Abstract
Per-Arnt-Sim (PAS) kinase (PASK, PASKIN, and PSK) is a member of the group of nutrient sensing protein kinases. These protein kinases sense the energy or nutrient status of the cell and regulate cellular metabolism appropriately. PAS kinase responds to glucose availability and regulates glucose homeostasis in yeast, mice, and man. Despite this pivotal role, the molecular mechanisms of PAS kinase regulation and function are largely unknown. This review focuses on what is known about PAS kinase, including its conservation from yeast to man, identified substrates, associated phenotypes and role in metabolic disease.
Keywords: PAS kinase, diabetes, glucose metabolism, signal transduction, obesity, nutrient sensing
Introduction
A cell’s ability to accurately coordinate its metabolism with nutrient availability is essential for proper health. Metabolic diseases are becoming a pandemic with more than one in 10 adults in the world being obese (World Health Organization), often leading to further disease such as diabetes, heart disease and cancer [National Cancer Institute, (1–3)]. Glucose metabolism is central to these metabolic diseases (4) making it crucial to understand the cellular mechanisms behind glucose allocation. Protein kinases are a key cellular mechanism for proper glucose allocation by enabling the simultaneous control of multiple proteins to direct glucose away from some pathways while stimulating others.
An individual cell may express hundreds of different protein kinases. It is estimated that there are 122 protein kinase homologs in yeast (5), 540 in mice (6), and 518 in humans (7), which phosphorylate over 30% of the proteome (8). Thus, it is not surprising that aberrant kinase function is associated with several human diseases and that protein kinases are one of the top drug targets for treating disease (9).
Per-Arnt-Sim (PAS) kinase (PASK, PASKIN, and PSK) has recently been shown to play pivotal roles in glucose homeostasis in yeast, mice, and man. This review will discuss what is known about PAS kinase activation and function, including its conservation from yeast to man, identified substrates, associated phenotypes and role in metabolic disease.
Conservation and Functional Domains of PAS Kinase
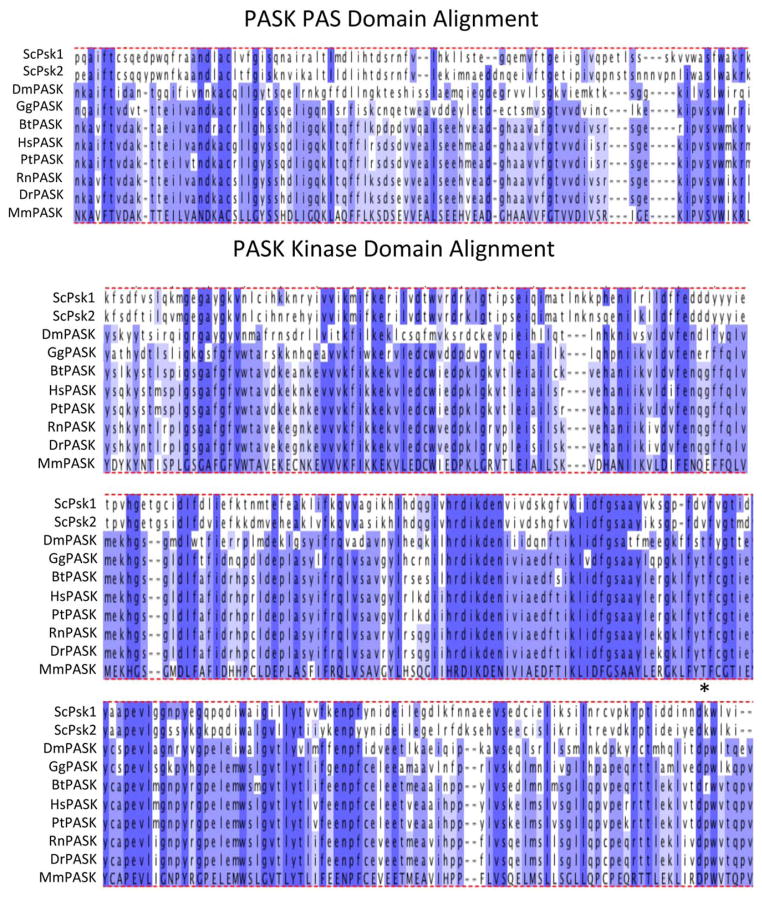
Nutrient sensing protein kinases regulate multiple targets in response to nutrient status, enabling a single enzyme to facilitate the appropriate cellular response. PAS kinase is a member of the group of nutrient sensing protein kinases and contains both a sensory PAS domain and a serine/threonine catalytic kinase domain (10,11). Both the PAS and kinase domains of PAS kinase are highly conserved from yeast to man (10,11), displaying significant amino acid conservation among organisms (Fig. 1). Saccharomyces cerevisiae is the only organism in which two orthologs of PAS kinase have been identified, namely Psk1 and Psk2 (10). Duplications of genes are common in yeast due to a whole genome duplication that occurred in an early ancestor (12–16). Most duplicate genes were eventually lost, while a few acquired accessory functions that allowed for their selection and maintenance, suggesting differential functions for the Psk1 and Psk2 proteins. Blast alignment of the yeast Psk1 protein with human PASK (hPASK) indicates 27% identity and 56% similarity for the PAS domain and 38% identity and 56% similarity for the kinase domain, arguing the evolutionary importance of these two domains. As the only reported mammalian protein to contain both a sensory PAS and protein kinase domain, it is no surprise that PAS kinase plays a key role in metabolic regulation in response to nutrient status.
