Abstract
Multidrug membrane transporters (efflux pumps) can selectively extrude a variety of structurally and functionally diverse substrates (e.g., chemotoxics, antibiotics), leading to multidrug resistance (MDR) and ineffective treatment of a wide variety of diseases. In this study, we have designed and constructed fusion gene (egfp-mexB) of N-terminal mexB with C-terminal egfp, inserted it into a plasmid vector (pMMB67EH), and successfully expressed it in ΔMexB (MexB deletion) strain of Pseudomonas aeruginosa to create a new strain that expresses MexA-(EGFP-MexB)-OprM. We characterized the fusion gene using gel electrophoresis and DNA sequencing, and determined their expression in live cells by measuring the fluorescence of EGFP in single live cells using fluorescence microscopy. Efflux function of the new strain was studied by measuring its accumulation kinetics of ethidium bromide (EtBr, a pump substrate) using fluorescence spectroscopy, which was compared with the cells (WT, ΔMexM, ΔABM, and nalB1) with various expression levels of MexAB-OprM. The new strain shows 6-fold lower accumulation rates of EtBr (15 μM) than ΔABM, 4-fold lower than ΔMexB, but only 1.1-fold higher than WT. As EtBr concentration increases to 40 μM, the new strain has nearly the same accumulation rate of EtBr as ΔMexB, but 1.4-fold higher than WT. We observed the nearly same level of inhibitory effect of CCCP (carbonyl cyanide-m-chlorophenylhydrazone) on the efflux of EtBr by the new strain and WT. Antibiotic susceptibility study shows that the minimum inhibitory concentrations (MICs) of aztreonam (AZT) and chloramphenicol (CP) for the new strain are 6-fold or 3-fold lower than WT, respectively, and 2-fold higher than those of ΔMexB. Taken together, the results suggest that the fusion protein partially retains the efflux function of MexAB-OprM. Modeled structure of the fusion protein shows that the position and orientation of the N-terminal fused EGFP domain may either partially block the translocation pore or restrict the movement of the individual pump domains, which leads to partially restrict efflux activity.
Keywords: EGPF-fused membrane transporter, efflux function, fluorescence spectroscopy, MexAB-OprM, multidrug resistance, Pseudomonas aeruginosa
Introduction
Multidrug membrane transporters (efflux pumps) can selectively extrude a wide range of substrates, which leads to multidrug resistance (MDR).1-2 These transporters exist in both prokaryotes and eukaryotes.3-4 Pseudomonas (P.) aeruginosa (a ubiquitous gram-negative bacterium) is a major opportunistic human pathogen and the leading cause of nosocomial infections in cancer, transplant, burn, and cystic fibrosis patients.5-7 Its efflux pumps can selectively extrude a wide variety of structurally and functionally diverse substrates, causing MDR. MDR is the primary cause of ineffective therapeutic treatments toward the infections, and the use of high doses of therapeutic agents that leads to severe side effects.
P. aeruginosa possesses several multidrug membrane transporters, including MexAB-OprM, that belongs to the resistance-nodulation-cell division (RND) family.2, 7-10 The MexAB-OprM is the primary membrane transporter in WT of P. aeruginosa. This tripartite drug efflux pump consists of two inner membrane proteins (MexA and MexB) and one outer membrane protein (OprM).11-13 This efflux pump extrudes a wide spectrum of structurally and functionally unrelated antibiotics (e.g., AZT, CP, tetracycline, and gentamicin) using the drive-force generated by proton gradients across the cellular membrane.14-15 The interplay between the MexAB-OprM efflux system and the outer membrane barrier plays an important role in MDR. 8, 16-17 In general, studies suggest that the transmembrane protein (MexB) specifically recognizes and captures its substrates either from the lipid bilayer of inner membrane or from cytoplasmic space, then transports them to the extracellular medium through OprM which forms a channel in the outer membrane.18 The cooperation between MexB and OprM is coordinated by the periplasmic membrane protein MexA.19-22 However, despite extensive studies, molecular mechanisms of multidrug membrane transporters remain not yet fully understood.6, 23 Therefore, direct visualization and study of MexB and its interaction with the pump substrates in live cells can shed new light on the roles of MexB in efflux function of MexAB-OprM.
Fluorescent proteins (e.g., GFP) have been widely used as reporter genes and non-invasive fluorescence probes to study molecular mechanisms and functions of proteins in live cells.24-26 Fluorescent proteins fused with membrane transporters have been used to determine the locations and topologies of ABC transporters.27-28 Our previous studies have showed that fusion of BmrA (ABC transporter) with EGFP retains its efflux function.29-30 However, other studies have shown that proteins (membrane transporters) fused with GFP may alter their functions.31-32 Notably, the effects of fusion of EGFP with tripartite efflux pumps (e.g., MexAB-OprM) on their efflux function have not yet been reported. Thus, it is crucial to first determine whether the MexA-(EGFP-MexB)-OprM retains the efflux function of MexAB-OprM, before we use the fusion protein to study the efflux mechanisms of MexAB-OprM.
Current methods for study of multidrug membrane transporters in bacterial and mammalian cells include the measurement of their accumulation of radioactively labeled substrates (14C and 3H) and the fluorescence dyes (e.g., EtBr).29-30, 33-36 Fluorescent dyes (EtBr) emit weak fluorescence in aqueous solution (outside cells) and become strongly fluorescent in non-polar and hydrophobic environments, especially as they enter the cells and intercalate with DNA.37 Thus, time-dependent (time-course) fluorescence intensity of EtBr is particularly suitable for real-time monitoring of efflux kinetics of multidrug membrane transporters in live cells. 29-30, 33-36, 38 Minimum inhibitory concentrations (MICs) of antibiotics that are specific substrates of the transporters and show the specific susceptibility toward given strains of bacterial cells have also been used to study efflux function of multidrug membrane transporters.39
In this study, we fused C-terminus of egfp with N-terminus of mexB to construct egfp-mexB fusion gene, and expressed it in MexB deleted strain of P. aeruginosa (ΔMexB) to create a new strain, MexA-(EGFP-MexB)-OprM. We characterized the effects of fusion EGFP on efflux function of MexAB-OprM by studying real-time accumulation of EtBr inside the cells, inhibitory effects of the proton ionophore (CCCP) on intracellular EtBr accumulation, and MICs of two representative antibiotics (AZT and CP) toward the cells. We used model structural analysis to further determine the potential effects of fusion EGFP on the efflux function of MexAB-OprM.
