A combination of real-time fluorescence imaging and high-temporal-resolution RNA sequencing reveals dynamic changes in growth, photosynthesis, and associated global gene expression during jasmonate signaling.
Abstract
Biotic stress constrains plant productivity in natural and agricultural ecosystems. Repression of photosynthetic genes is a conserved plant response to biotic attack, but how this transcriptional reprogramming is linked to changes in photosynthesis and the transition from growth- to defense-oriented metabolism is poorly understood. Here, we used a combination of noninvasive chlorophyll fluorescence imaging technology and RNA sequencing to determine the effect of the defense hormone jasmonate (JA) on the growth, photosynthetic efficiency, and gene expression of Arabidopsis (Arabidopsis thaliana) rosette leaves. High temporal resolution was achieved through treatment with coronatine (COR), a high-affinity agonist of the JA receptor. We show that leaf growth is rapidly arrested after COR treatment and that this effect is tightly correlated with changes in the expression of genes involved in growth, photosynthesis, and defense. Rapid COR-induced expression of defense genes occurred concomitantly with the repression of photosynthetic genes but was not associated with a reduced quantum efficiency of photosystem II. These findings support the view that photosynthetic capacity is maintained during the period in which stress-induced JA signaling redirects metabolism from growth to defense. Chlorophyll fluorescence images captured in a multiscale time series, however, revealed a transient COR-induced decrease in quantum efficiency of photosystem II at dawn of the day after treatment. Physiological studies suggest that this response results from delayed stomatal opening at the night-day transition. These collective results establish a high-resolution temporal view of how a major stress response pathway modulates plant growth and photosynthesis and highlight the utility of chlorophyll fluorescence imaging for revealing transient stress-induced perturbations in photosynthetic performance.
Plant productivity is dependent on the capture and conversion of solar energy and the subsequent allocation of reduced carbon into processes required for growth and reproduction. In natural environments, however, plants encounter stress conditions that negatively impact productivity by reducing photosynthesis and growth. Studies describing the negative effects of abiotic stress on photosynthetic efficiency and growth rate are well documented, including the effects of high light, water deficiency, and salinity. Biotic stress from pathogen infection and insect herbivory also decreases photosynthesis and growth (Zangerl et al., 2002; Bonfig et al., 2006; Berger et al., 2007; Ishiga et al., 2009; Nabity et al., 2009, 2013). That these effects can be greater than that attributed simply to the removal of leaf tissue from disease or herbivory suggests that signaling pathways activated by stress reduce photosynthetic efficiency in remaining leaves (Zvereva et al., 2010; Nabity et al., 2013). Biotic stress conditions typically divert resource allocation from growth processes to the production of defensive compounds whose biosynthesis is energetically demanding. Such growth-defense tradeoffs presumably evolved to increase plant fitness in rapidly changing environments (Herms and Mattson, 1992; Baldwin, 1998; Ballaré, 2009; Meldau et al., 2012; Huot et al., 2014). A greater understanding of how plant defense pathways modulate energy capture and conversion is an important goal of research aimed at improving biomass production through increased photosynthesis.
Plant defense responses to biotic stress are controlled by multiple hormone signaling pathways that interact in complex ways to regulate gene expression and metabolism (Pieterse et al., 2009; Erb et al., 2012). Down-regulation of photosynthetic genes and their corresponding proteins is a conserved feature of plant responses to many pathogens and herbivores (Reymond et al., 2004; Zou et al., 2005; Giri et al., 2006; Mitra and Baldwin, 2008; Bilgin et al., 2010). The physiological significance of this phenomenon, however, remains unclear. Reduced photosynthetic output may reflect a reduction in plant growth rate that is typically associated with biotic stress conditions and may also serve to limit the availability of nutrients to opportunistic plant parasites. On the other hand, stress-induced production of defense-related compounds depends on robust photosynthetic output and may in fact increase total photosynthetic demand (Bekaert et al., 2012; Rojas et al., 2014). The plant stress hormone jasmonate (JA) plays a key role in controlling resource allocation between the competing processes of growth and defense (Zhang and Turner, 2008; Hou et al., 2010; Yang et al., 2012; Nabity et al., 2013; Noir et al., 2013; Ullmann-Zeunert et al., 2013). JA perception and signaling is principally governed by intracellular levels of jasmonoyl-l-isoleucine (JA-Ile), which stimulates the formation of a coreceptor complex consisting of the CORONATINE INSENSITIVE1 (COI1) F-box protein and JASMONATE ZIM DOMAIN (JAZ) transcriptional repressor proteins (Xie et al., 1998; Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Katsir et al., 2008; Melotto et al., 2008; Fonseca et al., 2009; Sheard et al., 2010). Degradation of JAZ repressors by the ubiquitin-proteasome system activates transcription factors such as MYC2 that drive the expression of JA response genes in response to tissue injury and other forms of stress (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Chung et al., 2008; Koo et al., 2009). In comparison with detailed knowledge of how JA activates defense responses (Howe and Jander, 2008; Wu and Baldwin, 2010; Ballaré, 2011), relatively little is known about the mechanisms by which JA represses photosynthetic gene expression and growth processes.
