Abstract
The altered expression of microRNAs (miRNAs) is associated with a number of cancer types. The study of the association between the miRNA profile and cancer may be useful to identify potential biomarkers of certain types of cancer. In the present study, 19 miRNAs were identified by high-throughput sequencing in the serum of colorectal cancer (CRC) patients. A network analysis was performed based on a computational approach to identify associations between CRC and miRNAs. The present study may be useful to identify cancer-specific signatures and potentially useful biomarkers for the diagnosis of CRC. The network analysis of miRNA-target genes may aid in identifying altered miRNA regulatory networks that are involved in tumor pathogenesis.
Keywords: colorectal cancer, high-throughput sequencing, miRNA, biomarker
Introduction
Colorectal cancer (CRC) is an important contributor to cancer-related mortality and morbidity. Accumulated data has uncovered several critical genes and pathways important in the initiation and progression of CRC (1–3). Large-scale sequencing analyses have identified numerous recurrently mutated genes and chromosomal translocations (4–6). In addition, a number of microRNAs (miRNAs) have been previously reported to be associated with CRC (7,8). However, how miRNA changes contribute to colorectal tumorigenesis has not yet been defined. Further insight into these changes may identify potential biomarkers or therapeutic targets.
miRNAs are small non-protein coding RNA molecules that regulate gene expression (9,10). miRNAs are important in crucial cellular processes, including development, differentiation, proliferation, apoptosis and metabolism (11,12). miRNAs have been proven to interact with potential oncogenes or tumor suppressors, and a number of miRNAs are differentially expressed in normal and neoplastic tissues and in tumors. The differential expression of miRNA has been previously evaluated as a predictive signature of cancer (13–17).
The present study investigated the expression profile of miRNAs in the serum of CRC patients. In total, 19 miRNAs were identified by high-throughput sequencing. A network analysis was performed based on a computational approach to identify associations between CRC and miRNAs. The network analysis of miRNA-target genes may aid in identifying altered miRNA regulatory networks that are involved in tumor pathogenesis.
Materials and methods
Samples and RNA extraction
Primary tumor and neighboring non-tumorous tissues were obtained from five CRC patients. All samples were collected according to procedures approved by the Institutional Review Board of the Guangzhou Medical University (Guangzhou, China) and individuals can not be identified from data or images included in the present study.
Tissue samples were flash frozen in liquid nitrogen and stored at −80°C until nucleic acid extraction. In total, 200 mg of fresh frozen tissues were used to isolate total RNA by phenol extraction (TRIzol reagent; Invitrogen, Life Technologies, Carlsbad, CA, USA). RNA concentration and purity were controlled by A NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) was used to measure the quantity, integrity and purity of the small RNA. Patients provided written informed consent and the study was approved by the ethics committee of the Third Hospital of the Guangzhou Medical University.
miRNA sequencing and sequence analysis
The miRNA expression profile was determined using the Ion Torrent PGM™ sequencer (Life Technologies). Briefly, the Ion Total RNA-Seq kit v2 (Life Technologies) was used to make small RNA libraries that preserve strand information. The templates were prepared using the Ion OneTouch™ system (Life Technologies), and small RNA analysis was performed using the Ion PGM™ sequencer. The miRNA sequences were analyzed by the Torrent Suite software (Life Technologies), and by miRWalk (http://mirwalk.uni-hd.de) and miRBase (http://www.mirbase.org). In addition, gene ontology (GO) and pathway analyses were performed using several tools that identify pathways and GO based on data sets from sequencing with the intent to identify miRNA-related genes and pathways. The tools include Ingenuity Systems Bioinformatics Software (Ingenuity Systems, Inc., Redwood City, CA, USA), FatiGO (http://fatigo.bioinfo.cnio.es) and Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/gds).
Results
Altered miRNA expression in CRC patients
The miRNA profiles of five pairs of solid tumor and adjacent tissues were compared. In total, 16 miRNAs exhibited higher expression in the solid tumor tissues, while three miRNAs exhibited higher expression profiles in the normal tissues; these are listed in Table I.
Table I.
Identification of miRNAs with higher expression profiles in normal or solid tumor tissues.
| Solid tumor tissues | Normal adjacent tissues |
|---|---|
| hsa-miR-135b-5p | hsa-miR-100-5p |
| hsa-miR-146a-5p | hsa-miR-138-5p |
| hsa-miR-148b-3p | hsa-miR-191-5p |
| hsa-miR-17-5p | |
| hsa-miR-196a-5p | |
| hsa-miR-200a-3p | |
| hsa-miR-20a-5p | |
| hsa-miR-21-5p | |
| hsa-miR-223-3p | |
| hsa-miR-27a-5p | |
| hsa-miR-29b-3p | |
| hsa-miR-30e-5p | |
| hsa-miR-374b-5p | |
| hsa-miR-4787-5p | |
| hsa-miR-485-3p | |
| hsa-miR-660-5p |
miR/miRNA, microRNA; CRC, colorectal cancer.