Experimental Section
Reagents and Cell Strains
PCR polymerase (Agilent Technologies), plasmid isolation kit (Qiagen), DNA gel extraction kit (Qiagen), DNA ligation kit (Roche) which includes ligation enzyme (T4 DNA ligase) and ligation buffer, SalI/HindIII (NEB), CP (Calbiochem), IPTG (GBT), CCCP (≥ 97%, Sigma), carbenecillin (Sigma), AZT (Sigma), EtBr (Invitrogen), and E. coli DH5α (Invitrogen) were purchased and used as received. P. aeruginosa strains (WT, nalB1, ΔMexB, ΔABM) and plasmid (pMMB67EH) were provided by Hiroshi Yoneyama.40-41 All other reagents except indicated were purchased from Sigma and used as received. The nanopure deionized (DI) water (18 MΩ water, Barnstead) was used to prepare all solutions, including the L-broth (LB) medium (1% tryptone, 0.5% yeast extract and 0.5% NaCl in DI water, pH = 7.2).
Construction of egfp-mexB Fusion Gene and MexA-(EGFP-MexB)-OprM Strain
We fused N-terminal mexB gene with C-terminal egfp to prepare the egfp-mexB fusion gene by asymmetrical PCR amplification, and then inserted it into the vector (pMMB67EH) to prepare pMMB67EH-EGFP-MexB vector, as described in Figure S1 in online supplementary information.42 We first used one pair of egfp primers (egfp primer-1: 5′-CTTGTCGACAAGGGGATCCACCATGGTGAGCAAGG-3′ and egfp primer-2: 5′-CAATGAAAAACTTCGACATCTTGTACAGCTCGTCCATGC-3′), and EGFP plasmid as a template to amply egfp gene (Figure S1A-I). We then used only egfp primer-1 and the amplified double-strand (ds) egfp gene as the template to create the coding strand of egfp gene (single-strand, ss-egfp) (Figure S1A-II). Using the similar approaches, we used one pair of mexB primers (mexB primer-1: 5′-GCATGGACGAGCTGTACAAGATGTCGAAGTTTTTCATTG-3′, mexB primer-2: 5′-GATAAGCTTATCATTGCCCCTTTTCGAC-3′), and genomic DNA of P. aeruginosa as the template to generate and amplify the mexB gene (Figure S1B-I). We used mexB primer-2 and the amplified ds-mexB gene as the template to generate and amplify the template strand of mexB gene (ss-mexB) (Figure S1B-II), which has 20 bases that are complimentary to the ss-egfp at the 3′-end. By mixing the ss-egfp with ss-mexB, it creates 20 base-pair (bp) ds-DNA, that links the ss-egfp with ss-mexB, which is used as the template to generate and amplify egfp-mexB fusion gene using both egfp primer-1 and mexB primer-2 as primers (Figure S1AB-III). The PCR product of the egfp-mexB fusion gene was characterized by agarose gel electrophoresis (0.8%, E = 8-10 V/cm), and purified by the gel purification kit.
The purified PCR products of egfp-mexB gene and vector (pMMB67EH) were digested by a pair of restriction enzymes (SalI/HindIII), which were carried out by incubating the PCR products (10 ng) or the vector (5 ng) with two units of SalI/HindIII each in the presence of 10x digestion buffer (4 or 3 μL) in DI water (40 or 30 μL) at 37 °C for 3 h, respectively (Figure S1AB-IV). The digested products were characterized by the agarose gel electrophoresis and purified via the gel extraction kit. After purification, the digested and purified egfp-mexB gene and the vector (pMMB67EH) were ligated to produce pMMB67EH-EGFP-MexB (Figure S1C). The ligation was carried out by incubating digested and purified egfp-mexB fusion gene (20 nM) with digested and purified vector (5 nM) in the presence of one unit of T4 DNA ligase and 10× ligation buffer (2 μL) in DI water (20 μL) at 12°C overnight. The digestion, gel electrophoresis and ligation were carried out using the reported protocols.43
The pMMB67EH-EGFP-MexB was then transformed into DH5α competent cells (Invitrogen) using heat shock. Briefly, we mixed the pMMB67EH-EGFP-MexB (10 μL, 0.1 μg/μL) with the competent cell suspension (100 μL, 2×107 cells/mL), placed the mixture on ice for 30 min, followed by a heat shock at 42 °C for 90 s, then placed it back onto the ice for another 2 min. We then added the LB medium (1 mL) to the mixture, incubated it in a shaker (LabLine Orbit-Environ, 37°C, 200 rpm) for 1 h, and centrifuged the solution at 5000 rpm for 5 min. We removed the supernatant and suspended the pellets (transformed cells) in the fresh medium, inoculated them onto LB agar plates (0.5% yeast extract, 0.5% NaCl, 1% tryptone,1.5% agar and 100 μg/ml ampicillin), and incubated the plates at 37°C for 24 h. Positive ligations were verified by restriction enzyme digestion (SalI/HindIII), PCR amplification using the primers (egfp primer-1 and mexB primer-2), and DNA sequencing (Applied Biosystem, 3730xl DNA Analyzer).
Plasmids with the correct sequence were finally transformed into MexB deletion strain (ΔMexB) using electroporation.44-45 Briefly, the cells (ΔMexB) were cultured in the LB medium in the shaker (200 rpm, 37 °C) until they reach the early log phase (optical density at 600 nm, OD600nm = 0.3-0.5). The cells were then harvested using centrifugation (7000 g, 10 min, 4 °C), and washed with the sucrose (300 mM) three times, and resuspended in the sucrose. The cell suspension was placed on ice for 30 min. We then mixed the cells (80 μL) with the fusion plasmid (pMMB67EH-EGFP-MexB, 10 μL, 1 μg/μL), transferred them to a pre-chilled sterile electroporation cuvette (gap between two electrodes = 0.2 cm), and applied the electric pulse (2.5 kV, 25 μF, 5 ms, one pulse) (Biorad Gene Pulser). The cells were then cultured in the LB medium (3 mL) in the shaker (200 rpm, 37 °C) for 2 h, and harvested using centrifugation (5000 rpm, 5 min). The cells were streaked onto the LB agar plate containing 100 μg/ml carbenecillin, which was then incubated at 37 °C for 24 h. The clones were picked and cultured at 37 °C in the LB medium with 100 μg/mL carbenecillin to produce EGFP fused cells, MexA-(EGFP-MexB)-OprM.