Here, we combined RNA sequencing (RNA-seq) with a newly designed chlorophyll fluorescence imaging system to study the temporal relationship between JA-induced transcriptional reprogramming, leaf growth, and photosynthesis. To directly assess the effect of JA on growth and photosynthesis without potential secondary effects associated with tissue damage inflicted by insect herbivory or pathogen infection, we used exogenous coronatine (COR) as a chemical tool to achieve rapid, strong, and specific activation of the JA signaling pathway. COR is a polyketide effector molecule produced by Pseudomonas syringae pv tomato strain DC3000 (Pst DC3000). Whereas elicitation of JA responses with exogenous jasmonic acid or methyl jasmonate (MeJA) depends on biochemical conversion of these compounds to JA-Ile, COR is a structural mimic of JA-Ile and thus acts directly as an agonist of the COI1-JAZ coreceptor (Thines et al., 2007; Katsir et al., 2008; Melotto et al., 2008; Fonseca et al., 2009; Sheard et al., 2010). The relative potency of COR as an elicitor of JA responses may also reflect structural features of the molecule that render it resistant to catabolic pathways that degrade JA-Ile (Koo and Howe, 2012). Our integrated analyses of COR-treated Arabidopsis (Arabidopsis thaliana) showed that activation of JA signaling causes growth arrest but not concomitant loss of photosynthetic efficiency during the initial phase of the response. High-temporal-resolution measurements also revealed a transient reduction in photosynthesis at dawn (night-day transition) of the day following COR treatment. These collective findings highlight the utility of chlorophyll fluorescence imaging for studying the impact of dynamic environments on growth and photosynthetic performance.
RESULTS
COR Rapidly Arrests Growth without Immediate Effects on Photosynthesis
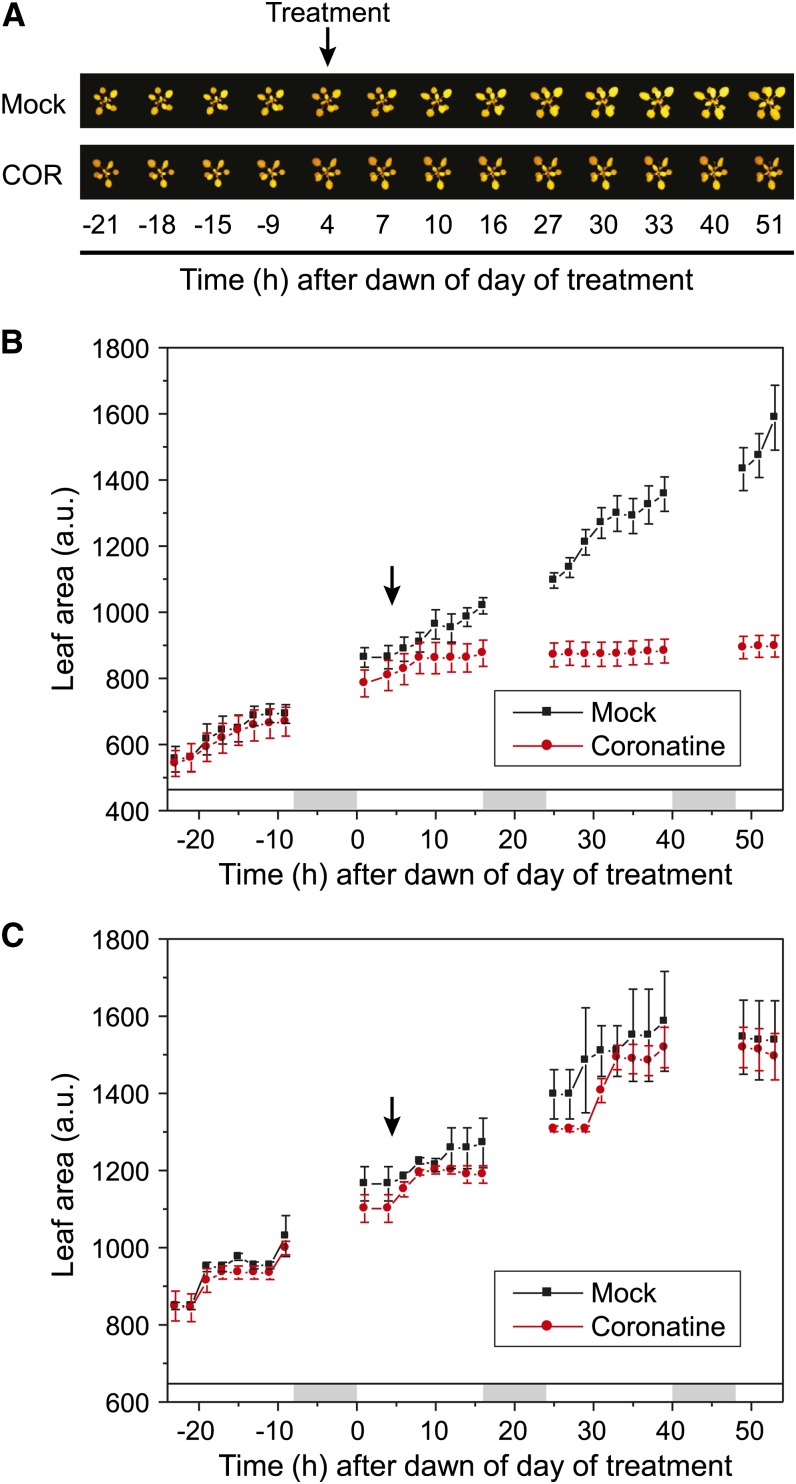
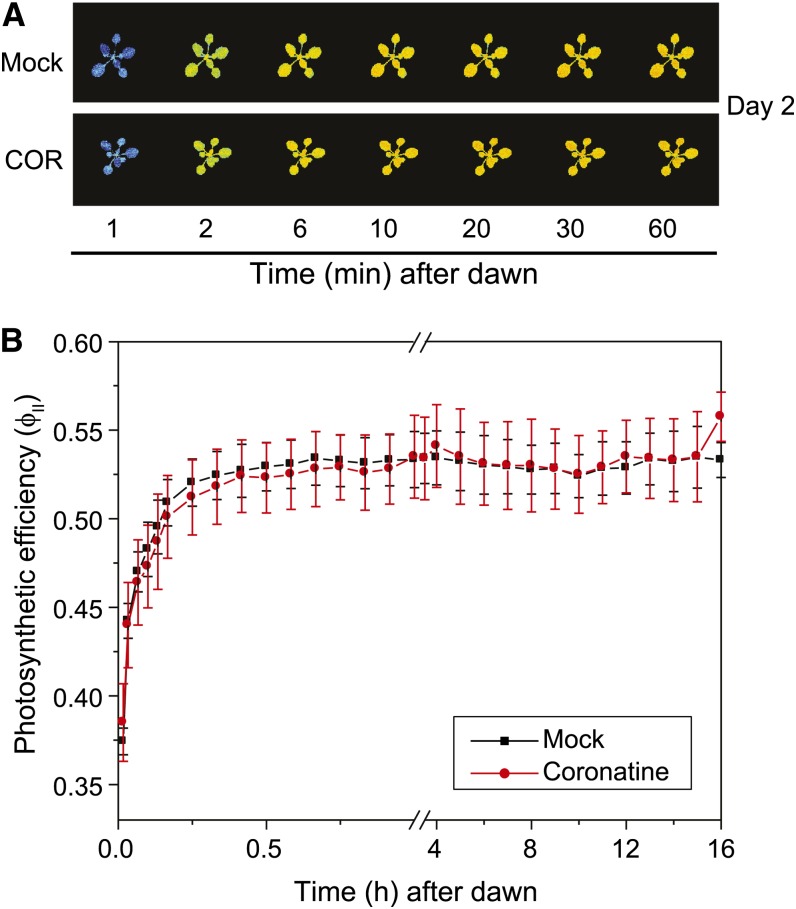
To profile the effects of JA signaling on growth and photosynthesis, we used a noninvasive, real-time fluorescence imaging system to measure leaf area and photosynthetic efficiency in response to COR treatment. Our imaging system was designed to continuously monitor the chlorophyll fluorescence of multiple plants, allowing analysis of mock- and COR-treated plants with high temporal resolution (Supplemental Fig. S1). We first assessed changes in growth by measuring leaf area at 2-h intervals spanning 1 d before and 2 d after COR treatment. Analysis of the resulting images showed that COR significantly inhibited (P < 0.001) growth within approximately 4 h of treatment and that this effect on leaf area persisted for the duration of the time course (Fig. 1, A and B). In contrast to wild-type (Columbia-0 [Col-0]) plants, COR treatment did not affect growth of the coi1-30 mutant, which lacks a functional receptor for JA-Ile and COR (P > 0.5; Fig. 1C). We next examined whether the rapid growth arrest by COR was accompanied by changes in photosynthetic capacity as determined by fluorescence imaging of the steady-state quantum efficiency of PSII (ΦII) in mock- and COR-treated plants. The results showed that COR does not have an immediate (i.e. day of treatment) effect on ΦII (Supplemental Fig. S2A). Thus, COR-induced arrest of leaf growth via the COI1 receptor system is not associated with short-term effects on photosynthetic efficiency. Imaging experiments performed over longer time frames showed that a single application of COR does not significantly affect ΦII at time points extending to 6 d after treatment (Supplemental Fig. S2B).
Figure 1.
The phytotoxin COR rapidly arrests plant growth. Arabidopsis (Col-0) plants were acclimated for 36 h in an imaging chamber and, 4 h after dawn of the following day (denoted by arrows), sprayed with either water (mock) or 5 μm COR. The 0-h time point corresponds to dawn of the day of treatment. A, False-color chlorophyll fluorescence images of representative plants at the indicated time points. B, Quantification of leaf area based on an analysis of the images shown in A. Data show means ± se (in arbitrary units [a.u.]) of three independent replicates. For each replicate, leaf area was determined for two to three actively growing leaves. The experiment was independently replicated three times. Photoperiod (white bars, light; gray bars, dark) is denoted above the x axis. C, Effect of COR treatment on growth of the JA signaling mutant coi1-30. Leaf area was quantified as described in B. Results were derived from two independent experiments. Photoperiod is denoted above the x axis.
Genes Associated with Photosynthesis and Growth Are Repressed by COR
To investigate the relationship between COR-induced growth arrest and changes in gene expression, we used RNA-seq to measure the fine-scale temporal dynamics of gene expression in the 24-h period following COR treatment. To control for diurnal changes in gene expression, a matched set of mock-treated plants was analyzed for each of the 20 time points within the time series (Supplemental Fig. S1). We assessed differential gene expression by calculating the difference in absolute expression between matched COR and mock samples (i.e. transcript levels in COR-treated minus mock-treated samples). This change in transcript level better reflected COR-induced expression patterns compared with fold change (i.e. transcript levels in COR-treated divided by mock-treated samples), particularly for those genes exhibiting a high absolute expression level or strong diurnal rhythm in the absence of COR treatment. The COR-induced temporal expression profile of all Arabidopsis genes is provided in Supplemental Table S1. Transcript levels measured by RNA-seq were highly correlated with quantitative PCR data for several selected genes, thereby validating the approach (Supplemental Fig. S3).
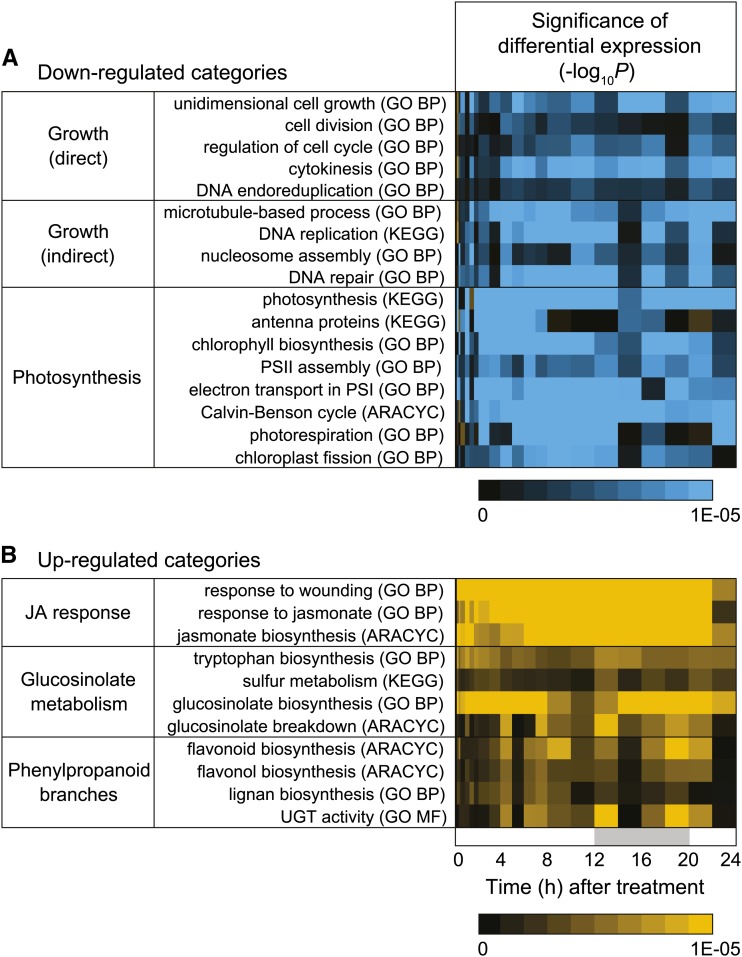
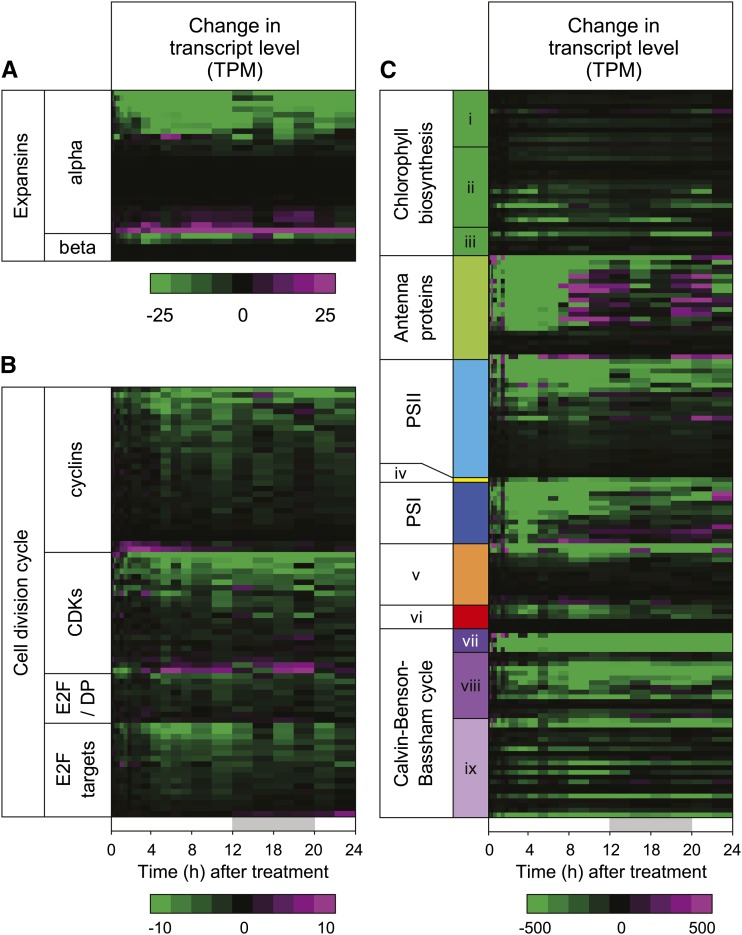
To obtain an unbiased analysis of processes affected by COR treatment, we performed a Mann-Whitney-Wilcoxon test of the change in transcript abundance for functional categories at each time point and expressed the results as a heat map of P values (Fig. 2). This analysis showed that gene functions associated with growth and photosynthesis were strongly repressed by COR treatment (Fig. 2A). Among the COR-repressed functional categories associated directly with plant growth were members of the expansin family and other genes associated with growth of the cell wall (Fig. 3A). Repressed expression of some expansin genes (e.g. EXPANSIN A8 [EXPA8]) was particularly striking, with transcript levels declining to the limit of detection after COR treatment. We also observed delayed repression of genes associated with cell division, including cyclins, cyclin-dependent kinases, E2 Promoter-Binding Factor (E2F)/DP transcription factors, and E2F target genes (Fig. 3B). Genes associated with DNA replication and microtubule processes were also repressed (Fig. 2A), which is a potential indirect effect of COR-induced growth arrest.
