SUMMARY
5-methylcytosine is a major epigenetic modification that is sometimes called “the fifth nucleotide.” However, our knowledge of how offspring inherit the DNA methylome from parents is limited. We generated nine single-base resolution DNA methylomes, including zebrafish gametes and early embryos. The oocyte methylome is significantly hypomethylated compared to sperm. Strikingly, the paternal DNA methylation pattern is maintained throughout early embryogenesis. The maternal DNA methylation pattern is maintained until the 16-cell stage. Then, the oocyte methylome is gradually discarded through cell division and is progressively reprogrammed to a pattern similar to that of the sperm methylome. The passive demethylation rate and the de novo methylation rate are similar in the maternal DNA. By the midblastula stage, the embryo’s methylome is virtually identical to the sperm methylome. Moreover, inheritance of the sperm methylome facilitates the epigenetic regulation of embryogenesis. Therefore, besides DNA sequences, sperm DNA methylome is also inherited in zebrafish early embryos.
INTRODUCTION
Epigenetic modifications such as DNA methylation and histone modifications play critical roles during embryogenesis (Li et al., 1992; Okano et al., 1999). However, knowledge of how much epigenetic information in gametes can be transferred to the offspring is limited. Recent studies show that epigenetic modifications from gametes in general are cleared and reestablished after fertilization (Blewitt et al., 2006; Daxinger and Whitelaw, 2010, 2012; Feng et al., 2010b; Henderson and Jacobsen, 2007; Wu and Zhang, 2010) except that a number of loci in some model organisms are resistant to the clearing (Arteaga-Vazquez and Chandler, 2010; Cavalli and Paro, 1998; Morgan et al., 1999). However, this theory lacks evidence in the form of high-resolution epigenetic maps in oocytes, sperm, and early embryos.
DNA methylation is one major epigenetic modification that is crucial for the development and differentiation of various cell types in an organism (Li et al., 1992; Okano et al., 1999). In mammals, DNA demethylation occurs in the whole-genome level after fertilization, but not in some loci, such as intracisternal A particle (IAP) (Daxinger and Whitelaw, 2010; Wu and Zhang, 2010). To further understand how offspring obtain DNA methylation information from parents, reduced representation bisulfite sequencing (RRBS) was used to achieve the most comprehensive genome-scale methylomes in mouse gametes and prespecified embryos (Smallwood and Kelsey, 2012; Smith et al., 2012), which explored the unique regulatory phase of DNA methylation in early mammalian embryos. Unfortunately, the RRBS method covers only 5% of the genome for the comparative analysis (Ball et al., 2009; Harris et al., 2010; Smith et al., 2012). The limited genome coverage in oocyte and early embryos prevents a full understanding of how much DNA methylation information is inherited and how it is transferred from sperm and oocyte to progenies, respectively. Moreover, the limited understanding extends to the biological purposes and outcomes of DNA methylation inheritance from gametes.
Although the genome-wide DNA demethylation is a hallmark of mammalian embryogenesis, a controversy surrounds the question of whether such a phenomenon is general for all vertebrates. Some studies report the absence of genome-wide demethylation in zebrafish (Danio rerio) and Xenopus (Macleod et al., 1999; Veenstra and Wolffe, 2001), but others argue for the existence of genome-wide demethylation in zebrafish embryos (MacKay et al., 2007; Mhanni and McGowan, 2004). Here, we chose zebrafish as the model to measure DNA methylomes at single-base resolution in gametes and early embryos. We reveal that zebrafish inherit the DNA methylome from sperm.
RESULTS
Single-Base Resolution DNA Methylomes of Zebrafish Gametes and Early Embryos
The zebrafish is a common model organism for vertebrate developmental studies. The annotated zebrafish genome is about 1.4 giga (G) bases, including 24.2 million CpG sites. Genetic polymorphisms (SNPs) would potentially interrupt the calling of methylation status of cytosines. Therefore, we performed whole-genome resequencing of the Tu¨ bingen (TU) strain (depth 22-fold) used in this study and identified about 11 million SNPs between our TU strain and the reference genome (Zv9, UCSC). Indeed, 1.2 million CpG sites are disrupted by SNPs in the TU genome. These sites were therefore excluded from further analyses.
To explore how progenies inherit the DNA methylation information from parents, we collected both sperm and oocytes, as well as cleavage-stage embryos at the 16-cell, 32-cell, and 64-cell stages, the early-blastula stage at 128 cell, the midblastula stage (MBT) at 1,000 cell (or 1k cell), the gastrula stage at the germ ring, and testis from inbred TU strain. We generated singlebase resolution methylomes in these samples with MethylC-seq (Lister et al., 2009). The average genomic depth among these nine samples was 13-fold per strand (Table 1). We did not observe significant methylation at non-CpG sites in any stage of embryos (data not shown). Therefore, all subsequent analyses were focused on the CpG sites.
Table 1.
Summary of Shotgun Bisulfite Sequencing
| Sample | Stage | Genome Depths |
BS Conversion Rate |
Number of CG (1×) |
CG (1×) Covered |
Number of CG (5×) |
CG (5×) Covered |
|---|---|---|---|---|---|---|---|
| Sperm | gamete | 31 | 99.53% | 22.0 M | 95.18% | 20.6 M | 89.25% |
| Egg | 35 | 99.73% | 22.1 M | 95.93% | 21.0 M | 90.89% | |
| 16 cell | cleavage | 19 | 99.44% | 21.6 M | 93.76% | 18.9 M | 82.07% |
| 32 cell | 32 | 99.89% | 22.1 M | 95.69% | 20.8 M | 90.19% | |
| 64 cell | 19 | 99.34% | 20.8 M | 90.19% | 17.8 M | 77.25% | |
| 128 cell | blastula | 22 | 99.51% | 21.9 M | 95.00% | 19.9 M | 86.14% |
| 1,000 cell | 38 | 99.53% | 22.0 M | 95.45% | 20.8 M | 90.15% | |
| Germ ring | gastrula | 20 | 99.57% | 21.7 M | 94.09% | 19.3 M | 83.41% |
| Testis | organ | 21 | 99.44% | 22.0 M | 95.27% | 20.1 M | 87.03% |
Paired reads were mapped uniquely to the reference genome (Zv9, UCSC) by Bismark. Number of CpG (1×) or number of CpG (5×) indicate total number of CpG sites mapped at least one read or five reads. Covered indicates proportion of mapped CpG sites over total CpG sites in genome. “M” indicates million.
First, we analyzed the genomic features of the zebrafish methylome and calculated the methylation level of each CpG site across the genome. In this study, we considered only CpG sites that were covered at least ten times. In all samples, the majority of CpG sites are either highly methylated (methylation level between 75% and 100%) or unmethylated (less than 25%) (Figure S1A available online) (Lister et al., 2009; Molaro et al., 2011). Then, we checked the methylation features in various functional elements such as promoters, exons, and CpG islands (CGI). The results show that the unmethylated CpG sites are highly enriched in promoters and CGIs (Figure S1B). In general, the pattern of DNA methylation in functional regulatory elements in zebrafish is similar to that observed in mammals (Lister et al., 2009; Molaro et al., 2011).