Cell Culture and Assays
We pre-cultured each of five strains of P. aeruginosa: WT, nalB1, ΔMexB, ΔABM, and MexA-(EGFP-MexB)-OprM, by inoculating a single clone of each strain from the LB agar plate into the LB medium and placed them in the shaker (37 °C, 200 rpm) for 12 h. We then cultured the cells in the fresh LB medium in the shaker (37 °C, 200 rpm) for another 8 h, except adding 2 mM IPTG and 100 μg/mL carbenecillin in the medium for the culture of MexA-(EGFP-MexB)-OprM cells to induce the expression of the fusion protein as the OD600 nm of the cell suspension reached 0.5. We harvested the cells using centrifugation (Beckman J2-21, JA-14 rotor, 6000 rpm, 23 °C, 10 min), washed them with the PBS buffer (50 mM phosphate, 100 mM NaCl, pH 7.0) three times, and re-suspended them in the buffer. The cell concentration in the buffer (OD600 nm = 0.1) was used to characterize the expression of the fusion protein (EGFP-MexB) in single live cells using fluorescence microscopy and to study the efflux function of membrane transporters by measuring accumulation kinetics of EtBr in the cells using fluorescence spectroscopy.
Characterization of Expression of MexA-(EGFP-MexB)-OprM in Single Live Cells
The cells suspended in the PBS buffer were imaged in a micro-chamber using dark-field optical microscopy and epi-fluorescence microscopy (Nikon, E-400) equipped with a CCD camera (Micromax, Roper Scientific). The design and construction of the micro-chamber and dark-field optical microscopy for imaging of single live cells were fully described in our previous studies.33, 35, 38, 46 In this study, the fluorescence filter cube (Chroma Tech) containing a band-pass excitation filter (455 ± 30 nm), band-pass emission filter (525 ± 30 nm) and a dichroic mirror (500 nm), was used for fluorescence imaging of expression of EGFP-MexB in single live cells. The dark-field optical microscope is equipped with a dark-field condenser (Oil 1.43-1.20, Nikon) and a 100x objective (Nikon Plan fluor 100× oil, iris, SL. N.A. 0.5-1.3, W.D. 0.20 mm).
Fluorescence Spectroscopic Study of Efflux Function of MexA-(EGFP-MexB)-OprM in Live Cells
Fluorescence intensity of EtBr (15 and 40 μM) incubated with the cells (OD 600 nm = 0.1) in the PBS buffer in the presence or absence of CCCP (0.1 mM) was acquired over time (1-2 h) with 3s time interval at room temperature using a fluorescence spectrometer (Perkin-Elmer, LS50B). The excitation and emission wavelengths were selected at 465 and 600 nm with a width of each excitation and emission slit at 10 nm, respectively.
Study of Minimum Inhibitory Concentration (MIC) of Antibiotics to the Cells
We measured the MICs of two representative antibiotics (AZT and CP) toward the cells: WT, MexA-(EGFP-MexB)-OprM, ΔMexB, ΔABM, and nalB1. Each strain was pre-cultured by inoculating a single clone from the LB agar plate into the LB medium (with 100 μg/mL carbenecillin for the EGFP fusion strain to maintain the plasmid) in the shaker (200 rpm, 37°C) overnight. The cell suspension for each strain (100 μl, ∼104 cells) was added into the refresh LB medium (2.9 mL) in the presence of dilution series of AZT (0-100 or 250 μg/mL) or CP (0-100 or 250 μg/mL) with 100 μg/mL carbenecillin and 2 mM IPTG for the fusion strain to induce the expression of fusion protein, in the shaker (200 rpm, 37°C) for another 16 h. The concentration and number of the cells in each cell cultured suspension were then measured using the UV-vis spectroscopy (OD600 nm) and dark-field optical microscopy, respectively.
Results and Discussion
Construction and Characterization of egfp-mexB Fusion Gene and its Expression
To directly visualize MexB and determine its roles in the efflux function of MexAB-OprM using fluorescence microscopy and spectroscopy, we designed and constructed a new strain of P. aeruginosa with MexB fused with EGFP (MexA-EGFP-MexB-OprM). We first designed and constructed the egfp-mexB fusion gene as shown in Figure 1 and as described in Methods (Figure S1). The amplified egfp-mexB gene and vector (pMMB67EH) were digested and ligated to produce fusion plasmid (pMMB67EH-EGFP-MexB) (Figure S1).
Figure 1.
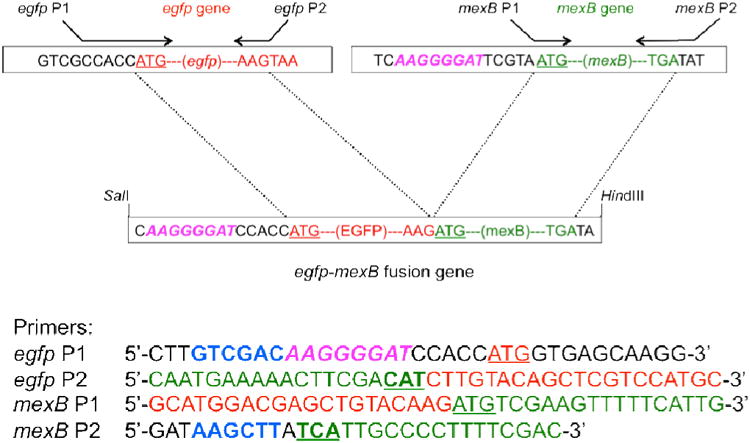
Design and construction of egfp-mexB fusion gene and their primers. Two egfp primers are used to amply the egfp gene marked in red using EGFP plasmid as a template. Two mexB primers are used to amply the mexB gene marked in green using the ds-mexB as a template. Hexanucleotides of egfp P1 and mexB P2 labeled by bold and blue are SalI and HindIII sties, respectively. These two primers are used to amply egfp-mexB fusion gene. The AAGGGGAT region labeled by pink and italic corresponds to Shine-Dalgarno (SD) sequence of the mexB gene. Underlined ATGs are initiation codons of egfp and mexB genes. CAT of egfp P2 primer as marked by bold and underlined is a complement triplet of the mexB initiation codon (ATG). TCA of mexB P2 labeled by bold and underlined is a complement triplet of the mexB termination codon (TGA). The detailed experimental design is described in Figure S1 in on-line supplementary information.
