Epigenetics and chromatin modification engage complex gene regulatory systems in plants.
Abstract
Chromatin modifications and epigenetics may play important roles in many plant processes, including developmental regulation, responses to environmental stimuli, and local adaptation. Chromatin modifications describe biochemical changes to chromatin state, such as alterations in the specific type or placement of histones, modifications of DNA or histones, or changes in the specific proteins or RNAs that associate with a genomic region. The term epigenetic is often used to describe a variety of unexpected patterns of gene regulation or inheritance. Here, we specifically define epigenetics to include the key aspects of heritability (stable transmission of gene expression states through mitotic or meiotic cell divisions) and independence from DNA sequence changes. We argue against generically equating chromatin and epigenetics; although many examples of epigenetics involve chromatin changes, those chromatin changes are not always heritable or may be influenced by genetic changes. Careful use of the terms chromatin modifications and epigenetics can help separate the biochemical mechanisms of regulation from the inheritance patterns of altered chromatin states. Here, we also highlight examples in which chromatin modifications and epigenetics affect important plant processes.
In current usage, the term epigenetics (for review, see Haig, 2004) is used to describe several distinct concepts: some researchers use epigenetics to describe heritable differences not caused by DNA sequence changes, whereas others use epigenetics to describe any changes in chromatin modifications or simply describe unusual patterns of inheritance. This use of the same term to describe different concepts can limit our ability to communicate accurately. In this article, we use the term epigenetics to describe heritable patterns of phenotypic variation that are not solely attributable to differences in DNA sequence. The term heritable in this definition indicates stable transmission of information through mitosis or meiosis in the absence of the original inducing signal. Chromatin state often plays a critical role in gene regulation, but alterations in chromatin state may not necessarily lead to heritable changes. For example, the chromatin state change may depend on particular genetic sequences or the continual presence of an endogenous cue. Studies on the potential role of epigenetics in plant development or response to the environment generally involve genetically identical cells but can struggle to provide strong evidence of heritability in the absence of the primary signal. In contrast, studies investigating the role of epigenetics in natural phenotypic variation can struggle to provide evidence that changes in phenotype, which correlate with variation in chromatin modifications, are not the result of underlying genetic changes. Thus, providing clear evidence for a role of epigenetics has proven challenging in many studies.
The relatively liberal usage of the term epigenetics to describe different concepts can interfere with our ability to clearly describe novel research findings. In some cases, epigenetics has been used to describe any situation of inheritance that does not follow simple Mendelian expectations or any example of gene regulation involving chromatin changes. By reserving the term epigenetic to describe heritability without direct involvement of DNA sequence, we can distinguish this concept of inheritance from the biochemical mechanisms of gene regulation involving chromatin states. In an attempt to better delineate confirmed epigenetic phenomena, we next describe situations that would not be considered epigenetic by this definition.
One common usage of epigenetics is as a catchall term to describe any unexpected, non-Mendelian pattern of inheritance. In some cases, researchers studying unusual patterns of inheritance have found evidence for epigenetic phenomena. For example, the basis for the variable phenotype condition by some alleles of the Agouti locus in mouse (Mus musculus; Morgan et al., 1999), the unstable patterns of transposon activity (discussed in McClintock, 1984), and paramutation (Chandler and Stam, 2004) all involve non-Mendelian inheritance and epigenetics. However, caution should be taken, because there are many examples of unusual patterns of inheritance that can be attributed to genetic changes. For example, transposon insertions can cause unstable phenotypes that behave in unexpected fashions. Studies on heritable changes of some flax (Linum usitatissimum) varieties in response to environmental stress also point to genetic rather than epigenetic changes (Johnson et al., 2011). Even examples of unusual inheritance in mutant plants defective in maintenance of chromatin modifications can reveal examples of genetic rather than epigenetic changes (Yi and Richards, 2008). Many non-Mendelian phenomena have genetic bases, such as quantitative traits, segregation distortion, and cytoplasmic inheritance. Although epigenetic phenomena can display unusual inheritance patterns, it is worth noting that genetic regulation can also lead to non-Mendelian patterns. The description of a phenomenon as epigenetic becomes particularly difficult in organisms that are not tractable to genetic studies. In species with complex polyploid genomes or crossing barriers, it is difficult to probe the actual mechanism of inheritance, and it can be easy to misapply the label epigenetic without showing the lack of primary sequence differences driving the phenomenon.
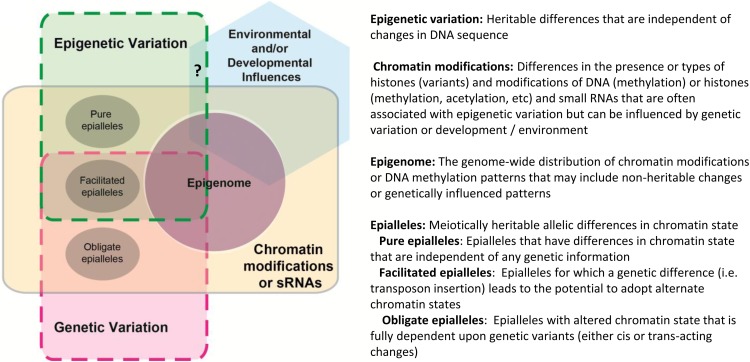
Another common usage of epigenetics is as a term to describe any gene regulation that involves chromatin modifications. The Latin prefix epi connotes over, and in one sense, chromatin modifications certainly provide potential information that may be over the genome sequence information. However, this definition of epigenetic does not have any requirement for transmission through mitosis or meiosis. Therefore, using epigenetic in this context may confound the description of chromatin state as opposed to inheritance patterns. In addition, using the term epigenetics to describe any chromatin changes would likely imply that all gene regulation is epigenetic, because chromatin modifications generally affect gene expression in eukaryotes. Many studies that find differences in chromatin modifications at a particular gene for two plants, tissues, or treatments will report epigenetic regulation of that gene. However, describing this as chromatin-based regulation of the gene would more precisely distinguish between chromatin changes and concepts of heritability. Chromatin modifications do function as an integral part of some epigenetic phenomena; however, some chromatin modifications are likely not heritable and therefore, might not be considered epigenetic (Fig. 1). For example, the phosphorylation of histone H3 Ser 10 (Nowak and Corces, 2004) is quite labile and changes during the cell cycle. There is little evidence that this particular modification provides heritable information that is transmitted through mitotic or meiotic cell divisions. Other chromatin modifications can exhibit stable inheritance, but this property varies substantially for different modifications. Next, we will discuss the potential avenues of heritability for a variety of chromatin modifications.
Figure 1.
Definitions and overlap of concepts often associated with epigenetics. sRNA, Small RNA.
DNA methylation can be heritable through cell divisions, because the methyl group is covalently linked to DNA. DNA replication adds unmethylated cytosines, resulting in hemimethylation at previously methylated sites in all contexts (CG, CHG, and CHH, where H = A, C, or T). Methylation of the newly synthesized strand can occur through context-specific mechanisms (Law and Jacobsen, 2010). For CG dinucleotides, enzymes, such as Methyltransferase1 (MET1), recognize the hemimethylated DNA and direct the maintenance methyltransferase to methylate the symmetrical unmethylated cytosine (Bostick et al., 2007). CHG methylation can be transmitted to newly replicated DNA in a similar fashion by chromomethylase enzymes, such as Chromomethylase3 (CMT3), and evidence also shows that CHG methylation has a self-reinforcing loop with histone H3 lysine9 dimethylation (Lindroth et al., 2004; Johnson et al., 2007; Du et al., 2012). For CHH sites, inherited or newly generated small interfering RNAs (siRNAs) can direct DNA methylation by the Domains Rearranged Methyltransferase2 (DRM2) methyltransferase through the RNA-directed DNA methylation pathway (Aufsatz et al., 2002; Cao et al., 2003; Kanno et al., 2005; Onodera et al., 2005; Law and Jacobsen, 2010), or in some cases, newly accessible DNA resulting from chromatin remodeling undergoes CHH methylation by the CMT2 methyltransferase (Zemach et al., 2013).
Demethylation also influences DNA methylation patterns. Passive demethylation occurs through the lack of active maintenance of DNA methylation, and active DNA demethylation occurs through catalytic removal of 5-methylcytosine (Zhang and Zhu, 2012). Plants encode a family of glycosylases that can actively remove DNA methylation. These enzymes can act in a developmentally programmed fashion to induce locus-specific loss of DNA methylation (Choi et al., 2002; Ibarra et al., 2012) but also play a role in pruning DNA methylation patterns in other tissues (Zhu et al., 2007). Thus, although DNA methylation patterns can be stably inherited, especially in CG and CHG contexts, a variety of natural mechanisms result in gain or loss of DNA methylation and thus, potentially interfere with the heritability of DNA methylation.
Mechanisms for heritability of histone modifications or variants are less obvious and less frequently described in the literature (Margueron and Reinberg, 2010). In some cases, particular histone variants, such as centromeric histone H3, seem to be stably maintained at consistent genomic positions (Allshire and Karpen, 2008). However, many of the nucleosomes that are deposited after replication do not initially contain the same modifications as the parental chromatin (Xu and Zhu, 2010; Abmayr and Workman, 2012; Budhavarapu et al., 2013). Plants contain a variety of enzymes that can add or remove histone modifications (Chen and Tian, 2007; Pfluger and Wagner, 2007; Chen et al., 2011; Deal and Henikoff, 2011; Thorstensen et al., 2011). The targeting of these enzymes to the proper locations and the turnover rate for different modifications will influence the ability of that modification to be stably inherited. The histone modification with the most evidence for stable inheritance in plants is histone H3 lysine9 dimethylation, which is coupled with CHG DNA methylation and acts in a self-reinforcing loop that likely provides a mechanism for stable inheritance of this chromatin modification (Johnson et al., 2007). Much remains unknown about the potential mechanisms for heritability of histone modifications, but evidence indicates that differences in histone modifications between two cells will not necessarily be transmitted through mitosis or meiosis.