The Dynamics of DNA Methylomes in Gametes and Early Embryos
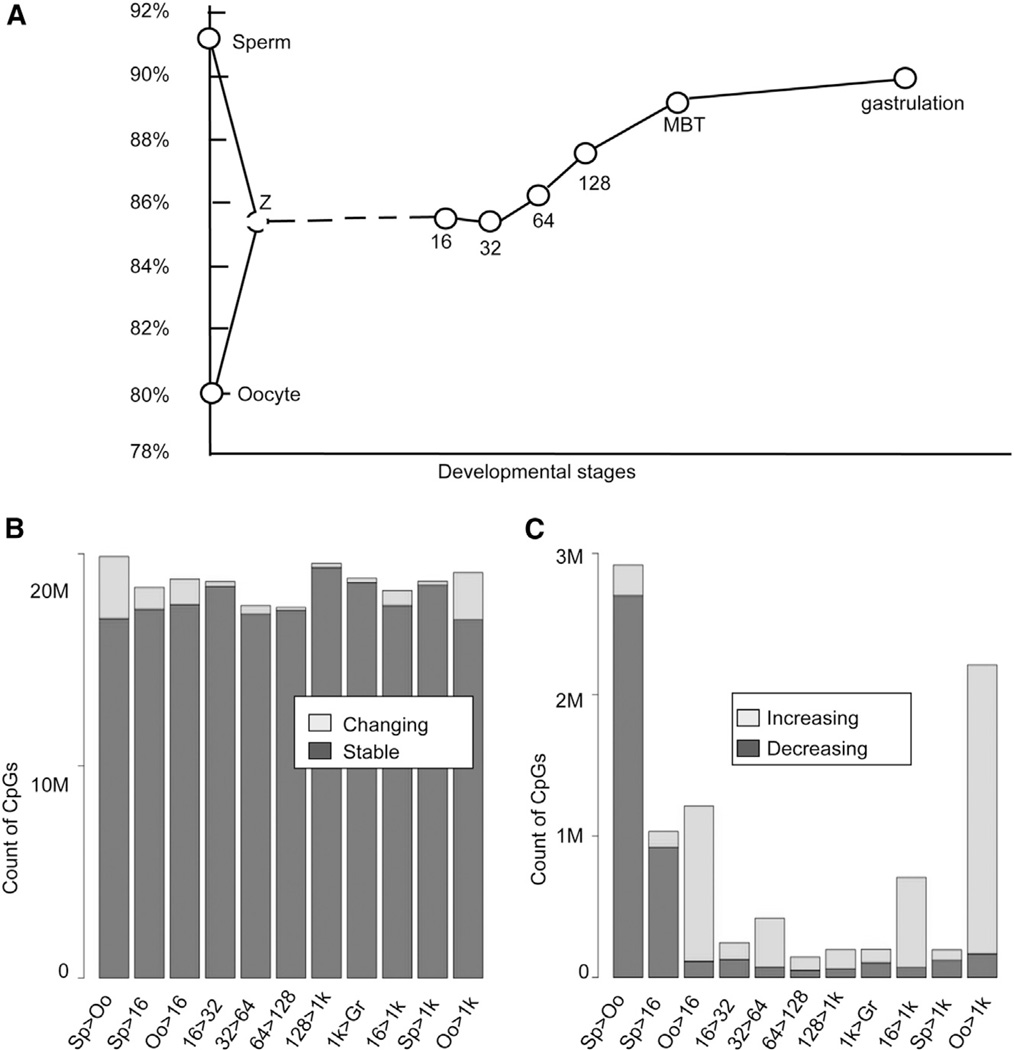
Next, we were interested in the dynamic change in the DNA methylome in zebrafish gametes and early embryos. We plotted the average DNA methylation level across the whole genome in gametes and early embryos. Surprisingly, the methylation level of oocytes is lowest among all examined samples, whereas sperm shows the highest methylation level (Figure 1A). Oocytes (average methylation level, 80%) are globally hypomethylated compared to sperm (91%). Interestingly, after fertilization, the methylation level appears to be stable in early cleavage stages of embryos compared to the predicted level in zygote, the mean level of sperm and oocyte. From 32 cell onward, the methylation level increases gradually to achieve a comparable level to sperm upon MBT (Figure 1A).
Figure 1. Dynamics of DNA Methylomes for Gametes and Early Embryos.
(A) The dynamics of the average DNA methylation level. The average methylation level is calculated as the percentage of methylated CpG in all CpG content. “Z” means the mean of methylation level between oocyte and sperm.
(B) Differentially methylated CpG sites between consecutive stages.
(C) Sites with increasing or decreasing methylation levels co-occur during each consecutive transition. The last two bars represent the comparisons between sperm or oocyte with the 1,000-cell stage, respectively.
See also Figure S1.
For each pair of consecutive stages, we compared the methylation level of each CpG site and called it as differentially methylated CpG if the difference in methylation level exceeded 0.2 with p value < 0.05 according to Fisher’s exact test. About 14% of CpG sites are differentially methylated CpGs between sperm and oocyte genome wide (Figure 1B). The methylation levels of most CpG sites are stable during the transitions of consecutive stages (Figure 1B), but we identified significant changes between oocyte and 1,000-cell embryos (Figure 1C).
Early Embryos Reset to a DNA Methylome Almost Identical to that in Sperm
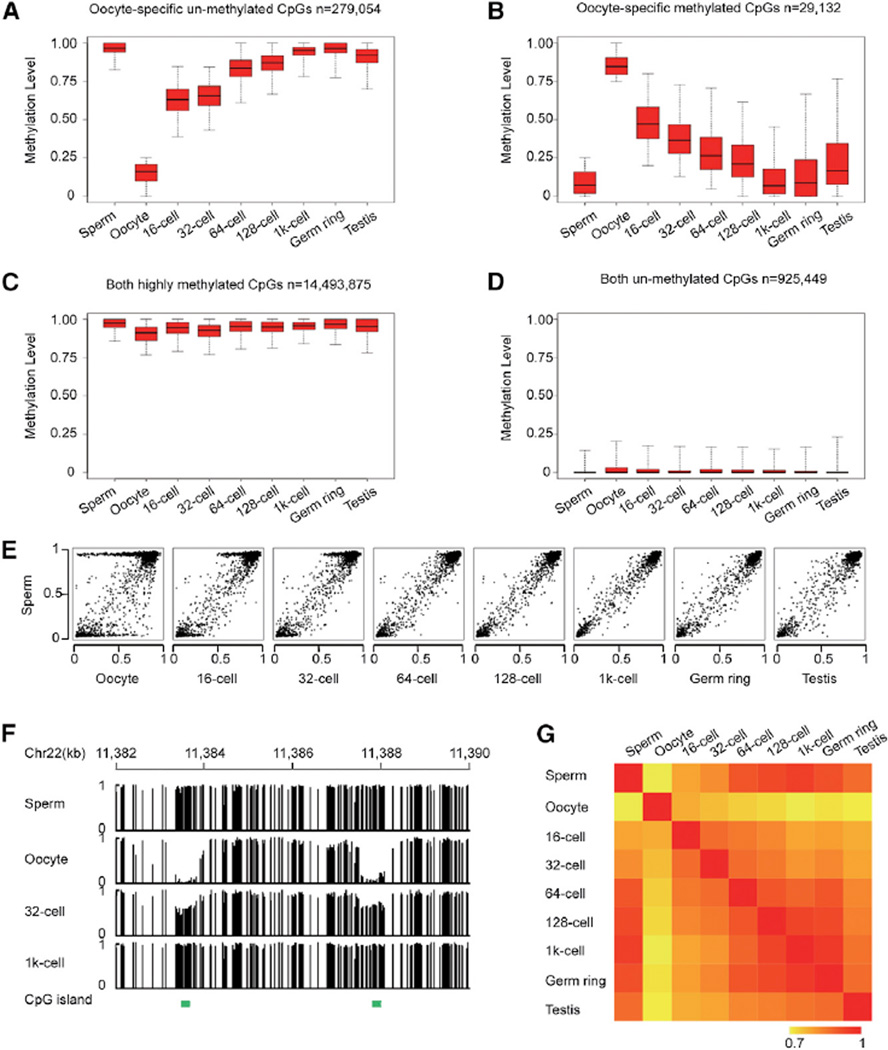
The methylomes of oocyte and sperm are significantly different (Figure 1A). But there is no significant difference between sperm and 1,000-cell embryo (Figure 1B). To investigate the potential similarities of the methylation landscape of sperm and 1,000-cell embryo, we began our analysis from oocyte-specific methylated sites (mCpG percentage ≥ 75% in oocyte and ≤ 25% in sperm) and oocyte-specific unmethylated sites (mCpG percentage ≥ 75% in sperm and ≤ 25% in oocyte). We plotted their dynamic changes across all examined stages of embryos. The methylation levels of both oocyte-specific unmethylated sites and oocyte-specific methylated sites become intermediately methylated in 16-cell embryos, which is consistent with the knowledge that the average methylation level in early embryos (16-cell embryos) is close to the mean value of oocyte and sperm (Figure 1A). Interestingly, those sites exhibit a gradual change to the similar levels seen in sperm upon MBT stage (Figures 2A and 2B). Similar results were also observed in the other differentially methylated sites between sperm and oocyte (Figures S2A and S2B). Sites with similar methylation levels between gametes do not exhibit significant changes during embryogenesis (Figures 2C, 2D, and S2C).