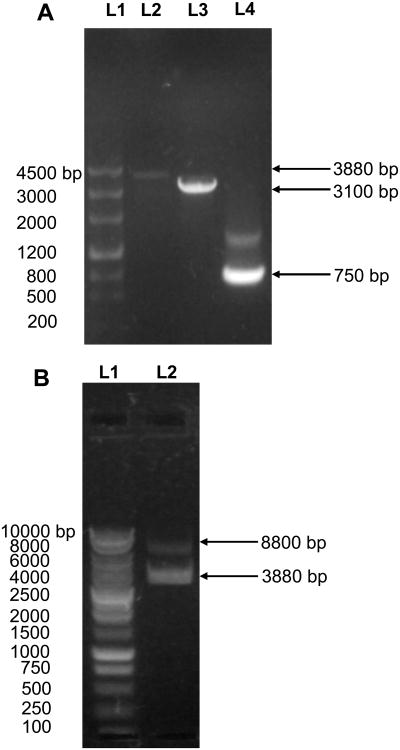
We characterized the fusion gene (egfp-mexB) using DNA sequencing (Figure S2 in on-supplementary information), and gel electrophoresis (Figure 2A). Note that PCR amplified egfp-mexB, mexB and egfp genes contain 3888, 3171 and 756 bp, respectively, and the fusion gene (egfp-mexB) was formed by overlapping (hybridizing) of the 39 bp of amplified mexB and egfp genes (20 bp each), which leads to 3888 bp, instead of 3927 bp. The results in Figure 2A show that the egfp-mexB gene (∼3880 bp in L2) includes mexB gene (∼3100 bp in L3), and egfp gene (∼750 bp in L4), indicating that the fusion gene (egfp-mexB) has been successfully constructed. We also characterized the fusion plasmid (pMMB67EH-EGFP-MexB) using DNA sequencing and gel electrophoresis (Figure 2B). The digested fusion plasmid shows two bands, the original plasmid (pMMB67EH, 8.8 kb) and the fusion gene (egfp-mexB, ∼3880 bp), which demonstrate that the fusion gene has been successfully inserted into the original plasmid and the fusion plasmid (pMMB67EH-EGFP-MexB) has been successfully constructed. The DNA sequences of both fusion gene and fusion plasmid show that the sequences of mexB are in excellent agreement with those reported previously.47
Figure 2.
Characterization of egfp-mexB fusion gene using agarose gel electrophoresis: (A): (L1) DNA markers/ladders in base pair (bp); (L2) The egfp-mexB fusion gene amplified using both egfp P1 and mexB P2 as primers (Figure S1A-B:III); (L3) The ds-mexB gene amplified using the genomic DNA of P. aeruginosa as a template, and mexB P1 and P2 as primers (Figure S1B-I); (L4) The ds-egfp gene amplified using the plasmid (pEGFP) as a template and egfp P1 and P2 as primers. Arrows point to the PCR products of 750, 3100 and 3880 bp, which agree well with the number of base pairs of egfp, mexB and egfp-mexB fusion gene. The sequence of egfp-mexB is characterized using DNA sequencer and shown in Figure S2 in on-line supplementary information. (B): (L1) DNA markers/ladders in bp; (L2) The pMMB67EH-EGFP-MexB vector digested by a pair of restriction enzymes, SalI/HindIII. Arrows point to the digested vector plasmids at 8800 bp and egfp-mexB fusion gene at 3880 bp.
We transformed the characterized fusion plasmid (pMMB67EH-EGFP-MexB) into MexB deletion strain (ΔMexB) and characterized their expression in single live cells by measuring fluorescence of EGFP in single live cells using fluorescence microscopy and spectroscopy. The results in Figure 3 show that all MexA-(EGFP-MexB)-OprM cells emit green fluorescence, while other strains of P. aeruginosa, WT (MexAB-OprM), nalB1 (over expression of MexAB-OprM), ΔABM (deletion of MexAB-OprM), and ΔMexB (deletion of MexB), do not.
Figure 3.
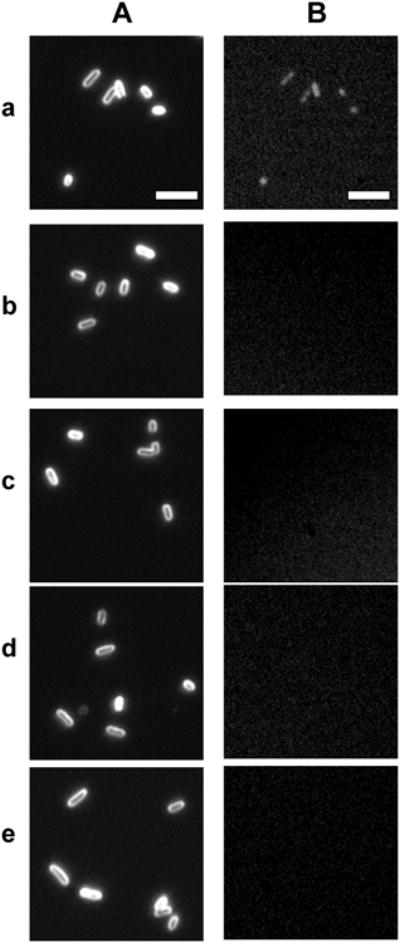
Characterization of expression of egfp-mexB fusion gene in single live cells. (A) Dark-field optical images and (B) green fluorescence images of single live cells: (a) MexA-(EGFP-MexB)-OprM; (b) ΔMexB; (c) WT; (d) nalB1; and (e) ΔABM, show green fluorescence of EGFP in (a) MexA-(EGFP-MexB)-OprM cells only, which indicates the expression of egfp-mexB fusion gene in the cells. Scale bar = 5 μm.