Another potential issue with equating chromatin modifications and epigenetics is that some chromatin modifications may result from genetic variation (Paszkowski and Grossniklaus, 2011; Pecinka et al., 2013). The identification of epialleles, alleles that vary in chromatin state, has increased with our ability to generate genome-wide maps of chromatin modifications, but understanding the mechanistic basis for epiallele formation remains challenging. Moreover, the epiallele designation can be misleading, because not all epialleles are epigenetic (Fig. 1). Obligate epialleles are completely dependent on genetic variation (Richards, 2006; Fig. 2) and persist, because the genome sequence encodes the instructions required to direct chromatin modifications. Therefore, the heritable information for this class of epialleles is genetic rather than epigenetic (Fig. 2).
Figure 2.
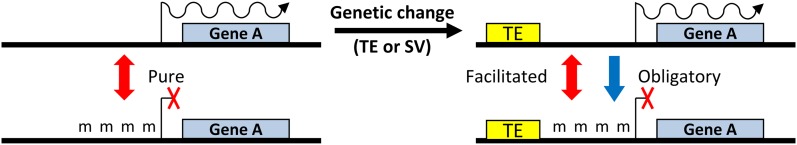
Classification of epialleles based on potential influence of genetic sequences. Pure epialleles shift between transmissible chromatin states (chromatin modification [m]) with no change in genetic sequence at linked or unlinked genomic positions. In other cases, a genetic change, such as a transposable element (TE) insertion or structural variant (SV), can produce directed chromatin changes, resulting in an obligatory allele. Alternatively, the genetic change can result in a poised allele that can exist in two potential chromatin states (facilitated).
The lines between genetic and epigenetic begin to blur when discussing the other two classes of epialleles, facilitated and pure, and any distinction between these epialleles may be very difficult to apply to natural situations. In both cases, the same genetic sequence can exhibit two potential chromatin states that show at least partial heritability. In some clear examples of facilitated epialleles, the genetic sequences at linked or unlinked genomic positions influence chromatin state. For example, in the Agouti spp. locus in mice, alleles that contain a transposon insertion upstream of the gene exhibit metastable inheritance of chromatin state (Morgan et al., 1999). Similarly, paramutation at the B locus of maize (Zea mays) requires certain sequences in the regulatory regions, but the presence of this genetic information does not always lead to one state or the other (Stam et al., 2002; Belele et al., 2013). In another scenario, a change in the DNA sequence can trigger a stable change to the chromatin environment that persists in the absence of the original genetic variant as observed at the Arabidopsis thaliana folate transporter1 (AtFOLT1) locus (Durand et al., 2012). Therefore, facilitated epialleles show a role for epigenetic influence and a role for genetic sequences that predispose an allele to epigenetic regulation (Fig. 2). Pure epialleles are completely independent from changes in the DNA sequence. Two apparent examples of pure epialleles, Linaria cycloidea (Lcyc; Cubas et al., 1999) and Clark Kent (clk; Jacobsen and Meyerowitz, 1997), exhibit stable differences in chromatin state without apparent genetic change. However, even these examples may have sequences that predispose these particular genes to being susceptible to altered chromatin state. These classifications attempt to provide a framework to consider the interaction between DNA sequences and epiallele formation, but it is clear in nature that the three classes exist in a continuum. These distinctions between epiallele types reveal the complexity in equating differing chromatin states at an allele with epigenetics.
Small RNAs are another example of a complex regulatory mechanism that can contribute to epigenetics but is not necessarily epigenetic itself (Bond and Baulcombe, 2014). Small RNAs, including siRNAs, microRNAs (miRNAs), and several other specific types, can influence transcription and RNA stability in plants. However, in most cases, the small RNA is produced from a genomic region and provides a sequence-specific genetic factor. Small RNAs can provide a mobile signal that affects distant genomic regions (Bender, 2004) or even distant cells (Melnyk et al., 2011). In some cases, the influence of the small RNA will only persist while the small RNA itself is present, but there are also mechanisms that can perpetuate the signal of small RNAs. Plant species have the potential to maintain small RNA signals, in some cases through RNA-dependent RNA polymerases; therefore, some small RNAs have the potential to create a self-perpetuating signal (Baulcombe, 2006). Some small RNAs can also trigger RNA-directed DNA methylation, thereby providing a signal that is later translated into a potentially heritable chromatin modification (Simon and Meyers, 2011). Although some small RNAs can exert heritable influences, many small RNAs exert only a transient influence on gene expression. For example, many miRNAs provide specific regulation that influences plant development or responses to the environment, but these responses would rarely fit the definition for epigenetics, because they are not heritable in the absence of the original inducing signal (Voinnet, 2009; Chen, 2012; Sunkar et al., 2012).
The term epigenomics complicates the distinction between chromatin modifications and epigenetics (Fig. 1). Originally, this term was used to describe studies that profile the genome-wide distribution of a particular chromatin modification, such as DNA methylation or histone modification (Callinan and Feinberg, 2006), that was often thought to contain epigenetic information. In recent years, epigenomics has come to encompass any study that profiles the genome-wide distribution of any chromatin-associated factors, such as histone modifications, histone variants, RNAs, and transcription factors (Bernstein et al., 2010), regardless of their heritability. Epigenomics studies provide comprehensive profiles of chromatin modifications, some of which may provide transmissible information. However, as discussed above, many of the chromatin modifications may be nonheritable or programmed by underlying DNA sequence. Therefore, it is important to distinguish between epigenetics and epigenomics, although the two words have similar connotations. Epigenomic studies improve our understanding of potential sources of epigenetic phenomena, and they can also reveal how genetic variation, development, or the environment influences the genome-wide distribution of chromatin modifications.
The above examples illustrate the complexity of gene regulation and some of the difficulties with invoking the term epigenetics. Before examining the potential contribution of epigenetics and chromatin modifications to plant processes, it is worthwhile to clearly distinguish the two types of heritable epigenetic information. Meiotic inheritance (or transgenerational) will often involve constitutive heterochromatin. Maintaining stable inheritance of information across generations requires this information to pass through many mitotic cell divisions. Given its stability, one would expect that this type of variation could play a role in natural variation, evolution, and polyploidy. By contrast, mitotic inheritance will often involve facultative heterochromatin that might vary among cells depending on their developmental history or environmental exposures. We will begin by considering the potential role for epigenetics and chromatin modifications in plant development and response to the environment. We will consider whether the line between mitotic and meiotic inheritance may sometimes be blurred. If some of the mitotically heritable changes in chromatin state that are influenced by development or the environment can be transmitted to offspring, this would create the potential for so-called soft inheritance, in which acquired characteristics could be transmitted to offspring (Richards, 2006). We will conclude by considering the potential role for epigenetics in natural variation and evolutionary processes.
EPIGENETICS AND PLANT DEVELOPMENT
Many open questions remain regarding the role of chromatin modifications and epigenetics in driving plant developmental processes. Development in multicellular organisms often involves differentiation of cells into specialized cell types that express different sets of genes. The silencing of certain genes in specific cell types could be considered to occur through facultative heterochromatin, with repressive chromatin in different locations based on cell type. The role of epigenetics in plant development is most likely limited to mitotic transmission of gene expression states. If the epigenetic memory of developmental decisions was inherited through meiosis, it would likely interfere with development of the subsequent generation. Epigenetics may contribute to plant development by stably maintaining gene expression states that are initially directed by sequence-specific factors, such as transcription factors or small RNAs. The Polycomb group (PcG) of genes provides a classic example for a role of epigenetics in developmental control of gene expression (Simon, 1995; Francis and Kingston, 2001; Ringrose and Paro, 2004; Grimaud et al., 2006). A set of well-characterized regulatory mechanisms leads to differential expression of key regulators in Drosophila melanogaster larvae. Initially, these regulatory mechanisms depend on sequence-specific transcriptional activators and repressors, but these gene expression states become locked in by the PcG complex. This process has limited sequence specificity and can regulate different sets of genes in different cell types. Homologs of some PcG genes in plant species may play related roles in developmental regulation of gene expression (Bemer and Grossniklaus, 2012; Holec and Berger, 2012). The PcG genes contribute to molecular memory of gene expression choices that are made during cellular differentiation and provide mitotically heritable information, but we have limited evidence of their contributions to meiotic inheritance (Orlando, 2003; Breiling et al., 2007).
