Abstract
The transcription factor Forkhead box protein M1 (FOXM1) is overexpressed in the majority of cancer patients. This overexpression is implicated to play a role in the pathogenesis, progression, and metastasis of cancer. This important role of FOXM1 demonstrates} its significance to cancer therapy. MicroRNAs (miRNAs) are small noncoding, endogenous, single-stranded RNAs that are pivotal posttranscriptional gene expression regulators. MiRNAs aberrantly expressed in cancer cells have important roles in tumorigenesis and progression. Currently, miRNAs are being studied as diagnostic and prognostic biomarkers and therapeutic tools for cancer. The rapid discovery of many target miRNAs and their relevant pathways has contributed to the development of miRNA-based therapeutics for cancer. In this review, we summarize the latest and most significant findings on FOXM1 and miRNA involvement in cancer development and describe the role/roles of miRNA/FOXM1 signaling pathways in cancer initiation and progression. Targeting FOXM1 via regulation of miRNA expression may have a role in cancer treatment, although the miRNA delivery method remains the key challenge to the establishment of this novel therapy.
Keywords: FOXM1, miRNA, transcription, invasion, metastasis, therapy
INTRODUCTION
Forkhead box protein M1 (FOXM1), also known as HFH11, MPP2, WIN, and Trident, is a member of the Forkhead superfamily of transcription factors, which are evolutionary conserved in the winged helix/Forkhead DNA-binding domain [1–5]. The term Forkhead is derived from the two head-like structures in Forkhead-mutant Drosophila embryos, which have exhibited defects during formation of the anterior and posterior gut [6]. The human FOXM1 gene, which consists of 10 exons, is mapped to chromosome 12p13-3 [1]. Among these 10 exons, exon Va and exon VIIa are alternatively spliced, giving rise to three distinct FOXM1 variants: FOXM1a, FOXM1b, and FOXM1c. FOXM1a is considered to be a dominant-negative regulator of other FOXM1 isoforms, as it harbors both exon Va and exon VIIa, resulting in disruption of the transactivation domain by exon VIIa. However, both FOXM1b, which contains neither exon Va nor exon VIIa, and FOXM1c, which contains exon Va only, are transcriptionally active and can directly promote target gene expression in an isoform-specific manner [7].
Genome-wide gene expression profiling of cancers has independently and consistently identified FOXM1 as one of the genes whose expression is most commonly upregulated in human solid tumors, including liver, prostate, brain, breast, lung, colon, pancreatic, skin, cervical, ovarian, oral, hematological, and nervous system tumors [8–10]. Several reports indicated that FOXM1 can regulate many aspects of cancer biology, including cell proliferation, cell-cycle progression, cell differentiation, DNA damage repair, tissue hemostasis, angiogenesis, and apoptosis. These findings point to a principal role for FOXM1 in tumorigenesis. In addition, a recent study linked FOXM1 deregulation with cancer progression and metastasis [11].
MicroRNAs (miRNAs) are short noncoding RNAs 20–24 nucleotides in length that posttranscriptionally regulate gene expression [12]. They guide the binding of RNA-induced silencing complexes (RISCs) to regions of partial complementarity located mainly within 3' untranslated regions of target messenger RNAs (mRNAs), resulting in mRNA degradation and/or translational inhibition [13]. Importantly, miRNAs play critical roles in a broad range of biological processes, including proliferation, differentiation, apoptosis, and stress response, linking them with the pathogenesis of numerous human diseases, including cancer [14].
In this review, we provide a brief introduction of miRNAs and FOXM1 and describe their roles in cancer development and progression and the possibility of modulating miRNA and FOXM1 functions for therapeutic gain in cancer patients. Furthermore, we highlight the importance of the miRNA/FOXM1 signaling pathway in cancer development and progression, the targeting of which may be a preventive and therapeutic strategy for cancer.
FOXM1 IN TUMORIGENESIS AND PROGRESSION
Cell-Cycle Progression and Cell Proliferation
The most basic property of cancer cells is their infinite capacity for proliferation. Normal cells are guided by regulatory mechanisms to enter, advance through, and exit the cell cycle in an orderly manner. In contrast, cancer cells disregard these regulatory signals and operate independently [15]. The involvement of FOXM1 in tumorigenesis is largely related to its central role in cell-cycle progression and proliferation. FOXM1 is a key regulator of G1/S and G2/M transition and M-phase progression. Besides regulating Cdc25A expression at the G1/S checkpoint, FOXM1 controls the transcription of S-phase kinase-associated protein 2 (Skp2) and Cks1, which are specific subunits of the Skp1/Cullin 1/F-box complex essential for the ubiquitinylation and degradation of the cyclin-dependent kinase inhibitor proteins p21Cip1 and p27Kip1. Furthermore, FOXM1 expression is essential for transcription of the mitotic regulatory genes, such as Cdc25B, Aurora B kinase, survivin, and centromere proteins A and B, which are involved in G2/M transition, maintenance of proper chromosomal stability, and segregation of chromosomes during mitosis [16]. Accordingly, the majority of FOXM1-depleted cells experience delays in transition to (or from) G2 phase and severe mitotic abnormalities upon entry into mitosis. Frequently, FOXM1-deficient cells also have mitotic spindle defects, chromosome misalignment, mitotic spindle checkpoint dysfunction, and cytokinesis failure. As a result, the cells undergo centrosome amplification and even endoreduplication, eventually becoming aneuploid and polyploid [17, 18].