Fluorescence Study of Efflux Function of MexA-(EGFP-MexB)-OprM in Live Cells
To determine whether EGFP-MexB protein expressed in ΔMexB cells maintains the efflux function of MexAB-OprM and whether EGFP interfere the efflux function of the transporter, we studied the efflux kinetics of EtBr by MexAB-OprM in live cells in real time by measuring accumulation rates of intracellular EtBr using fluorescence spectroscopy. It is worth noting that EtBr emits week fluorescence in aqueous solution (outside the cells), and its fluorescence intensity increases substantially (up to 10-fold) as they enter into the cells and intercalate with DNA.37 Therefore, EtBr has been used to monitor and characterize the efflux kinetics of membrane transporters in live cells in real time.33, 35-36, 38, 40
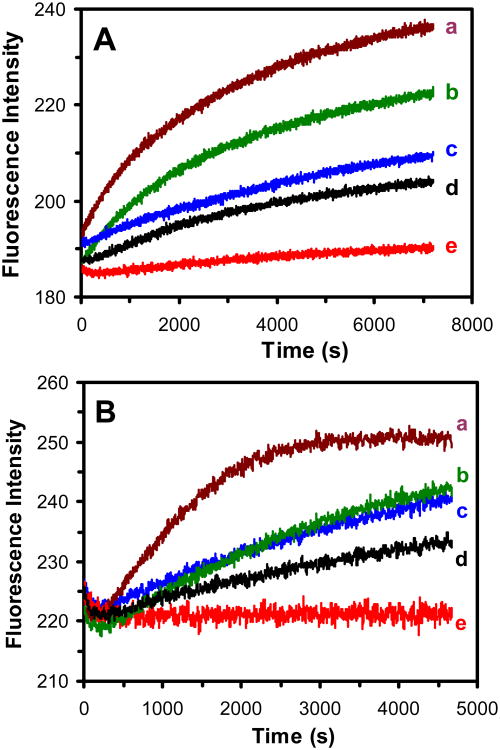
Time-dependent fluorescence intensity of EtBr (15 and 40 μM) incubated with the live cells (ΔABM, ΔMexB, MexA-EGFP-MexB-OprM, WT, and nalB-1) in PBS buffer show that fluorescence intensity of EtBr increases with time and the rates highly depend upon the expression level of MexAB-OprM in the cells (Figure 4). The highest rates and highest fluorescence intensity at each given time were observed for EtBr incubated with the ΔABM cells (Figure 4a), while the lowest rates and lowest fluorescence intensity were found for nalB-1 cells with over-expression of MexAB-OprM (Figure 4e). The results show that the MexAB-OprM extrudes the EtBr out of the cells, which leads to the lowest accumulation of EtBr in nalB1. The results also demonstrate that we can use the accumulation rates of EtBr in the cells to characterize the efflux function of MexAB-OprM.
Figure 4.
Study of the dependence of accumulation and efflux kinetics of the intracellular EtBr on the expression of MexAB-OprM and MexA-(EGFP-MexB)-OprM transporters in live cells. Time-dependent fluorescence intensity of EtBr: (A) 15 and (B) 40 μM, incubated with the cells (OD600 nm = 0.1 in PBS buffer, pH 7.2): (a) ΔABM, (b) ΔMexB, (c) MexA-(EGFP-MexB)-OprM, (d) WT, and (e) nalB1 strains.
The highest accumulation rate and highest fluorescence intensity of EtBr in the ΔABM cells are followed by those of ΔMexB (Figure 4b), MexA-(EGFP-MexB)-OprM (Figure 4c), WT (Figure 4d), and nalB1 cells (Figure 4e). Interestingly, the rates of MexA-(EGFP-MexB)-OprM are only 1.1-fold higher than those of WT (nearly the same as WT), but 4-fold lower than ΔMexB in the presence of 15 μM EtBr (Figure 4A). The results suggest that the fusion strain extrudes the intracellular EtBr out of the cells nearly as effectively as WT cells, and it is much more effectively than ΔMexB.
As concentration of EtBr increases from 15 μM (Figure 4A) to 40 μM (Figure 4B), the accumulation rates of EtBr in MexA-(EGFP-MexB)-OprM increase and they are nearly the same as ΔMexB, but 1.4-fold higher than WT, showing that the ineffectiveness of efflux function of fusion pump is magnified upon the over-flowing (higher concentration) of intracellular EtBr. The results (Figure 4) suggest that the fused EGFP may create steric effect upon the efflux function of MexB, and hence the fusion pump (MexA-EGFP-MexB-OprM) cannot extrude the substrate as effectively as WT (MexAB-OprM). The results in Figure 4 further demonstrate that EGFP is fused with MexB, and MexA-(EGFP-MexB)-OprM is expressed. Should EGFP be cleaved from MexB, we would have observed the same efflux function of the fusion strain as that of WT.
Study of Inhibitory Effect of CCCP upon Efflux Function of MexA-(EGFP-MexB)-OprM in Live Cells
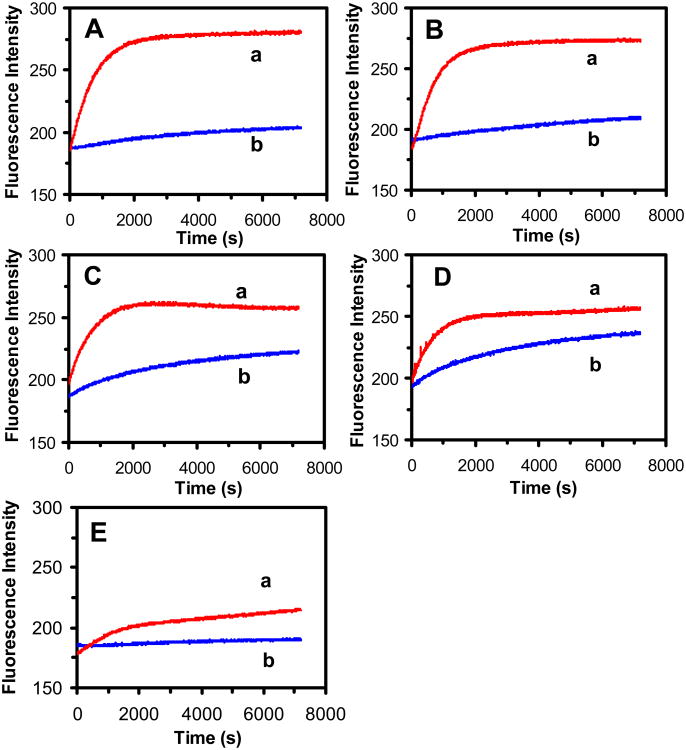
We further characterized the efflux function of fusion pump by study of inhibitory effect of CCCP upon the efflux function of the pump (Figure 5). Note that the CCCP can effectively decrease the proton gradients across the cellular membrane, remove the energy source of the transporters and hence disable the efflux function of the proton-motive transporters. The results (Figure 5) show that the fluorescence intensity of intracellular EtBr increases with time much more rapidly in the presence of CCCP than in the absence of CCCP, indicating the inhibitory effect of CCCP upon the efflux functions of the transporters, and the accumulation rates of intracellular EtBr by the cells are much more rapidly in the presence of CCCP than its absence. Among five strains, the CCCP affects the efflux function of WT most (Figure 5A), closely followed by fusion strain (MexA-EGFP-MexB-OprM) (Figure 5B), then ΔMexB (Figure 5C), ΔABM (Figure 5D), and nalB1 (Figure 5E). The lowest effect of CCCP upon the efflux function of nalB1 could be attributed to the insufficient dose of CCCP to disable over-expression of nalB1. Interestingly, the CCCP also affects the accumulation rates of EtBr by ΔABM, which could be attributed to the presence of other proton-motive transporters (e.g., MexCD-OprJ, MexEF-OprN) in ΔABM.48-49
Figure 5.