Figure 2. Early Embryos Reset the Methylation Landscape to a Pattern that Is Almost Identical to Sperm.
(A and B) Box plots of methylation values of CpG sites highlight the dynamic patterns of oocyte-specific unmethylated sites (methylation level ≥ 0.75 in sperm and ≤ 0.25 in oocyte) and oocyte-specific methylated sites (methylation level ≥ 0.75 in oocyte and ≤ 0.25 in sperm) across early embryogenesis. Black line indicates median, edges stand for the 25th/75th percentile, and whiskers stand for the 2.5th/97.5th percentile.
(C and D) Box plots of DNA methylation level for sites with similar methylation level in both gametes across early embryogenesis.
(E) Dot plots show pairwise comparisons of average methylation level in CGIs.
(F) Graphical representation of methylation pattern at two CGIs within a locus in gametes, 32-cell, and 1,000-cell embryos. Green bars highlight the positions of CGI. Vertical line height indicates the methylation level.
(G) Pearson correlations coefficient heatmap among methylomes. Correlation coefficients are colored yellow to red to indicate low to high, respectively.
See also Figure S2 and Table S1.
Analyses of the dynamics of CGIs further support this finding. CGIs are known as important functional genomic regions in the regulation of gene expression. The DNA methylation state of CGIs is also reprogrammed to the sperm methylation state upon MBT specification (Figures 2E, 2F, and S2D). Further analysis shows that genome-wide correlation of methylomes between sperm and early embryos becomes increasingly higher upon specification to the MBT stage (Figure 2G). In contrast, the change of correlation between methylomes of oocytes and early embryos presents an entirely reversed trend (Figure 2G). Additionally, six oocyte-specific methylated regions and ten oocyte-specific unmethylated regions were independently validated by bisulfite-sequencing PCR, which provided further proof that all the examined loci reset to sperm methylation state upon specification to MBT stage (Table S1). In summary, evidence of CpG sites, CGIs, and whole-genome correlation all support the idea that sperm defines the DNA methylome of early embryos upon specification to MBT.
Upon specification to MBT, differentially methylated sites between sperm and oocyte are reset gradually to sperm methylation pattern, whereas from MBT onward, the methylome is further programmed (Figures 2A, 2B, 2E, and 2G) and becomes more and more different from sperm. But all the reprogramming to the somatic cell is based on the sperm methylation pattern, which is limited to specific regions. As a result, the methylome of somatic cells is still close to the sperm pattern. These data suggest that MBT is a transitional stage during DNA methylation reprogramming in zebrafish early embryogenesis.
5hmC Is Not Involved in the DNA Methylation Reprogramming in Zebrafish Early Embryos
Next, we were interested in how the early embryo methylome becomes similar to sperm methylome upon the MBT stage. Our previous results show that there is no genome-wide DNA demethylation between gametes and early cleavage stages of embryos but that a significant number of sites are demethylated during early embryogenesis (Figure 1C). Previous reports show that the AID/Gadd45 enzyme mediates DNA demethylation in zebrafish (Rai et al., 2008). But, AID/Gadd45 has no activity before 4 hr postfertilization (Rai et al., 2008). Given that oocyte-specific methylated regions become totally unmethylated by the MBT stage (around 3 hr postfertilization), we chose not to pursue the potential role of AID/Gadd45 further in this work. Several recent studies have suggested that 5hmC may be involved in paternal DNA demethylation after fertilization (Inoue and Zhang, 2011; Iqbal et al., 2011). To explore whether 5hmC mediates DNA demethylation in zebrafish early embryos, we used a glycosylation-mediated enrichment method (Robertson et al., 2012) to detect the genomic distribution of 5hmC in sperm, 2-cell, and 16-cell embryos. Our data show a very limited number of 5hmC-enriched regions (Table S2), none of which are associated with DNA demethylation regions, thereby suggesting that 5hmC does not mediate DNA demethylation in early embryos. To further confirm this result, we measured 5hmC in 32-cell embryos with singlenucleotide resolution using Tet-assisted bisulfite sequencing (TAB-seq) (Yu et al., 2012). The data demonstrate a small number of 5hmC sites and no 5hmC enriched regions in 32-cell embryos (Table S3). Additionally, 5hmC is not detectable by immunofluorescence staining in zebrafish early embryos (Figure S3). Taken together, 5hmC does not mediate the DNA demethylation in zebrafish early embryos.