FIG 1.
Alignment of the PAS kinase PAS domain and kinase domain amino acid sequences from various model organisms. The amino acid sequence from S. cerevisiae Psk1 (ScPsk1), S. cerevisiae Psk2 (ScPsk2), D. melanogaster (DmPASK), G. gallus (GgPASK), B. taurus (BtPASK), H. sapiens (HsPASK), P. troglodytes (PtPASK), R. norvegicus (RtPASK), D. rerio (DrPASK), and M. musculus (MmPASK) were obtained from GenBank and aligned using the Clustal Omega and Jalview software (49,50). The semiconserved phosphorylation loop threonine is indicated by an asterisk. No discernable PASK homologs were found for the model organisms A. thaliana, O. sativa, C. elegans, or A. mellifera. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The mechanism by which PAS kinase activity is regulated is unknown, but it is likely through its N-terminal PAS domain. PAS domains often play important roles in mediating protein-protein interactions, signal transfer, and subcellular localization by regulating an attached functional domain (17). They are known to react to a variety of intracellular stimuli including light, oxygen, redox state, or metabolites and can bind small ligands to trigger appropriate downstream responses (18). As expected for a regulatory domain, removal of the hPASK PAS domain increases catalytic activity (10) while addition of purified PAS domain inhibits the activity in trans (10,19). The NMR structure of the hPASK PAS domain is similar to the oxygen sensor FixL (11), which is able to sense oxygen through a heme ligand. Amezcua et al. screened over 750 organic compounds and found that the hPASK PAS domain selectively bound nine related but nonbiologically relevant small molecules with high affinity within its hydrophobic core (19). They provide evidence that hPASK PAS domain binds directly to the kinase domain and that ligand binding disrupts this interaction. Together, these results suggest small organic molecules bind to the inhibitory PAS domain, releasing it from the kinase domain. However, the exact biologically relevant ligand is yet to be determined.
The C-terminal end of PAS kinase contains a catalytic serine/threonine kinase domain that belongs to the CAMK family based on both amino acid sequence and protein structure (20). Most protein kinases require phosphorylation of at least one amino acid within the activation loop of their kinase domain to be activated (21). hPASK contains an activation loop threonine (T1116) that is not conserved in yeast (see Fig. 1). In addition, biochemical assays and crystallographic evidence indicate that activation loop phosphorylation is not necessary for hPASK activation (20). Together this data led to an investigation of the structural features of PAS kinase that enable activation in the absence of activation loop phosphorylation (20). The kinase domain of hPASK adopts the classical two-lobe structure typical of eukaryotic protein kinases but contains a unique additional β-hairpin replacing part of the αC helix. Other kinases that do not require activation loop phosphorylation typically have a negatively charged or nonpolar residue in their activation loop, whereas hPASK has a phosphorylatable threonine residue. Although not necessary for activation, hPASK auto-phosphorylation within the activation loop has been shown to increase catalytic activity (10). Hence, activation loop phosphorylation may regulate activity in certain cellular contexts or in response to particular substrates.
These biochemical and structural investigations into the role of the PAS and kinase domains have led to the current model for PAS kinase regulation summarized in Fig. 2. The PAS domain binds to the kinase domain and inhibits catalytic activity. Under activating conditions, a small metabolite binds to the PAS domain causing a conformational change, releasing PAS domain binding and activating the kinase domain. In addition, autophosphorylation or transphosphorylation of PAS kinase could lead to stable activation by disrupting the PAS and kinase domain interaction.
FIG 2.