Study of inhibitory effects of CCCP (0.1 mM) on efflux function of MexAB-OprM and MexA-(EGFP-MexB)-OprM transporters in live cells. Time-dependent fluorescence intensity of EtBr (15 μM) incubated with the cells (OD600 nm = 0.1 in PBS buffer, pH 7.2): (A) WT; (B) MexA-(EGFP-MexB)-OprM; (C) ΔMexB; (D) ΔABM; and (E) nalB1 strains, in (a) the presence and (b) absence of CCCP.
Taken together, the results show that the fusion transporter (MexA-EGFP-MexB-OprM) retains significant level of efflux function of MexAB-OprM. The fusion pump displays similar efflux kinetics to WT in response to the lower dose of EtBr (Figure 4A), suggesting that the fusion strain may be suitable for the study of efflux function of the transporter in low dose substrates. Nonetheless, fusion of EGFP with MexB may create steric effects upon its folding and/or interactions with MexA, OprM or pump substrates, and affect the efflux function of MexAB-OprM (Figure 4B).
Antibiotic Susceptibility Study of Efflux Function of MexA-(EGFP-MexB)-OprM
To further investigate the efflux function of the fusion pump (MexA-EGFP-MexB-OprM), we determined the susceptibility of all five strains of P. aeruginosa (WT, ΔMexB, MexA-EGFP-MexB-OprM, ΔABM, and nalB1) toward two representative antibiotics (AZT and CP). The results (Table 1) show that MIC of AZT for WT (3.13 μg/mL) is about 5-fold lower than nalB1 (15.7 μg/mL), and 16-fold higher than ΔABM (0.2 μg/mL). The MIC of CP for WT (25 μg/mL) is 4-fold lower than nalB1 (100 μg/mL), and 16-fold higher than ΔABM (1.56 μg/mL). The results in Table 1 show that MICs of AZT and CP for each strain highly depend upon the expression level of MexAB-OprM and they are the effective and reliable parameters to characterize the efflux function of the transporter.
Table I. Study of Minimum Inhibitory Concentration (MIC) of Antibiotics, Aztreonam (AZT) and Chloramphenicol (CP) toward Various Strains of Cells.
| Strains | MIC (μg/mL) | |
|---|---|---|
| AZT | CP | |
| WT (PAO4290: normal expression of MexAB-OprM) | 3.13 | 25 |
| MexA-(EGFP-MexB)-OprM (N-terminus of MexB fused with EGFP) | 0.5 | 8 |
| ΔMexB (TNP071: deletion of MexB, derivation of PAO4290) | 0.25 | 4 |
| ΔABM (TNP076: deletion of MexAB-OprM, derivative of PAO4290) | 0.2 | 1.56 |
| nalB1 (TNP030#1: a mutant that over-expression of MexAB-OprM) | 15.7 | 100 |
The results in Table 1 show that MIC of AZT for fusion strain (0.5 μg/mL) is about 6-fold lower than WT (3.13 μg/mL), and 2-fold higher than ΔMexB (0.25 μg/mL). The MIC of CP for fusion strain (8 μg/mL) is about 3-fold lower than WT (25 μg/mL), and 2-fold higher than ΔMexB (4 μg/mL). The results further demonstrate the expression of fusion transporter (MexA-EGFP-MexB-OprM). Should EGFP be cleaved from MexB, we would have observed the same MICs for the fusion strain as that of WT. The results also further affirm our hypothesis that the expression of EGFP-MexB in ΔMexB partially restores the efflux function of MexAB-OprM, and fusion of EGFP with MexB may affect the efflux function of the transporter.
Structural Modeling and Analysis of MexA-(EGFP-MexB)-OprM Transporter
To understand how fusion of C-terminal EGFP with N-terminal MexB could create steric effects upon efflux function of MexAB-OprM, we constructed secondary structure of MexAB-OprM model based on the coordinates of the data-driven docking model of the multidrug efflux pump (AcrA-AcrB-TolC) from P. aeruginosa (Figure 6A).50 We then fused the EGFP domains to the N-termini of MexB in its trimeric assemblage (Figure 6B)
Figure 6.
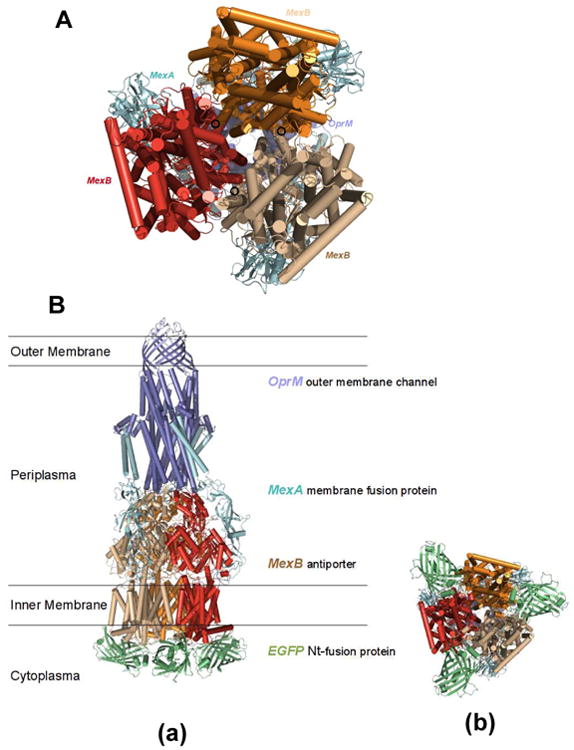
The modeled structures of MexA-(EGFP-MexB)-OprM expressed in the cells (ΔMexB): (A) Secondary structure of MexAB-OprM model is constructed based on the coordinates of the data-driven docking model of the multidrug efflux pump (AcrA-AcrB-TolC) from P. aeruginosa. The model shows the location of the MexB N-termini at the cytosolic end of the translocation pore of the transporter with the unstructured loop of the N-termini pointing inward towards the trimeric pore. (B) The EGFP domains were fused to the N-termini of MexB in its trimer assemblage: (a) stereo side view and (b) bottom view of the structures. Spatial placement of the EGFP domains in the cytosol is limited by potential EGFP to EGFP, or EGFP to MexB clashes in the trimeric configuration. The position and orientation of the N-terminal fused EGFP domains may either partially block the translocation pore or restrict the movement of the individual pump domains, thereby restricting efflux activity.