GO and pathway analyses of miRNA target genes
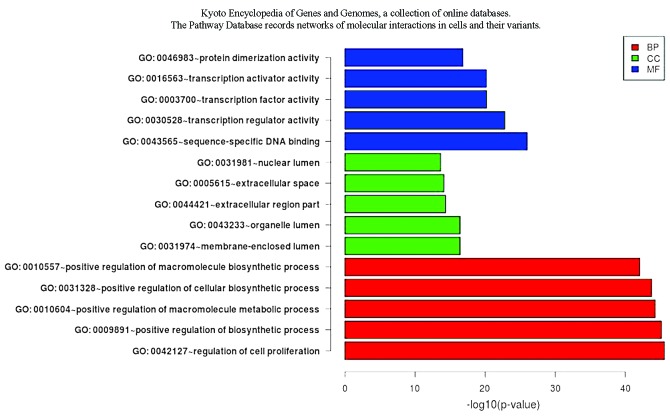
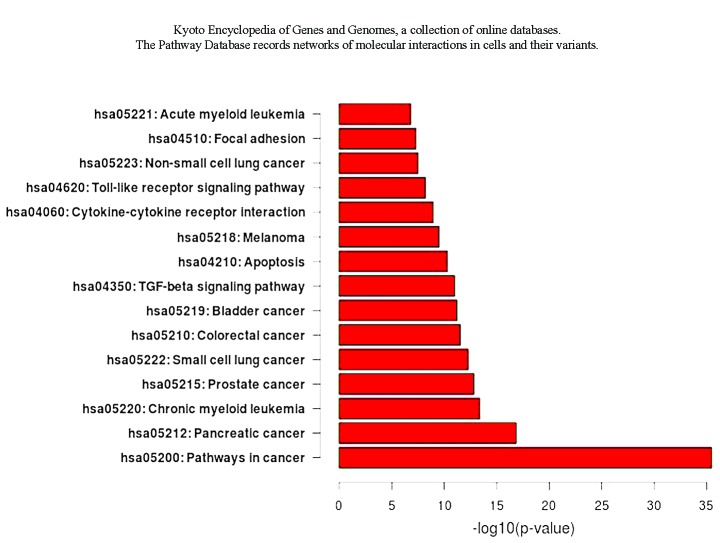
The results of the GO analysis showed that a number of target genes are involved in cell proliferation and apoptosis. The pathway analysis showed that the transforming growth factor β and Toll-like signal pathways are also involved (Fig. 1 and 2).
Figure 1.
GO analysis. A number of target genes were found to be involved in cell proliferation and apoptosis. BP, biological process; CC, celluar component; MF, molecular function; GO, gene ontology.
Figure 2.
Pathway analysis. Transforming growth factor β and Toll-like signal pathways were found to be involved in cell proliferation and apoptosis.
Discussion
miRNAs are small non-coding RNAs that enhance the cleavage or translational repression of specific mRNAs with recognition site(s) in the 3′-untranslated region. Since the identification of the miRNAs, several large-scale studies have compared the profiles of miRNA expression patterns between non-tumor and tumor tissues (18,19). A number of lines of evidence have shown that miRNA expression is predictive of outcome in patients with solid tumors. In lung cancer, low levels of let-7a have been associated with a short survival time following surgery. In addition, previous miRNA microarray expression profiling of tumors and paired non-tumorous tissues has been performed in colon cancer patients to identify miRNA expression patterns associated with outcome, and high levels of miR-21 have been found to be associated with a short overall survival time, independent of other factors (19,20).
In the current study, 19 miRNAs were identified by high-throughput sequencing. A network analysis was performed based on a computational approach to identify associations between CRC and miRNAs. The current study may be useful to identify cancer-specific signatures and potentially useful biomarkers for the diagnosis of CRC. The network analysis of miRNA-target genes may aid in identifying altered miRNA regulatory networks that are involved in tumor pathogenesis.
Acknowledgements
The present study was supported by the Science and Information Technology Foundation of Guangzhou (grant nos. 2010J-E141 and 2011J4100051), the Guangdong Provincial Science and Technology Department (grant no. 2011B0904005260) and the Natural Science Foundation of Guangdong Province (grant no. S2012040006707).
References
- 1.Tjandra JJ, Kilkenny JW, Buie WD, et al. Standards Practice Task Force; American Society of Colon and Rectal Surgeons: Practice parameters for the management of rectal cancer (revised) Dis Colon Rectum. 2005;48:411–423. doi: 10.1007/s10350-004-0937-9. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 3.Aaltonen LA, Peltomäki P, Leach FS, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 4.Bass AJ, Lawrence MS, Brace LE, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 6.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 7.Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 8.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 10.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 12.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Perera RJ, Ray A. MicroRNAs in the search for understanding human diseases. BioDrugs. 2007;21:97–104. doi: 10.2165/00063030-200721020-00004. [DOI] [PubMed] [Google Scholar]
- 14.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 15.Caldas C, Brenton JD. Sizing up miRNAs as cancer genes. Nat Med. 2005;11:712–714. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- 16.Jones KB, Salah Z, Del Mare S, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volinia S, Galasso M, Costinean S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 19.Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20:1272–1286. doi: 10.1158/1055-9965.EPI-11-0035. [DOI] [PubMed] [Google Scholar]
- 20.Jeong HC, Kim EK, Lee JH, Lee JM, Yoo HN, Kim JK. Aberrant expression of let-7a miRNA in the blood of non-small cell lung cancer patients. Mol Med Rep. 2011;4:383–387. doi: 10.3892/mmr.2011.430. [DOI] [PubMed] [Google Scholar]