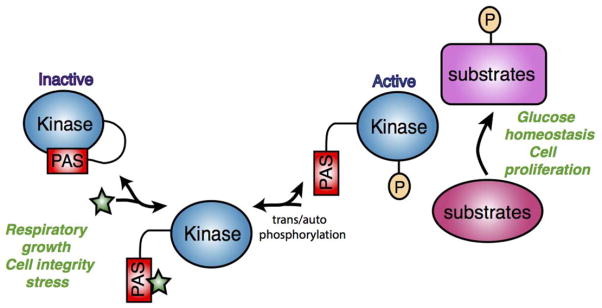
A current model for PAS kinase regulation and function. The PAS domain binds to and inhibits the kinase domain. Under activating growth conditions, such as growth that stimulates respiration or cell integrity stress, a small metabolite (indicated by the star) may bind the PAS domain, releasing it from the kinase domain. Auto or transphosphorylation may also activate PAS kinase or stabilize transient metabolite activation. PAS kinase then phosphorylates downstream targets to regulate glucose metabolism as well as cell proliferation. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Activation and Regulation of PAS Kinase
While the underlying molecular mechanisms behind PAS kinase regulation are yet to be determined, growth conditions that activate both yeast and mammalian PAS kinase have been reported (Table 1). In yeast, PAS kinase is activated by two different pathways, the cell integrity stress pathway or the glucose repression pathway (22). Although the precise mechanisms of activation by the cell integrity pathway are unknown, both Psk1 and Psk2 are activated by cell wall or membrane stress as well as overexpression of the cell integrity sensor-transducer WSC1. In contrast, only one of the two PAS kinase homologs, Psk1, is activated by the glucose repression pathway. The yeast homolog of (AMP)-activated protein kinase (AMPK), otherwise known as SNF1, is the master commander of the glucose repression pathway and controls the shift from fermentation to respiration when glucose levels are low. SNF1 is necessary and sufficient for Psk1 activation by nonglucose carbon sources that either require or favor respiration. This activation is both transcriptional as well as post-transcriptional. The second yeast homolog, Psk2, is transcriptionally downregulated and thus inactivated by carbon sources that stimulate respiration, suggesting differential roles for Psk1 and Psk2.
TABLE 1.
Cellular conditions known to activate PAS kinase in yeast and mammalian cells
| Organism (cells) | Activation pathway | Cellular growth conditions | Genetic modifiers | Study |
|---|---|---|---|---|
| S. cerevisiae | Cell integrity stress | CW, CPZ, SDS, high temp (37 °C) | WSC1OE | (22) |
| Glucose repression | Nonglucose carbon sources |
reg1 snf1 |
(22) | |
|
| ||||
| M. musculus (Min-6) | High glucose | High glucose | (23) | |
|
| ||||
| hPASK In vitro | Phospholipids | In vitro assay | (27) | |
Calcofluor white (CW) elicits cell wall stress, chlorpromazine (CPZ) elicits cell membrane stress while sodium dodecyl sulfate (SDS) elicits a more general cell stress. Nonglucose carbon sources tested include ethanol, glycerol, galactose, and raffinose. Growth on these nonglucose carbon sources as sole carbon source either requires or favors respiration in yeast, whereas high glucose favors high respiration rates in Min-6 cells. OE stands for overexpression.
Similar to yeast PAS kinase, hPASK is activated by conditions that stimulate high respiration in Min-6 pancreatic β-cells suggesting a conserved pathway of activation. In contrast to yeast, high glucose levels trigger high respiration rates in these cells, with an increase in both hPASK mRNA and protein (23). Further support is provided in a recent study by Hurtado-Carneiro et al., in which PASK is activated by glucose and glucagon-like peptide-1 (GLP-1), a peptide whose levels increase after a meal to signal nutrient availability to the central nervous system (24). Conversely, hPASK decreased in pancreatic islets of type 2 diabetics where high glucose may be expected (25). These differences could be due to differing genetic backgrounds including the low level of hPASK mRNA in normal tissue verses cell lines (26) or altered hPASK levels in a disease state.
In addition to activation by growth conditions that stimulate respiration, mammalian PASK has been shown to be activated through the binding of different phospholipids (27). Monophosphorylated phosphatidylinositols proved to be the strongest binding phospholipids and caused the highest auto-phosphorylation of PASK. In contrast, diphosphorylated and triphosphorylated phosphatidylinositols inhibited PASK auto-phosphorylation. Also important to note is that rather than binding to the sensory PAS domain, monophosphorylated phosphatidlyinositols bind to the kinase domain. These results suggest a multi-ligand regulation of PASK activity through the interaction of phospholipids with the kinase domain and an unidentified metabolite with the PAS domain.
PAS Kinase Phenotypes and Substrates
Since the discovery of PAS kinase in 2001 (10,11) studies in humans, mice, and yeast have been performed to better understand its regulation and function. Both phenotypic analysis (summarized in Table 2) and substrate discovery (summarized in Table 3) have been used to elucidate its role and cellular importance, however, the underlying molecular mechanisms behind its function are largely unknown.
TABLE 2.