Figure 2.
Temporal dynamics of gene expression in response to COR treatment. Plants were treated with either COR or a mock control as described in the gene expression analysis section of Supplemental Figure S1. Heat maps depict functional categories of genes that were down-regulated (A; blue) or up-regulated (B; yellow) at the indicated times (h) after COR treatment. The significance of changes in transcript levels was determined at each time point by the Mann-Whitney-Wilcoxon test with Benjamini-Hochberg correction, with −log10-transformed P values displayed in the heat map. Functional categories from GO (BP, Biological Process; MF, Molecular Function), AraCyc, and KEGG were analyzed together using the AraPath annotation set. UGT, UDP-Glycosyltransferase.
Figure 3.
Global repression of growth- and photosynthesis-associated genes in response to COR treatment. Plants were treated with COR or a mock control as described in Supplemental Figure S1. Transcript levels were measured by RNA-seq. Heat maps depict the change in absolute expression, as calculated from TPM in COR-treated sample minus TPM of the time-matched mock sample. A, Expression pattern of α- and β-expansin genes in Arabidopsis. B, Expression pattern of genes associated with the cell division cycle. CDKs, Cyclin-dependent kinases, including CDK subunits and inhibitors; E2F and DP are transcription factors. C, Expression pattern of genes involved in the following photosynthetic processes: i, tetrapyrrole biosynthesis; ii, chlorophyllide a biosynthesis; iii, chlorophyll a biosynthesis; iv, cytochrome b6/f complex; v, photosynthetic electron transport; vi, ATP synthase; vii, carbon fixation; viii, reduction; and ix, regeneration.
COR treatment also strongly reduced the abundance of many PHOTOSYNTHESIS-ASSOCIATED GENE (PAG) transcripts, which dominate the leaf transcriptome (Baerenfaller et al., 2008). Among this group of repressed genes were those encoding components of the light-harvesting complex, photosystem subunits, electron transport chain, chlorophyll biosynthesis pathway, and the Calvin-Benson-Bassham cycle (Fig. 2A). Two general patterns of PAG repression were apparent: a strong, transient repression 2 to 10 h after COR treatment and a more rapid (1 h) repression that was sustained for the duration of the time course (Fig. 3C). Genes associated with the light-harvesting complexes (e.g. CHLOROPHYLL A/B BINDING PROTEIN3), photosystems (e.g. PHOTOSYSTEM II SUBUNIT P-1), and chlorophyll biosynthesis (e.g. PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A), for example, exhibited strong and transient repression. In contrast, genes encoding components of the Calvin-Benson-Bassham cycle (e.g. RUBISCO ACTIVASE [RCA]) exhibited a more sustained repression pattern (Fig. 3; Supplemental Fig. S4). We used the coi1-30 mutant to test whether gene repression by COR is dependent on the JA receptor. Control experiments showed that induction of the JA-responsive gene ALLENE OXIDE SYNTHEASE was abolished in coi1-30 plants (Supplemental Fig. S5). We also found that transcripts associated with photosynthesis and growth, including RCA and EXPA8, were repressed by COR in Col-0 but not in coi1-30 plants. Collectively, these results indicate that repression of growth-related genes by COR correlates with reduced growth as determined by leaf area measurements, whereas repression of PAG expression by COR is not associated with reduced photosynthesis under these experimental conditions.
Repression of PAG Transcript Abundance Correlates with the Induction of Defense Genes
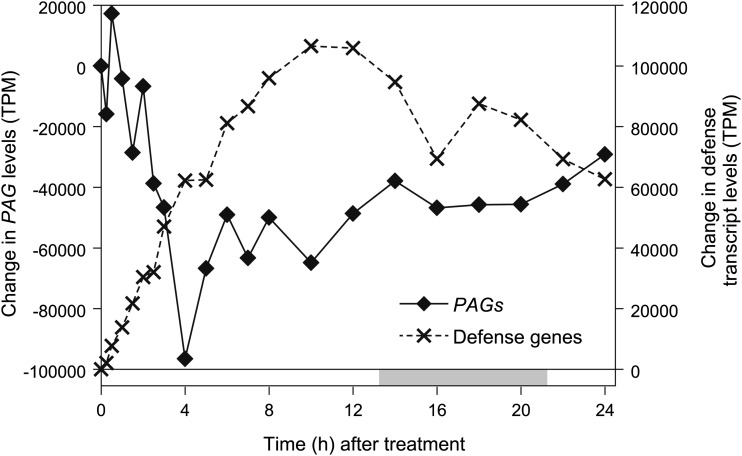
We analyzed the RNA-seq data to determine how COR-induced changes in the abundance of PAG transcripts relate to the expression dynamics of defense-related genes. As expected, functional categories associated with JA-triggered defense responses were strongly induced by COR (Fig. 2B). Among these categories were responses to wounding and JA stimulus as well as the defense-related glucosinolate and phenylpropanoid metabolic pathways. These sets of defense genes were induced as early as 15 min after COR treatment and, in many cases, were up-regulated for the duration of the time course. To quantitatively compare PAG repression with defense gene induction, we determined the cumulative change in levels of PAG and defense-related transcripts over time. Summing of the expression values for the 50 most strongly repressed PAG genes showed a sharp peak in repression at the 4-h time point (Fig. 4; Supplemental Table S2). This cumulative decrease of approximately 97,000 transcripts per million (TPM) accounted for 9.7% of the entire leaf transcriptome. By comparison, the 50 most strongly induced defense genes increased cumulatively by approximately 62,000 TPM at the 4-h time point, with a later peak of 107,000 TPM 10 h after treatment (Fig. 4). Direct comparison of the cumulative changes in PAG and defense transcripts showed a good inverse correlation (r = −0.87) within the first 4 h after COR treatment. These results show that JA-triggered expression of defense genes correlates temporally with PAG repression and that this reprogramming of the leaf transcriptome can occur without significant reduction in photosynthetic efficiency.
Figure 4.
Comparison of COR-induced changes in PAG and defense-related transcript levels. Plants were treated with either COR or a mock control as described in Supplemental Figure S1. Cumulative changes in transcript levels are shown for the 50 most strongly repressed PAGs (solid line, left axis) and the 50 most strongly induced defense-related genes (dashed line, right axis). Photoperiod is denoted above the x axis as described for Figure 1.