The contribution of FOXM1 to the proliferation of human non-small cell lung cancer is well defined. Researchers used an interferon-inducible Mx-Cre recombinase transgene to delete the mouse FOXM1 fl/fl-targeted allele before inducing formation of lung tumors with urethane. Mx-Cre FOXM1−/− mice exhibited diminished proliferation of lung tumor cells, causing a marked reduction in the number and size of lung adenomas. Transient transfection experiments with A549 lung adenocarcinoma cells demonstrated that depletion of FOXM1 expression by small interfering RNA caused diminished DNA replication and mitosis and reduced anchorage-independent growth of cell colonies on soft agar [19]. In a study of hepatocellular carcinoma (HCC), conditionally deleted FOXM1b-expressing mouse hepatocytes failed to proliferate and were highly resistant to HCC development in response to a diethylnitrosamine/phenobarbital liver tumor induction protocol. The mechanism of resistance to HCC proliferation was associated with nuclear accumulation of the cell-cycle inhibitor p27Kip1 and reduced expression of the Cdk1 activator Cdc25B phosphatase [20]. In another study, FOXM1 was overexpressed in malignant glioma cells, and FOXM1b was the predominant FOXM1 isoform expressed in human glioma cells but not in normal brain cells. Enforced FOXM1B expression caused SW1783 and Hs683 glioma cells, which do not form tumor xenografts, to regain tumorigenicity in nude mouse model systems. Inhibition of FOXM1 expression in U-87MG glioma cells suppressed their anchorage-independent growth in vitro and tumorigenicity in vivo. The mechanism of FOXM1 that contributed to glioma tumorigenicity was FOXM1's regulation of the expression of Skp2, which is known to promote degradation of p27Kip1 [21].
In colorectal tumor studies, investigators used either Rosa26-FOXM1b transgenic mice with ubiquitous expression of human FOXM1b complementary DNA or mice in which the FOXM1fl/fl-targeted allele was deleted from colonic epithelial cells using the gut-specific Villin-Cre recombinase transgene. After 12 weeks of treatment with azoxymethane and dextran sodium sulfate, Rosa26-FOXM1b transgenic mice had more and larger colorectal tumors than did wild-type Rosa26-FOXM1b+/+ mice. Likewise, the researchers observed much less development and growth of colorectal tumors in Villin-Cre FOXM1−/− mice than in FOXM1 fl/fl mice after treatment with azoxymethane and dextran sodium sulfate, which was associated with decreased expression of cyclin A2, cyclin B1, survivin, and T-cell factor 4 genes. Moreover, FOXM1-depleted colon cancer cell lines exhibited reduced DNA replication and anchorage-independent growth [22]. Similarly, increased expression of FOXM1b accelerated the development, proliferation, and growth of prostatic tumors in both TRAMP and LADY double transgenic mice. Furthermore, development of prostate carcinomas in TRAMP/Rosa26-FOXM1 double transgenic mice required high levels of expression of FOXM1 protein to overcome sustained expression of the alternative reading frame tumor suppressor, a potent inhibitor of the transcriptional activity of FOXM1. Depletion of FOXM1 expression in the prostate cancer cell lines PC-3, LNCaP, and DU-145 by small interfering RNA transfection greatly reduced their proliferation and anchorage-independent growth on soft agar. This phenotype was associated with increased nuclear expression of the cyclin-dependent kinase inhibitor p27Kip1 and diminished expression of the S-phase–promoting protein cyclin A2 and M-phase–promoting protein cyclin B1 [23].
Cellular Senescence and Apoptosis
Cellular senescence is a tumor-suppressing mechanism that arrests cell proliferation in potential tumors. Recent studies demonstrated that FOXM1 contributes to cell transformation by antagonizing cellular senescence. For example, FOXM1 depletion can trigger the onset of p53- and p16INK4A-independent senescence of gastric cancer cells by inducing p27Kip1 expression [24]. Furthermore, researchers have observed cellular senescence in mouse embryonic fibroblasts derived from FOXM1-deficient mice, which express markers such as irreversible cell-cycle arrest, increased expression of the cellular senescence regulator p19Arf, and increased senescence-associated β-galactosidase activity [16]. In contrast, FOXM1 overexpression can suppress oxidative stress-induced cellular senescence and accumulation of senescence markers such as p53, p21Cip1, and p19Arf.
Cancer cells can survive and expand not only via proliferation but also by resisting cell death signals [25]. Apoptosis, the most common form of cell elimination, is a strictly regulated stepwise process, and a variety of signals can trigger it. However, cancer cells have developed multiple mechanisms to avoid cell destruction. Investigators have shown that proteasome inhibitors suppress FOXM1 expression and simultaneously induce apoptosis in human cancer cell lines [26]. In another study, FOXM1 knockdown sensitized human cancer cells to apoptosis induced by proteasome inhibitors such as MG132, bortezomib, and thiostrepton, but it did not affect autophagy levels following treatment with these drugs [27]. Also, researchers observed that downregulation of FOXM1 expression sensitized human cancer cells of different origins to DNA damage-induced apoptosis, which suppressed activation of proapoptotic c-Jun N-terminal kinase and positively regulated expression of antiapoptotic Bcl-2. This suggested that c-Jun N-terminal kinase activation and Bcl-2 downregulation mediate the sensitivity of cancer cells to DNA-damaging agent-induced apoptosis after targeting of FOXM1 [28].
Angiogenesis, Invasion, Metastasis, and Epithelial-Mesenchymal Transition
Tumor growth and progression are highly dependent on adequate blood supply for acquiring nutrients and oxygen as well as disposing of metabolic waste and carbon dioxide. Angiogenesis fulfills these requirements. After completion of embryonic development, angiogenesis is downregulated. However, at the onset of tumor development, an angiogenic switch is turned on and remains activated to facilitate tumor growth and progression [15]. FOXM1b is overexpressed in human glioblastoma cells, and forced FOXM1b expression in anaplastic astrocytoma cells leads to the formation of highly angiogenic glioblastomas in nude mice. Similarly, promotion of gastric tumorigenesis by FOXM1b is directly and highly correlated with transactivation of vascular endothelial growth factor (VEGF) and increased angiogenesis. FOXM1 depletion is correlated with reduced expression and activity of VEGF, leading to decreased angiogenesis, whereas FOXM1 overexpression has resulted in transactivation of VEGF and increased angiogenesis in vitro. Suppression of FOXM1 expression has compromised the tumorigenicity of glioma and gastric cancer cells in orthotopic mouse models and led to decreased tumor vascularization. Researchers identified that VEGF is a direct transcriptional target of FOXM1b [29, 30].