Paternal DNA Copies Maintain Sperm Methylome, and Maternal DNA Copies Maintain Oocyte Methylome in 16-Cell Embryos
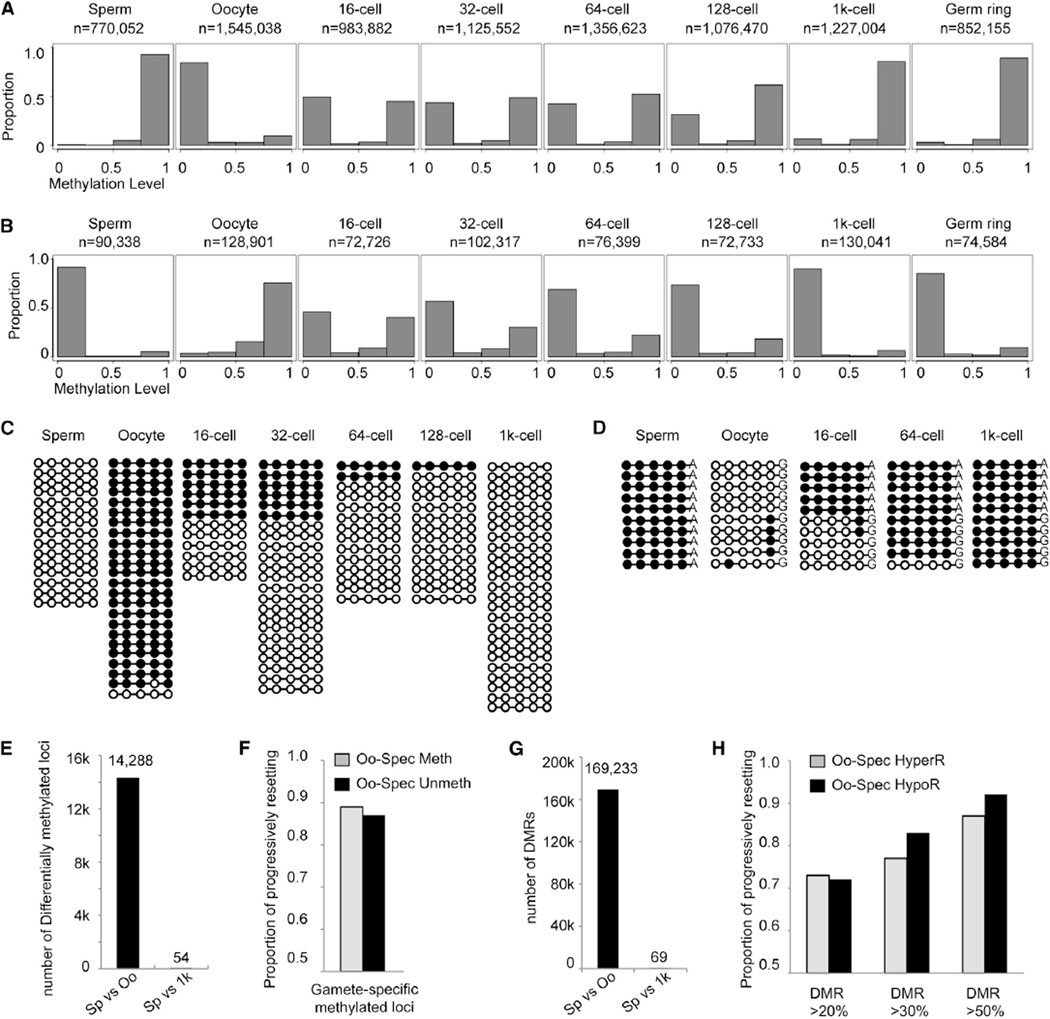
Sperm and oocyte methylome are significantly different, but embryos reset to a methylome very similar to sperm upon MBT. To understand how differentially methylated regions (DMRs) between oocyte and sperm reprogram to the same state, we tracked the dynamic changes of oocyte-specific unmethylated loci.In this analysis, we picked up the paired reads covering at least four consecutive oocyte-specific unmethylated sites (mCpG percentage ≥ 75% in sperm and ≤ 25% in oocyte) from each stage and then calculated the methylation level of these reads. As expected, almost all paired reads in oocyte-specific unmethylated regions are highly methylated in sperm (Figure 3A, first panel) and are unmethylated in oocytes (Figure 3A, second panel). While in the cleavage 16-cell embryos, about half of the paired reads are unmethylated, and the other half are highly methylated (Figure 3A, third panel). This result is consistent with the previous finding that the average methylation level in early embryos (16-cell embryos) is close to the mean value of oocyte and sperm (Figure 1A). Similar findings were also revealed for paired reads, which covered at least four oocyte-specific methylated sites (mCpG percentage ≤ 25% in sperm and ≥ 75% in oocyte) in the 16-cell stage (Figure 3B first, second, and third panels). Two representative loci are shown in Figures 3C and S4A. Additionally, results of bisulfite-sequencing PCR show that the DNA methylation levels of both oocyte-specific methylated loci and unmethylated loci in 8-cell embryos are around 50% (Table S1). These results indicate that the DNA methylomes of 8-cell and 16-cell embryos result from the addition of sperm and oocyte methylome, thus suggesting that paternal DNA may maintain sperm methylome and maternal DNA may maintain oocyte methylome.
Figure 3. The Methylome of Paternal DNA Is Maintained, and the Methylome of Maternal DNA Is Gradually Reset to Sperm Pattern.
(A) Distribution of the average methylation values of paired reads, which include at least four consecutive oocyte-specific unmethylated CpG sites (mCpG percentage ≥ 75% in sperm and ≤ 25% in oocyte) in gametes and early embryos. n is the number of paired reads in each stage.
(B) Distribution of average methylation values of paired reads covering at least four consecutive oocyte-specific methylated sites (mCpG percentage ≥ 75% in oocyte and ≤ 25% in sperm). x axis represents methylation level. Bins are at 0.25 intervals (bins at 0.05 are shown in Figures S1C and S1D).
(C) Dynamic changes of DNA methylation for a representative locus located in chr7:21,315,648-21,315,662. Paired reads from whole-genome data covering all five CpGs are shown. Open circles represent unmethylated CpGs, and filled circles represent methylated CpGs.
(D) Dynamic changes of DNA methylation for a representative locus in chr6: 27,393,375-27,393,560 tracked with SNPs to distinguish maternal and paternal DNA. The data are validated using bisulfite PCR. Ten reads were randomly picked for sperm and oocyte. Five reads from paternal DNA and five reads from maternal DNA are randomly picked in 16-cell, 64-cell, and 1,000-cell stages of embryos. See also Table S4.
(E) Number of differentially methylated loci covered by oocyte-specific methylated and unmethylated paired reads.
(F) Genome-wide analysis of the proportion of differentially methylated loci covered by paired reads progressively resetting to sperm methylation state. Gray bars represent oocyte-specific hypermethylated loci, and black bars represent oocyte-specific hypomethylated loci.
(G) Number of DMRs genome wide, methylation difference > 0.2.
(H) Genome-wide analysis of the proportion of DMRs progressively resetting to sperm methylation state. Methylation level difference of the DMRs was set higher than 0.2, 0.3, and 0.5, respectively.
See also Figures S3 and S4 and Tables S2 and S3.
To validate this, we tracked DNA methylation states of paternal and maternal DNA copies according to SNPs from crossed zebrafish, TU strain female with Tupfel long fin (TL) strain male. About two million homogenous SNPs were identified between TL and TU strains. The bisulfite-sequencing PCR method was used to examine the methylation states of 22 DMRs (Table S4). All of these loci contain SNPs, which can distinguish maternal DNA from paternal DNA. As expected, paternal DNA copies maintain the sperm methylation state, and maternal DNA copies maintain the oocyte methylation state in the 16-cell embryos. A representative locus is presented in Figure 3D.
Collectively, our data demonstrate that paternal DNA copies maintain the sperm methylome and maternal DNA copies maintain the oocyte methylome, indicating that both paternal and maternal DNA use a DNA maintenance system to maintain the methylome until the 16-cell stage. Our mRNA expression data also demonstrate that maintenance methyltransferase DNMT1 is highly expressed across all of these early cleavage stages (Table S5), which is consistent with a previous report (Rai et al., 2006). Our data show that no whole-genome DNA demethylation occurs after zebrafish fertilization, a finding that goes against previous reports that have demonstrated whole-genome DNA demethylation (MacKay et al., 2007; Mhanni and McGowan, 2004).
For DMRs between Gametes, the Methylation Pattern of Maternal DNA Gradually Resets to Sperm Methylation State after the 16-Cell Stage
Next, we were interested in how the DNA methylome is established after the 16-cell stage. Similar to the 16-cell stage, the majority of paired-reads are still either highly methylated or unmethylated in later stages (Figures 3A and 3B). Notably, both oocyte-specific unmethylated reads (Figure 3A) and oocyte-specific methylated reads (Figure 3B) genome wide gradually decrease after the 16-cell stage, and the majority of them are changed to the sperm methylation state upon MBT.
For an oocyte-specific methylated locus, the proportion of the methylated reads also gradually decreases, and the locus becomes the sperm unmethylated state in 1,000-cell stage (Figure 3C). This result indicates that demethylation of this oocyte-specific highly methylated locus occurs through passive demethylation. For an oocyte-specific unmethylated locus, the proportion of the unmethylated reads also gradually decreases, and the locus is methylated in the 1,000-cell stage (Figure S4A), indicating that de novo methylation occurs in this locus. Our messenger RNA sequencing (mRNA-seq) results show that de novo methyltransferases DNMT3, 4, 5, and 7 (mammalian homology DNMT3b) are highly expressed at these stages (Table S5).