Real-Time Fluorescence Imaging Reveals a COR-Induced Transient Decrease in Photosynthesis
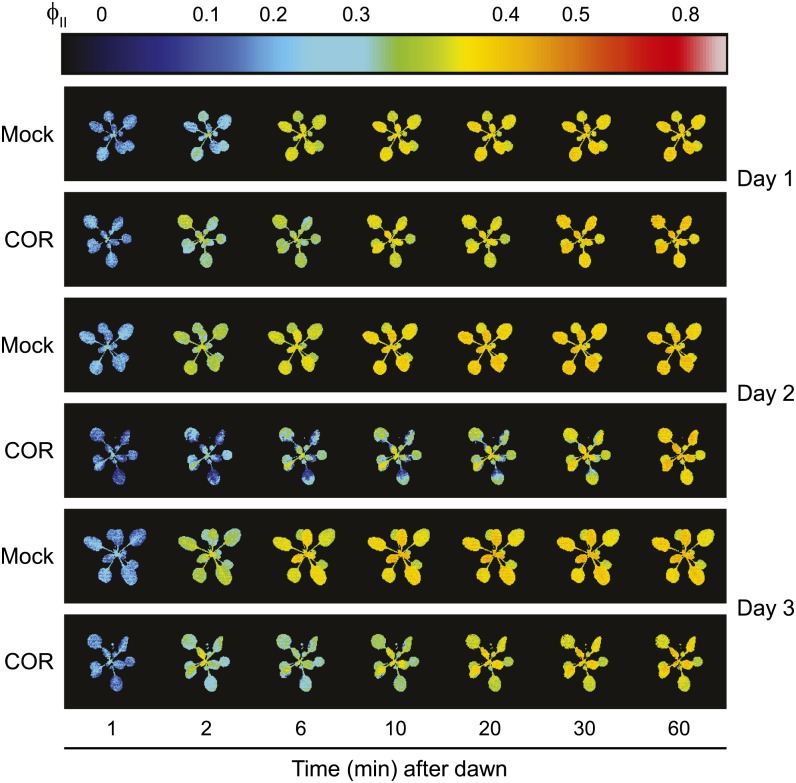
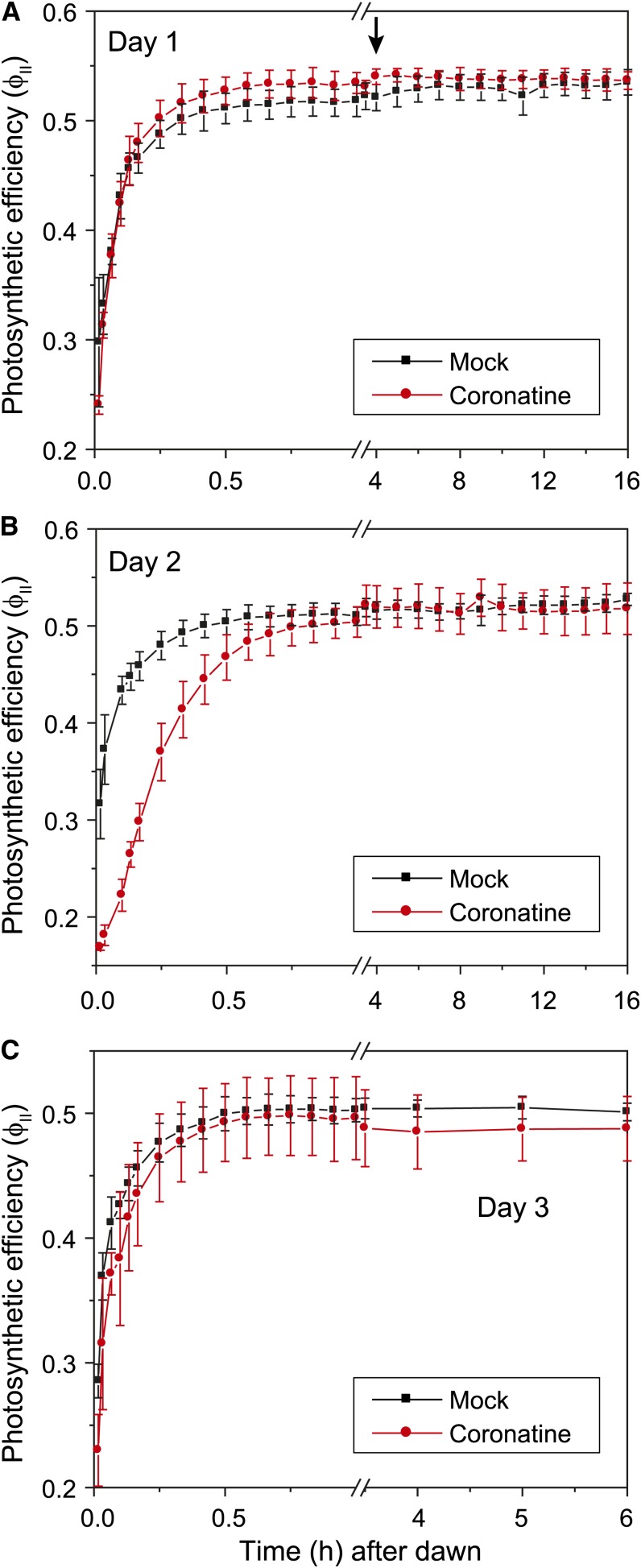
In our initial studies to determine how COR treatment affects photosynthetic parameters, hourly measurements of steady-state ΦII provided evidence for a transient decrease in photosynthetic efficiency at dawn (dark-light transition) of the day after COR treatment (data not shown). To further investigate the timing of this effect, we increased the frequency of fluorescence measurements at the dark-light transition of each day (Supplemental Fig. S1). Images were acquired for 1 d prior to treatment (day 0), the day of treatment (day 1), and the following 2 d after treatment (days 2 and 3). Changes in ΦII were not detected on day 0 or day 1. However, ΦII images collected at high temporal resolution revealed a significant reduction in PSII efficiency during pre-steady-state photosynthesis at dawn of day 2 (Fig. 5). Quantification of ΦII data showed that mock-treated plants reached steady-state photosynthetic rates within approximately 20 min of the dark-to-light transition, whereas the establishment of steady-state photosynthesis was significantly delayed in COR-treated plants (Fig. 6). Within 1 h of exposure to light, ΦII levels in COR- and mock-treated plants were indistinguishable, indicating that the effect of COR is transient. The transient reduction in ΦII at dawn of day 2 was much less pronounced on day 3 (Fig. 6C), suggesting a photosynthetic acclimation response to COR. We also found that nonphotochemical exciton quenching (NPQ) was unaffected by COR except during the transient response at dawn of day 2, where NPQ was reversibly elevated, likely reflecting the onset of the photoprotective energy-dependent exciton quenching response (Supplemental Fig. S6). High-resolution ΦII measurements performed with the coi1-30 mutant demonstrated that the COR-induced decrease in ΦII on day 2 was dependent on the COI1 receptor (Fig. 7).
Figure 5.
COR treatment reduces photosynthetic efficiency in a delayed but transient manner. Col-0 plants were acclimated in the imaging chamber for 36 h and, 4 h after dawn of the following day (day 1), were treated with either water (mock) or COR. False-color chlorophyll fluorescence images of ΦII (scale bar at top) are shown from a representative experiment at selected times after dawn (dark-light transition) of day 1 (the day of treatment), day 2 (1 d after treatment), and day 3 (2 d after treatment). The experiment was independently replicated three times.
Figure 6.
Quantification of the effect of COR on photosynthetic efficiency. Col-0 plants were treated with COR or a mock control as described in the legend to Figure 5. The time of treatment is denoted by the arrow in A. ΦII values (mean ± se, n = 3 replicates) were calculated from chlorophyll fluorescence images captured at the indicated times after the onset of dawn (dark-light transition) of day 1 (A), day 2 (B), and day 3 (C).
Figure 7.
COR-induced perturbation of photosynthetic efficiency is dependent on COI1. coi1-30 plants were treated with COR or a mock control and imaged for chlorophyll fluorescence as described in the legend to Figure 5. A, Representative false-color images of ΦII at selected times after dawn (dark-light transition) of day 2 (1 d after treatment). B, ΦII values calculated from chlorophyll fluorescence images denote means ± se of three independent replicates. For each replicate, ΦII was quantified for two to three actively growing leaves per plant.
Early Morning Decrease in Photosynthesis Is Associated with Altered Stomatal Behavior
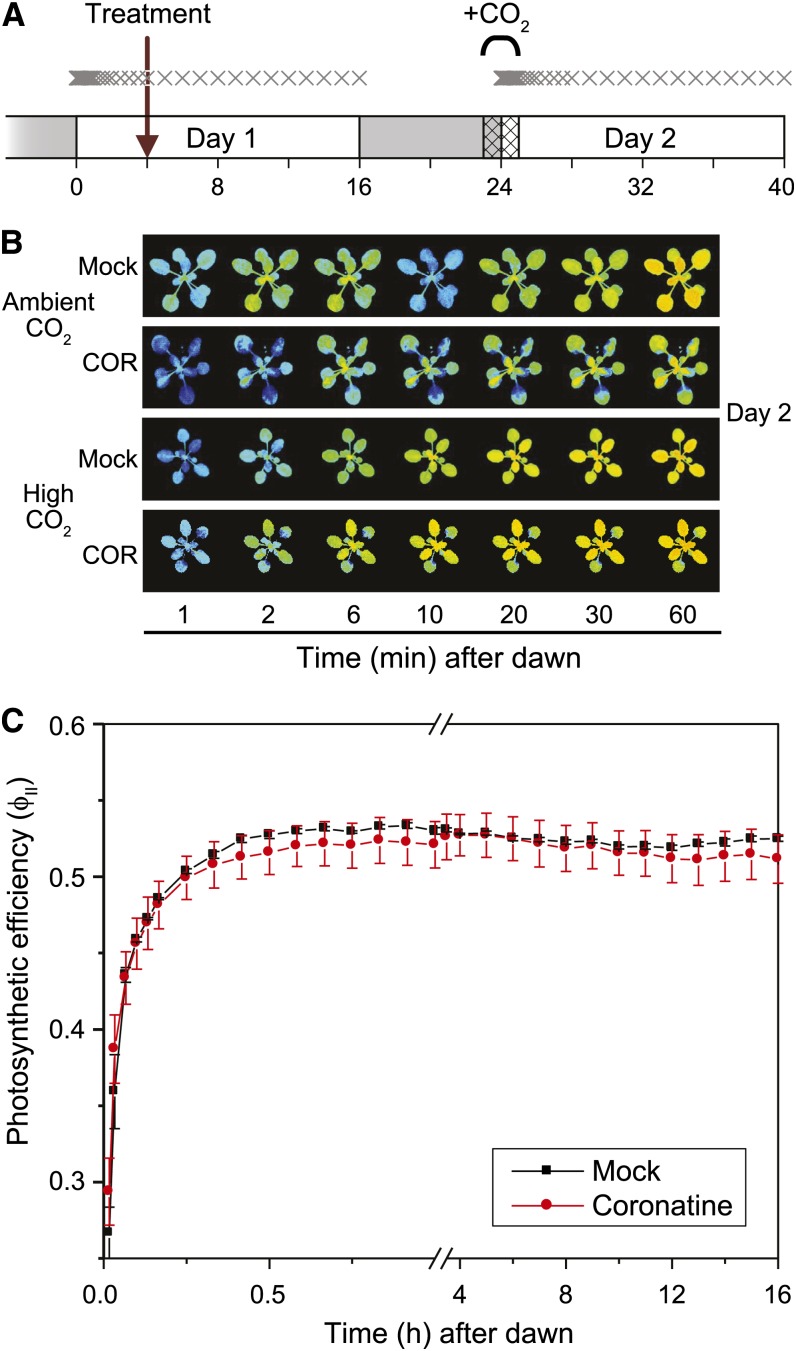
Photosynthetic induction in response to light involves a circadian-regulated opening of stomata to increase gas exchange. We hypothesized that the early morning (day 2) effect of COR on photosynthesis may be related to altered stomatal behavior. It was previously reported that exogenous MeJA causes stomatal closure via a process that depends on abscisic acid and reactive oxygen species (ROS; Hossain et al., 2011; Daszkowska-Golec and Szarejko, 2013). That we did not observe a COR-induced decrease of photosynthesis on day 1 is consistent with the observation that COR does not immediately induce stomatal closure (Melotto et al., 2006; Montillet et al., 2013). We thus considered the possibility that COR might delay stomatal opening at the dawn (dark-light transition) of day 2. Based on the fact that high CO2 concentrations can overcome stomatal limitations in photosynthesis (Farquhar and Sharkey, 1982), we tested whether the exposure of COR-treated plants to elevated CO2 levels could complement the ΦII decrease (Fig. 8A). Indeed, COR elicited the early morning ΦII decrease at ambient CO2, but upon supplementation of plants with high CO2 (2,000 µL L−1) for a 2-h period (beginning 1 h before dawn), this effect on ΦII was alleviated (Fig. 8, B and C).