Many of the key developmental regulators identified by forward genetics encode transcription factors (Ramachandran et al., 1994). However, there is strong evidence that chromatin modifications and complex gene regulatory mechanisms, such as small RNAs, play important roles in plant development. Forward genetics experiments have identified a number of genes that contribute to chromatin remodeling or chromatin modifications that can lead to developmental abnormalities (Goodrich and Tweedie, 2002; Reyes et al., 2002; Wagner, 2003; Reyes, 2006; Köhler and Hennig, 2010; Bemer and Grossniklaus, 2012; Holec and Berger, 2012). There is also very strong evidence for a role of miRNAs in controlling developmental transitions in many plant species, largely through their control of transcription factors (Carrington and Ambros, 2003; Jones-Rhoades et al., 2006; Chen, 2009). Direct analyses of chromatin modifications in different cell types have provided evidence that the profiles of some histone modifications change substantially during development (Roudier et al., 2009). For example, H3K27me3 contributes to regulation of important transcription factors in some cell types and shows clear tissue-specific patterns that are often associated with tissue-specific expression of the target genes (Lafos et al., 2011; Zheng and Chen, 2011; Makarevitch et al., 2013). Other marks, such as histone acetylation and H3K4me3, show tissue-specific patterns (Berr et al., 2010) but may be an effect, rather than a cause, of tissue-specific gene expression.
The role of DNA methylation in plant development has not been fully resolved. Plants that have lost functions for some components of the DNA methylation machinery can exhibit altered morphology and development (Finnegan et al., 1996; Ronemus et al., 1996; Richards, 1997). However, this may simply reflect ectopic expression of some genes that influence morphology and development as opposed to indicating a normal role for DNA methylation in regulation of development. For example, Arabidopsis plants compromised in certain key components of the DNA methylation machinery will express SUPPRESSOR OF drm1 drm2 cmt3(SDC). SDC is not normally expressed in vegetative tissues, but when ectopically expressed in certain DNA methylation mutants, it leads to altered development (Henderson and Jacobsen, 2008). Genome-wide analyses of DNA methylation do find examples of tissue-specific differences in vegetative tissues, but the frequency of these changes is relatively small compared to differences among ecotypes or the frequency of tissue-specific gene expression (Zemach et al., 2010; Zhang et al., 2011; Eichten et al., 2013a; Schmitz et al., 2013a). It remains possible that tissue-specific changes in DNA methylation play an important role in the regulation of certain aspects of development (Li et al., 2011; Zhong et al., 2013), but it seems that the bulk of DNA methylation in plants remains relatively stable during vegetative development.
In contrast to the generally stable patterns of DNA methylation in vegetative tissues, DNA methylation in reproductive and endosperm tissues shows dynamic changes (Lauria et al., 2004; Gehring et al., 2009; Hsieh et al., 2009; Zemach et al., 2010; Bauer and Fischer, 2011; Zhang et al., 2011; Gutierrez-Marcos and Dickinson, 2012). During male gametogenesis, the vegetative nucleus undergoes major alterations to DNA methylation patterns (Slotkin et al., 2009; Borges et al., 2012; Calarco et al., 2012; Ibarra et al., 2012; Jullien et al., 2012). Similarly, the central cell of the female gamete also experiences substantial changes in DNA methylation (Choi et al., 2002; Gehring et al., 2006; Hsieh et al., 2009; Ibarra et al., 2012). Neither of these cells will contribute genetic information to the vegetative tissues in the next generation. However, it has been hypothesized that the loss of epigenetic silencing in these cells allows the production of siRNAs that can reinforce silencing of transposons in adjacent reproductive cells that will contribute to the next generation (Slotkin et al., 2009; Baroux et al., 2011; Ibarra et al., 2012; Wollmann and Berger, 2012). This likely contributes to imprinted gene expression in endosperm tissue and the evidence that some targets of imprinting are key regulators of endosperm development (Li and Berger, 2012; Gehring, 2013).
Although Waddington originally used the term epigenetics in a developmental context (Waddington, 1942; Haig, 2004), here, we make the case that a definition of epigenetics requiring heritability makes it difficult to establish an epigenetic component for many of the chromatin modifications that occur during development. This is not to say that epigenetics does not play an important role in development but instead, highlights the complexities in truly assigning a role for epigenetics in the developmental regulation of gene expression. It is hard to show that the chromatin differences among cells would be heritable in the absence of ongoing signals or positional cues. A key exception is imprinting, in which allele-specific expression is controlled by the parent of origin. The maternal and paternal alleles share the same DNA sequence and are present in the same nucleus, but they exhibit different expression levels (Gehring, 2013).
Plants and animals seem to have differences in how chromatin modifications and DNA methylation are reprogrammed before the next generation (Feng et al., 2010). In addition, they have differences in the ease of generating pluripotent cells. In many animal systems, generating pluripotent cells requires substantial treatments to remodel chromatin (Meissner, 2010; Adachi and Schöler, 2012). In contrast, in most plant systems, generating stable tissue culture lines often requires only treatment with combinations of plant hormones. However, tissue culture often causes chromatin changes (Kaeppler et al., 2000; Tanurdzic et al., 2008; He et al., 2012; Stroud et al., 2013). In summary, strong evidence shows that chromatin varies among different cell types and that some of these modifications play important roles in plant development, but whether specific examples are epigenetic (transmissible) during plant development remains less clear.
EPIGENETICS AND RESPONSE TO THE ENVIRONMENT
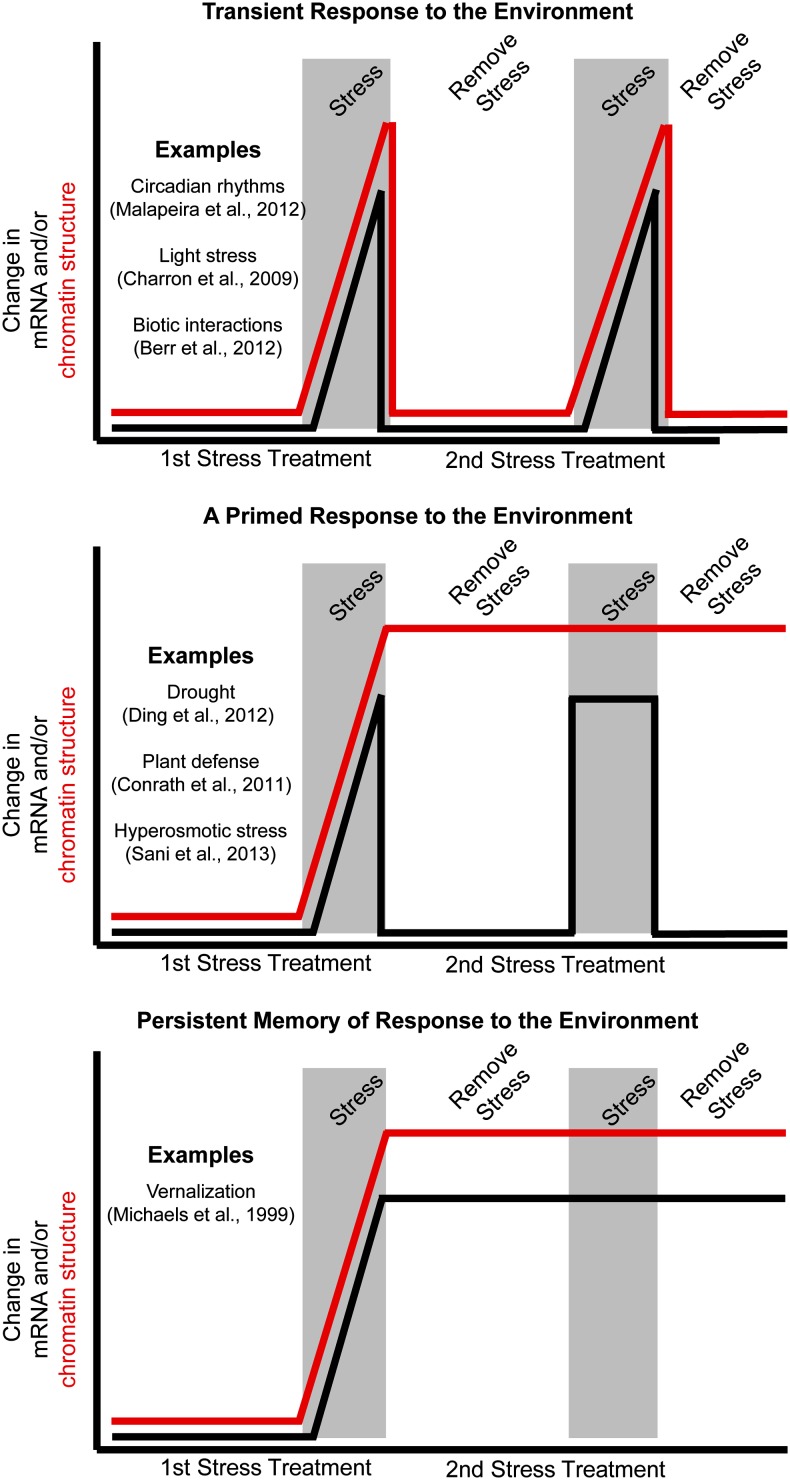
The term epigenetics is often invoked to describe plant responses to environmental cues (Finnegan, 2002; Boyko and Kovalchuk, 2011; Mirouze and Paszkowski, 2011; Paszkowski and Grossniklaus, 2011; Gutzat and Mittelsten Scheid, 2012; Pecinka and Mittelsten Scheid, 2012). The concept of environmental responses as epigenetic is often used to describe a range of findings from heritable phenotypic consequences to the observation of chromatin changes after environmental treatments. Stable phenotypic consequences after an environmental treatment may reflect physiological or morphological changes triggered by the initial environmental cues that are maintained without epigenetic contributions. However, the observation of chromatin changes does not necessarily imply epigenetics, because these changes may be transient effects that require the ongoing presence of the stimulus. For example, drought conditions may result in altered leaf wax deposition, and this morphological change could influence phenotypic responses to subsequent environmental conditions but would not reflect cellular inheritance of the memory of the stress. Conceptually, mitotically transmissible memory that programs responses to environmental cues may provide part of the mechanism that plants use to alter gene expression in response to the environment. There are some clear examples of epigenetic mitotic memory of environmental conditions, such as vernalization (see below), but there are many more examples in which dynamic changes in chromatin modifications provide a portion of the mechanism for transient gene expression changes in response to the environment (Fig. 3). We will begin by discussing the immediate and short-term effects of environmental stress. In the next section, we will touch on the more contentious issue of whether environmental exposures can lead to heritable changes in offspring reminiscent of Lamarckian inheritance (Daxinger and Whitelaw, 2010).