Metastasis is a complex multistep process involving local invasion, intravasation, extravasation, micrometastasis formation, and colonization [15]. During cancer cell invasion, degradation of the basement membrane and various components of the extracellular matrix is facilitated by a group of zinc-dependent endopeptidases, the matrix metalloproteinases (MMPs). In particular, MMP-2 and MMP-9 are consistently implicated to have roles in tumor invasion, metastasis, and angiogenesis [31, 32]. Increasing evidences suggest that FOXM1 actively participates in the metastatic process. Specifically, FOXM1 contributes to glioma progression by directly targeting MMP-2 gene transcription and thus enhancing tumor cell invasion [33]. In pancreatic cancer cells, downregulation of FOXM1 expression has reduced MMP-2, MMP-9, and VEGF expression, resulting in inhibition of migration, invasion, and angiogenesis [34]. Also, overexpression of FOXM1 in SUM102 and SKBR3 breast cancer cells, which express FOXM1 at low levels, has resulted in increased migration and invasion. Moreover, downregulation of FOXM1 expression has inhibited the expression of many factors involved in degradation of the extracellular matrix and angiogenesis, such as plasminogen activator, urokinase plasminogen activator, urokinase receptor, MMP-2, MMP-9, and VEGF, as well as the activity of MMP-9 and VEGF [35].
Epithelial-mesenchymal transition (EMT), a process by which epithelial cells acquire mesenchymal characteristics, leading to increased migratory and invasive potential of cancer cells, has a critical role in metastasis [15]. Overexpression of FOXM1 may lead to acquisition of the EMT phenotype in pancreatic cancer cells via activation of mesenchymal cell markers such as ZEB1, ZEB2, Snail2, E-cadherin, and vimentin [36]. FOXM1 also promotes EMT, migration, invasion, and metastasis in pancreatic cancer cells via direct transcriptional activation of caveolin-1, a key structural protein in the caveolae that is strongly implicated to have a role in the development of cancer [37]. In HCC cells, FOXM1b activates the Akt/Snail1 pathway and stimulates expression of stathmin, lysyl oxidase, lysyl oxidase like-2, and several other genes involved in metastasis. Furthermore, an Arf-derived peptide that inhibits FOXM1b expression impedes metastasis of FOXM1b-expressing HCC cells [38]. From this report, we can conclude that FOXM1b is a potent activator of tumor metastasis and that Arf-mediated inhibition of FOXM1b may play an important role in suppression of metastasis.
MIRNAS REGULATE SIGNALING PATHWAYS OF CANCER CELLS
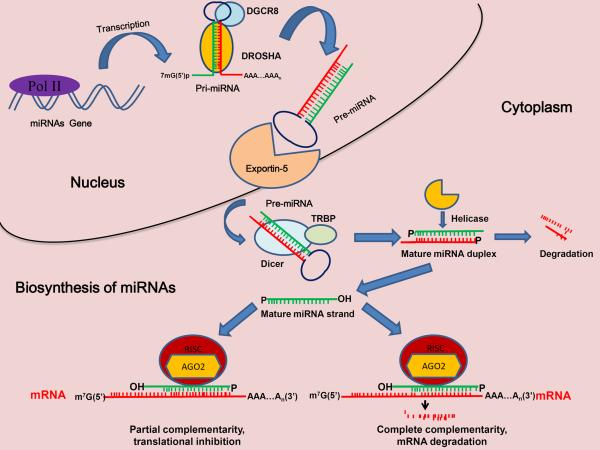
More than 1500 mature human miRNA sequences are listed in the miRNA database. These sequences play important roles in virtually all biological pathways in mammals and other multicellular organisms. The miRNA maturation process consists of several posttranscriptional processing steps (Fig. [1]) [12]. For most miRNAs, transcription is carried out by RNA polymerase II, yielding a 5'-capped, 3'-polyadenylated primary miRNA (pri-miRNA) that can be longer than 1000 bases and contain one or more hairpin stem-loop structures (miRNA clusters), with mature miRNAs locatedon one arm of each stem loop. Following transcription, the pri-miRNA is processed by the nuclear RNase III endonuclease DROSHA, yielding a 60- to 70-nucleotide double-stranded hairpin structure, precursor miRNA (pre-miRNA). In turn, pre-miRNAs are actively exported to the cytoplasm via the exportin-5 complex, where they are further processed by the RNase III endonuclease DICER. This enzyme cleaves at the opposite end from DROSHA near the terminal loop of the pre-miRNA hairpin, leading to the production of approximately 22-nucleotide double-stranded molecules (miRNA duplexes). The resulting duplex is then incorporated into the miRNA-induced silencing complex (RISC), where the miRNA duplex is unwound, facilitating the separation of mature antisense miRNA (guide strand) and transfer of it to argonaute proteins within the miRISC. Meanwhile, the passenger strand is released and, usually, rapidly degraded. Following complex assembly, the mature single-stranded miRNA guides the miRISC to target mRNAs. Target recognition and hybridization in animals occur primarily via incomplete base-pairing complementarity between the miRNA and the target mRNA, resulting in mismatches and bulges that, in turn, lead to target gene silencing, which can occur via translational repression and/or mRNA degradation. In the rare cases of nearly perfect miRNA complementarity with the target mRNA in animals, the mRNA can be endonucleolytically cleaved. Whether translational repression occurs at or after the initiation of translation is unclear. Conversely, degradation of the target mRNA, which is induced by miRNA without cleavage by argonaute, may lead to deadenylation, decapping, and exonucleolytic degradation of the mRNA [12, 13].
Fig. (1).
The process of miRNA maturation. In the nucleus, RNA polymerase II (Pol II) transcribes pri-miRNA and is cleaved by the DROSHA/DGCR8 complex to form premiRNA. The pre-miRNA is then transported to the cytoplasm by exportin-5, where it is further processed by the RNase III endonuclease DICER to form mature single-stranded miRNA. Next, the strand enters the RISC and suppresses the target mRNA via either translational repression (partial complementarity) or target degradation (complete complementarity).
Despite their subtle effects on individual targets, miRNAs are responsible for the modulation of multiple signaling pathways involved in cell growth, proliferation, differentiation, motility, and apoptosis [39]. In fact, a number of miRNAs are located in fragile regions of the human genome that are associated with cancer development, and dysregulated miRNAs play crucial roles in tumor initiation, progression, and metastasis and are often associated with diagnosis, prognosis, and response to therapy (Table 1).