Furthermore, the oocyte-specific differentially methylated loci tracked by SNPs show that paternal DNA copies maintain the sperm methylation state in the 64-cell and the 1,000-cell stages. At the 64-cell stage, however, the number of maternal DNA copies with oocyte-specific methylation state (OSMS) decreases, and some maternal DNA copies transform to the sperm methylation state (Figure 3D and Table S4). In the 1,000-cell stage, the methylation state of maternal DNA is similar to the methylation state of sperm (Figure 3D and Table S4). In summary, for all of these loci, paternal DNA copies maintain the sperm methylation state until the MBT stage and maternal DNA copies with OSMS gradually decrease during cell division and reprogram to a sperm methylation pattern upon MBT stage.
We also examined how many oocyte-specific unmethylated loci and oocyte-specific methylated loci progressively reset to sperm pattern through cell division. Our whole-genome analyses between sperm and oocyte show there are 14,288 differentially methylated loci covered by paired reads, including 13,594 oocyte-specific unmethylated loci and 694 oocyte-specific methylated loci (Figure 3E). When comparing sperm and MBT, only 54 (0.38%) loci remain as differentially methylated loci (adjusted t test, p < 0.05, methylation level difference > 0.2), indicating that almost all of the gamete-specific methylated or unmethylated loci reset to sperm methylation state. Then, we calculated how many of the loci progressively reset to sperm methylation state through cell division. Our data include the methylomes of 16-cell, 32-cell, 64-cell, 128-cell, and 1,000-cell stages. Therefore, we can calculate the methylation level changes of all the specific loci in four consecutive transitions, including 16-cell to 32-cell, 32-cell to 64-cell, 64-cell to 128-cell, and 128-cell to 1,000-cell stages. We defined a locus that was progressively reset to sperm methylation state through cell division if the locus was transformed into sperm methylation state upon the MBT stage, and the DNA methylation level of this specific locus gradually increased (for oocyte-specific unmethylated locus) or decreased (for oocyte-specific methylated locus) in at least three of four consecutive transitions. We found that about 89% of oocyte-specific methylated loci and 87% oocyte-specific unmethylated loci follow our definition of progressive resetting (Figure 3F).
Moreover, we applied the analysis to DMRs (methylation level difference > 0.2). Between sperm and oocyte, there are 162,196 oocyte hypomethylated regions (HypoRs) and 7,037 oocyte-specific hypermethylated regions (HyperRs) (Figure 3G). When sperm are compared to MBT, only 69 of them (0.041%) are still DMRs, indicating that almost all DMRs between gametes reset to sperm methylation state. For all DMRs, 73% of oocyte-specific HyperRs and 72% of HypoRs follow our definition of progressive resetting (Figure 3H). To reduce the effect of sequencing noises, we focused on the DMRs with a methylation difference higher than 0.5. As expected, the percentages of HyperRs and HypoRs following our definition of progressive resetting increase to 87% and 92%, respectively (Figure 3H). Representative regions of all six large HOX genes clusters progressively reset to sperm methylation state (Figures S4B–S4G).
In summary, paternal DNA maintains sperm methylome until MBT stage. For maternal DNA, the oocyte-specific methylation pattern is gradually discarded during cell division and progressively resets to a sperm methylome pattern. Our data indicate that maternal DNA involves passive DNA demethylation and de novo methylation simultaneously.
The Methylome of Maternal DNA Could Reset to the Sperm Pattern as One Unit
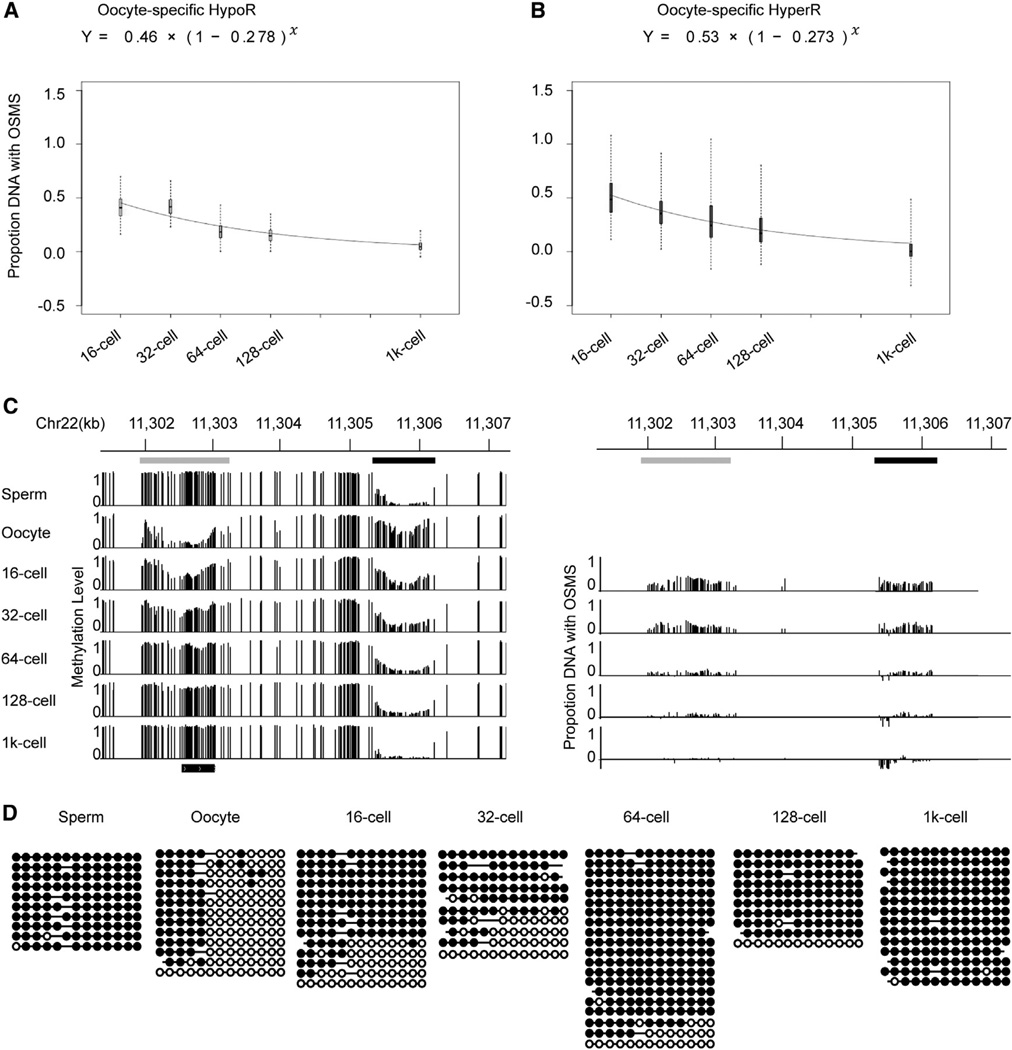
Next, we were interested in the progressively resetting rates of the oocyte-specific HyperRs and HypoRs. To achieve the rate, we needed to know the proportion of DNA copies with OSMS to the total DNA copies in each stage. Because DNA presents either sperm methylome state or oocyte methylome state (Figures 3A–3C), we can estimate the proportion of DNA copies with OSMS in a developmental stage according to the methylation level of sperm, oocyte, and this specific stage. Therefore, we got the estimated proportion of DNA copies with OSMS for all DMRs (methylation level difference > 0.2) in each stage (Experimental Procedures). Then, an exponential model was used to fit the proportion-time curve. We calculated the fitting curves of the de novo methylation rate for oocyte-specific HypoRs and the demethylation rate for HyperRs across early embryogenesis, respectively (Figures 4A and 4B). Excitingly, the average de novo methylation rate of HypoRs is 0.278, and the demethylation rate of HyperRs is 0.273. The two rates are almost exactly the same. A representative locus is shown, which includes one oocyte-specific HyperR, one HypoR, and unchanged methylation regions. The proportion of DNA copies with OSMS for each CpG site within the oocyte-specific HyperR and HypoR is also calculated. In general, the CpG sites within the HyperR and HypoR are reset at a similar rate (Figure 4C). Unfortunately, for the unchanged regions,we cannot track whether maternal DNA copies with oocyte methylation states are reset because their methylation states show no difference among gametes and remain stable across development. Given that the resetting rates for the HyperR and HypoR are the same, this 5 kb region should reset to sperm methylation state as one unit. Additionally, a representative locus (covered by paired reads) includes both common methylated CpG sites and differentially methylated sites between sperm and oocyte. In this locus, the paired reads with the oocyte methylation state decrease as one unit and reset to sperm methylation state (Figure 4D). These data suggest that the whole oocyte methylome could reset to sperm pattern as one unit during cell division.