Figure 3.
Differential expression and chromatin alterations in responses to the environment. Top, Genes in this class respond transiently and have no memory to the environment by undergoing changes to both the local chromatin state and mRNA levels. Middle, Genes in this class respond to a change in the environment by altering chromatin states and changing mRNA levels. This alteration in chromatin states can persist, which enables a more rapid mRNA response after a subsequent exposure to that environment. Bottom, A memory of the environment is enabled by persistent changes in both gene expression and local chromatin states after exposure to the environment.
Many published reports have documented changes in chromatin or siRNA abundance in response to environmental conditions (Gutzat and Mittelsten Scheid, 2012; Khraiwesh et al., 2012). Histone modification patterns can vary after environmental exposure and often are associated with altered expression at some loci (Kwon et al., 2009; Kim et al., 2010; van Dijk et al., 2010; Luo et al., 2012; Zong et al., 2013). In some cases, environmental treatments, such as heat, can affect heterochromatin (Lang-Mladek et al., 2010; Pecinka et al., 2010; Tittel-Elmer et al., 2010). Environmental stress can also affect the abundance of miRNAs or siRNAs in plants (Ruiz-Ferrer and Voinnet, 2009; Khraiwesh et al., 2012). Transposons can also respond to changes in environmental conditions (Grandbastien et al., 2005; Pecinka et al., 2010; Ito et al., 2011) and may influence the responsiveness of nearby genes to environmental cues (Feschotte, 2008; Naito et al., 2009; Ito et al., 2011; Bucher et al., 2012; Wheeler, 2013; Cavrak et al., 2014). Together, these reports provide clear evidence for an important role of chromatin and siRNAs in plant responses to the environment. Although chromatin changes or small RNAs have the potential to contribute to heritable changes, they can also lead to transient changes in gene expression. Few examples have shown actual mitotic or meiotic memory of environmental treatments.
Vernalization provides an example of mitotic epigenetic memory during the lifetime of a plant. Certain plant varieties germinate in the fall, overwinter as a vegetative rosette, and subsequently, flower early in the spring season when the days lengthen. Vernalization is the exposure to long-term cold that occurs during this overwintering period and renders plants competent to flower early in the spring (Amasino and Michaels, 2010). Plants with this lifecycle can take advantage of an ecological niche that enables successful reproduction in the early spring, when many other plant varieties have just begun to germinate. A phenotypic analysis of the impact of temperature and day length on flowering in henbane (Hyoscyamus niger) elegantly showed the epigenetic basis for vernalization (Lang and Melchers, 1947). Although the specific mechanisms of vernalization vary between species, clear evidence shows an epigenetic basis of vernalization in Arabidopsis (Schmitz and Amasino, 2007). Before the extended period of cold, the floral repressor FLOWERING LOCUS C (FLC) is expressed and suppresses the transition from vegetative growth to flowering. During extended periods of cold, FLC is silenced, in part, by changes to chromatin, including histone modifications (Bastow et al., 2004; Sung and Amasino, 2004; Song et al., 2012). This altered chromatin and expression state is then stably transmitted mitotically and renders plants competent to flower, even in the absence of the original stimulus (cold temperature). Vernalization is not meiotically heritable, because it resets every generation. Failure to reset the requirement for vernalization would be detrimental, because it would lead plants to flower rapidly before the onset of winter, reducing overall reproductive success.
Another potential example of epigenetic contributions to environmental response is the priming or training response to environmental conditions. Priming refers to changes in phenotype or gene expression after repeated exposures to an environmental stress. A phenotypic response or gene expression change will be trained by initial exposures to stress and will exhibit more pronounced or rapid responses to subsequent treatments of the same environmental condition (Fig. 3). There are examples of priming in response to abiotic (Sung et al., 2003; Ding et al., 2012; Sani et al., 2013) and biotic (Conrath, 2011) environmental cues. It is important to note that the observations of priming are often rooted in phenotypic observations of how responses to subsequent treatments differ from initial treatments. In some cases, the priming response has been associated with alterations in chromatin modifications (Jaskiewicz et al., 2011; Sani et al., 2013) or DNA methylation (Dowen et al., 2012; Yu et al., 2013). However, it is difficult to fully document the role of epigenetics in this phenomenon (Sani et al., 2013). The initial treatment of an environmental stress often results in numerous physiological and morphological changes. These differences may provide a different response to subsequent treatments in the absence of a true epigenetic memory.
EPIGENETICS AND TRANSMISSION OF ENVIRONMENTAL EFFECTS TO OFFSPRING
The section above focused on how plants respond to environmental cues with chromatin changes, including mitotically transmissible changes. In this section, we will evaluate the evidence that environmental exposures of the parents may transmit altered expression states to offspring. It is possible that environmental stresses during the parental generation could promote changes, leading to offspring being more prepared to deal with a similar environment. This may be especially true in plant systems with limited seed dispersal, leading to very similar environmental conditions for both parent and offspring. There are some hints that this might occur (Boyko and Kovalchuk, 2011; Bilichak et al., 2012; Luna et al., 2012; Rasmann et al., 2012; Slaughter et al., 2012), but it is still unclear if this soft inheritance occurs (Pecinka and Mittelsten Scheid, 2012) and how large an impact on phenotype it may have.
There are clear examples of maternal effects in which the environment of the maternal parent can influence seed or seedling traits of the offspring (Galloway, 2005). In many cases, these are most pronounced if the environmental stresses occur during seed development and maturation. However, the observation of maternal effects does not necessarily translate to epigenetics per se. In an extreme case, in which the maternal parent is subjected to severe environmental stress, the seeds will often be smaller and have reduced viability, likely because of direct physiological changes during exposure rather than inheritance of expression states altered by stress. The separation of seed viability or physiology from true inherited epigenetic changes is complex. Imprinting provides a clear example of epigenetic parental effects that influence the relative expression of the maternal and paternal alleles in offspring endosperm tissue. However, there is little evidence that imprinting is sensitive to environmental conditions.
What are the hints of epigenetic memory of environmental stresses that could be transmitted to offspring? Reports indicate that somatic homologous recombination rates in offspring can be affected by parental environmental exposures (Molinier et al., 2006; Yao and Kovalchuk, 2011; Puchta and Hohn, 2012), but this may not be a highly reproducible response (Pecinka et al., 2009; Ülker et al., 2012). Several recent articles provide evidence for heritable phenotypic changes or chromatin changes during development or in the offspring of plants exposed to biotic stress (Boyko and Kovalchuk, 2011; Dowen et al., 2012; Luna et al., 2012; Rasmann et al., 2012; Slaughter et al., 2012). Groups have also reported heritable changes in DNA methylation or other chromatin modifications in the progeny of plants subjected to abiotic stresses (Verhoeven et al., 2010; Bilichak et al., 2012; Verhoeven and van Gurp, 2012; Colaneri and Jones, 2013). There are also reports that tissue culture, a very severe environmental condition, results in heritable changes in DNA methylation levels (Stroud et al., 2013). Despite these reports, there are still concerns about whether they represent bona fide examples of epigenetics that are induced by the environment (Pecinka and Mittelsten Scheid, 2012).
Parental environmental exposures certainly can affect the phenotype of offspring, but this might have little effect on phenotype. For most agronomic species, there is limited evidence that performing selection using environmental variation as opposed to genetic variation can lead to improved performance. It is clear that using particular environments for selecting ideal genetic combinations has resulted in more locally adapted varieties. However, there is limited evidence that breeders have been successful in performing local adaptation of a variety using repeated growth of materials in a particular environment without allowing for genetic diversity or recombination of diverse alleles. That being said, this fascinating topic deserves additional attention and will likely reveal nuances in how plants adjust to environmental conditions.
A POTENTIAL ROLE FOR EPIGENETICS IN EVOLUTION AND PHENOTYPIC VARIATION WITHIN SPECIES
The above sections focused on the potential for epigenetics and chromatin modifications to contribute to changes in gene expression associated with development or environmental responses. Epigenetics could also contribute to natural variation within species and potentially, local adaptation. Epigenetics could theoretically impact natural variation as a faster acting and less permanent method of gene regulation compared with genetic variation. The potential reversibility of epiallelic states opens the possibility of temporary adaptation or exploration of cryptic genomic information (Richards, 2006, 2011; Weigel and Colot, 2012). In a similar vein, the potential instability of epiallelic states and the requirement for continued active silencing may suggest that longer term evolutionary changes in plants would often use genetics as opposed to epigenetics. This may leave a role for epigenetics in natural variation within a species but suggests that stable differences between species likely involve primarily genetic changes.