Table 1.
MiRNAs and Their Targets in Cancer Cells
| MiRNA | Tumor type | Expression | Diagnostic/prognostic biomarker | Target | Mechanism | Reference |
|---|---|---|---|---|---|---|
| let-7 | Breast cancer, HCC, lung cancer, HNSCC, oral cancer, gastroenterop ancreatic neuroendocrine tumors | Reduced | Combined low expression of let-7d and miR-205 in HNSCC cells (poor prognosis); an let-7 miRNA-binding site (KRAS-LCS6) polymorphism in the KRAS 3'untranslated region correlated with reduced survival durations in oral cavity cancer cases (poor prognosis) | HMGA1 | Inhibits proliferation of cancer cells and stem cell self-renewal; promotes terminal differentiation at the larval to adult transition in both nematode stem cells and fly wing imaginal discs | [76–79] |
| lin-28/lin-28b | HCC, ovarian cancer | Increased | High expression of lin-28b correlates with risk of ovarian cancer progression and death (poor prognosis) | IGF-2 | Blocks maturation and affects the differentiation and proliferation of embryonic stem cells | [78, 80] |
| miR-9 | CRC, medulloblastoma | Reduced | Methylation of miR-9-1 correlates with lymph node metastasis in CRC cases (poor prognosis) | TrkC | Promotes cancer cell growth arrest and apoptosis while targeting the proliferative truncated TrkC isoform | [81, 82] |
| miR-21 | Breast cancer, bladder cancer, laryngeal carcinoma, tongue squamous cell carcinoma | Increased | High expression of miR-21 is an independent prognostic factor indicating poor survival of tongue squamous cell carcinoma (poor prognosis) | BTG2/TPM1 | Contributes to the malignant phenotype by maintaining a low level of BTG2 expressioninhi bits apoptosis partly via TPM1 silencing, affects the expression of its protein targets via translational inhibition | [83–87] |
| miR-34 family | CLL, acute myeloid leukemia, NPC | Reduced | Low expression of miR-34a correlates with chemotherapy-refractory disease, impaired DNA damage response, and apoptosis resistance in CLL cases (poor prognosis) | CREB/p53 | MiR-34b causes cell-cycle abnormalities and reduces anchorage-independent growth | [88–90] |
| miR-92 | CLL, CRC | Increased | Different levels of miR-92 expression discriminate CRC patients and control subjects (diagnostic) | pVHL | Aberrant regulation of pVHL levels by miR-92-1 promotes the HIF/VEGF axis | [91, 92] |
| miR-661 | Breast cancer | Reduced | -- | MTA1 | Inhibits the motility, invasiveness, anchorage-independent growth, and tumorigenicity of invasive cancer cells | [93] |
| miR-15a/miR-16 | Multiple myeloma, NPC | Reduced | -- | AKT3/BRCA1/MAP kinases/MAP3K IP3/RPS6 | Regulates proliferation and growth of cancer cells | [94, 95] |
| miR-101 | Bladder transitional cell carcinoma, HCC | Reduced | -- | EZH2/FOS | Inhibits cell proliferation, colony formation, invasion, and migration | [96, 97] |
| miR-129 | Bladder cancer, CRC | Reduced | Low expression of miR-129 correlates with poor outcome of bladder cancer (poor prognosis) | GALNT1/SOX4 | Inhibits growth and induces cell death | [81, 83] |
| miR-122 | HCC | Reduced | Low expression of miR-122 segregates with specific gene expression profiles linked with HCC progression (poor prognosis) | ADAM17 | Reduces migration, invasion, anchorage-independent growth, tumorigenesis, angiogenesis, and intrahepatic metastasis | [98, 99] |
| miR-205 | HNSCC, prostate cancer | Reduced | Low expression of miR-205 correlates with locoregional recurrence independently of disease severity at diagnosis and treatment of HNSCC (poor prognosis), combined low expression of let-7d and miR-205 in HNSCC cases (poor prognosis) | E2F1/E2F5/ERBB3 | Counteracts EMT and reduces cell migration/invasion | [76, 100] |
| miR-449a | Prostate cancer | Reduced | -- | HDAC1 | Induces cell-cycle arrest, apoptosis, and a senescent-like phenotype | [101] |
| miR-141 | Clear cell renal cell cancer | Reduced | Combination of upregulated miR-155 and downregulated miR-141 expression results in 97% overall correct classification of matched malignant and nonmalignant clear cell renal tissue samples (diagnostic) | -- | -- | [102] |
| miR-451 | Gastric cancer | Reduced | Low levels of miR-451 expression in gastric cancer cases (poor prognosis) | MIF | Reduces cell proliferation and increases sensitivity to radiotherapy | [103] |
| miR-206 | Rhabdomyos arcoma | Reduced | -- | MET | Promotes myogenic differentiation and blocks tumor growth by switching the global mRNA expression profile to one that resembles that of mature muscle | [104] |
| miR-155 | Breast cancer, Hodgkin lymphoma, NPC, pancreatic cancer, clear cell renal cell cancer | Increased | Combination of upregulated miR-155 and downregulated miR-141 expression results in 97% overall correct classification of matched malignant and nonmalignant clear cell renal tissue samples (diagnostic), high expression of miR-155 correlates with poor survival of pancreatic cancer (poor prognosis) | AGTR1/FGF7/IKBKE/ZIC3/ZNF537 | -- | [84, 88, 102, 105, 106] |
| miR-196a/b | Breast cancer, mixed-lineage leukemia, esophageal adenocarcinoma | Increased | High expression of miR-196a correlates with high-grade dysplasia in Barrett esophagus cases (poor prognosis) | KRT5/S100A9/SPRR2C | MiR-196b increases proliferative capacity and survival and partially blocks differentiation | [84, 107, 108] |
| miR-29a/c | Breast cancer | Increased | Low expression of miR-29c in CLL cases (poor prognosis) | B7-H3/TTP | MiR-29a modulates expression of the immunoinhibitory molecule B7-H3;miR-29a suppresses the expression of TTP, leading to epithelial polarity and metastasis | [109–111] |
HNSCC, head and neck squamous cell carcinoma; CRC, colorectal cancer; CLL, chronic lymphocytic leukemia; HIF, hypoxia-inducible factor; TTP.