Phenotypes associated with PAS kinase in yeast, mice, and man
| Organism | Genotype | Growth conditions | Phenotypes | Study |
|---|---|---|---|---|
| S. cerevisiae | psk1− psk2− | Galactose, 39 °C Glucose (log and SP) |
Growth inhibition Glycogen hyperaccumulation Trehalose hyperaccumulation |
(37) |
| psk2− | Galactose, 39 °C Glucose (log and SP) |
Limited growth Intermediate glycogen hyperaccumulation Intermediate trehalose hyperaccumulation |
(37) | |
| psk1− | Glucose (log and SP) | Intermediate glycogen hyperaccumulation Intermediate trehalose hyperaccumulation |
(37) | |
| PSK2OEstm1− | Glucose, 37 °C | Rescues growth of stm1− mutant | (37) | |
| psk2−rom2− | Glucose, 37 °C | Exacerbates growth of rom2ts mutant | (22) | |
| PSK2OEtor2ts | Glucose, 37 °C | Overexpression rescues tor2ts | (42) | |
| PSK2OEUGP1OE | Galactose, 37 °C | Overexpression rescues UGP1OE toxicity | (39,48) | |
|
| ||||
| M. musculus | pask−/− | 16 H fasting Glucose (10 mM) Insulin (20 mM) |
Increased fasting plasma glucose and glucagon Impaired inhibition of glucagon secretion Decreased islet and whole pancreas insulin Increased glucagon secretion |
(25) |
| hPASKOE | Glucose (1 mM) | Inhibited glucagon release in TC1–9 islets | (25) | |
| pask−/− | Glucose (1 mM) | Constitutive glucagon release in TC1–9 islets | (25) | |
| pask−/− | Glucose (1mM) High fat diet (45%) |
Decreased insulin levels Glucose tolerant Insulin sensitive Less weight gain Hypermetabolic (higher O2 consumption, CO2 production and heat generation) Reduced liver triglyceride accumulation Increased ATP production |
(30) | |
| pask−/− | Acute hypoxia | Higher hypoxic ventilatory response | (31) | |
| hPASK(KD)OE | Decreased intracellular insulin in TC1–9 cells | (25) | ||
| hPASK G1117E | Hyperinsulin secretion in CD1 islets | (28) | ||
|
| ||||
| H. Sapiens | hPASK G1117E | MODY associated | (28) | |
Log is log/exponential phase, SP is stationary phase, OE is overexpression and MODY is Maturity-Onset Diabetes of the Young.
TABLE 3.
Summary of PAS kinase substrates reported from studies of yeast and hPASK
| Organism | Substrate | Evidence | Study |
|---|---|---|---|
| S. cerevisiae | Ugp1 UDP-glucose Pyrophosphorylase |
In vitro kinase assay with Psk2 PSK-dependent in vivo phosphorylation UGP1-S11A mutant mimics psk1−psk2− mutant |
(22,37,39) |
| Caf20 Cap Associated Factor |
In vitro kinase assay with Psk2 | (39) | |
| Tif11 (eIF1A) Translation Initiation Factor |
In vitro kinase assay with Psk2 | (39) | |
| Sro9 Suppressor of Rho3 |
In vitro kinase assay with Psk2 | (39) | |
| Gsy2 Glycogen Synthase |
In vitro kinase assay with Psk2 | (39) | |
|
| |||
| H. sapiens | Gsy Glycogen Synthase |
In vitro kinase assay with hPASK Copurification |
(38) |
| eEF1A1 Eukaryotic Translation Elongation Factor |
Yeast and mammalian two-hybrid Copurification In vitro kinase assay with hPASK Colocalization |
(40) | |
| S6 Ribosomal Protein S6 |
In vitro kinase assay with hPASK In vivo phosphorylation assay |
(27) | |
| Pdx-1 Pancreatic Duodenal Homeobox-1 |
In vitro kinase assay with hPASK | (29) | |
The protein abbreviation and description are given for each substrate.
In humans, a rare mutation in hPASK (hPASK G1117E) may be a genetic modifier of Maturity-Onset Diabetes of the Young (MODY) since it was found in one of eighteen patients with MODY (28). This mutation was found to cause increased basal insulin secretion from pancreatic cells when transfected into mouse islets. In addition, activated hPASK is involved in the regulation of glucose induced preproinsulin and pancreatic duodenum homeobox-1 (PDX1) gene expression, a key transcription factor involved in both pancreatic development and insulin gene expression (23,29). In human islets, overexpression of hPASK also causes an inhibition of glucagon secretion at various glucose concentrations (25). Glucagon is a hormone that is secreted by the pancreas to stimulate glucose secretion when blood glucose is low. Together these findings suggest that hPASK may play a key role in the pathophysiology of type 2 diabetes.