Initial coordinates of the complete trimeric pump were obtained from the data-driven docking of AcrA-TolC onto AcrB.50 The individual components of the tripartite multidrug efflux pump from P. aeruginosa are highly homologous to the corresponding components in the MexAB-OprM transporter with up to 69% sequence identity between the antiporter AcrB and MexB.51
Three dimensional structure alignment shows low average root-mean-square deviations (RMSDs) for coordinate displacements of 2.053 Å for AcrA/MexA (PDB codes: 2F1M and 1VF7), 2.355 Å for AcrB/MexB (PDB codes: 2J8S and 2V50) and 2.064 Å for TolC/OprM (PDB codes: 2VDD and 1WP1).52-53 The working model was assembled by secondary structure matching of MexB on AcrB in its trimeric configuration.53-54 The EGFP model was assembled separately using SWISS-MODEL,55 based on 3D structure of the GFP (PDB code: 3OGO) with a sequence identity of 98.7%. The prediction of secondary structures shows the linker region connecting the C-terminus of the EGFP domain to the N-terminus of MexB to be unstructured.56 The connecting residues were built manually using program Coot.54 The EGFP domains were positioned in the cytoplasmic space with no clashes to the MexB domains and the membrane region. The structural models (Figure 6) were generated using PyMOL.57
The structural model shows the location of the MexB N-termini at the cytosolic end of the translocation pore of the transporter with the unstructured loop of the N-termini pointing inward towards the trimeric pore (Figure 6A). Spatial placement of the EGFP domains in the cytosol is limited by potential EGFP to EGFP, or EGFP to MexB clashes in the trimeric configuration. The model structure shows that the position and orientation of the N-terminal fused EGFP domains may either partially block the translocation pore or restrict the movement of the individual pump domains, thereby partially restricting efflux activity (Figure 6B).
Summary
In summary, we have designed and constructed the fusion gene (egfp-mexB), inserted it into the plasmid vector (pMMB67EH) and successfully expressed it in ΔMexB strain of P. aeruginosa. This approach allows us to create the new strain of P. aeruginosa that expresses MexA-(EGFP-MexB)-OprM. We characterized the efflux function of the new strain by measuring their accumulation kinetics of intracellular EtBr (a pump substrate), inhibitory effects of CCCP upon its efflux function, and MICs of two representative antibiotics (AZT and CP). We compared these results with the cells with various expression levels of MexAB-OprM (WT, ΔABM, ΔMexB, and nalB1), and found that the new strain shows 4-fold lower accumulation rates of EtBr (15 μM) than ΔMexB (deletion of MexB), but only 1.1-fold higher than WT (nearly the same as WT). As EtBr concentration increases to 40 μM, the new strain has nearly the same accumulation rate of EtBr as ΔMexB, but 1.4-fold higher than WT. Interestingly, the CCCP creates nearly the same level of inhibitory effects upon the efflux function of the new strain as the WT, which are 2-3 times higher than their effects upon ΔMexB and ΔABM. Antibiotic susceptibility study shows that the fusion strain has 6-fold lower MIC of AZT and 3-folder lower MIC of CP than WT, and only 2-fold higher than ΔMexB. Taken together, these results suggest that the fusion protein, MexA-(EGFP-MexB)-OprM, partially retains the efflux function of MexAB-OprM. Modeled structural study of the fusion pump shows that the position and orientation of the N-terminal fused EGFP domains may either partially block the translocation pore or restrict the movement of the individual pump domains, which lead to partially restrict efflux activity. The work is in progress to characterize topologies and translocation mechanisms of fusion pump and develop new approaches to generate the labeled MexB without affecting their efflux functions.
Supplementary Material
Supplementary Information Available: Figure S1: Construction of fusion egfp-mexB
Figure S2: DNA sequence of egfp-mexB
Acknowledgments
This work is supported in part by NSF (NIRT: CBET 0507036) and NIH (R01 GM0764401) and 3R01 (GM0764401-04S1). Ding and Lee are grateful for the support of Dominion Scholar Fellowship and NSF-GRAS (CBET 0507036), respectively.
References
- 1.Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Curr Opin Struct Biol. 2004;14:741–747. doi: 10.1016/j.sbi.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Poole K. Annu Med. 2007;39:162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 3.Blair JM, Piddock LJ. Curr Opin Microbiol. 2009;12:512–519. doi: 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Pfister O, Liao R. Circ Res. 2008;102:998–1001. doi: 10.1161/CIRCRESAHA.108.176453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alibert-Franco S, Pradines B, Mahamoud A, Davin-Regli A, Pagès JM. Curr Med Chem. 2009;16:301–317. doi: 10.2174/092986709787002619. [DOI] [PubMed] [Google Scholar]
- 6.Mesaros N, Nordmann P, Plésiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A, Tulkens PM, Van Bambeke F. Clin Microbiol Infect. 2007;13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 7.Pietras Z, Bavro VN, Furnham N, Pellegrini-Calace M, Milner-White EJ, Luisi BF. Curr Drug Targets. 2008;9:719–728. doi: 10.2174/138945008785747743. [DOI] [PubMed] [Google Scholar]
- 8.Germ M, Yoshihara E, Yoneyama H, Nakae T. Biochem Biophys Res Commun. 1999;261:452–455. doi: 10.1006/bbrc.1999.1045. [DOI] [PubMed] [Google Scholar]