Figure 4. DNA Demethylation Rate of Oocyte-Specific HypoRs and De Novo Methylation Rate of HyperRs.
(A and B) Fitting curves of the DNA demethylation rate of oocyte-specific HypoRs and de novo methylation rate of HyperRs. The methylation level difference of HypoRs and HyperRs is greater than 0.2. HypoRs and HyperRs represent hypomethylated and hypermethylated regions versus sperm, respectively. OSMS represents oocyte-specific methylation state, which is compared with sperm. Box plot of proportion DNA with OSMS was drawn for each stage. Black line indicates median, edges stand for the 25th/75th percentile, and whiskers represent the 2.5th/97.5th percentile.
(C) Graphical representation of methylation dynamics of one region containing one oocyte-specific HyperR (demethylation locus, black bar) and one oocyte-specific HypoR (de novo methylation locus, gray bar). The left panel shows the dynamics of the methylation level. The right panel shows the relative proportion of DNA copies with oocyte methylation state for each CG site within the DMRs.
(D) Dynamics of a representative locus in chr16: 46,387,770-46,387,944 covered by paired reads containing common methylated sites and differentially methylated sites (sperm versus oocyte). Paired reads with oocyte methylation landscape decrease and reprogram to sperm methylation state.
See also Table S5.
DNA Methylome Reprogramming Associates with Early Embryonic Developmental Transitions
DNA methylation is critical for proper embryonic and tissue-specific development (De Carvalho et al., 2010; Jaenisch and Bird, 2003; Kim et al., 2010). Therefore, we pursued the potential functions of DNA methylation reprogramming during early embryogenesis. Given that the DNA methylation level at promoters is highly related to gene expression (Feng et al., 2010a; Lister et al., 2009; Zemach et al., 2010), we pursued how the DNA methylation pattern in promoter is reprogrammed among gametes and early embryos. As shown in Figure 5A, the DNA methylation pattern of promoters in sperm is significantly different than oocyte but is very similar to MBT. Abundant differences begin to emerge again during the transition from MBT to gastrula. Moreover, genes with differentially methylated promoters between oocyte and sperm are almost identical to those between oocyte and MBT (Table S6), which is consistent with the MBT methylome being virtually identical to sperm.
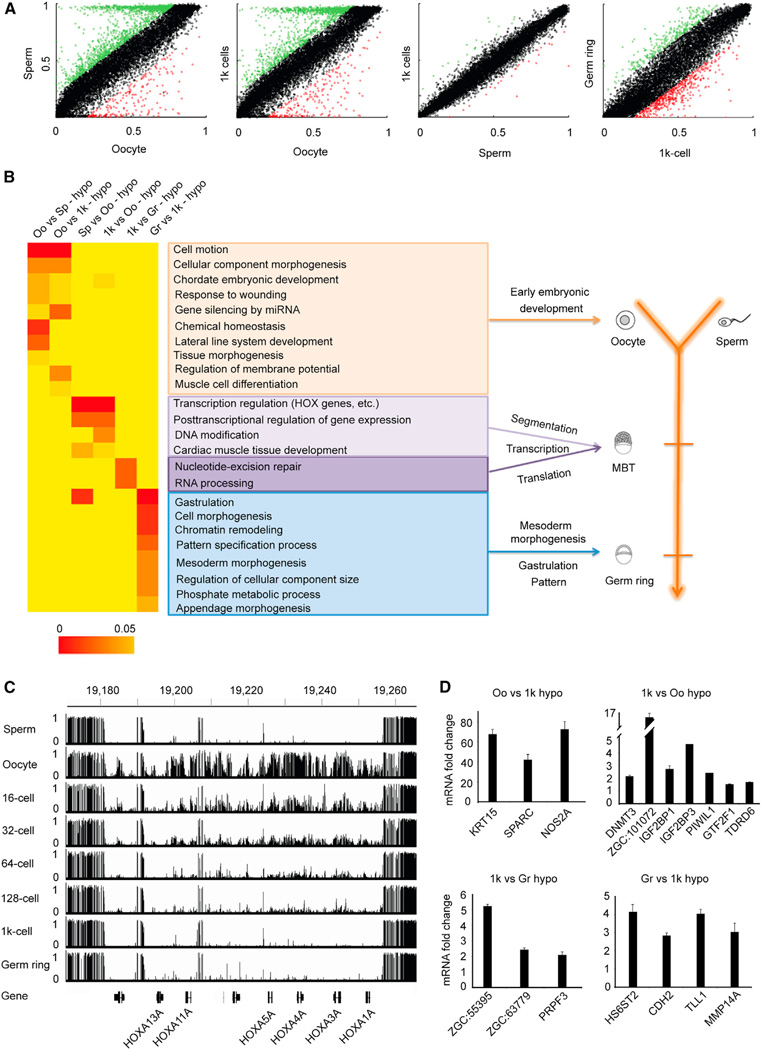
Figure 5. Functional Significance of Sperm Methylome Pattern.
(A) Dot plots show pairwise comparisons of average methylation level in promoters (2 kb up TSS) based on RefSeq gene annotations. Differentially methylated promoters are highlighted in green and red. See also Table S6.
(B) The heatmap represents the combination of GO term enrichment in genes with oocyte-specific, sperm-specific, MBT-specific, and gastrula-specific hypomethylated promoters. The statistical significance of the heatmap is colored red to orange to indicate high to low, respectively. Yellow is background.
See also Tables S6 and S7.
(C) Graph represents methylation at one HOXa gene cluster region in gametes and early embryos. Vertical line height indicates the methylation level.
(D) RT-qPCR validates the expression changes for genes with methylation level significantly changed in promoters among developmental stages transition. Data show mean ± SD from one representative experiment in triplicate; at least two independent experiments were performed with similar results.
See also Figure S5.
In order to explore the biological significance of inheriting sperm methylome, we used gene ontology analysis to examine the functional enrichments of genes with differentially methylated promoters (Table S6). All of the enriched categories are listed (Figure 5B). Interestingly, consistent with the fact that maternally stored mRNAs direct early embryonic development prior to MBT (Korzh, 2009; Yasuda and Schubiger, 1992), genes with oocyte-specific hypomethylated promoters (oocyte versus sperm and oocyte versus MBT) show strong enrichment in the processes that regulate very early embryogenesis such as cellular component morphogenesis, cell motion, and lateral line system development (Figure 5B, orange window).