In addition to the rare hPASK G1117E mutation being associated with diabetes, several PASK-deficiency phenotypes have been associated with symptoms of metabolic syndrome and diabetes in mice (25,30). Initial studies indicate no abnormalities in development, growth, or reproductive functions in PASK knockout mice (26). However, when placed on a high fat diet, PASK knockout mice gain less weight, are hypermetabolic, and display reduced insulin and triglyceride levels when compared to their wildtype littermates (30). Furthermore, they are more insulin sensitive and glucose tolerant. Additional studies found increased glucose and glucagon levels following 16 H of fasting, as well as decreased insulin levels in knockout mice (25). Besides these phenotypes associated with diabetes, a recent study showed that female PASK-deficient mice have an increased ventilatory response to acute hypoxia treatment; however, they are unable to reach ventilatory acclimatization after chronic hypoxia exposure (31). A major regulator of glucose consumption is the availability of oxygen, with glucose consumption being higher in hypoxic tissues (32–34). This suggests a potential link between oxygen sensing and PASK, resulting in altered regulation of glucose metabolism.
In addition to knockout phenotypes, the identification of bona fide PAS kinase substrates is critical to understanding its cellular role as well as contribution to metabolic disease. Four mammalian PASK substrates have been reported, namely pancreatic duodenal homeobox-1 (Pdx-1), glycogen synthase (Gsy), eukaryotic translation elongation factor 1A1 (eEF1A1), and ribosomal protein S6 (S6).
The mammalian substrate, Pdx-1, was identified by da Silva et al. as a direct in vitro substrate for hPASK and there is evidence that hPASK also controls Pdx-1 expression (23,29). Pdx-1 is an essential transcription factor found in β-cells that plays a key role in regulating genes crucial for glucose sensing and insulin gene expression (35). Accordingly, mutations in Pdx-1 have been associated with the development of MODY (36).
Glycogen metabolism is an important process that is tightly regulated to maintain proper glucose and energy homeostasis. PAS kinase has been shown to negatively regulate Gsy, the enzyme responsible for glycogen synthesis, in both yeast (37) and mammals (38). Rutter et al. provided the first evidence by showing direct in vitro phosphorylation of yeast Gsy2, and decreased Gsy activity in vivo (37). Wilson et al. then demonstrated the in vitro phosphorylation of Gsy by mammalian PASK and provided in vivo evidence for its negative regulation (38). Finally, Gsy was identified in a large scale peptide microarray assay for hPASK substrates, along with other proteins central to glycogen metabolism and glycolysis (27). Together, these results provide compelling evidence for conserved PASK-dependent phosphorylation and regulation of Gsy.
The role of PAS kinase in glucose homeostasis is expanded in yeast through its well-characterized substrate Ugp1 (UDP-glucose pyrophosphorylase). Ugp1 is responsible for the production of UDP-glucose, which is the primary glucose donor for most glucose-dependent cellular reactions. In yeast, the two major storage forms of glucose are glycogen and treha-lose. Phenotypes associated with PAS kinase deficiency are centered on hyperaccumulation of each of these storage molecules. Surprisingly, PAS kinase-dependent phosphorylation does not affect the enzymatic activity of Ugp1 (39) but rather partitions UDP-glucose to be used towards structural components at the expense of glycogen storage (22). PAS kinase-deficient yeast accumulates excess glycogen in a phospho-Ugp1 dependent manner (37). In addition, they are unable to grow on nonglucose carbon sources at high temperatures (i.e., Galactose, 39 °C) due to a defect in cell wall structural components (37,39). To determine potential pathways that might be involved with the PAS kinase-deficient growth phenotype, a high-copy suppressor screen was performed (37). A number of different genetic suppressors were found including genes involved in glucose metabolism such as phosphoglucomutase (PGM1, PGM2) and the glucose derepression gene SIP1, as well as genes involved in translational control.
A role for PAS kinase in translational control is also suggested by both mammalian and yeast substrates. Yeast and mammalian two-hybrid screening as well as copurification studies identified translation elongation factor 1 alpha 1 (eEF1A1) as a binding partner of hPASK (40). hPASK phosphorylates eEF1A1, colocalizes in HeLa and sperm cells, and increases translation efficiency. Additionally, Schlafali et al. show ribosomal protein S6 as an in vitro and in vivo target of PAS kinase (27). Finally, several ribosomal proteins appeared to be phosphorylated by PAS kinase in a high throughput screen [website publication only (41)], further substantiating a role of PAS kinase in translational regulation.
S. cerevisiae has proved to be a valuable model organism in searching for PAS kinase substrates, providing additional support for the role of PAS kinase in translation. The Caf20, Tif11, and Sro9 proteins, all of which are involved in translational regulation, were shown to be phosphorylated by Psk2 in vitro (37). In addition, when overexpressed, Psk2 is able to rescue growth of a stm1 mutant, which encodes for translation initiation factor eIF4B (37). The abundance of evidence from both yeast and mammalian studies strongly supports a role for PAS kinase in the regulation of translation.