THE MIRNA/FOXM1 SIGNALING PATHWAY IN CANCER CELLS
Given the importance of miRNA and FOXM1 expression to cancer initiation and progression, modulation of miRNA and FOXM1 signaling pathways may play a critical role in cancer biology, and investigation of this modulation is urgently needed. Authors reported that expression of the miRNAhsa-miR-370 was decreased and that FOXM1 expression was directly downregulated by hsa-miR-370 in gastric cancer cell lines [40]. The presence of Helicobacter pylori and CagA inhibited hsa-miR-370 expression, which led to FOXM1 overexpression and cell proliferation. In a study of non-small cell lung cancer cases, miR-134 expression correlated with invasive potential and the EMT phenotype, and the researchers identified FOXM1, a potential metastasis promoter, as a direct functional target of miR-134, demonstrating that miR-134 inhibited EMT via FOXM1 [41]. Furthermore, investigators found that downregulation of miR-370 expression occurred frequently in leukemia cell lines and primary leukemic cells obtained from patients with de novo acute myeloid leukemia [42]. Ectopic expression of miR-370 in HL60 and K562 leukemia cells led to cell growth arrest and senescence. In contrast, downregulation of miR-370 expression enhanced the proliferation of these cells. Mechanistically, miR-370 may function as a tumor suppressor by targeting FOXM1. Thus, epigenetic silencing of miR-370 leads to derepression of FOXM1 expression and consequently contributes to acute myeloid leukemia development and progression. In a pancreatic cancer study, overexpression of FOXM1 led to acquisition of the EMT phenotype via activation of the mesenchymal cell markers ZEB1, ZEB2, Snail2, and vimentin, which is consistent with increased sphere-forming (pancreatospheres) capacity and cancer stem cell surface marker (CD44 and EpCAM) expression in pancreatic cancer cells [36]. Furthermore, overexpression of FOXM1 led to decreased expression of miRNAs (let-7a, let-7b, let-7c, miR-200b, and miR-200c). However, re-expression of miR-200b inhibited the expression of ZEB1, ZEB2, and vimentin as well as FOXM1 and induced the expression of E-cadherin, leading to reversal of the EMT phenotype. This demonstrated that FOXM1 overexpression is responsible for acquisition of the EMT and cancer stem cell phenotypes, which is mediated in part by regulation of miR-200b expression.
THE MIRNA/FOXM1 SIGNALING PATHWAY IN CANCER TREATMENT
As described above, the oncogenic transcription factor FOXM1 is overexpressed in the majority of human cancers and is implicated to have a role in cell migration, invasion, angiogenesis, and metastasis. This important role of FOXM1 related to cancer affirms its importance cancer therapy [43]. Recently reported data suggest that targeting FOXM1 in single-agent or combination therapy for cancer is promising[44]. After being extensively demonstrated to be deregulated in cancer cells, miRNAs are now being studied as targets and tools for cancer treatment. In particular, Reversal of miRNA expression to normal levels holds remarkable potential for therapeutic interventions [45, 46]. Of note, miRNAs delivered to tumors that specifically inhibit FOXM1 expression may be developed as feasible anticancer drugs.
However, the miRNA delivery method is the most troublesome issue in miRNA-based therapy. In 2007, Valadi et al. [47] found that miRNAs are contained in exosomes. They showed that miRNAs are not only intracellular gene expression regulators but also humoral factors, suggesting that miRNAs can act as tools for facilitation of cell-cell communication. Also, Johnstone and colleagues discovered exosomes in 1987 when they observed that sheep reticulocytes secrete exosomes to remove the transferring receptor during maturation [48]. Exosomes are homogenous in shape, range from 50 to 100 nm in diameter, and float on a sucrose gradient at a density of 1.13–1.19 g/ml [49]. They are formed by invagination of the plasma membrane into endosomes followed by inward vesiculation and vesicular release into the extracellular space [50, 51]. Exosomes are characterized by their enrichment in Alix, the tumor susceptibility gene TSG101, heat shock protein 70, and the tetraspanins CD63, CD81, and CD9 [51, 52]. Exosome secretion is observed in most cell types under both physiological and pathological conditions, especially tumor cells and hematopoietic cells, including reticulocytes [53–56], dendritic cells [57], B and T lymphocytes [58–60], platelets [61], mast cells [62, 63], macrophages [64], epithelial cells [65], fibroblasts [66], astrocytes, and neurons [67]. The extent of exosome secretion can be affected by not only Ca2+ levels [68–70] but also amino acid availability, as nutrient starvation, stress, rapamycin-based treatment, and hormone-based treatment induce autophagy, thus blocking exosome secretion [71].
Resolving the problems with miRNA delivery is essential for cancer treatment with these molecules”?}. Therefore, examination of the use of exosomal miRNAs for cancer treatment is natural. For example, the exosomal miRNA miR-223 from tumor-associated macrophages is transported to breast cancer cells, supporting that macrophages regulate the invasiveness of breast cancer cells via exosome-mediated delivery of oncogenic miRNAs [72]. In addition, exosomal miRNAs derived from HCC cells can be taken up by other cells and target transforming growth factor-β–activated kinase 1, resulting in enhancement of the growth of transformed recipient cells [73]. It The researchers in that studyalso showed that proliferation of a prostate carcinoma cell line was inhibited by the addition of an exosomal fraction isolated from a noncancerous prostate epithelial cell line. These observations suggest that transfer of exosomal miRNAs derived from noncancerous cells to cancerous cells inhibits proliferation of the latter. Indeed, expression of some tumor-suppressive miRNAs, such as miR-16, miR-205, and miR-143, was downregulated in prostate cancer cell lines at the cellular and extracellular levels. This supports the idea that secretory tumor-suppressive miRNAs should be transferred from noncancerous to cancerous cells in accordance with the miRNA concentration gradient.