Figure 8.
COR-induced decrease in photosynthetic efficiency is eliminated by high CO2. A, Diagram of the experimental setup. Following the initial acclimation in the imaging chamber, plants were treated (arrow) with water (mock) or COR 4 h after the dawn of day 1. One set of plants (high CO2) was subsequently treated with 2,000 µL L−1 CO2 for 2 h, beginning 1 h before dawn (hashed region) of day 2 (1 d after COR treatment). A second set of control plants (ambient CO2) was maintained at ambient CO2 levels for the duration of the experiment. Chlorophyll fluorescence images were taken at the time points denoted by X. B, Representative false-color images of ΦII taken at the indicated times after dawn of day 2. C, ΦII values calculated from chlorophyll fluorescence images of plants treated with high CO2. Data show means ± sd of three independent replicates. For each replicate, ΦII was quantified for two to three actively growing leaves per plant.
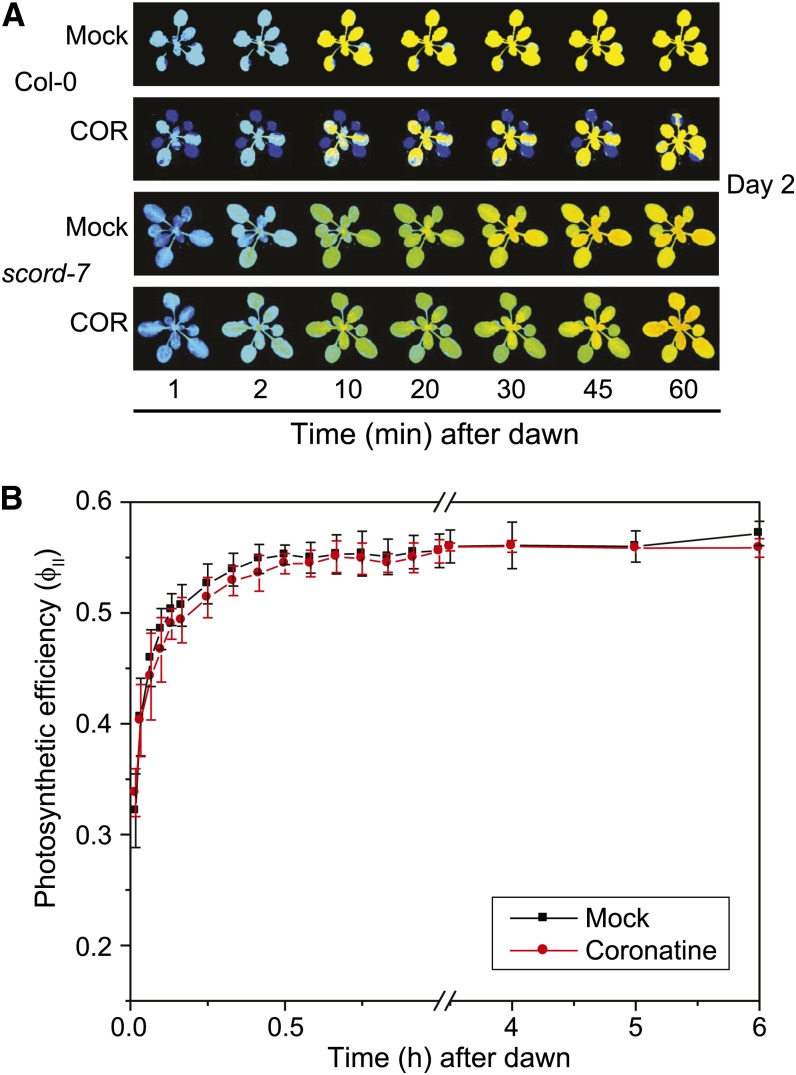
We next examined the response of the susceptible to coronatine-deficient Pst DC3000 (scord7) mutant, which is compromised in stomatal closure (Zeng et al., 2011). Application of COR resulted in the expected decrease in ΦII in control Col-0 plants, but this effect was not observed in the scord7 mutant (Fig. 9A). Quantitative analysis of the data showed that ΦII values in mock- and COR-treated scord7 plants were indistinguishable (Fig. 9B). Together, these results suggest that COR treatment delays the opening of stomata at dawn of the following day (day 2), temporarily limiting CO2 assimilation and the establishment of steady-state photosynthesis. Given the reported effects of JA signaling on ROS production and the inhibitory effect of ROS on stomatal opening (McAinsh et al., 1996; Suhita et al., 2004), we also tested the possibility that elevated ROS levels might be responsible for the COR-induced reduction of ΦII. We detected a significant increase in hydrogen peroxide (H2O2) levels 48 h after COR treatment but not at time points preceding or coinciding with the transient ΦII effect (Supplemental Fig. S7). Therefore, it is unlikely that the accumulation of ROS is responsible for the decrease in ΦII.
Figure 9.
The scord7 mutant does not exhibit a COR-induced decrease in photosynthetic efficiency. Wild-type (Col-0) and scord7 mutant plants were treated with COR (or mock control) as described in the legend to Figure 5. A, Representative false-color images of ΦII at selected times after dawn (dark-light transition) of day 2 (1 d after treatment). B, ΦII values calculated from chlorophyll fluorescence images denote means ± sd of two independent replicates. For each replicate, ΦII was quantified for two to three actively growing leaves per plant.
DISCUSSION
In this study, we exploited a potent agonist of the JA receptor system as a chemical tool to address the question of how a major branch of plant immunity simultaneously represses growth and activates defense and to determine how the rapid activation of JA signaling modulates photosynthesis during the transition from growth- to defense-oriented metabolism. This approach is based on the premise that COR activates JA responses with high specificity and, unlike biotic challenges, does so without the potential complicating effects of tissue damage or attacker-derived effector molecules that target other physiological processes in the host (Zhao et al., 2003; Melotto et al., 2006; Boller and He, 2009; Koo and Howe, 2009; Wu and Baldwin, 2010). Our results show that COR treatment rapidly arrests leaf growth as determined by leaf area measurements and that this effect correlates with the repression of genes involved in cell division and expansion. We found, for example, that genes encoding A- and D-type cyclins, which are involved in the control of the G1/S transition (Gutierrez, 2009), are among the most strongly repressed genes associated with cell cycle regulation. These findings are consistent with previous studies showing that exogenous MeJA inhibits cell expansion and also arrests cells in the G1 phase prior to the S transition (Światek et al., 2002; Pauwels et al., 2008; Zhang and Turner, 2008; Noir et al., 2013). Noir et al. (2013) reported that genes encoding specific A- and D-type cyclins (CYCA3 and CYCD3) are induced by MeJA during the developmental switch from cell proliferation to endoreduplication. These particular cyclin subtypes are negative regulators of endoreduplication and thus may play dual roles in JA-mediated growth repression by arresting the cell cycle in proliferating cells and inhibiting the switch to endoreduplication in expanding cells. It is possible that we did not detect the up-regulation of these genes because our analysis was focused on a relatively short time period (24 h) following COR treatment.
Other JA-signaled processes may also play a role in growth suppression, for example, by inhibiting the growth-promoting effects of GAs. Recent studies indicate that JA-triggered degradation of JAZ proteins serves to increase the abundance of growth-repressing DELLA proteins through a mechanism involving direct JAZ-DELLA interaction (Hou et al., 2010; Yang et al., 2012). In addition, the Repressor of Ga1-3-Like Protein (RGL3) member of the DELLA family of proteins in Arabidopsis is strongly induced at the transcriptional level by JA (Wild et al., 2012), and we found that this gene is also strongly expressed in response to COR treatment (Supplemental Table S1). Although it is clear that JA-induced expression of RGL3 modulates host defense responses (Wild et al., 2012), a direct role for RGL3 in growth repression remains to be determined.