Besides glucose and translational regulation, recent studies have suggested the involvement of yeast PAS kinase in the Target of Rapamycin TOR2 pathway (42). Tor2 is a well-studied, essential nutrient responsive protein kinase that regulates growth and cell-cycle dependent polarization of the actin cytoskeleton. A tor2ts mutant can be rescued by overexpression of its downstream substrate Rho1, or Rho1 activating GDP/GTP exchange factors such as Rom2 (43). In addition, overexpression of yeast PAS kinase, either Psk1 or Psk2, is also able to suppress the tor2ts mutant (42). This suppression is dependent on the phosphorylation of Ugp1, which forms a complex with Rom2. Although the downstream pathways activated through this association are unknown, this suppression of the tor2ts mutant supports novel roles for PAS kinase in the regulation of cell proliferation. Previous studies indicate that activation of AMPK or mTOR signaling pathways was not dependent on PASK function in mammalian cells (30). In contrast, recent studies show that knockdown of PASK mRNA impairs AMPK and mTOR/S6K1 response to glucose in neuroblastoma cells (24), supporting an overlap of PASK and TOR function.
Although numerous PAS kinase substrates have been identified, the in vivo effects of phosphorylation of these proteins are largely unknown. In addition, these proteins are not likely to explain the pleiotropic effects seen in PASK-deficient mice, including reduced triglyceride accumulation and weight gain as well as increased metabolism and insulin sensitivity. In looking for a consensus sequence for PAS kinase phosphorylation targets, studies show that hPASK prefers basic residues at the −3 and −5 positions upstream of the serine or threonine phosphoacceptor in substrate peptides (20), and shows a strong preference for arginine at −3 position (20,27). The Ugp1 serine 11 phosphorylation site, which is the one bona fide in vivo phosphorylation target of yeast PAS Kinase, matches this consensus sequence found for hPASK, providing a basis for predicting in vivo phosphorylation site preferences 20).
Conclusions
Nutrient sensing kinases play a critical role in controlling glucose metabolism by simultaneously regulating interrelated metabolic pathways in response to glucose. Three nutrient sensing kinases, AMPK, TOR, and PASK play key roles in proper glucose regulation. AMPK responds to cellular energy levels and upregulates energy producing pathways while inhibiting energy consumption pathways [for a recent review see (44)]. TOR responds to amino acids and other nutrients to regulate growth and proliferation [for a recent review see (45)]. PAS kinase, the focus of this review, is regulated by glucose levels and is necessary for glucose homeostasis. Integration of the AMPK, PASK, and TOR pathways would link cellular energy status with glucose allocation and cell proliferation.
Recently, PAS kinase has been shown to integrate with the AMPK and TOR pathways in both yeast and mammalian cells. In yeast, AMPK (SNF1) is necessary and sufficient for Psk1 activation (22), and overexpression of either PSK1 or PSK2 rescues the tor2ts growth defect (42). In mammalian cells, PAS kinase may integrate with the mTOR pathway through phosphorylation of S6, a target of mTOR (27). The well-studied AMPK and mTOR pathways are primary drug targets for the treatment of diabetes and cancer. Metformin, an AMPK activator, has been the most commonly prescribed drug for the treatment of type 2 diabetes for almost 20 years and is currently in 60 clinical trials for treatment of a wide variety of cancers [for a recent review see (46)]. mTOR inhibitors have also been shown to be an effective treatment against a wide variety of cancers [for a recent review see (47)]. Since PAS kinase is regulated by AMPK and affects targets downstream of TOR in both yeast and mammalian cells, it may prove to be a valuable nonessential target for the treatment of diabetes and cancer. In support of PAS kinase as a therapeutic target, PAS Kinase is implicated in the development of MODY (28), and phenotypes seen in the PASK-deficient mice are directly related to development of obesity, diabetes, and cardiovascular disease (30).
Despite its obvious importance, the molecular mechanisms behind PAS kinase function are largely unknown, with few verified substrates reported. A better understanding of PAS kinase regulation and function will not only increase our understanding of conserved, basic pathways of metabolism, but may lead to novel therapeutic drug targets for the treatment of metabolic disease.