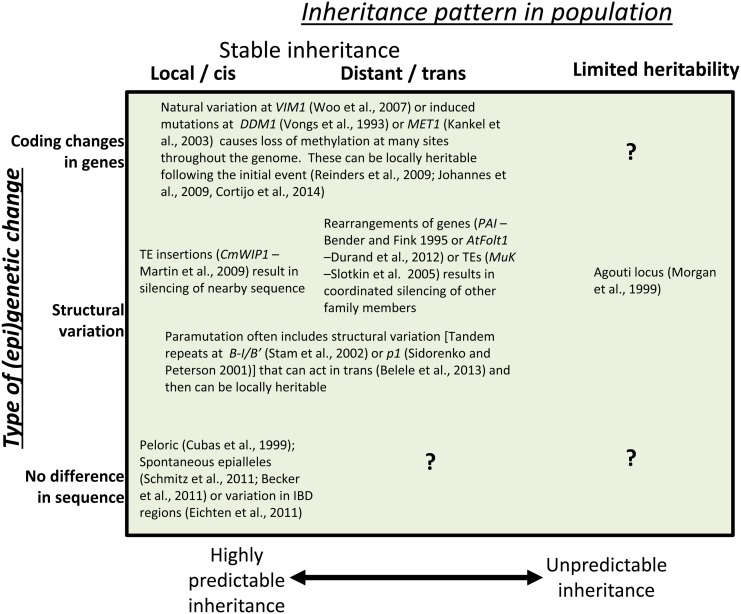
There are many examples of natural variation for DNA methylation or other chromatin modifications. The main difficulty in assigning natural variation for chromatin modifications as epigenetic is the problem of separating chromatin changes from genetic changes that also occur between individuals of the same species. There are some well-characterized examples of epigenetic natural variation that may contribute to phenotypic variation within plant species (Fig. 4). In recent years, there has been substantial progress in characterizing natural variation for DNA methylation or other chromatin modifications. However, it has become clear that this variation can be driven by both genetic and epigenetic mechanisms. The development of epigenetic recombinant inbred lines (epiRILs), populations that segregate for variation in DNA methylation patterns with limited genetic variation, has provided a tool for showing the effects of epigenetics on natural variation (Johannes et al., 2009; Reinders et al., 2009).
Figure 4.
Classification of examples of variable DNA methylation levels in populations. The inheritance patterns for the DNA methylation patterns can include examples of local (cis) inheritance, examples of distant (trans) inheritance, or highly unstable examples that have limited or no heritability. These examples also can be classified based on whether a genetic change is involved and if so, what type of genetic change occurs. PAI, Phosphoribosylanthranilate isomerase; TE, transposable element; VIM1, Variant in methylation1; CmWIP1, Cucumis melo WPP-domain interacting protein1; MuK, Mu killer; IBD, identity by descent.
There are examples in which natural phenotypic variation within a species seems to be regulated by epigenetics (Fig. 4). Linaria vulgaris, a snapdragon plant named after Carl Linnaeus, shows natural variation in flower symmetry (Cubas et al., 1999). The peloric variant displays radial symmetry and has increased levels of DNA methylation in the promoter and coding region of Lcyc. The underlying sequence of the Lcyc alleles and surrounding regions is identical, suggesting an epigenetic basis for this phenotype. The symmetry phenotype is heritable through generations, and occasional reversion to wild-type symmetry can occur on branches because of the loss of promoter methylation (Cubas et al., 1999). Similar examples at the colorless nonripening locus in tomato (Solanum lycopersicum; Manning et al., 2006), the Clark Kent alleles of the SUPERMAN locus (Jacobsen and Meyerowitz, 1997), and the qui-quinine starch locus (Silveira et al., 2013) in Arabidopsis display aberrant DNA methylation patterns with no apparent dependence on changes in genetic sequence. There is also evidence for natural variation for DNA methylation levels at ribosomal DNA (Riddle and Richards, 2002) and some repetitive elements (Rangwala et al., 2006; Rangwala and Richards, 2007) in Arabidopsis; however, it is not clear whether these differences contribute to morphological changes.
Paramutation also provides an example of epiallelic variation within populations (Chandler, 2007; Hollick, 2012). Paramutation describes a directed allelic interaction that can occur in heterozygous individuals and results in differences in expression levels without affecting primary sequence (Chandler, 2007; Hollick, 2012). Paramutation can clearly result in epigenetic variation, but only certain alleles at a particular locus are subject to regulation by paramutation (Chandler and Stam, 2004), which may reflect the need for certain genetic elements to create an environment that is permissive for a role of epigenetics for the regulation of facilitated epialleles. Similarly, there are many examples of potential chromatin-based regulation that may be linked to genetic changes, such as transposon insertions (Kinoshita et al., 2007; Saze and Kakutani, 2007; Martin et al., 2009; Robbins et al., 2009).
Technological advances have provided the opportunity to study genome-wide changes in DNA methylation (Schmitz and Ecker, 2012), histone modifications (Dong et al., 2012; Makarevitch et al., 2013), or small RNAs (Zhai et al., 2008; Shivaprasad et al., 2012) in different individuals of the same species. Genome-wide studies of DNA methylation have provided evidence for natural variation in DNA methylation patterns (Vaughn et al., 2007; Eichten et al., 2011, 2013b; Regulski et al., 2013; Schmitz et al., 2013a, 2013b). Each of these studies identified numerous examples of differentially methylated regions between different individuals of the same species. However, identification of a differentially methylated region does not simply imply that this variation is epigenetic in nature, because genetic variation can lead to variation in DNA methylation, which is seen from an obligate epiallele within the phosphoribosylanthranilate isomerase (PAI) gene family in Arabidopsis (Bender and Fink, 1995). Some accessions of Arabidopsis have an inverted repeat of the PAI1 locus that leads to the production of small RNAs and silencing of the entire gene family by small RNAs that direct DNA methylation (Melquist and Bender, 2004). Variation for transposable elements may contribute to DNA methylation of nearby low-copy sequences (Hollister and Gaut, 2009; Eichten et al., 2012). The effects of genetic changes on DNA methylation state can be confounded by examples of DNA methylation variants that may have arisen as a result of a genetic variant but are no longer dependent on that genetic variant for their maintenance over generational time. This is exemplified by the AtFOLT copy number variants of Arabidopsis (Durand et al., 2012). AtFOLT1 exists as a single copy locus in some Arabidopsis accessions, but a complex rearrangement and duplication of this locus, AtFOLT2, exists in other accessions. The nature of this duplication leads to silencing by small RNAs and DNA methylation of the AtFOLT1 locus. Therefore, segregating populations from two parents that differ in the AtFOLT copy numbers results in rare individuals that contain only the silenced AtFOLT1 locus. Even more fascinating is that this silenced locus can be maintained over at least six generations in the absence of the inducing trigger (the complex rearrangement and duplication). This finding suggests that the DNA methylation patterns present in one individual in a population may reflect both the exposures to other alleles that occurred in past generations.
Although much of the natural variation in DNA methylation in plant populations may reflect contributions of genetic variation, evidence indicates that at least a portion of this variation reflects pure or facilitated epialleles. Identification of pure or facilitated epialleles is challenging, because any dependence on genotype needs to be ruled out as a direct causal factor. This is especially difficult in plant populations that contain abundant genetic variation. Therefore, the strongest evidence for pure epialleles has arisen in controlled plant populations with known pedigrees and limited genetic variation. Recent studies in Arabidopsis and maize that combined epigenomic profiling methods with populations of plants that were derived from multiple generations of successive growth revealed evidence for these pure epialleles (Becker et al., 2011; Eichten et al., 2011; Schmitz et al., 2011, 2013b). In these experiments, the variation in genotype for whole genomes or specific regions of the genome was reduced to spontaneous mutations. The frequency at which these epialleles naturally appear is greater than the known spontaneous genetic mutation rate, indicating their independence from genotype. Moreover, one of the identified epialleles from Arabidopsis reverted to the wild-type methylation state after one additional generation of growth (Becker et al., 2011). These pure epialleles may arise within inbred populations and contribute to spontaneous variation (Havecker et al., 2012).
Epialleles have the potential to exhibit unexpected patterns of inheritance, such as paramutation or high levels of instability (Cubas et al., 1999; Becker et al., 2011; Schmitz et al., 2011). Several recent studies have attempted to document the patterns of inheritance for differential methylation using association mapping with natural populations (Eichten et al., 2013b; Schmitz et al., 2013a) or biparental populations (Eichten et al., 2013b, Regulski et al., 2013; Schmitz et al., 2013b). Although a minority of the loci exhibits patterns that would suggest paramutation-like patterns or unstable inheritance (Greaves et al., 2014), the majority of DNA methylation variation seems to be under local (cis) control and inherited in a relatively faithful manner (Schmitz et al., 2013a). In some cases, the inheritance of DNA methylation patterns is locally controlled, even in the absence of DNA sequence variation (Eichten et al., 2011), providing evidence for stable inheritance of pure epialleles.
