied with high temporal resolution: comparison of wild type and auxin-response mutants. Planta 194: 215–222 [Google Scholar]
- Hagen G, Guilfoyle TJ. (1985) Rapid induction of selective transcription by auxins. Mol Cell Biol 5: 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall PJ, Bandurski RS. (1986) [3H]Indole-3-acetyl-myo-inositol hydrolysis by extracts of Zea mays L. vegetative tissue. Plant Physiol 80: 374–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G, Somerville C. (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. (2011) A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK. (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowska A, Kowalczyk S. (2005) A specific enzyme hydrolyzing 6-O(4-O)-indole-3-ylacetyl-β-D-glucose in immature kernels of Zea mays. J Plant Physiol 162: 207–213 [DOI] [PubMed] [Google Scholar]
- Jakubowska A, Kowalczyk S, Leźnicki AJ. (1993) Enzymatic hydrolysis of 4-O and 6-O-indol-3-ylacetyl-β-D-glucose in plant tissues. J Plant Physiol 142: 61–66 [Google Scholar]
- Kinoshita N, Wang H, Kasahara H, Liu J, Macpherson C, Machida Y, Kamiya Y, Hannah MA, Chua NH. (2012) IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 24: 3590–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Enders TA, Strader LC. (2013) Auxin biosynthesis and storage forms. J Exp Bot 64: 2541–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M, Sandberg G. (2001) Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol 127: 1845–1853 [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk S, Bandurski RS. (1990) Isomerization of 1-O-indol-3-ylacetyl-β-D-glucose. Enzymatic hydrolysis of 1-O, 4-O, and 6-O-indol-3-ylacetyl-β-D-glucose and the enzymatic synthesis of indole-3-acetyl glycerol by a hormone metabolizing complex. Plant Physiol 94: 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk S, Jakubowska A, Zielińska E, Bandurski RS. (2003) Bifunctional indole-3-acetyl transferase catalyses synthesis and hydrolysis of indole-3-acetyl-myo-inositol in immature endosperm of Zea mays. Physiol Plant 119: 165–174 [Google Scholar]
- LeClere S, Tellez R, Rampey RA, Matsuda SP, Bartel B. (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem 277: 20446–20452 [DOI] [PubMed] [Google Scholar]
- Liu X, Cohen JD, Gardner G. (2011) Low-fluence red light increases the transport and biosynthesis of auxin. Plant Physiol 157: 891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hegeman AD, Gardner G, Cohen JD. (2012) Protocol: high-throughput and quantitative assays of auxin and auxin precursors from minute tissue samples. Plant Methods 8: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K. (2013) Auxin metabolism and homeostasis during plant development. Development 140: 943–950 [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull A, Kowalczyk M, Marchant A, Celenza J, Cohen J, Sandberg G. (2002) Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. In Perrot-Rechenmann C, Hagen G, eds, Auxin Molecular Biology. Springer, Dordrecht, The Netherlands, pp 249–272 [PubMed] [Google Scholar]
- Ludwig-Müller J. (2000) Indole-3-butyric acid in plant growth and development. Plant Growth Regul 32: 219–230 [Google Scholar]
- Ludwig-Müller J, Jülke S, Bierfreund NM, Decker EL, Reski R. (2009) Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol 181: 323–338 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J, Vertocnik A, Town CD. (2005) Analysis of indole-3-butyric acid-induced adventitious root formation on Arabidopsis stem segments. J Exp Bot 56: 2095–2105 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan K, Basheer C, Bajic VB, Swarup S. (2003) Enhancement of plant-microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol 132: 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. (1997) Auxin metabolism. Physiol Plant 100: 431–442 [Google Scholar]
- Normanly J, Slovin J, Cohen J. (2010) Auxin biosynthesis and metabolism. In Davies P, ed, Plant Hormones. Springer, Dordrecht, The Netherlands, pp 36–62 [Google Scholar]
- Normanly J, Slovin JP, Cohen JD. (1995) Rethinking auxin biosynthesis and metabolism. Plant Physiol 107: 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K. (2012) Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J 72: 523–536 [DOI] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, Palme K. (2008) Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1: 321–337 [DOI] [PubMed] [Google Scholar]
- Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM. (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282: 10036–10046 [DOI] [PubMed] [Google Scholar]