References
- 1.Sciacca L, Vigneri R, Tumminia A, Frasca F, Squatrito S, et al. Clinical and molecular mechanisms favoring cancer initiation and progression in diabetic patients. Nutr Metab Cardiovasc Dis. Sep;23(9):808–815. doi: 10.1016/j.numecd.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Palomer X, Salvado L, Barroso E, Vazquez-Carrera M. An overview of the crosstalk between inflammatory processes and metabolic dysregulation during diabetic cardiomyopathy. Int J Cardiol. Aug 6; doi: 10.1016/j.ijcard.2013.07.150. pii:S0167-5273(13)01361-2. [DOI] [PubMed] [Google Scholar]
- 3.Van de Voorde J, Pauwels B, Boydens C, Decaluwe K. Adipocytokines in relation to cardiovascular disease. Metabolism. Jul 15; doi: 10.1016/j.metabol.2013.06.004. pii:S0026-0495(13)00184-4. [DOI] [PubMed] [Google Scholar]
- 4.Gillies RJ, Robey I, Gatenby RA. Causes and consequences of increased glucose metabolism of cancers. J Nucl Med. 2008;49(Suppl 2):24S–42S. doi: 10.2967/jnumed.107.047258. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H, Klemic JF, Chang S, Bertone P, Casamayor A, et al. Analysis of yeast protein kinases using protein chips. Nat Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- 6.Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc Natl Acad Sci USA. 2004;101:11707–11712. doi: 10.1073/pnas.0306880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 8.Cohen P. The regulation of protein function by multisite phosphorylation–a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 9.Melnikova I, Golden J. Targeting protein kinases. Nat Rev Drug Discov. 2004;3:993–994. doi: 10.1038/nrd1600. [DOI] [PubMed] [Google Scholar]
- 10.Rutter J, Michnoff CH, Harper SM, Gardner KH, McKnight SL. PAS kinase: an evolutionarily conserved PAS domain-regulated serine/threonine kinase. Proc Natl Acad Sci USA. 2001;98:8991–8996. doi: 10.1073/pnas.161284798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofer T, Spielmann P, Stengel P, Stier B, Katschinski DM, et al. Mammalian PASKIN, a PAS-serine/threonine kinase related to bacterial oxygen sensors. Biochem Biophys Res Commun. 2001;288:757–764. doi: 10.1006/bbrc.2001.5840. [DOI] [PubMed] [Google Scholar]
- 12.Byrne KP, Wolfe KH. Consistent patterns of rate asymmetry and gene loss indicate widespread neofunctionalization of yeast genes after whole-genome duplication. Genetics. 2007;175:1341–1350. doi: 10.1534/genetics.106.066951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conant GC, Wolfe KH. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol. 2007;3:129. doi: 10.1038/msb4100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grassi L, Fusco D, Sellerio A, Cora D, Bassetti B, et al. Identity and divergence of protein domain architectures after the yeast whole-genome duplication event. Mol Bio Syst. 2010;6:2305–2315. doi: 10.1039/c003507f. [DOI] [PubMed] [Google Scholar]
- 15.Maclean CJ, Greig D. Reciprocal gene loss following experimental whole-genome duplication causes reproductive isolation in yeast. Evolution. 2011;65:932–945. doi: 10.1111/j.1558-5646.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- 16.Sugino RP, Innan H. Estimating the time to the whole-genome duplication and the duration of concerted evolution via gene conversion in yeast. Genetics. 2005;171:63–69. doi: 10.1534/genetics.105.043869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amezcua CA, Harper SM, Rutter J, Gardner KH. Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation. Structure. 2002;10:1349–1361. doi: 10.1016/s0969-2126(02)00857-2. [DOI] [PubMed] [Google Scholar]
- 20.Kikani CK, Antonysamy SA, Bonanno JB, Romero R, Zhang FF, et al. Structural bases of PAS domain-regulated kinase (PASK) activation in the absence of activation loop phosphorylation. J Biol Chem. 2010;285:41034–41043. doi: 10.1074/jbc.M110.157594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 22.Grose JH, Smith TL, Sabic H, Rutter J. Yeast PAS kinase coordinates glucose partitioning in response to metabolic and cell integrity signaling. EMBO J. 2007;26:4824–4830. doi: 10.1038/sj.emboj.7601914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.da Silva Xavier G, Rutter J, Rutter GA. Involvement of Per-Arnt-Sim (PAS) kinase in the stimulation of preproinsulin and pancreatic duodenum homeobox 1 gene expression by glucose. Proc Natl Acad Sci USA. 2004;101:8319–8324. doi: 10.1073/pnas.0307737101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurtado-Carneiro V, Roncero I, Blazquez E, Alvarez E, Sanz C. PAS kinase as a nutrient sensor in neuroblastoma and hypothalamic cells required for the normal expression and activity of other cellular nutrient and energy sensors. Mol Neurobiol. doi: 10.1007/s12035-013-8476-9. in press. [DOI] [PubMed] [Google Scholar]