It is well established that JA signaling represses the expression of PAGs and, depending on the treatment and plant species under study, the abundance of the corresponding proteins (Giri et al., 2006; Mitra and Baldwin, 2008; Nabity et al., 2009; Bilgin et al., 2010; Chen et al., 2011; Gfeller et al., 2011; Shan et al., 2011). Long-term exposure of aerial plant tissues to COR (or MeJA) also results in the loss of chlorophyll accumulation and decreased photosynthesis (Kenyon and Turner, 1990; Jung, 2004; Uppalapati et al., 2005; Ishiga et al., 2009; Shan et al., 2011). Whereas our transcriptome analysis revealed strong repression of PAGs in response to COR treatment, the effect on photosynthesis during maximal PAG repression was negligible. This finding suggests that photosynthesis in the Arabidopsis leaf can tolerate major fluctuations in the expression of components of the photosynthetic apparatus without immediately impacting energy capture and conversion. In support of this idea, we observed that the decrease in RCA transcript levels in COR-treated leaves was not accompanied by significant reduction in RCA protein content (data not shown). Photosynthetic robustness, which is the capacity to produce photosynthetic products in the face of genetic or environmental perturbation, may thus be an important part of the plant’s strategy to ensure an adequate production of defense compounds during critical early stages of the defense response (Kitano, 2004; Luo et al., 2009).
As the primary photosynthetic organ in plants, leaves are the major source of reduced carbon skeletons that fuel the biosynthesis of energy-rich macromolecules. Accordingly, the vast majority of biosynthetic resources within the leaf are dedicated to photosynthesis (Baerenfaller et al., 2008). We found that 72% of the top 100 expressed genes in mock-treated Col-0 leaves have a photosynthesis-related function. Consistent with many previous studies (Schaffer et al., 2001), our high-density time series showed that most, if not all, PAGs exhibit diurnal expression (Supplemental Fig. S4). Such temporal patterns of expression highlight the importance of including time-matched mock controls when assessing the effect of stress treatments on gene expression and for considering the timing of treatment with respect to diurnal and circadian cycles. Although the effect of COR treatment on fold repression of PAG expression was relatively modest (less than 2-fold on average) in comparison with the fold induction of defense genes, it is important to note that this effect reflects a large absolute decrease in the size of the PAG transcript pool. It is possible that highly abundant PAG transcripts in the leaf provide a buffering capacity required for the rapid induction of defense-related genes during stress. Such a mechanism would allow immediate redirection of biosynthetic capacity, including the cellular machinery for transcription and translation, from growth to defense without short-term losses in photosynthesis (Bilgin et al., 2010). The relatively slow turnover time of many photosynthetic proteins may also allow for the maintenance of photosynthetic capacity under conditions where increased JA signaling reduces the abundance of PAG transcripts.
It is possible that we did not observe sustained negative effects of COR on photosynthesis because, unlike pathogen infection or insect herbivory, the treatment does not cause physical damage to tissue. Other studies that employed chlorophyll fluorescence imaging reported spatial heterogeneity in the reduction of photosynthesis in response to insect and pathogen attack, with the strongest effects localized to the site of leaf damage (Zangerl et al., 2002; Bonfig et al., 2006; Berger et al., 2007; Nabity et al., 2013). There is evidence to indicate that these spatially restricted effects result from changes in hydraulic conductance and water stress rather than increased JA signaling per se (Reymond et al., 2000; Nabity et al., 2009). Other studies describing a negative effect of COR on photosynthesis used Pst DC3000 infection assays (Ishiga et al., 2009) in which host responses are modulated not only by COR but also by the action of numerous type III effectors and programmed cell death responses (Zhao et al., 2003; Ishiga et al., 2009).
Although we did not observe sustained reduction of ΦII in response to COR treatment, our newly developed chlorophyll fluorescence imaging technology revealed a previously unreported transient decrease of ΦII at dawn on the morning after treatment. This effect was dependent on the COI1 receptor and occurred well after the onset of PAG repression. That the early morning decrease in ΦII did not correlate temporally with changes in bulk H2O2 levels suggests that reduced photosynthesis at this phase of the response is not caused by COR-induced ROS accumulation, which has been observed in tomato (Solanum lycopersicum) leaves (Ishiga et al., 2009). CO2 supplementation experiments and analysis of the scord7 mutant provided evidence that COR indirectly controls light-induced photosynthesis by affecting stomatal opening at dawn. Because the rate of stomatal opening is expected to strongly affect photosynthetic productivity under fluctuating environmental conditions, the defense-related transient effect described here could have a significant impact on the productivity of field-grown plants (Lawson et al., 2012). Further work is needed to understand how the JA pathway modulates stomata opening at the dark-to-light transition and to determine whether the early morning decrease in ΦII occurs under natural stress conditions. It is possible that the effect of a single application of COR on photosynthesis differs from that elicited by grazing insect herbivores that repeatedly wound leaf tissue and thereby continuously stimulate JA-Ile production. Likewise, photoperiod, light intensity, and humidity may also affect the extent to which JA signaling impacts photosynthetic parameters.
Under growth conditions in which water and nutrients are not limiting, the rate of plant growth and biomass accumulation is directly related to photosynthetic efficiency. Our results show, however, that light energy capture by photosynthesis remains largely unaffected in leaves whose growth is rapidly arrested by COR treatment. This finding suggests that JA signaling effectively uncouples growth from photosynthesis and is consistent with the view of JA as a signal to redirect biosynthetic capacity from growth to defense (Ballaré, 2009; Meldau et al., 2012). Recent studies of nitrogen flux dynamics in response to JA elicitation and insect herbivory support this idea (Ullmann-Zeunert et al., 2013). Other studies have shown that simulated herbivory can redirect the allocation of fixed carbon from leaves to roots (Schwachtje et al., 2006; Ferrieri et al., 2013). Carbon partitioning to belowground tissues may represent a plant strategy to protect resources from consumption and to better tolerate herbivory (Schwachtje et al., 2006). Additional work is needed to determine how JA-induced inhibition of leaf growth is related to resource partitioning and biomass accumulation and to better understand the genetic mechanisms that control growth-defense tradeoffs in dynamic environments.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used as the wild-type genetic background for all experiments. Soil-grown plants were maintained in a growth chamber with a 16-h day (100 µE m−2 s−1 cool-white fluorescent light, 22°C) and 8-h night (18°C) and fertilized weekly with 0.5× Hoagland solution. Soil-grown plants were 3 to 4 weeks old when treated for experiments. For studies with the coi1-30 mutant (SALK_035548, obtained from the Arabidopsis Biological Resource Center and described by Yang et al. [2012]), homozygous coi1-30 seedlings were selected on the basis of their JA-insensitive root growth phenotype. Seedlings were grown on vertically oriented square petri plates containing solid medium (1× Linsmaier and Skoog [Caisson Laboratories], 0.7% [w/v] phytoblend agar [Caisson Laboratories], and 0.8% [w/v] Suc) supplemented with 20 µm MeJA (Sigma-Aldrich). In parallel, Col-0 seedlings were grown on solid medium without MeJA (only for coi1-30 experiments), and homozygous coi1-30 and Col-0 seedlings were transplanted to soil after 8 d. For fine-scale analysis of gene expression by RNA-seq, plants were grown on 10 cm × 10 cm square petri plates containing solid medium (1× Linsmaier and Skoog and 0.5% [w/v] phytoblend agar). The medium did not contain Suc to ensure photoautotrophic growth. Nine high-quality Col-0 seeds (minimum size of 300 µm) were sown per plate with equidistant spacing and were maintained in a growth chamber (Percival Scientific) at 22°C with a 16-h day (100 µE m−2 s−1) and 8-h night. To minimize within-chamber variation, plates were randomly rotated twice per week. Before sowing on solid medium, seeds were surface sterilized with 40% (v/v) commercial bleach for 10 min and washed 10 times with sterile water. All seeds were stratified for 3 to 4 d at 4°C prior to germination.
RNA Extraction and Quantitative PCR
For RNA-seq analysis, plants grown on solid medium were sprayed with sterile water (mock) or 5 µm COR (prepared in sterile water), as described in Supplemental Figure S1. Five plants (including roots) from the same plate were pooled for each sample, with two biological replicates collected per sample. For quantitative PCR (qPCR) analysis, soil-grown plants were sprayed with mock or 5 µm COR solution at 3.5 h after dawn, and at harvest two plants were pooled for each sample, with three biological replicates collected per sample. For both RNA-seq and qPCR analyses, the 16-h-light/8-h-dark photoperiod was maintained during the course of the experiment, and mock-treated samples were collected for each time point to account for changes caused by diurnal rhythms. Harvested tissue was immediately frozen in liquid nitrogen and stored at −80°C until processing. Frozen tissue was homogenized using a TissueLyser II (Qiagen) and 2-mm stainless steel beads. RNA was extracted using the RNeasy kit (Qiagen) with on-column DNase (Qiagen) treatment to remove genomic DNA, as per the manufacturer’s protocols. RNA quality was assessed by A260-A280 ratios (typically, 2.1–2.2) using an ND-1000 UV Nanodrop spectrophotometer (Thermo Scientific) and by RNA integrity (greater than 7.0) determined with a Bioanalyzer (Agilent). For qPCR analyses, complementary DNA (cDNA) was reverse transcribed from 100 ng of total RNA with random primers using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) as per the manufacturer’s instructions. The resulting cDNA was diluted to 0.5 ng µL−1 with RNase-free water.