- 9.Maseda H, Yoneyama H, Nakae T. Antimicrob Agents Chemother. 2000;44:658–664. doi: 10.1128/aac.44.3.658-664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Antimicrob Agents Chemother. 2000;44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwagi T, Yoneyama H, Narita S, Nakagawa A, Nakae T. J Biol Chem. 2004;279:52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 12.Akama H, Matsuura T, Kashiwagi S, Yoneyama H, Narita S, Tsukihara T, Nakagawa A, Nakae T. J Biol Chem. 2004;279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 13.Wilke MS, Heller M, Creagh AL, Haynes CA, McIntosh LP, Poole K, Strynadka NC. Proc Natl Acad Sci U S A. 2008;105:14832–14837. doi: 10.1073/pnas.0805489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakae T. Microbiologia. 1997;13:273–284. [PubMed] [Google Scholar]
- 15.Paulsen IT, Brown MH, Skurray RA. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakajima A, Sugimoto Y, Yoneyama H, Nakae T. Microbiol Immunol. 2002;46:391–395. doi: 10.1111/j.1348-0421.2002.tb02711.x. [DOI] [PubMed] [Google Scholar]
- 17.Poole K. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 18.Mokhonov V, Mokhonova E, Yoshihara E, Masui R, Sakai M, Akama H, Nakae T. Protein Expr Purif. 2005;40:91–100. doi: 10.1016/j.pep.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Morita Y, Kimura N, Mima T, Mizushima T, Tsuchiya T. J Gen Appl Microbiol. 2001;47:27–32. doi: 10.2323/jgam.47.27. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima A, Sugimoto Y, Yoneyama H, Nakae T. J Biol Chem. 2000;275:30064–30068. doi: 10.1074/jbc.M005742200. [DOI] [PubMed] [Google Scholar]
- 21.Nehme D, Li XZ, Elliot R, Poole K. J Bacteriol. 2004;186:2973–2983. doi: 10.1128/JB.186.10.2973-2983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piddock LJ. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 23.Uwate M, Ichise YK, Shirai A, Omasa T, Nakae T, Maseda H. Microbiol Immunol. 2013;57:263–272. doi: 10.1111/1348-0421.12032. [DOI] [PubMed] [Google Scholar]
- 24.Gerdes HH, Rudolf R. Protoplasma. 1999;209:1–8. doi: 10.1007/BF01415695. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi A, Maeda T, Maeda M. Biol Pharm Bull. 2004;27:1916–1922. doi: 10.1248/bpb.27.1916. [DOI] [PubMed] [Google Scholar]
- 26.Orford M, Mean R, Lapathitis G, Genethliou N, Panayiotou E, Panayi H, Malas S. Biochem Biophys Res Commun. 2009;384:199–203. doi: 10.1016/j.bbrc.2009.04.089. [DOI] [PubMed] [Google Scholar]
- 27.Drew D, Sjostrand D, Nilsson J, Urbig T, Chin CN, de Gier JW, von Heijne G. Proc Natl Acad Sci U S A. 2002;99:2690–2695. doi: 10.1073/pnas.052018199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandlur SM, Wei L, Levine J, Russell J, Kaur P. J Biol Chem. 2004;279:27799–27806. doi: 10.1074/jbc.M402898200. [DOI] [PubMed] [Google Scholar]
- 29.Ding F, Lee K, Vahedi-Faridi A, Huang T, Xu XHN. Anal Bioanal Chem. 2011;400:223–235. doi: 10.1007/s00216-011-4727-7. [DOI] [PubMed] [Google Scholar]
- 30.Lee KJ, Browning LM, Huang T, Ding F, Nallathamby PD, Xu XHN. Anal Bioanal Chem. 2010;397:3317–3328. doi: 10.1007/s00216-010-3864-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shukla S, Saini P, Smriti JS, Ambudkar SV, Prasad R. Eukaryot Cell. 2003;2:1361–1375. doi: 10.1128/EC.2.6.1361-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abe-Dohmae S, Ikeda Y, Matsuo M, Hayashi M, Okuhira K, Ueda K, Yokoyama S. J Biol Chem. 2004;279:604–611. doi: 10.1074/jbc.M309888200. [DOI] [PubMed] [Google Scholar]
- 33.Kyriacou SV, Nowak ME, Brownlow WJ, Xu XHN. J Biomed Opt. 2002;7:576–586. doi: 10.1117/1.1506707. [DOI] [PubMed] [Google Scholar]
- 34.Mortimer PG, Piddock LJ. J Antimicrob Chemother. 1991;28:639–653. doi: 10.1093/jac/28.5.639. [DOI] [PubMed] [Google Scholar]
- 35.Xu XHN, Brownlow WJ, Kyriacou SV, Wan Q, Viola JJ. Biochemistry. 2004;43:10400–10413. doi: 10.1021/bi036231a. [DOI] [PubMed] [Google Scholar]
- 36.Nallathamby PD, Lee KJ, Desai T, Xu XHN. Biochemistry. 2010;49:5942–5953. doi: 10.1021/bi100268k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cosa G, Focsaneanu KS, McLean JRN, McNamee JP, Scaiano JC. Photochemistry and Photobiology. 2001;73:585–599. doi: 10.1562/0031-8655(2001)073<0585:PPOFDD>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 38.Xu XHN, Brownlow WJ, Huang S, Chen J. Biochem Biophys Res Commun. 2003;305:79–86. doi: 10.1016/s0006-291x(03)00692-2. [DOI] [PubMed] [Google Scholar]
- 39.Al-Hamad A, Upton M, Burnie J. J Antimicrob Chemother. 2009;64:731–734. doi: 10.1093/jac/dkp271. [DOI] [PubMed] [Google Scholar]
- 40.Ocaktan A, Yoneyama H, Nakae T. J Biol Chem. 1997;272:21964–21969. doi: 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- 41.Yoneyama H, Ocaktan A, Tsuda M, Nakae T. Biochem Biophys Res Commun. 1997;233:611–618. doi: 10.1006/bbrc.1997.6506. [DOI] [PubMed] [Google Scholar]
- 42.Mather MW, McReynolds LM, Yu CA. Gene. 1995;156:85–88. doi: 10.1016/0378-1119(95)00074-g. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook JF, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- 44.Diver JM, Bryan LE, Sokol PA. Anal Biochem. 1990;189:75–79. doi: 10.1016/0003-2697(90)90046-c. [DOI] [PubMed] [Google Scholar]
- 45.Choi KH, Kumar A, Schweizer HP, Microbiol J. Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Xu XHN, Chen J, Jeffers RB, Kyriacou SV. Nano Lett. 2002;2:175–182. [Google Scholar]
- 47.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik D, Lagrou JM, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 48.Poole K, Srikumar R. Curr Top Med Chem. 2001;1:59–71. doi: 10.2174/1568026013395605. [DOI] [PubMed] [Google Scholar]
- 49.Paulsen IT. Curr Opin Microbiol. 2003;6:446–451. doi: 10.1016/j.mib.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. Proc Natl Acad Sci U S A. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.CCP4. Acta Crystallogr D Biol Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 53.Krissinel E, Henrick K. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Schwede T, Kopp J, Guex N, Peitsch MC. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones DT. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 57.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information Available: Figure S1: Construction of fusion egfp-mexB
Figure S2: DNA sequence of egfp-mexB