To examine the contribution of secretory tumor-suppressive miRNAs to cancer initiation in depth, researchers generated miR-143–overexpressing HEK293 cells and observed an approximately 50% decrease in proliferation of a prostate cancer cell line that was induced by suppression of the miR-143 target gene KRAS after the addition to the cell cultures of an exosome derived from the HEK293 cells [74]. Importantly, the decrease in proliferation was reversed by transfection of an anti-miR-143 antibody into the prostate cancer cell line. These data indicated that the cell-proliferation inhibition was attributable to the secretory miR-143 in the exosomes of miR-143–overexpressing HEK293 cells. In addition, exogenously transduced miR-143 did not suppress the proliferation of noncancerous cells, suggesting that excessive amounts of tumor-suppressive miRNAs did not provide an additional growth-inhibitory effect on normal cells, in which expression of tumor-suppressive miRNAs was maintained at physiological levels. The investigators observed no overt side effects of dendritic cell-derived exosomes on exosome-mediated gene delivery in vivo.
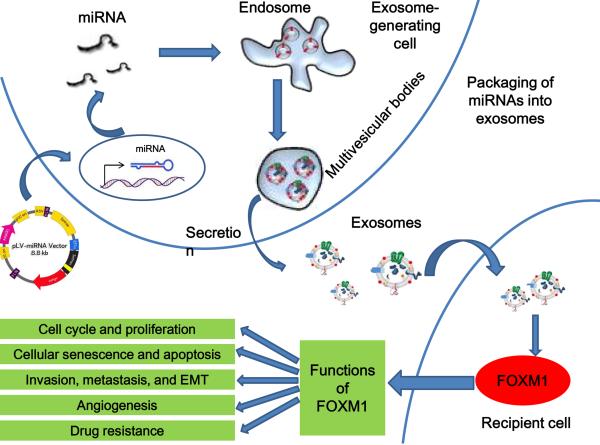
Taken together, the results of these studies demonstrate that miRNAs that specifically target FOXM1 can be loaded into exosomes and then delivered to cancer cells, leading to the development of miRNAs feasible anticancer drugs (Fig. [2]).
Fig. (2).
Schematic of miRNA transfer by exosomes. MiRNA overexpression is induced via transfection with an miRNA expression vector. MiRNAs are then selectively incorporated into the intraluminal vesicles of multivesicular bodies (MVBs). Exosomes containing miRNAs are derived from the MVBs and may be released into the extracellular environment via either fusion of MVBs with the cell surface or a budding pathway. Exosomes also may bind to the plasma membrane of a target cell. Recruited exosomes may either fuse directly with the plasma membrane or be endocytosed and then fuse with the delimiting membrane of an endocytic compartment. Both of these fusion pathways result in the delivery of exosomal miRNA to the cytoplasm of the target cell, where it may associate with and silence corresponding mRNA.
CONCLUSIONS AND PERSPECTIVES
Cancer seems to be the plague of the 21st century, as with each passing year, an increasing number of people die of it. Originally, FOXM1 caught the attention of investigators as a critical regulator of cell proliferation and cell-cycle progression [44]. Understanding of FOXM1's function and how it is regulated will undoubtedly provide new clues in identifying the mechanisms of tumorigenesis and cancer progression. All of the research findings described herein indicate that this oncogene may be the key to making significant progress in cancer treatment. For example, FOXM1 is a potential therapeutic target in patients with cancer, as its expression is upregulated in cases of numerous human malignancies, but it is not expressed in normal nondividing cells, thus indicating promising specificity for cancer cells in a targeted therapeutic approach [17]. The discovery of miRNAs and development of their study methods have raised an enormous number of questions concerning details regarding regulation of miRNA expression and posttranscriptional regulation of gene expression the roles of miRNAs in signaling pathways in cell proliferation, apoptosis, and invasion; and improvement of study methods. The most promising field of miRNA research would be that focusing on the delineation of tumorigenic pathways and implementation of cancer therapeutics [75]. The miRNA/FOXM1 signaling pathway may have a promising role in cancer initiation and progression, and miRNAs that specifically target FOXM1 may be investigated as anticancer drugs. However, we must make to identify the miRNAs that can effectively inhibit FOXM1 expression and investigate simple, efficient methods of delivering miRNAs to cancer cells.
Acknowledgments
Funding This work was supported in part by grants R01-CA129956, R01-CA148954, R01-CA152309, and R01-CA172233 from the National Institutes of Health (to K.X.).