Additionally, compared to oocyte, genes with MBT-specific (also sperm-specific) hypomethylated promoters are strongly enriched in posttranscriptional regulation, DNA modification, and transcription factors (Figure 5B, purple window). These data are consistent with the activation of zygotic genome transcription at MBT stage (Kimmel et al., 1995). In sperm, many developmental regulated genes, such as all 48 HOX genes—which are known to be critical for the patterning of the animal embryo’s anteroposterior (AP) axis (Prince et al., 1998)—do not express (data not shown), even with unmethylated promoters (Figures 5C and S4B–S4G). Moreover, the majority of these genes still cannot express before the MBT stage because zygotic genome transcription is absent during that period. These data suggest that depositing those DNA methylation patterns in sperm will direct later embryonic development and will meanwhile prevent these genes from overex-pressing too early to disturb the early embryogenesis.
We also explored the functional enrichment of genes with differentially methylated promoters between MBT and gastrula stage (germ ring). Interestingly, genes with MBT-specific hypomethylated promoters are highly enriched in RNA processing (Figure 5B, purple window), which is consistent with the activation of zygotic genome transcription at the MBT stage. In addition, genes with gastrulation-specific hypomethylated promoters versus MBT are highly enriched in gastrulation, pattern specification, and mesoderm morphogenesis (Figure 5B, blue window).
Collectively, our data indicate that DNA methylome reprogramming associates with early embryonic developmental transitions.
Inheriting Sperm Methylome Can Facilitate the Epigenetic Regulation of Embryogenesis
Given that DNA methylation can regulate gene expression (Feng et al., 2010a; Lister et al., 2009; Zemach et al., 2010), we analyzed mRNA expressions of sperm, oocytes, 1,000-cell embryos, and germ-ring embryos generated by mRNA-seq. Combining analysis between mRNA expression and DNA methylation shows that gene expression is anticorrelated with methylated CpG density in promoter for each stage (Figure S5A). Further analyses clearly show that gene expression varies inversely with methylation levels of the TSS-proximal region (Figures S5B and S5C). Taken together, these data indicate that the DNA methylation pattern associates with gene expressions in early embryos.
MBT is the stage for the activation of zygotic genome transcription (Kimmel et al., 1995). Because the majority of genes are biallelically expressed except the imprinted ones, the methylomes of maternal and paternal DNAs becoming identical should be important for the activation of zygotic genome transcription. Our data show that the maternal and paternal DNA methylomes become the same as sperm pattern by the MBT stage. Although the differentially expressed genes among various stages are not significantly correlated with the change of DNA methylation genome wide, 14 genes (Table S7) with sperm-specific HypoRs and overexpressed in the MBT embryos versus the oocyte are still enriched in the categories of RNA metabolism and regulation of transcription. Expression of these genes would be important for the zygotic genome transcription. Therefore, the MBT embryos using sperm methylome pattern can facilitate the activation of zygotic genome transcription in the MBT stage.
Moreover, genes with both hypomethylated promoters and that are overexpressed in the germ-ring embryos versus the 1,000-cell embryos are enriched in appendage development, cell adhesion, and pattern specification process (Table S7). These data agree with the understanding that gastrulation is a morphogenetic process in the formation of embryonic germ layers and the segmentation pattern established in gastrula stage. The expressions of a number of genes associated with embryo developmental transition were further confirmed via quantitative RT-PCR (Figure 5D). Taken together, the associations between DNA methylation and mRNA expressions indicate that inheriting sperm methylome is important for embryogenesis.
To further validate the function of sperm methylome pattern, we performed androgenetic and gynogenetic assays. By transferring sperm nucleus into enucleated oocyte, we found that 7 embryos from 27 transplanted eggs reached the dome stage, 5 of them further reached the bud stage, and only 3 of them finally survived beyond 18 hr (Figure S5D). Injecting another oocyte’s nuclear to an enucleated oocyte does not yield any developing embryos (0/63). Meanwhile, dechorionated oocytes cannot show any gynogenetic cell cleavage event using SrCl2, which has been shown to activate gynogenetic development in mammals (O’Neill et al., 1991). Additionally, consistent with the published data (Huang et al., 2003), nuclear transfer using somatic cell nuclear can easily produce zebrafish larva (data not shown), suggesting that unsuccessfully gynogenetic development should not result from lacking sperm fertilization process. Considering that somatic cells’ DNA methylome is close to the sperm pattern (Figure 2G, germ ring), those results suggest that the sperm methylation pattern should be critical for embryogenesis.
DISCUSSION
It is believed that epigenetic information such as DNA methylation and histone modifications is cleared and re-established after fertilization with the exception of a number of loci (Arteaga-Vazquez and Chandler, 2010; Cavalli and Paro, 1998; Morgan et al., 1999). However, with high-coverage DNA methylomes, we found that the sperm DNA methylation landscape is not cleared after fertilization; instead, sperm methylome is inherited in zebrafish. Compared to oocyte, sperm has quite smaller cellular volume with limited RNAs and proteins. Therefore, it is commonly believed that the majority of information for the embryogenesis of offspring is carried by egg, and sperm carries just one set of DNA. However, our study illustrates that besides DNA that can be inherited, sperm DNA methylome can also be inherited in zebrafish. And the inheriting sperm methylome can facilitate the embryogenesis.
In mammals, active DNA demethylation in paternal DNA and passive DNA demethylation in maternal DNA occurs after fertilization, followed by de novo methylation after inner cell mass (ICM) (Gu et al., 2011; Inoue and Zhang, 2011; Iqbal et al., 2011). In zebrafish, however, paternal DNA methylome maintained stable upon MBT stage, and maternal DNA methylome was maintained before the 16-cell stage but then gradually discarded, a process that involves passive DNA demethylation and de novo methylation simultaneously. These data indicate that zebrafish and mammals use a different mechanism for DNA reprogramming after fertilization. Our data show that 5hmC is not involved in the DNA demethylation during zebrafish early embryogenesis. The proportion of oocyte-specific hyper-methylated reads is gradually decreased, suggesting that demethylation is passive rather than active. Interestingly, the demethylation rate for these regions is about 27%, but not 50%, suggesting a mechanism of asymmetric division that establishes the DNA methylation pattern during embryogenesis. Perhaps new synthesized maternal DNAs maintain the oocyte methylation pattern by DNMTs in some cells, but not in all of them.
Zebrafish inherit sperm DNA methylome, but the underlying molecular mechanism remains to be elucidated. It has been shown that small RNA from germ-line cells mediates DNA methylation reprogramming to guide epigenetic inheritance in plant (Calarco et al., 2012). Previous studies also show that Piwi-interacting RNA (piRNA) mediates DNA methylation of transposon elements during germ cell maturation in mouse (Aravin et al., 2007, 2008). Perhaps zebrafish early embryos use a similar mechanism to reprogram the DNA methylation as mediated by piRNAs, which is abundant in zebrafish sperm (Houwing et al., 2007). Our data show that SrCl2-treated oocytes cannot initiate gynogenetic development, whereas previous data show that gynogenetic zebrafish can be generated for oocytes fertilized by UV-light-irradiated sperm (Walker et al., 2009). UV can break DNA into fragments, but UV-irradiated sperm should still be able to carry piRNAs into oocyte, which may guide the DNA methylation reprogramming. We cannot rule out that other important factors from sperm play a crucial role in DNA methylation inheritance.
Our data suggest that the whole oocyte methylome can act as one unit to be discarded. The possibility remains that the whole maternal DNA occurs through a de novo methylation mechanism to re-establish the methylome. How embryos control the switch from maintaining oocyte methylome to replace it with sperm methylome later on also requires further exploration. Additionally, future research should focus on how early embryos distinguish maternal DNA from paternal DNA.