The creation of epiRILs provides another tool for studying the potential contribution of epigenetics to natural variation (Johannes et al., 2009; Reinders et al., 2009). Populations of Arabidopsis were generated that contained perturbed epigenomes, but genetic variation was largely held constant. These populations segregate for genomic regions that have been stripped of DNA methylation by passage through a mutant background (decrease in DNA methylation1 [ddm1] or met1). The resulting lines segregate for altered DNA methylation state as well as some novel sequence changes caused by the reactivation of some transposon families in these mutant backgrounds (Miura et al., 2001; Singer et al., 2001; Kato et al., 2004). The analysis of the epiRILs reveals evidence for phenotypic variation within these populations (Johannes et al., 2009; Reinders et al., 2009; Roux et al., 2011; Latzel et al., 2012, 2013; Zhang et al., 2013). This result suggests that the loss of DNA methylation releases cryptic information in the Arabidopsis genome, and the segregation for these changes can impact morphology. To support this hypothesis, differentially methylated regions (DMRs) in the epiRILs were recently identified, and a subset of DMRs was used as markers to identify quantitative trait loci associated with phenotypic variation. The results indicated that the altered DNA methylation levels in the epiRILs lead to phenotypic changes (Cortijo et al., 2014). The phenotypic variation produced by the epiRILs resembles that found in natural strains of Arabidopsis (Roux et al., 2011), and therefore, much of the variation captured in epiRILs may also be segregating in natural plant populations that have not had intentional perturbation of epiallelic states. In fact, many of the DMRs identified in the epiRILs overlap with DMRs present in natural strains (Cortijo et al., 2014). Although not all variation observed in epiRILs is fully epigenetic, they are powerful in that they reveal the potential for epigenetic variation to affect phenotypic variation in plant genomes.
Understanding the role of epialleles in evolution is still in its infancy (Finnegan, 2002; Rapp and Wendel, 2005). Given that there are examples of pure epialleles that affect phenotype, it should be possible for natural selection to act on epigenetic variation and drive changes in epiallele frequency within populations. However, unresolved questions remain. First, given the overall longer periods of time that are relevant for evolutionary change among species, it is likely that potentially unstable inheritance of epialleles could be supplanted by genetic variation. If it is advantageous to silence expression of a gene throughout development, it is likely that, over time, mutations will arise that will abolish function of that gene. Second, a major question about the potential role of epigenetics in evolutionary processes is whether the environment influences the rate and nature of epiallelic variation. The evidence for directed, stable, meiotically heritable epialleles induced by environmental conditions is somewhat rare and may actually be unexpected. If epigenetic information can be influenced by directional environmental conditions, it might be expected that it could fluctuate back if the environment changes again. These types of changes would be unlikely to provide long-term stability that would contribute to differences between species but instead, might contribute to shorter term evolutionary processes, such as local adaptation. However, the potential for environmental stress to increase the rate of epigenetic variation may provide an alternative role for epigenetics in evolution. If environmental stress results in frequent alterations in chromatin modifications that are then heritable, it could provide a source of increased morphological variation that could be subject to natural selection. Depending on the stability of these changes, they would have the potential to contribute to the origin of novel epialleles that could be driven to fixation.
Epigenetics may also contribute to evolutionary processes that occur in the formation of polyploids or the accommodation of transposons. There is evidence that the combination of genomes in polyploids can be accompanied by changes in chromatin modifications (Chen, 2007; Doyle et al., 2008; Ng et al., 2012; Hegarty et al., 2013; Madlung and Wendel, 2013). These novel chromatin changes may provide the variation needed to identify individuals that can balance the contributions of the different genomes and may be necessary for the successful stabilization of polyploids. There is abundant evidence that many epigenetic processes affect transposons. Many epigenetic mechanisms silence transposons and prevent their proliferation (Slotkin and Martienssen, 2007; Lisch, 2009; Bucher et al., 2012). Evolutionary pressures have likely driven a balance between silencing of transposons and allowing for expression of nearby genes (Hollister and Gaut, 2009; Tenaillon et al., 2010; Levin and Moran, 2011; Lisch and Bennetzen, 2011).
CONCLUSION
In this article, we considered the role for chromatin modification and epigenetics in contributing to various plant processes. The distinction between chromatin modifications and epigenetics is useful, because it allows the term epigenetics to connote stable transmission or inheritance of information. Although by our more restricted definition, chromatin-based regulation of gene expression is not necessarily epigenetic, we do not wish to diminish the importance of this fascinating type of gene regulation. A careful use of these terms can help delineate the mechanisms for gene regulation versus the type of inheritance for gene regulation. Epigenetics has become a widely used term, and the word is in some danger of losing any real meaning if it is applied to too many distinct types of concepts. Although there are clear examples for a role for epigenetics in plant evolution, development, response to the environment, and parental effects, there are also many other interesting mechanisms that contribute to these processes. We have attempted to highlight the potential role for both epigenetics and chromatin-based processes in contributing to each of these areas of plant research and hope that we have conveyed our excitement about the future of these research areas.
Acknowledgments
We apologize for the many important works that were not cited because of space limitations; there is a vibrant community of researchers studying complex mechanisms of gene regulation in plants, and we are grateful to this full community. We thank Peter Hermanson, Kit Leffler, Rhiannon Macrae, Kelly Lane, Qing Li, and Amanda Waters for assistance in editing and manuscript preparation.
Glossary
- DMR
differentially methylated region
- epiRIL
epigenetic recombinant inbred line
- miRNA
microRNA
- PcG
Polycomb group
- siRNA
small interfering RNA
Footnotes
This work was supported by the National Science Foundation (grant nos. IOS–1339194 to R.J.S. and IOS–1237931 to N.M.S.), the National Institutes of Health (grant no. R00 GM100000 to R.J.S.), and the U.S. Department of Agriculture-National Institute of Food and Agriculture (grant no. 2010–04122 to N.M.S.).
References
- Abmayr SM, Workman JL. (2012) Holding on through DNA replication: histone modification or modifier? Cell 150: 875–877 [DOI] [PubMed] [Google Scholar]
- Adachi K, Schöler HR. (2012) Directing reprogramming to pluripotency by transcription factors. Curr Opin Genet Dev 22: 416–422 [DOI] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH. (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino RM, Michaels SD. (2010) The timing of flowering. Plant Physiol 154: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, van der Winden J, Matzke AJ, Matzke M. (2002) RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA (Suppl 4) 99: 16499–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Raissig MT, Grossniklaus U. (2011) Epigenetic regulation and reprogramming during gamete formation in plants. Curr Opin Genet Dev 21: 124–133 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Bauer MJ, Fischer RL. (2011) Genome demethylation and imprinting in the endosperm. Curr Opin Plant Biol 14: 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC. (2006) Short silencing RNA: the dark matter of genetics? Cold Spring Harb Symp Quant Biol 71: 13–20 [DOI] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D. (2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249 [DOI] [PubMed] [Google Scholar]
- Belele CL, Sidorenko L, Stam M, Bader R, Arteaga-Vazquez MA, Chandler VL. (2013) Specific tandem repeats are sufficient for paramutation-induced trans-generational silencing. PLoS Genet 9: e1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Grossniklaus U. (2012) Dynamic regulation of Polycomb group activity during plant development. Curr Opin Plant Biol 15: 523–529 [DOI] [PubMed] [Google Scholar]
- Bender J. (2004) DNA methylation of the endogenous PAI genes in Arabidopsis. Cold Spring Harb Symp Quant Biol 69: 145–153 [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR. (1995) Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83: 725–734 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. (2010) The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, McCallum EJ, Ménard R, Meyer D, Fuchs J, Dong A, Shen WH. (2010) Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22: 3232–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilichak A, Ilnystkyy Y, Hollunder J, Kovalchuk I. (2012) The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS ONE 7: e30515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond DM, Baulcombe DC. (2014) Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol 24: 100–107 [DOI] [PubMed] [Google Scholar]
- Borges F, Calarco JP, Martienssen RA. (2012) Reprogramming the epigenome in Arabidopsis pollen. Cold Spring Harb Symp Quant Biol 77: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Boyko A, Kovalchuk I. (2011) Genome instability and epigenetic modification—heritable responses to environmental stress? Curr Opin Plant Biol 14: 260–266 [DOI] [PubMed] [Google Scholar]
- Breiling A, Sessa L, Orlando V. (2007) Biology of polycomb and trithorax group proteins. Int Rev Cytol 258: 83–136 [DOI] [PubMed] [Google Scholar]
- Bucher E, Reinders J, Mirouze M. (2012) Epigenetic control of transposon transcription and mobility in Arabidopsis. Curr Opin Plant Biol 15: 503–510 [DOI] [PubMed] [Google Scholar]
- Budhavarapu VN, Chavez M, Tyler JK. (2013) How is epigenetic information maintained through DNA replication? Epigenetics Chromatin 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijó JA, Becker JD, et al. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, Feinberg AP. (2006) The emerging science of epigenomics. Hum Mol Genet 15: R95–R101 [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE. (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Mittelsten Scheid O. (2014) How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet 10: e1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL. (2007) Paramutation: from maize to mice. Cell 128: 641–645 [DOI] [PubMed] [Google Scholar]
- Chandler VL, Stam M. (2004) Chromatin conversations: mechanisms and implications of paramutation. Nat Rev Genet 5: 532–544 [DOI] [PubMed] [Google Scholar]
- Chen X. (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2012) Small RNAs in development—insights from plants. Curr Opin Genet Dev 22: 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hu Y, Zhou DX. (2011) Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim Biophys Acta 1809: 421–426 [DOI] [PubMed] [Google Scholar]
- Chen ZJ. (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Tian L. (2007) Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta 1769: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33–42 [DOI] [PubMed] [Google Scholar]