REFERENCES
- [1].Korver W, Roose J, Heinen K, et al. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics. 1997;46:435–42. doi: 10.1006/geno.1997.5065. [DOI] [PubMed] [Google Scholar]
- [2].Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- [3].Kaestner KH, Lee KH, Schlondorff J, et al. Six members of the mouse forkhead gene family are developmentally regulated. Proc Natl Acad Sci U S A. 1993;90:7628–31. doi: 10.1073/pnas.90.16.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–20. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- [5].Clevidence DE, Overdier DG, Tao W, et al. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc Natl Acad Sci U S A. 1993;90:3948–52. doi: 10.1073/pnas.90.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weigel D, Jurgens G, Kuttner F, Seifert E, Jackle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–58. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- [7].Ye H, Kelly TF, Samadani U, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–41. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–50. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Uddin S, Ahmed M, Hussain A, et al. Genome-wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am J Pathol. 2011;178:537–47. doi: 10.1016/j.ajpath.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Okabe H, Satoh S, Kato T, et al. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129–37. [PubMed] [Google Scholar]
- [11].Halasi M, Gartel AL. FOX(M1) News--It Is Cancer. Mol Cancer Ther. 2013;12:245–54. doi: 10.1158/1535-7163.MCT-12-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- [13].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [16].Wang IC, Chen YJ, Hughes D, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- [18].Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–9. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- [19].Kim IM, Ackerson T, Ramakrishna S, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–61. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- [20].Kalinichenko VV, Major ML, Wang X, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–50. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu M, Dai B, Kang SH, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- [22].Yoshida Y, Wang IC, Yoder HM, Davidson NO, Costa RH. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–31. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- [23].Kalin TV, Wang IC, Ackerson TJ, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–20. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zeng J, Wang L, Li Q, et al. FoxM1 is up-regulated in gastric cancer and its inhibition leads to cellular senescence, partially dependent on p27 kip1. J Pathol. 2009;218:419–27. doi: 10.1002/path.2530. [DOI] [PubMed] [Google Scholar]
- [25].Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- [26].Gartel AL. A new target for proteasome inhibitors: FoxM1. Expert Opin Investig Drugs. 2010;19:235–42. doi: 10.1517/13543780903563364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pandit B, Gartel AL. FoxM1 knockdown sensitizes human cancer cells to proteasome inhibitor-induced apoptosis but not to autophagy. Cell Cycle. 2011;10:3269–73. doi: 10.4161/cc.10.19.17735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Halasi M, Gartel AL. Suppression of FOXM1 sensitizes human cancer cells to cell death induced by DNA-damage. PLoS One. 2012;7:e31761. doi: 10.1371/journal.pone.0031761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang Y, Zhang N, Dai B, et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68:8733–42. doi: 10.1158/0008-5472.CAN-08-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li Q, Zhang N, Jia Z, et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–9. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Overall CM, Dean RA. Degradomics: systems biology of the protease web. Pleiotropic roles of MMPs in cancer. Cancer Metastasis Rev. 2006;25:69–75. doi: 10.1007/s10555-006-7890-0. [DOI] [PubMed] [Google Scholar]
- [32].van Kempen LC, Coussens LM. MMP9 potentiates pulmonary metastasis formation. Cancer Cell. 2002;2:251–2. doi: 10.1016/s1535-6108(02)00157-5. [DOI] [PubMed] [Google Scholar]
- [33].Dai B, Kang SH, Gong W, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–9. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- [34].Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67:8293–300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- [35].Ahmad A, Wang Z, Kong D, et al. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat. 2010;122:337–46. doi: 10.1007/s10549-009-0572-1. [DOI] [PubMed] [Google Scholar]
- [36].Bao B, Wang Z, Ali S, et al. Over-expression of FoxM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J Cell Biochem. 2011;112:2296–306. doi: 10.1002/jcb.23150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [37].Huang C, Qiu Z, Wang L, et al. A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res. 2012;72:655–65. doi: 10.1158/0008-5472.CAN-11-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Park HJ, Gusarova G, Wang Z, et al. Deregulation of FoxM1b leads to tumour metastasis. EMBO Mol Med. 2011;3:21–34. doi: 10.1002/emmm.201000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- [40].Feng Y, Wang L, Zeng J, et al. Fork head box M1 is overexpressed in Helicobacter pylori-induced gastric carcinogenesis and is negatively regulated by hsa-miR-370. Mol Cancer Res. 2013 May 10; doi: 10.1158/1541-7786.MCR-13-0007. Epub. [DOI] [PubMed] [Google Scholar]
- [41].Li J, Wang Y, Luo J, et al. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells. FEBS Lett. 2012;586:3761–5. doi: 10.1016/j.febslet.2012.09.016. [DOI] [PubMed] [Google Scholar]
- [42].Zhang X, Zeng J, Zhou M, et al. The tumor suppressive role of miRNA-370 by targeting FoxM1 in acute myeloid leukemia. Mol Cancer. 2012;11:56. doi: 10.1186/1476-4598-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang Z, Ahmad A, Li Y, et al. Forkhead box M1 transcription factor: a novel target for cancer therapy. Cancer Treat Rev. 2010;36:151–6. doi: 10.1016/j.ctrv.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Halasi M, Gartel AL. Targeting FOXM1 in cancer. Biochem Pharmacol. 2013;85:644–52. doi: 10.1016/j.bcp.2012.10.013. [DOI] [PubMed] [Google Scholar]
- [45].Borralho PM, Kren BT, Castro RE, et al. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689–700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]
- [46].Borralho PM, Simoes AE, Gomes SE, et al. miR-143 overexpression impairs growth of human colon carcinoma xenografts in mice with induction of apoptosis and inhibition of proliferation. PLoS One. 2011;6:e23787. doi: 10.1371/journal.pone.0023787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- [48].Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–20. [PubMed] [Google Scholar]
- [49].Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- [50].Bedoui S, Prato S, Mintern J, et al. Characterization of an immediate splenic precursor of CD8+ dendritic cells capable of inducing antiviral T cell responses. J Immunol. 2009;182:4200–7. doi: 10.4049/jimmunol.0802286. [DOI] [PubMed] [Google Scholar]
- [51].Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- [52].Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- [53].Johnstone RM, Bianchini A, Teng K. Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood. 1989;74:1844–51. [PubMed] [Google Scholar]
- [54].Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–8. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Fader CM, Savina A, Sanchez D, Colombo MI. Exosome secretion and red cell maturation: Exploring molecular components involved in the docking and fusion of multivesicular bodies in K562 cells. Blood Cells Mol Dis. 2005;35:153–7. doi: 10.1016/j.bcmd.2005.07.002. [DOI] [PubMed] [Google Scholar]
- [56].Geminard C, de Gassart A, Vidal M. Reticulocyte maturation: mitoptosis and exosome release. Biocell. 2002;26:205–15. [PubMed] [Google Scholar]
- [57].Zitvogel L, Regnault A, Lozier A, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- [58].Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–72. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Clayton A, Turkes A, Navabi H, Mason MD, Tabi Z. Induction of heat shock proteins in B-cell exosomes. J Cell Sci. 2005;118:3631–8. doi: 10.1242/jcs.02494. [DOI] [PubMed] [Google Scholar]