- 25.da Silva Xavier G, Farhan H, Kim H, Caxaria S, Johnson P, et al. Per-arnt-sim (PAS) domain-containing protein kinase is downregulated in human islets in type 2 diabetes and regulates glucagon secretion. Diabetologia. 2011;54:819–827. doi: 10.1007/s00125-010-2010-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katschinski DM, Marti HH, Wagner KF, Shibata J, Eckhardt K, et al. Targeted disruption of the mouse PAS domain serine/threonine kinase PASKIN. Mol Cell Biol. 2003;23:6780–6789. doi: 10.1128/MCB.23.19.6780-6789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlafli P, Troger J, Eckhardt K, Borter E, Spielmann P, et al. Substrate preference and phosphatidylinositol monophosphate inhibition of the catalytic domain of the Per-Arnt-Sim domain kinase PASKIN. FEBS J. 2011;278:1757–1768. doi: 10.1111/j.1742-4658.2011.08100.x. [DOI] [PubMed] [Google Scholar]
- 28.Semplici F, Vaxillaire M, Fogarty S, Semache M, Bonnefond A, et al. Human mutation within Per-Arnt-Sim (PAS) domain-containing protein kinase (PASK) causes basal insulin hypersecretion. J Biol Chem. 2011;286:44005–44014. doi: 10.1074/jbc.M111.254995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An R, da Silva Xavier G, Hao HX, Semplici F, Rutter J, et al. Regulation by Per-Arnt-Sim (PAS) kinase of pancreatic duodenal homeobox-1 nuclear import in pancreatic beta-cells. Biochem Soc Trans. 2006;34(Pt 5):791–793. doi: 10.1042/BST0340791. [DOI] [PubMed] [Google Scholar]
- 30.Hao HX, Cardon CM, Swiatek W, Cooksey RC, Smith TL, et al. PAS kinase is required for normal cellular energy balance. Proc Natl Acad Sci USA. 2007;104:15466–15471. doi: 10.1073/pnas.0705407104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soliz J, Soulage C, Borter E, van Patot MT, Gassmann M. Ventilatory responses to acute and chronic hypoxia are altered in female but not male Paskin-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2008;295:R649–658. doi: 10.1152/ajpregu.00876.2007. [DOI] [PubMed] [Google Scholar]
- 32.Krebs HA. The Pasteur effect and the relations between respiration and fermentation. Essays Biochem. 1972;8:1–34. [PubMed] [Google Scholar]
- 33.Racker E, Wu R. Limiting factors in glycolysis of ascites tumour cells and the pasteur effect. Regl Cell Metab. 1958:205–229. [Google Scholar]
- 34.Wu R. Regulatory mechanisms in carbohydrate metabolism. V Limiting factors of glycolysis in HeLa cells. J Biol Chem. 1959;234:2806–2810. [PubMed] [Google Scholar]
- 35.Boucher MJ, Selander L, Carlsson L, Edlund H. Phosphorylation marks IPF1/PDX1 protein for degradation by glycogen synthase kinase 3-dependent mechanisms. J Biol Chem. 2006;281:6395–6403. doi: 10.1074/jbc.M511597200. [DOI] [PubMed] [Google Scholar]
- 36.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 37.Rutter J, Probst BL, McKnight SL. Coordinate regulation of sugar flux and translation by PAS kinase. Cell. 2002;111:17–28. doi: 10.1016/s0092-8674(02)00974-1. [DOI] [PubMed] [Google Scholar]
- 38.Wilson WA, Skurat AV, Probst B, de Paoli-Roach A, Roach PJ, et al. Control of mammalian glycogen synthase by PAS kinase. Proc Natl Acad Sci USA. 2005;102:16596–16601. doi: 10.1073/pnas.0508481102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith TL, Rutter J. Regulation of glucose partitioning by PAS kinase and Ugp1 phosphorylation. Mol Cell. 2007;26:491–499. doi: 10.1016/j.molcel.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 40.Eckhardt K, Troger J, Reissmann J, Katschinski DM, Wagner KF, et al. Male germ cell expression of the PAS domain kinase PASKIN and its novel target eukaryotic translation elongation factor eEF1A1. Cell Physiol Biochem. 2007;20:227–240. doi: 10.1159/000104169. [DOI] [PubMed] [Google Scholar]
- 41.Brandon L, Probst SX, Leeju Wu, Carolyn H, Michnoff, et al. Two Distinct High Throughput Screens of PAS Kinase Yield Convergent Insight to Enzyme Function. Available at: http://www.mcknightlab.com/Documents/publications/ons/PASK_HTS.pdf.
- 42.Cardon CM, Beck T, Hall MN, Rutter J. PAS kinase promotes cell survival and growth through activation of Rho1. Sci Signal. 2012;5:ra9. doi: 10.1126/scisignal.2002435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt A, Bickle M, Beck T, Hall MN. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 44.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinn BJ, Kitagawa H, Memmott RM, Gills JJ, Dennis PA. Repositioning metformin for cancer prevention and treatment. Trends Endocrinol Metab. Sep;24(9):469–80. doi: 10.1016/j.tem.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Khan KH, Yap TA, Yan L, Cunningham D. Targeting the PI3K-AKT-mTOR signaling network in cancer. Chin J Cancer. 2013;32:253–265. doi: 10.5732/cjc.013.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grose JH, Sundwall E, Rutter J. Regulation and function of yeast PAS kinase: a role in the maintenance of cellular integrity. Cell Cycle. 2009;8:1824–1832. doi: 10.4161/cc.8.12.8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]