- Colaneri AC, Jones AM. (2013) Genome-wide quantitative identification of DNA differentially methylated sites in Arabidopsis seedlings growing at different water potential. PLoS ONE 8: e59878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U. (2011) Molecular aspects of defence priming. Trends Plant Sci 16: 524–531 [DOI] [PubMed] [Google Scholar]
- Cortijo S, Wardenaar R, Colomé-Tatché M, Gilly A, Etcheverry M, Labadie K, Caillieux E, Hospital F, Aury JM, Wincker P, et al. (2014) Mapping the epigenetic basis of complex traits. Science 343: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E. (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161 [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. (2010) Transgenerational epigenetic inheritance: more questions than answers. Genome Res 20: 1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. (2011) Histone variants and modifications in plant gene regulation. Curr Opin Plant Biol 14: 116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z. (2012) Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat Commun 3: 740. [DOI] [PubMed] [Google Scholar]
- Dong X, Reimer J, Göbel U, Engelhorn J, He F, Schoof H, Turck F. (2012) Natural variation of H3K27me3 distribution between two Arabidopsis accessions and its association with flanking transposable elements. Genome Biol 13: R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR. (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109: E2183–E2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. (2008) Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42: 443–461 [DOI] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. (2012) Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Bouché N, Perez Strand E, Loudet O, Camilleri C. (2012) Rapid establishment of genetic incompatibility through natural epigenetic variation. Curr Biol 22: 326–331 [DOI] [PubMed] [Google Scholar]
- Eichten SR, Briskine R, Song J, Li Q, Swanson-Wagner R, Hermanson PJ, Waters AJ, Starr E, West PT, Tiffin P, et al. (2013a) Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Ellis NA, Makarevitch I, Yeh CT, Gent JI, Guo L, McGinnis KM, Zhang X, Schnable PS, Vaughn MW, et al. (2012) Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet 8: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Swanson-Wagner RA, Schnable JC, Waters AJ, Hermanson PJ, Liu S, Yeh CT, Jia Y, Gendler K, Freeling M, et al. (2011) Heritable epigenetic variation among maize inbreds. PLoS Genet 7: e1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Vaughn MW, Hermanson PJ, Springer NM. (2013b) Variation in DNA methylation patterns is more common among maize inbreds than among tissues. Plant Genome 6: 2 [Google Scholar]
- Feng S, Jacobsen SE, Reik W. (2010) Epigenetic reprogramming in plant and animal development. Science 330: 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. (2008) Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ. (2002) Epialleles: a source of random variation in times of stress. Curr Opin Plant Biol 5: 101–106 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE. (2001) Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol 2: 409–421 [DOI] [PubMed] [Google Scholar]
- Galloway LF. (2005) Maternal effects provide phenotypic adaptation to local environmental conditions. New Phytol 166: 93–99 [DOI] [PubMed] [Google Scholar]
- Gehring M. (2013) Genomic imprinting: insights from plants. Annu Rev Genet 47: 187–208 [DOI] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Tweedie S. (2002) Remembrance of things past: chromatin remodeling in plant development. Annu Rev Cell Dev Biol 18: 707–746 [DOI] [PubMed] [Google Scholar]
- Grandbastien MA, Audeon C, Bonnivard E, Casacuberta JM, Chalhoub B, Costa AP, Le QH, Melayah D, Petit M, Poncet C, et al. (2005) Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet Genome Res 110: 229–241 [DOI] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES. (2014) Inheritance of Trans Chromosomal Methylation patterns from Arabidopsis F1 hybrids. Proc Natl Acad Sci USA 111: 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Nègre N, Cavalli G. (2006) From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res 14: 363–375 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Dickinson HG. (2012) Epigenetic reprogramming in plant reproductive lineages. Plant Cell Physiol 53: 817–823 [DOI] [PubMed] [Google Scholar]
- Gutzat R, Mittelsten Scheid O. (2012) Epigenetic responses to stress: triple defense? Curr Opin Plant Biol 15: 568–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. (2004) The (dual) origin of epigenetics. Cold Spring Harb Symp Quant Biol 69: 67–70 [DOI] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Fedito P, Hardcastle TJ, Baulcombe DC. (2012) Metastable differentially methylated regions within Arabidopsis inbred populations are associated with modified expression of non-coding transcripts. PLoS ONE 7: e45242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Chen X, Huang H, Xu L. (2012) Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet 8: e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty M, Coate J, Sherman-Broyles S, Abbott R, Hiscock S, Doyle J. (2013) Lessons from natural and artificial polyploids in higher plants. Cytogenet Genome Res 140: 204–225 [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. (2008) Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev 22: 1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holec S, Berger F. (2012) Polycomb group complexes mediate developmental transitions in plants. Plant Physiol 158: 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JB. (2012) Paramutation: a trans-homolog interaction affecting heritable gene regulation. Curr Opin Plant Biol 15: 536–543 [DOI] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. (2009) Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res 19: 1419–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Meyerowitz EM. (1997) Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277: 1100–1103 [DOI] [PubMed] [Google Scholar]
- Jaskiewicz M, Conrath U, Peterhänsel C. (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, Agier N, Bulski A, Albuisson J, Heredia F, Audigier P, et al. (2009) Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet 5: e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Moss T, Cullis C. (2011) Environmentally induced heritable changes in flax. J Vis Exp 47: 2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. (2007) The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. (2012) DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22: 1825–1830 [DOI] [PubMed] [Google Scholar]
- Kaeppler SM, Kaeppler HF, Rhee Y. (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43: 179–188 [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kato M, Takashima K, Kakutani T. (2004) Epigenetic control of CACTA transposon mobility in Arabidopsis thaliana. Genetics 168: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu JK, Zhu J. (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, To TK, Nishioka T, Seki M. (2010) Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ 33: 604–611 [DOI] [PubMed] [Google Scholar]
- Kinoshita Y, Saze H, Kinoshita T, Miura A, Soppe WJ, Koornneef M, Kakutani T. (2007) Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J 49: 38–45 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L. (2010) Regulation of cell identity by plant Polycomb and trithorax group proteins. Curr Opin Genet Dev 20: 541–547 [DOI] [PubMed] [Google Scholar]
- Kwon CS, Lee D, Choi G, Chung WI. (2009) Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J 60: 112–121 [DOI] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A, Melchers G. (1947) Vernalisation and devernalisation bei einer zweijahrigen Pflanze. Z Naturforsch B 2b: 444–449 [Google Scholar]
- Lang-Mladek C, Popova O, Kiok K, Berlinger M, Rakic B, Aufsatz W, Jonak C, Hauser MT, Luschnig C. (2010) Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol Plant 3: 594–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latzel V, Allan E, Bortolini Silveira A, Colot V, Fischer M, Bossdorf O. (2013) Epigenetic diversity increases the productivity and stability of plant populations. Nat Commun 4: 2875. [DOI] [PubMed] [Google Scholar]
- Latzel V, Zhang Y, Karlsson Moritz K, Fischer M, Bossdorf O. (2012) Epigenetic variation in plant responses to defence hormones. Ann Bot (Lond) 110: 1423–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria M, Rupe M, Guo M, Kranz E, Pirona R, Viotti A, Lund G. (2004) Extensive maternal DNA hypomethylation in the endosperm of Zea mays. Plant Cell 16: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL, Moran JV. (2011) Dynamic interactions between transposable elements and their hosts. Nat Rev Genet 12: 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Berger F. (2012) Endosperm: food for humankind and fodder for scientific discoveries. New Phytol 195: 290–305 [DOI] [PubMed] [Google Scholar]
- Li W, Liu H, Cheng ZJ, Su YH, Han HN, Zhang Y, Zhang XS. (2011) DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet 7: e1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, et al. (2004) Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23: 4286–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. (2009) Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 60: 43–66 [DOI] [PubMed] [Google Scholar]
- Lisch D, Bennetzen JL. (2011) Transposable element origins of epigenetic gene regulation. Curr Opin Plant Biol 14: 156–161 [DOI] [PubMed] [Google Scholar]
- Luna E, Bruce TJ, Roberts MR, Flors V, Ton J. (2012) Next-generation systemic acquired resistance. Plant Physiol 158: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K. (2012) Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta 1819: 129–136 [DOI] [PubMed] [Google Scholar]
- Madlung A, Wendel JF. (2013) Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet Genome Res 140: 270–285 [DOI] [PubMed] [Google Scholar]
- Makarevitch I, Eichten SR, Briskine R, Waters AJ, Danilevskaya ON, Meeley RB, Myers CL, Vaughn MW, Springer NM. (2013) Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25: 780–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. (2010) Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11: 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, Morin H, Pitrat M, Dogimont C, Bendahmane A. (2009) A transposon-induced epigenetic change leads to sex determination in melon. Nature 461: 1135–1138 [DOI] [PubMed] [Google Scholar]
- McClintock B. (1984) The significance of responses of the genome to challenge. Science 226: 792–801 [DOI] [PubMed] [Google Scholar]
- Meissner A. (2010) Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol 28: 1079–1088 [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Molnar A, Baulcombe DC. (2011) Intercellular and systemic movement of RNA silencing signals. EMBO J 30: 3553–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melquist S, Bender J. (2004) An internal rearrangement in an Arabidopsis inverted repeat locus impairs DNA methylation triggered by the locus. Genetics 166: 437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, Paszkowski J. (2011) Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol 14: 267–274 [DOI] [PubMed] [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. (2001) Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214 [DOI] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. (2006) Transgeneration memory of stress in plants. Nature 442: 1046–1049 [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. (1999) Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 23: 314–318 [DOI] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR. (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Ng DW, Lu J, Chen ZJ. (2012) Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr Opin Plant Biol 15: 154–161 [DOI] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG. (2004) Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet 20: 214–220 [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Orlando V. (2003) Polycomb, epigenomes, and control of cell identity. Cell 112: 599–606 [DOI] [PubMed] [Google Scholar]