Primers (Integrated DNA Technologies) were designed for each gene (Supplemental Table S3) using Oligo Explorer (Gene Link) or PerlPrimer (O. Marshall) with the following guidelines: 19 to 30 bp per primer with an amplicon of 100 to 160 bp, melting temperature of 65°C to 70°C (Integrated DNA Technologies Oligoanalyzer; settings of 0.25 µm oligonucleotide concentration and 50 mm Na+ and Mg2+ salt concentrations) with less than 2°C difference between primer pairs, and minimal intraprimer and interprimer complementarity. Primer efficiency was calculated for each primer pair as the mean efficiency of all genuine amplifications determined from the log-linear phase of each amplification plot using LinRegPCR version 2012.0 (Ruijter et al., 2009). qPCR was performed on an ABI 7500 Fast qPCR instrument (Applied Biosystems) on Fast Optical 96-well plates (Applied Biosystems) using Power SYBR Green (Applied Biosystems). Reactions consisted of 2 µL of diluted cDNA template (1 ng total), 1 μL of 5 µm forward and reverse primers (0.5 µm reaction concentration), 5 µL of 2× Power SYBR master mix, and 2 µL of nuclease-free water for a final reaction volume of 10 µL. Standard reactions were run with the following conditions: 50°C for 2 min, 95°C for 10 min, then 40 cycles of 15 s at 95°C for denaturation and 60 s at 60°C for annealing and polymerization. A dissociation curve was performed at the end of each reaction using default parameters (15 s at 95°C, 60 s at 60°C–95°C in 1°C increments, and 15 s at 95°C), which confirmed a single peak for each set of primers. Primer specificity was further assessed by separating multiple reactions per primer set from different runs on agarose gels, which confirmed the expected length of the amplicons. No-reverse transcription controls were run for each cDNA sample to confirm the absence of genomic DNA contamination. No-template controls were included for each primer set per run to confirm the absence of contamination and primer dimers. The no-template control wells consistently recorded no signal or were 10 or more cycle threshold above the target signal. All reactions were run with two technical replicates, which typically did not differ by more than 0.2 to 0.5 cycle threshold. Four reference genes (Protein Phosphatase 2A, Yellow Leaf Specific Gene8, Elongation Factor 1a, and F-Box Family Gene) reported previously (Vandesompele et al., 2002) to have stable expression in Arabidopsis were profiled for the entire time course. These reference genes were used to calculate a normalization factor for each sample, to which the expression of all other genes was normalized as described by Vandesompele et al. (2002). The efficiencies for each primer set were determined by LinRegPCR.
RNA-seq Analysis
The time points for fine-scale analysis of gene expression were selected with a bias for early responses: a 0-h control, then every 15 min for the first 30 min after treatment, then every 30 min until 3 h, every 1 h until 8 h, and finally every 2 h until 24 h after treatment, resulting in 21 time points including the 0 h (Supplemental Fig. S1). Two biological replicates were sequenced for each time point and treatment pair, except the 0-h control, for which three replicates were sequenced, resulting in 83 RNA samples. Tissue was handled and RNA extracted as described above, and the integrity of RNA samples was assessed with a Bioanalyzer (Agilent) to ensure an RNA integrity score of at least 7 for RNA sequencing. Barcoded sequencing libraries were created from high-quality total RNA using the Illumina RNAseq kit following the manufacturer’s instructions. Normalized libraries were run on the Illumina HiSeq 2000 sequencer with multiplexing of six libraries per lane on two flow cells, producing an average of 22.9 ± 2.8 million reads per sample. The number of reads sequenced and the number of open reading frames detected per sample were similar between each cell, indicating that sequencing from the two flow cells was comparable. One RNA sample (0-h control replicate) was sequenced on both flow cells, and the gene expression levels were highly correlated (r2 = 0.997), demonstrating directly that variability between the flow cells was very low. Quality control of reads was assessed with the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), using the artifacts filter to remove sequencing artifacts, the clipper to discard sequences with unknown nucleotides, and the quality trimmer to trim nucleotides below a quality score of 30 and discard sequences shorter than 40 nucleotides. Overall, this quality control discarded 1.3% ± 0.3% of reads. Reads were mapped to the Arabidopsis genome using the Illumina iGenomes The Arabidopsis Information Resource 10 index with RSEM (Li and Dewey, 2011) running default parameters and are expressed as TPM (reads per kilobase of model per million mapped reads normalized to transcript coverage). One sample (mock, 6 h) was poorly correlated with its biological replicate and had a higher duplicate read rate relative to other samples, likely due to low input concentration. This sample was discarded, and as such the 6-h mock time point was represented by one biological replicate. Differential gene expression was assessed by subtracting the number of transcripts (TPM) in COR-treated samples from that in the time-matched, mock-treated sample. Differentially expressed functional categories were determined by a Mann-Whitney-Wilcoxon test (Mann and Whitney, 1947) with Benjamini-Hochberg correction that was performed for each time point using the change in transcript level (COR minus mock). Functional gene categories were defined by the AraPath knowledgebase, which combines several annotation sets including the Arabidopsis Gene Ontology (GO), AraCyc pathway, and Kyoto Encyclopedia of Genes and Genomes (KEGG) functions (Lai et al., 2012). For targeted comparison of PAG and defense gene expression patterns, we defined a list of PAGs from the GO categories Thylakoid and Photosynthesis plus the KEGG and AraCyc lists for Photosynthesis, Carbon Fixation, and Chlorophyll Biosynthesis and defined a list of defense genes from the GO categories Response to Wounding and Response to JA Stimulus plus the KEGG and AraCyc lists for Jasmonic Acid Biosynthesis (Supplemental Table S4.).
Growth and Chlorophyll Fluorescence Measurements
Chlorophyll fluorescence images of intact plants were obtained from a custom-designed plant imaging system to be described elsewhere (J. Cruz, L. Savage, R. Zegarac, W.K. Kovac, C.C. Hall, J. Chen, and D.M. Kramer, unpublished data). This system uses a white light source to deliver actinic light (100 μmol photons m−2 s−1), providing light conditions that closely resemble those of the growth chamber and permitting noninvasive, continuous monitoring over an extended period. Chlorophyll fluorescence was probed with pulses of red light from a monochromatic red light source, and images were acquired with a CCD camera outfitted with an infrared band-pass filter. Images were acquired for steady-state fluorescence (Fs) and maximum fluorescence (Fm′; determined during a pulse of saturating light), and ΦII was estimated as (Fm′ – Fs)/Fm′. Images for the dark-adapted fluorescence maximum (Fm) were collected before dawn (during a pulse of saturating light), and NPQ was calculated as (Fm – Fm′)/Fm′ (Baker and Oxborough, 2004). Soil-grown plants were transferred to the imaging chamber (with a photoperiod synchronized to the growth chamber) approximately 36 h before treatment for acclimation, as described in Supplemental Figure S1. Images were acquired at increasing time intervals following the night-day transition at dawn: every 2 min for the initial 10-min interval after dawn; every 5 min for the following 1-h interval after dawn; every 30 min for the following 6 h after dawn; and finally at hourly intervals until dusk. Image processing was performed by Visual Phenomics software (Tessmer et al., 2013). Growth and ΦII measurements were averaged from two to three actively growing leaves of one plant per treatment per experiment, and experiments were independently replicated at least three times, unless indicated otherwise. All reported measurements are means of the independent replicates. ImageJ software (Schneider et al., 2012) was used for the analysis of maximum fluorescence images. The area enclosed by a perimeter outlining the edges of two to three actively growing leaves was used as a measure of growth. For CO2 supplementation experiments, the imaging system was supplemented with CO2 to 2,000 µL L−1 1 h before dawn for 2 h (i.e. CO2 supplemented from 5 am until 7 am, with dawn at 6 am) on the day after treatment. For statistical comparison of growth between mock- and COR-treated plants, we compared the slopes (growth rate) from linear regression analysis of leaf area and calculated a P value (two-tailed) to test the null hypothesis that the growth rate is unchanged by COR treatment.
Measurement of ROS
Soil-grown plants were sprayed with mock or 5 µm COR solution at 3.5 h after dawn. At harvest, two plants were pooled for each sample, with four biological replicates per sample, and were immediately frozen in liquid nitrogen and stored at −80°C until processing. H2O2 production was measured using the Amplex Red H2O2/peroxidase assay kit (Invitrogen) following the manufacturer’s instructions. Homogenized frozen tissue was extracted in 500 μL of 25 mm sodium phosphate buffer (pH 7.4). Extracts were clarified by centrifugation (12,000g) at 4°C. Fifty microliters of the resulting supernatant was incubated with 0.2 units mL−1 horseradish peroxidase in the dark for 30 min at room temperature. Fluorescence was measured with a fluorescence microplate reader (Perkin-Elmer) using excitation at 530 nm and emission at 590 nm.
The raw RNA-seq read data are deposited in the Short Read Archive (http://www.ncbi.nlm.nih.gov/sra/) and are accessible through accession number PRJNA245231.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Design of experiments for analysis of growth, photosynthetic efficiency, and gene expression profiling.
Supplemental Figure S2. COR treatment does not have an immediate effect on ΦII.
Supplemental Figure S3. Validation of RNA-seq data by quantitative PCR.
Supplemental Figure S4. COR treatment decreases PAG transcript levels.
Supplemental Figure S5. COR-induced changes in gene expression are dependent on COI1.
Supplemental Figure S6. COR treatment elevates NPQ at dawn of the day after treatment.
Supplemental Figure S7. Effect of COR treatment on H2O2 production.
Supplemental Table S1. High-resolution temporal profiling of the Arabidopsis transcriptome in response to COR treatment.
Supplemental Table S2. Top 50 repressed and top 50 induced defense genes.
Supplemental Table S3. Primers used for qPCR analysis.
Supplemental Table S4. List of photosynthesis- and defense-associated genes used in this study.
Supplementary Material
Acknowledgments
We thank Tom Sharkey and Sean Weise for constructive discussions during the course of this project, Marco Herde for help with RNA-seq analysis, Lalita Patel and Marcelo Campos for help with the experiments, and Yuki Yoshida for providing seeds of the coi1-30 mutant.