- [60].Peters PJ, Geuze HJ, Van der Donk HA, et al. Molecules relevant for T cell-target cell interaction are present in cytolytic granules of human T lymphocytes. Eur J Immunol. 1989;19:1469–75. doi: 10.1002/eji.1830190819. [DOI] [PubMed] [Google Scholar]
- [61].Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–9. [PubMed] [Google Scholar]
- [62].Skokos D, Botros HG, Demeure C, et al. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J Immunol. 2003;170:3037–45. doi: 10.4049/jimmunol.170.6.3037. [DOI] [PubMed] [Google Scholar]
- [63].Raposo G, Tenza D, Mecheri S, et al. Accumulation of major histocompatibility complex class II molecules in mast cell secretory granules and their release upon degranulation. Mol Biol Cell. 1997;8:2631–45. doi: 10.1091/mbc.8.12.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110:3234–44. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].van Niel G, Raposo G, Candalh C, et al. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121:337–49. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- [66].Lespagnol A, Duflaut D, Beekman C, et al. Exosome secretion, including the DNA damage-induced p53-dependent secretory pathway, is severely compromised in TSAP6/Steap3-null mice. Cell Death Differ. 2008;15:1723–33. doi: 10.1038/cdd.2008.104. [DOI] [PubMed] [Google Scholar]
- [67].Faure J, Lachenal G, Court M, et al. Exosomes are released by cultured cortical neurones. Mol Cell Neurosci. 2006;31:642–8. doi: 10.1016/j.mcn.2005.12.003. [DOI] [PubMed] [Google Scholar]
- [68].Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–90. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- [69].Merendino AM, Bucchieri F, Campanella C, et al. Hsp60 is actively secreted by human tumor cells. PLoS One. 2010;5:e9247. doi: 10.1371/journal.pone.0009247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Fader CM, Sanchez D, Furlan M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–50. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- [71].Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–8. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- [72].Yang M, Chen J, Su F, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–48. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kosaka N, Iguchi H, Yoshioka Y, et al. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J Biol Chem. 2012;287:1397–405. doi: 10.1074/jbc.M111.288662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Papaconstantinou IG, Lykoudis PM, Gazouli M, et al. A review on the role of microRNA in biology, diagnosis, and treatment of pancreatic adenocarcinoma. Pancreas. 2012;41:671–7. doi: 10.1097/MPA.0b013e31823c9d21. [DOI] [PubMed] [Google Scholar]
- [76].Childs G, Fazzari M, Kung G, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–45. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Christensen BC, Moyer BJ, Avissar M, et al. A let-7 microRNA-binding site polymorphism in the KRAS 3' UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003–7. doi: 10.1093/carcin/bgp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–18. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Rahman MM, Qian ZR, Wang EL, et al. Frequent overexpression of HMGA1 and 2 in gastroenteropancreatic neuroendocrine tumours and its relationship to let-7 downregulation. Br J Cancer. 2009;100:501–10. doi: 10.1038/sj.bjc.6604883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lu L, Katsaros D, Shaverdashvili K, et al. Pluripotent factor lin-28 and its homologue lin-28b in epithelial ovarian cancer and their associations with disease outcomes and expression of let-7a and IGF-II. Eur J Cancer. 2009;45:2212–8. doi: 10.1016/j.ejca.2009.05.003. [DOI] [PubMed] [Google Scholar]
- [81].Bandres E, Agirre X, Bitarte N, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–43. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- [82].Ferretti E, De Smaele E, Po A, et al. MicroRNA profiling in human medulloblastoma. Int J Cancer. 2009;124:568–77. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- [83].Dyrskjot L, Ostenfeld MS, Bramsen JB, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–60. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- [84].Hui AB, Shi W, Boutros PC, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- [85].Li J, Huang H, Sun L, et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- [86].Liu M, Wu H, Liu T, et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009;19:828–37. doi: 10.1038/cr.2009.72. [DOI] [PubMed] [Google Scholar]
- [87].Yang Y, Chaerkady R, Beer MA, Mendell JT, Pandey A. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics. 2009;9:1374–84. doi: 10.1002/pmic.200800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Chen HC, Chen GH, Chen YH, et al. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer. 2009;100:1002–11. doi: 10.1038/sj.bjc.6604948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Pigazzi M, Manara E, Baron E, Basso G. miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 2009;69:2471–8. doi: 10.1158/0008-5472.CAN-08-3404. [DOI] [PubMed] [Google Scholar]
- [90].Zenz T, Mohr J, Eldering E, et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood. 2009;113:3801–8. doi: 10.1182/blood-2008-08-172254. [DOI] [PubMed] [Google Scholar]
- [91].Ghosh AK, Shanafelt TD, Cimmino A, et al. Aberrant regulation of pVHL levels by microRNA promotes the HIF/VEGF axis in CLL B cells. Blood. 2009;113:5568–74. doi: 10.1182/blood-2008-10-185686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–81. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- [93].Reddy SD, Pakala SB, Ohshiro K, Rayala SK, Kumar R. MicroRNA-661, a c/EBPalpha target, inhibits metastatic tumor antigen 1 and regulates its functions. Cancer Res. 2009;69:5639–42. doi: 10.1158/0008-5472.CAN-09-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Roccaro AM, Sacco A, Thompson B, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–80. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhu JY, Pfuhl T, Motsch N, et al. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–41. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Friedman JM, Liang G, Liu CC, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–9. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- [97].Li S, Fu H, Wang Y, et al. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194–202. doi: 10.1002/hep.22757. [DOI] [PubMed] [Google Scholar]
- [98].Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–36. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Tsai WC, Hsu PW, Lai TC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–82. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- [100].Gandellini P, Folini M, Longoni N, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–95. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- [101].Noonan EJ, Place RF, Pookot D, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–24. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- [102].Jung M, Mollenkopf HJ, Grimm C, et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13:3918–28. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Bandres E, Bitarte N, Arias F, et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281–90. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- [104].Taulli R, Bersani F, Foglizzo V, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–78. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Gibcus JH, Tan LP, Harms G, et al. Hodgkin lymphoma cell lines are characterized by a specific miRNA expression profile. Neoplasia. 2009;11:167–76. doi: 10.1593/neo.08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Greither T, Grochola LF, Udelnow A, et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- [107].Maru DM, Singh RR, Hannah C, et al. MicroRNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–8. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Popovic R, Riesbeck LE, Velu CS, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–22. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–5. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Stamatopoulos B, Meuleman N, Haibe-Kains B, et al. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113:5237–45. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- [111].Xu H, Cheung IY, Guo HF, Cheung NK. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res. 2009;69:6275–81. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]