A great number of genes are highly expressed in oocytes, in which the hypomethylated methylome can promote these genes expressions during the oocyte maturation. But, in sperm, the majority of genes expressions are silent even with the unmethylated promoters, probably because the highly packed chromosome of sperm can prevent the DNA methylation information from being accessed by transcriptional machinery. Therefore, the DNA methylome blueprint for regulation of embryogenesis can be deposited in the sperm, which will not disturb genes’ expression in the sperm. Upon the MBT stage, the blueprint is successfully transferred to the embryos. Then the DNA methylome information is accessible, and the sperm methylome pattern can facilitate both the activation of the zygotic genome transcription and the epigenetic regulation of embryogenesis. At the same time, sperm methylome in MBT can help silence or suppress the expression of oocyte-specific genes.
In summary, our study shows that zebrafish inherit DNA methylome from sperm during early embryogenesis, which sets the stage for further research on how progenies inherit epigenetic information from their parents.
EXPERIMENTAL PROCEDURES
Zebrafish Stock and Collection of Gametes and Early Embryos
The wild-type zebrafish lines TU and TL were raised under standard conditions. Sperm were released by gently but repeatedly disrupting the testis with a pipette tip in Hank’s balanced salt solution (HBSS). Unfertilized oocytes were collected by squeezing the abdomen of females about 5 min after spawning. Embryos were grown in embryo medium at 28°C and were staged according to standard morphological criteria.
MethylC-Seq, Reads Filtering, and Alignment
Genomic DNA (≥ 100 ng) spiked with 0.5% unmethylated cl857 Sam7 Lambda DNA (Promega) was used to construct the DNA library. Bisulfite treatment was performed according to EZ DNA Methylation-Gold kit (Zymo Research) instruction manual. The library was sequenced by Hiseq 2000. Reads trimming was performed by Trimmomatic with default parameters. Filtered paired-end methyC-seq reads were mapped against the reference (Zv9) by Bismark_v0.6.4 (Krueger and Andrews, 2011). A custom script was used to examine whether paired-end reads were overlapped, and the overlapped part was trimmed from one end to prevent double counting in the same observation (see also Extended Experimental Procedures).
Resequencing
The inbred TU and TL strains were used to extract DNA for resequencing. Reads were aligned to the reference (Zv9, UCSC) by BWA with default parameters. SNP calling was performed by SAMtools (http://samtools.sourceforge.net), and SNP quality was set to be equal or greater than 20.
Estimated Proportion of DNA Strands with Maternal Methylation Pattern
For each DMR j, between sperm and oocyte, we calculated the average methylation level of CpGs within the region for each stage. For embryo stage i (i means cell division times since 16-cell stage, thus, i = 0, 1, 2, 3, and 6 for 16-cell, 32-cell, 64-cell, 128-cell, and 1,000-cell stages, respectively), the proportion of DNA strands with maternal methylation pattern was estimated as Pi,j. The methylation level for stage i in region j is formulated:
Therefore, the formula above could be rewritten as:
MLoocyte j means methylation level of oocyte in the region j. Then, an exponential model y = a*exp(b*x) is used for decoding the functional relationship of P~i, with sperm-specific HypoR data set and egg-specific HypoR data set, respectively. The analyses were implemented with R software.
Identification of Differentially Methylated Promoters and GO Annotation
Only promoters (2 kb upstream from the transcriptional start site) containing at least five CG sites that were each covered by at least five reads were considered. The methylation level of each promoter was determined as the ratio of the number of alignments with C (methylated) over the number of alignments with either C or T for all CG sites in the promoter. Promoters with differences in methylation levels larger than 20% between any two developmental stages (two-tailed Fisher’s exact test, p value < 0.05) were finally defined as differentially methylated promoters. Gene ontology (GO) analysis of genes with differentially methylated promoters was performed using DAVID (http://david.abcc.ncifcrf.gov/). GO terms with p value of less than 0.05 were determined to be statistically significant.
5hmC Sequencing
5-hmC DNA enrichment (hMeDIP-seq) sequence libraries were constructed according to Quest 5-hmC DNA Enrichment Kit (Zymo Research). TAB-seq of 5-hmC sequencing was performed as described (Yu et al., 2012) (see also Extended Experimental Procedures).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants to J.L. from the 973 Program of China (2011CB510101), National Natural Science Foundation of China (NSFC) (91219104 and 81171902), CAS (XDA01040407), and 100 Talents Project. W.C. is supported by NSFC (91231112 and 31171244) and CAS Excellent Youth Scholars (KSCX2-EW-Q-5-2). J.Z. is supported by NSFC (31200958). C.H. is supported by National Institutes of Health (HG006827). Sarah F. Reichard and Li Lan contributed to the editing.
Footnotes
ACCESSION NUMBERS
The GEO accession number for the sequencing data reported in this paper is GSE44075.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, five figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.04.041.
REFERENCES
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI intransposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga-Vazquez MA, Chandler VL. Paramutation in maize: RNA mediated trans-generational gene silencing. Curr. Opin. Genet. Dev. 2010;20:156–163. doi: 10.1016/j.gde.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2006;2:e49. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijo´ JA, Becker JD, Martienssen RA. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: more questions than answers. Genome Res. 2010;20:1623–1628. doi: 10.1101/gr.106138.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA. 2010a;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010b;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Huang H, Ju B, Lee KY, Lin S. Protocol for nuclear transfer in zebrafish. Cloning Stem Cells. 2003;5:333–337. doi: 10.1089/153623003772032835. [DOI] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo´ PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Korzh V. Before maternal-zygotic transition … There was morphogenetic function of nuclei. Zebrafish. 2009;6:295–302. doi: 10.1089/zeb.2008.0573. [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay AB, Mhanni AA, McGowan RA, Krone PH. Immunological detection of changes in genomic DNA methylation during early zebrafish development. Genome. 2007;50:778–785. doi: 10.1139/g07-055. [DOI] [PubMed] [Google Scholar]
- Macleod D, Clark VH, Bird A. Absence of genome-wide changes in DNA methylation during development of the zebrafish. Nat. Genet. 1999;23:139–140. doi: 10.1038/13767. [DOI] [PubMed] [Google Scholar]
- Mhanni AA, McGowan RA. Global changes in genomic methylation levels during early development of the zebrafish embryo. Dev. Genes Evol. 2004;214:412–417. doi: 10.1007/s00427-004-0418-0. [DOI] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- O’Neill GT, Rolfe LR, Kaufman MH. Developmental potential and chromosome constitution of strontium-induced mouse parthenogenones. Mol. Reprod. Dev. 1991;30:214–219. doi: 10.1002/mrd.1080300308. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, Cairns BR, Jones DA. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol. Cell. Biol. 2006;26:7077–7085. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AB, Dahl JA, Ougland R, Klungland A. Pull-down of 5-hydroxymethylcytosine DNA using JBP1-coated magnetic beads. Nat. Protoc. 2012;7:340–350. doi: 10.1038/nprot.2011.443. [DOI] [PubMed] [Google Scholar]
- Smallwood SA, Kelsey G. De novo DNA methylation: a germ cell perspective. Trends Genet. 2012;28:33–42. doi: 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Wolffe AP. Constitutive genomic methylation during embryonic development of Xenopus. Biochim. Biophys. Acta. 2001;1521:39–44. doi: 10.1016/s0167-4781(01)00280-9. [DOI] [PubMed] [Google Scholar]
- Walker C, Walsh GS, Moens C. Making gynogenetic diploid zebrafish by early pressure. J. Vis. Exp. 2009;28:e1396. doi: 10.3791/1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda GK, Schubiger G. Temporal regulation in the early embryo: is MBT too good to be true? Trends Genet. 1992;8:124–127. doi: 10.1016/0168-9525(92)90369-F. [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D, et al. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.