Glossary
- JA
jasmonate
- JA-Ile
jasmonoyl-l-isoleucine
- RNA-seq
RNA sequencing
- COR
coronatine
- Pst DC3000
Pseudomonas syringae pv tomato strain DC3000
- MeJA
methyl jasmonate
- Col-0
Columbia-0
- ΦII
quantum efficiency of PSII
- TPM
transcripts per million
- NPQ
nonphotochemical exciton quenching
- ROS
reactive oxygen species
- H2O2
hydrogen peroxide
- qPCR
quantitative PCR
- cDNA
complementary DNA
- GO
Gene Ontology
Footnotes
This work was supported by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (grant no. DE–FG02–91ER20021) and by the Natural Sciences and Engineering Research Council of Canada (fellowships to I.T.M. and B.A.R.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Baker NR, Oxborough K (2004) Chlorophyll fluorescence as a probe of photosynthetic productivity. In GC Papageorgiou, Govindjee, eds, Chlorophyll a Fluorescence. Springer, Dordrecht, The Netherlands, pp 65–82 [Google Scholar]
- Baldwin IT. (1998) Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA 95: 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL. (2009) Illuminated behaviour: phytochrome as a key regulator of light foraging and plant anti-herbivore defence. Plant Cell Environ 32: 713–725 [DOI] [PubMed] [Google Scholar]
- Ballaré CL. (2011) Jasmonate-induced defenses: a tale of intelligence, collaborators and rascals. Trends Plant Sci 16: 249–257 [DOI] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Hudson CM, Pires JC, Conant GC. (2012) Metabolic and evolutionary costs of herbivory defense: systems biology of glucosinolate synthesis. New Phytol 196: 596–605 [DOI] [PubMed] [Google Scholar]
- Berger S, Benediktyová Z, Matouš K, Bonfig K, Mueller MJ, Nedbal L, Roitsch T. (2007) Visualization of dynamics of plant-pathogen interaction by novel combination of chlorophyll fluorescence imaging and statistical analysis: differential effects of virulent and avirulent strains of P. syringae and of oxylipins on A. thaliana. J Exp Bot 58: 797–806 [DOI] [PubMed] [Google Scholar]
- Bilgin DD, Zavala JA, Zhu J, Clough SJ, Ort DR, DeLucia EH. (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33: 1597–1613 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfig KB, Schreiber U, Gabler A, Roitsch T, Berger S. (2006) Infection with virulent and avirulent P. syringae strains differentially affects photosynthesis and sink metabolism in Arabidopsis leaves. Planta 225: 1–12 [DOI] [PubMed] [Google Scholar]
- Chen Y, Pang Q, Dai S, Wang Y, Chen S, Yan X. (2011) Proteomic identification of differentially expressed proteins in Arabidopsis in response to methyl jasmonate. J Plant Physiol 168: 995–1008 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszkowska-Golec A, Szarejko I. (2013) Open or close the gate: stomata action under the control of phytohormones in drought stress conditions. Front Plant Sci 4: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M, Meldau S, Howe GA. (2012) Role of phytohormones in insect-specific plant reactions. Trends Plant Sci 17: 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar GD, Sharkey TD. (1982) Stomatal conductance and photosynthesis. Annu Rev Plant Physiol 33: 317–345 [Google Scholar]
- Ferrieri AP, Agtuca B, Appel HM, Ferrieri RA, Schultz JC. (2013) Temporal changes in allocation and partitioning of new carbon as 11C elicited by simulated herbivory suggest that roots shape aboveground responses in Arabidopsis. Plant Physiol 161: 692–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R. (2009) The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Gfeller A, Baerenfaller K, Loscos J, Chételat A, Baginsky S, Farmer EE. (2011) Jasmonate controls polypeptide patterning in undamaged tissue in wounded Arabidopsis leaves. Plant Physiol 156: 1797–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri AP, Wünsche H, Mitra S, Zavala JA, Muck A, Svatos A, Baldwin IT. (2006) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: VII. Changes in the plant’s proteome. Plant Physiol 142: 1621–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. (2009) The Arabidopsis cell division cycle. The Arabidopsis Book 7: e0120, /10.1199/tab.0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms DA, Mattson WJ. (1992) The dilemma of plants: to grow or defend. Q Rev Biol 67: 283–335 [Google Scholar]
- Hossain MA, Munemasa S, Uraji M, Nakamura Y, Mori IC, Murata Y. (2011) Involvement of endogenous abscisic acid in methyl jasmonate-induced stomatal closure in Arabidopsis. Plant Physiol 156: 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Lee LYC, Xia K, Yan Y, Yu H. (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. (April 27, 2014) Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant http://doi/10.1093/mp/ssu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y, Uppalapati SR, Ishiga T, Elavarthi S, Martin B, Bender CL. (2009) The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol 181: 147–160 [DOI] [PubMed] [Google Scholar]
- Jung S. (2004) Effect of chlorophyll reduction in Arabidopsis thaliana by methyl jasmonate or norflurazon on antioxidant systems. Plant Physiol Biochem 42: 225–231 [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon J, Turner JG. (1990) Physiological changes in Nicotiana tabacum leaves during development of chlorosis caused by coronatine. Physiol Mol Plant Pathol 37: 463–477 [Google Scholar]
- Kitano H. (2004) Biological robustness. Nat Rev Genet 5: 826–837 [DOI] [PubMed] [Google Scholar]
- Koo AJ, Howe GA. (2009) The wound hormone jasmonate. Phytochemistry 70: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Gao X, Jones AD, Howe GA. (2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J 59: 974–986 [DOI] [PubMed] [Google Scholar]
- Koo AJK, Howe GA. (2012) Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front Plant Sci 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L, Liberzon A, Hennessey J, Jiang G, Qi J, Mesirov JP, Ge SX. (2012) AraPath: a knowledgebase for pathway analysis in Arabidopsis. Bioinformatics 28: 2291–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T, Kramer DM, Raines CA. (2012) Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Curr Opin Biotechnol 23: 215–220 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Wei H, Ye L, Wang K, Chen F, Luo L, Liu L, Li Y, Crabbe MJC, Jin L, et al. (2009) Photosynthetic metabolism of C3 plants shows highly cooperative regulation under changing environments: a systems biological analysis. Proc Natl Acad Sci USA 106: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR. (1947) On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat 18: 50–60 [Google Scholar]
- McAinsh MR, Clayton H, Mansfield TA, Hetherington AM. (1996) Changes in stomatal behavior and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiol 111: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldau S, Erb M, Baldwin IT. (2012) Defence on demand: mechanisms behind optimal defence patterns. Ann Bot (Lond) 110: 1503–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, et al. (2008) A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Mitra S, Baldwin IT. (2008) Independently silencing two photosynthetic proteins in Nicotiana attenuata has different effects on herbivore resistance. Plant Physiol 148: 1128–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montillet JL, Leonhardt N, Mondy S, Tranchimand S, Rumeau D, Boudsocq M, Garcia AV, Douki T, Bigeard J, Laurière C, et al. (2013) An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol 11: e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabity PD, Zavala JA, DeLucia EH. (2009) Indirect suppression of photosynthesis on individual leaves by arthropod herbivory. Ann Bot (Lond) 103: 655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabity PD, Zavala JA, DeLucia EH. (2013) Herbivore induction of jasmonic acid and chemical defences reduce photosynthesis in Nicotiana attenuata. J Exp Bot 64: 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noir S, Bömer M, Takahashi N, Ishida T, Tsui TL, Balbi V, Shanahan H, Sugimoto K, Devoto A. (2013) Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol 161: 1930–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A. (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RM, Krishnamurthy V, Dicke M, Farmer EE. (2004) A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Tzin V, Mysore KS. (2014) Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front Plant Sci 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PEH, Jahnke S, van Dongen JT, Schittko U, Baldwin IT. (2006) SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci USA 103: 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Wang J, Chua L, Jiang D, Peng W, Xie D. (2011) The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol 155: 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhita D, Raghavendra AS, Kwak JM, Vavasseur A. (2004) Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol 134: 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Światek A, Lenjou M, Van Bockstaele D, Inzé D, Van Onckelen H. (2002) Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol 128: 201–211 [PMC free article] [PubMed] [Google Scholar]
- Tessmer OL, Jiao Y, Cruz JA, Kramer DM, Chen J. (2013) Functional approach to high-throughput plant growth analysis. BMC Syst Biol (Suppl 6) 7: S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Ullmann-Zeunert L, Stanton MA, Wielsch N, Bartram S, Hummert C, Svatoš A, Baldwin IT, Groten K. (2013) Quantification of growth-defense trade-offs in a common currency: nitrogen required for phenolamide biosynthesis is not derived from ribulose-1,5-bisphosphate carboxylase/oxygenase turnover. Plant J 75: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, Jones W, Bender CL. (2005) The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J 42: 201–217 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P. (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44: 1–24 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangerl AR, Hamilton JG, Miller TJ, Crofts AR, Oxborough K, Berenbaum MR, de Lucia EH. (2002) Impact of folivory on photosynthesis is greater than the sum of its holes. Proc Natl Acad Sci USA 99: 1088–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Brutus A, Kremer JM, Withers JC, Gao X, Jones AD, He SY. (2011) A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog 7: e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Turner JG. (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA. (2003) Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J 36: 485–499 [DOI] [PubMed] [Google Scholar]
- Zou J, Rodriguez-Zas S, Aldea M, Li M, Zhu J, Gonzalez DO, Vodkin LO, DeLucia E, Clough SJ. (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol Plant Microbe Interact 18: 1161–1174 [DOI] [PubMed] [Google Scholar]
- Zvereva EL, Lanta V, Kozlov MV. (2010) Effects of sap-feeding insect herbivores on growth and reproduction of woody plants: a meta-analysis of experimental studies. Oecologia 163: 949–960 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.