- Pencík A, Rolcík J, Novák O, Magnus V, Barták P, Buchtík R, Salopek-Sondi B, Strnad M. (2009) Isolation of novel indole-3-acetic acid conjugates by immunoaffinity extraction. Talanta 80: 651–655 [DOI] [PubMed] [Google Scholar]
- Péret B, Swarup K, Ferguson A, Seth M, Yang Y, Dhondt S, James N, Casimiro I, Perry P, Syed A, et al. (2012) AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Candela H, Robles P, López-Torrejón G, del Pozo JC, Micol JL. (2010) A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol 51: 1661–1673 [DOI] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. (2010) Cellular responses to auxin: division versus expansion. Cold Spring Harb Perspect Biol 2: a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Prasad K, Grigg SP, Sanchez-Perez GF, Scheres B. (2013) Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci USA 110: 1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Gu H, Zhao Y, Ma Z, Shi G, Yang Y, Pichersky E, Chen H, Liu M, Chen Z, et al. (2005) An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. (2004) A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol 135: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi K, Landberg K, Nilsson L, Ljung K, Sundås-Larsson A. (2011) TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J 65: 897–906 [DOI] [PubMed] [Google Scholar]
- Salopek-Sondi B, Šamec D, Mihaljević S, Smolko A, Pavlović I, Janković I, Ludwig-Müller J. (2013) Influence of stress hormones on the auxin homeostasis in Brassica rapa seedlings. Plant Cell Rep 32: 1031–1042 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Barkoulas M, Tsiantis M. (2010) Control of leaf and vein development by auxin. Cold Spring Harb Perspect Biol 2: a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE. (1999) Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol 121: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Petrášek J. (2011) Why plants need more than one type of auxin. Plant Sci 180: 454–460 [DOI] [PubMed] [Google Scholar]
- Slovin J, Bandurski R, Cohen J. (1999) Auxin. In Hooykaas PJJ, Hall MA, Libbenga KR, eds, New Comprehensive Biochemistry, Vol 33 Elsevier, Amsterdam, pp 115–140 [Google Scholar]
- Spiess GM, Zolman BK. (2013) Peroxisomes as a source of auxin signaling molecules. In del Río LA, ed, Peroxisomes and their Key Role in Cellular Signaling and Metabolism, Vol 69 Springer, Dordrecht, The Netherlands, pp 257–281 [DOI] [PubMed] [Google Scholar]
- Staswick PE. (2009) The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol 150: 1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Strader LC, Bartel B. (2011) Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B. (2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23: 984–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YY, Epstein E, Normanly J. (2000) Characterization of auxin conjugates in Arabidopsis. Low steady-state levels of indole-3-acetyl-aspartate, indole-3-acetyl-glutamate, and indole-3-acetyl-glucose. Plant Physiol 123: 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E, Rojo E, León J, Sánchez-Serrano JJ. (1997) Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol 115: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, et al. (2010) Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Petrásek J, Zádníková P, Hoyerová K, Pešek B, Raz V, Swarup R, Bennett M, Zazímalová E, Benková E, et al. (2010) The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606 [DOI] [PubMed] [Google Scholar]
- Walz A, Park S, Slovin JP, Ludwig-Müller J, Momonoki YS, Cohen JD. (2002) A gene encoding a protein modified by the phytohormone indoleacetic acid. Proc Natl Acad Sci USA 99: 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemariam MG, Onkokesung N, Baldwin IT, Galis I. (2012) Jasmonoyl-L-isoleucine hydrolase 1 (JIH1) regulates jasmonoyl-L-isoleucine levels and attenuates plant defenses against herbivores. Plant J 72: 758–767 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xu R, Ma CJ, Vlot AC, Klessig DF, Pichersky E. (2008) Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol 147: 1034–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Dai X, Zhao Y, Ljung K, Noel JP, Chory J. (2013) Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol 9: 244–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B. (2008) Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Nyberg M, Bartel B. (2007) IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol 64: 59–72 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C, Ross JR, Koscheski P, Yang Y, Pichersky E, Noel JP. (2003) Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15: 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.