- Paszkowski J, Grossniklaus U. (2011) Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr Opin Plant Biol 14: 195–203 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Abdelsamad A, Vu GT. (2013) Hidden genetic nature of epigenetic natural variation in plants. Trends Plant Sci 18: 625–632 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O. (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Mittelsten Scheid O. (2012) Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol 53: 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Rosa M, Schikora A, Berlinger M, Hirt H, Luschnig C, Mittelsten Scheid O. (2009) Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE 4: e5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J, Wagner D. (2007) Histone modifications and dynamic regulation of genome accessibility in plants. Curr Opin Plant Biol 10: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H, Hohn B. (2012) In planta somatic homologous recombination assay revisited: a successful and versatile, but delicate tool. Plant Cell 24: 4324–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Hiratsuka K, Chua NH. (1994) Transcription factors in plant growth and development. Curr Opin Genet Dev 4: 642–646 [DOI] [PubMed] [Google Scholar]
- Rangwala SH, Elumalai R, Vanier C, Ozkan H, Galbraith DW, Richards EJ. (2006) Meiotically stable natural epialleles of Sadhu, a novel Arabidopsis retroposon. PLoS Genet 2: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SH, Richards EJ. (2007) Differential epigenetic regulation within an Arabidopsis retroposon family. Genetics 176: 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp RA, Wendel JF. (2005) Epigenetics and plant evolution. New Phytol 168: 81–91 [DOI] [PubMed] [Google Scholar]
- Rasmann S, De Vos M, Casteel CL, Tian D, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G. (2012) Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol 158: 854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MT, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res 23: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wulff BB, Mirouze M, Marí-Ordóñez A, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J. (2009) Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev 23: 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC. (2006) Chromatin modifiers that control plant development. Curr Opin Plant Biol 9: 21–27 [DOI] [PubMed] [Google Scholar]
- Reyes JC, Hennig L, Gruissem W. (2002) Chromatin-remodeling and memory factors. New regulators of plant development. Plant Physiol 130: 1090–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. (1997) DNA methylation and plant development. Trends Genet 13: 319–323 [DOI] [PubMed] [Google Scholar]
- Richards EJ. (2006) Inherited epigenetic variation—revisiting soft inheritance. Nat Rev Genet 7: 395–401 [DOI] [PubMed] [Google Scholar]
- Richards EJ. (2011) Natural epigenetic variation in plant species: a view from the field. Curr Opin Plant Biol 14: 204–209 [DOI] [PubMed] [Google Scholar]
- Riddle NC, Richards EJ. (2002) The control of natural variation in cytosine methylation in Arabidopsis. Genetics 162: 355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Robbins ML, Wang P, Sekhon RS, Chopra S. (2009) Gene structure induced epigenetic modifications of pericarp color1 alleles of maize result in tissue-specific mosaicism. PLoS ONE 4: e8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657 [DOI] [PubMed] [Google Scholar]
- Roudier F, Teixeira FK, Colot V. (2009) Chromatin indexing in Arabidopsis: an epigenomic tale of tails and more. Trends Genet 25: 511–517 [DOI] [PubMed] [Google Scholar]
- Roux F, Colomé-Tatché M, Edelist C, Wardenaar R, Guerche P, Hospital F, Colot V, Jansen RC, Johannes F. (2011) Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics 188: 1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O. (2009) Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60: 485–510 [DOI] [PubMed] [Google Scholar]
- Sani E, Herzyk P, Perrella G, Colot V, Amtmann A. (2013) Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol 14: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Kakutani T. (2007) Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J 26: 3641–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Amasino RM. (2007) Vernalization: a model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim Biophys Acta 1769: 269–275 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Ecker JR. (2012) Epigenetic and epigenomic variation in Arabidopsis thaliana. Trends Plant Sci 17: 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, He Y, Valdés-López O, Khan SM, Joshi T, Urich MA, Nery JR, Diers B, Xu D, Stacey G, et al. (2013a) Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res 23: 1663–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. (2011) Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ, et al. (2013b) Patterns of population epigenomic diversity. Nature 495: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC. (2012) Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J 31: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko LV, Peterson T. (2001) Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize p1 gene. Plant Cell 13: 319–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira AB, Trontin C, Cortijo S, Barau J, Del Bem LE, Loudet O, Colot V, Vincentz M. (2013) Extensive natural epigenetic variation at a de novo originated gene. PLoS Genet 9: e1003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. (1995) Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol 7: 376–385 [DOI] [PubMed] [Google Scholar]
- Simon SA, Meyers BC. (2011) Small RNA-mediated epigenetic modifications in plants. Curr Opin Plant Biol 14: 148–155 [DOI] [PubMed] [Google Scholar]
- Singer T, Yordan C, Martienssen RA. (2001) Robertson’s Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev 15: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter A, Daniel X, Flors V, Luna E, Hohn B, Mauch-Mani B. (2012) Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol 158: 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Freeling M, Lisch D. (2005) Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet 37: 641–644 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA. (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Angel A, Howard M, Dean C. (2012) Vernalization—a cold-induced epigenetic switch. J Cell Sci 125: 3723–3731 [DOI] [PubMed] [Google Scholar]
- Stam M, Belele C, Ramakrishna W, Dorweiler JE, Bennetzen JL, Chandler VL. (2002) The regulatory regions required for B’ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics 162: 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Ding B, Simon SA, Feng S, Bellizzi M, Pellegrini M, Wang GL, Meyers BC, Jacobsen SE. (2013) Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2: e00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Kaplan F, Lee KJ, Guy CL. (2003) Acquired tolerance to temperature extremes. Trends Plant Sci 8: 179–187 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM. (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G. (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17: 196–203 [DOI] [PubMed] [Google Scholar]
- Tanurdzic M, Vaughn MW, Jiang H, Lee TJ, Slotkin RK, Sosinski B, Thompson WF, Doerge RW, Martienssen RA. (2008) Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol 6: 2880–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Hollister JD, Gaut BS. (2010) A triptych of the evolution of plant transposable elements. Trends Plant Sci 15: 471–478 [DOI] [PubMed] [Google Scholar]
- Thorstensen T, Grini PE, Aalen RB. (2011) SET domain proteins in plant development. Biochim Biophys Acta 1809: 407–420 [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I. (2010) Stress-induced activation of heterochromatic transcription. PLoS Genet 6: e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Hommelsheim CM, Berson T, Thomas S, Chandrasekar B, Olcay AC, Berendzen KW, Frantzeskakis L. (2012) Reevaluation of the reliability and usefulness of the somatic homologous recombination reporter lines. Plant Cell 24: 4314–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk K, Ding Y, Malkaram S, Riethoven JJ, Liu R, Yang J, Laczko P, Chen H, Xia Y, Ladunga I, et al. (2010) Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol 10: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn MW, Tanurdzić M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD, Dedhia N, McCombie WR, Agier N, Bulski A, et al. (2007) Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol 5: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven KJ, Jansen JJ, van Dijk PJ, Biere A. (2010) Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol 185: 1108–1118 [DOI] [PubMed] [Google Scholar]
- Verhoeven KJ, van Gurp TP. (2012) Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS ONE 7: e38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. (1993) Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Waddington C. (1942) The epigenotype. Endeavour 1: 18–20 [Google Scholar]
- Wagner D. (2003) Chromatin regulation of plant development. Curr Opin Plant Biol 6: 20–28 [DOI] [PubMed] [Google Scholar]
- Weigel D, Colot V. (2012) Epialleles in plant evolution. Genome Biol 13: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler BS. (2013) Small RNAs, big impact: small RNA pathways in transposon control and their effect on the host stress response. Chromosome Res 21: 587–600 [DOI] [PubMed] [Google Scholar]
- Wollmann H, Berger F. (2012) Epigenetic reprogramming during plant reproduction and seed development. Curr Opin Plant Biol 15: 63–69 [DOI] [PubMed] [Google Scholar]
- Woo HR, Pontes O, Pikaard CS, Richards EJ. (2007) VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev 21: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Zhu B. (2010) Nucleosome assembly and epigenetic inheritance. Protein Cell 1: 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Kovalchuk I. (2011) Abiotic stress leads to somatic and heritable changes in homologous recombination frequency, point mutation frequency and microsatellite stability in Arabidopsis plants. Mutat Res 707: 61–66 [DOI] [PubMed] [Google Scholar]
- Yi H, Richards EJ. (2008) Phenotypic instability of Arabidopsis alleles affecting a disease Resistance gene cluster. BMC Plant Biol 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Lepère G, Jay F, Wang J, Bapaume L, Wang Y, Abraham AL, Penterman J, Fischer RL, Voinnet O, et al. (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA 110: 2389–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D. (2010) Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA 107: 18729–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Liu J, Liu B, Li P, Meyers BC, Chen X, Cao X. (2008) Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet 4: e1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu JK. (2012) Active DNA demethylation in plants and animals. Cold Spring Harb Symp Quant Biol 77: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Xu C, von Wettstein D, Liu B. (2011) Tissue-specific differences in cytosine methylation and their association with differential gene expression in sorghum. Plant Physiol 156: 1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Fischer M, Colot V, Bossdorf O. (2013) Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol 197: 314–322 [DOI] [PubMed] [Google Scholar]
- Zheng B, Chen X. (2011) Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr Opin Plant Biol 14: 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. (2007) The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol 17: 54–59 [DOI] [PubMed] [Google Scholar]
- Zong W, Zhong X, You J, Xiong L. (2013) Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol Biol 81: 175–188 [DOI] [PubMed] [Google Scholar]