Abstract
MicroRNAs (miRNAs) belonging to the evolutionary conserved miR-302 family play important functions in Embryonic Stem Cells (ESCs). The expression of some members, such as the human miR-302 and mouse miR-290 clusters, is regulated by ESC core transcription factors. However, whether miRNAs act downstream of signaling pathways involved in human ESC pluripotency remains unknown. The maintenance of pluripotency in hESCs is under the control of the TGFβ pathway. Here, we show that inhibition of the Activin/Nodal branch of this pathway affects the expression of a subset of miRNAs in hESCs. Among them, we found miR-373, a member of the miR-302 family. Proper levels of miR-373 are crucial for the maintenance of hESC pluripotency, since its overexpression leads to differentiation towards the mesendodermal lineage. Among miR-373 predicted targets, involved in TGFβ signaling, we validated the Nodal inhibitor Lefty. Our work suggests a crucial role for the interplay between miRNAs and signaling pathways in ESCs.
Keywords: human embryonic stem cells, microRNAs, TGFβ signaling, mesendoderm
Introduction
In human Embryonic Stem Cells (hESCs), the Activin/Nodal branch of the TGFβ pathway is both necessary and sufficient to sustain pluripotency [1, 2], since hESCs treated with Smad2/3 inhibitors exit pluripotency and engage in differentiation [3, 4]. Components of the hESC core transcriptional regulatory circuitry [5], such as Nanog, are among the genes that are activated by Smad2/3 [6, 7]. However, the full repertoire of Smad2/3 targets remains largely unexplored.
microRNAs (miRNAs) have lately emerged as important regulators in ESCs [8], where members of the conserved miR-302 family represent the vast majority of the total number of miRNAs [9, 10]. We previously showed that in hESCs, miR-302 both controls mesendodermal fate specification by targeting Lefty [11] and regulates their exit from pluripotency and neuroectodemal specification by targeting NR2F2 [12]. Despite the well-established role of miRNAs in hESCs, their link to signaling pathways remains largely elusive. In particular, whether hESC miRNAs are downstream targets of the Activin/Nodal branch is currently unknown.
Here, among nine miRNAs that respond to perturbations of TGFβ signaling in hESCs, we focus on miR-373/373*, a miR-302 family member [13, 14]. We find that miR-373/373* overexpression leads to exit from pluripotency and differentiation of hESCs towards mesendodermal fates. Mechanistically, we show that these effects could be mediated, at least partially, by the repression of Lefty. Given that Lefty genes are activated by Nodal/Activin signaling [15], our results suggest that a network of positive/negative feedback loops, involving signaling pathways and miRNAs of the miR-302 family, function cooperatively in order to maintain hESCs in a delicate balance between pluripotency and differentiation.
Materials and Methods
hESC culture and transgenesis
hESC line RUES2 was cultured as previously described [16]. Stable transgenic RUES2 lines were generated by nucleofection of a plasmid encoding an optimized transposase [17] together with the ePB-puro-TT-intRFP-miR-373 piggyBac vector, containing the human mir-373 stem loop flanked by 80 nucleotides on each side in the synthetic intron of the ePB-puro-TT-intRFP vector [18]. Single-cell derived colonies were clonally passaged and selected with puromycin. Where indicated, cells were induced with 1μg/ml doxycycline (Sigma).
microRNA expression profiling and Real-Time qPCR analysis
RUES2 cells were treated with either 10μM of SB431542 (Tocris Bioscience) or an equivalent volume of DMSO (Sigma) and total RNA was extracted using Trizol (Invitrogen). miRNA expression profiling was performed on a biological triplicate using the Illumina human v2 miRNA panel, which detects 1,146 human miRNAs. We considered significant a change in expression with p<0.01 and FDR <0.25. For miRNA Real-Time qPCR analysis, cDNA was generated using the miScript Reverse Transcription Kit (Qiagen) and analyzed with the miScript SYBR Green PCR kit and Primer Assays (Qiagen). mRNA Real-Time qPCR analysis was performed as described [12] with primers listed in Table S2. Results were analyzed with the REST-MCS2 software [19].
Immunostaining and Western blot
Cells were stained with primary anti-BRA (R&D), anti-GATA6 (SC) and anti-Sox17 (R&D), and secondary anti-goat- or anti-rabbit-Alexa488 (Molecular Probes) antibodies. Images were acquired on an Olympus IX71 inverted microscope and processed in LSM Viewer or ImageJ. Cell counting was performed using Matlab.
Western blot was performed as described [12] with anti-phospho-SMAD2/3 (Cell Signaling), anti-SMAD2/3 (Transduction Laboratories), anti-α/β-tubulin (Cell Signaling) and secondary HRP antibodies.
Chromatin immunoprecipitation Assay
Chromatin from RUES2 cells was immunoprecipitated with the MAGnify Chromatin Immunoprecipitation System (Invitrogen) with anti-SMAD2/3 (AF3797, R&D Systems) or with a control rabbit IgG antibody. Primers for Real-Time qPCR analysis are listed in Table S2.
Luciferase reporter Assays
HeLa cells were transfected as described [11] with 400ng/well of pRL-derivative, 40ng/well of pGL3-Control vector (Promega) and 10nM miR-373, miR-373* mimic (Qiagen) or negative control (AllStars Negative ct siRNA, Qiagen). Each condition was performed in triplicate. After 48 hours, cells were analyzed with the Dual-Luciferase Reporter kit (Promega).
Results
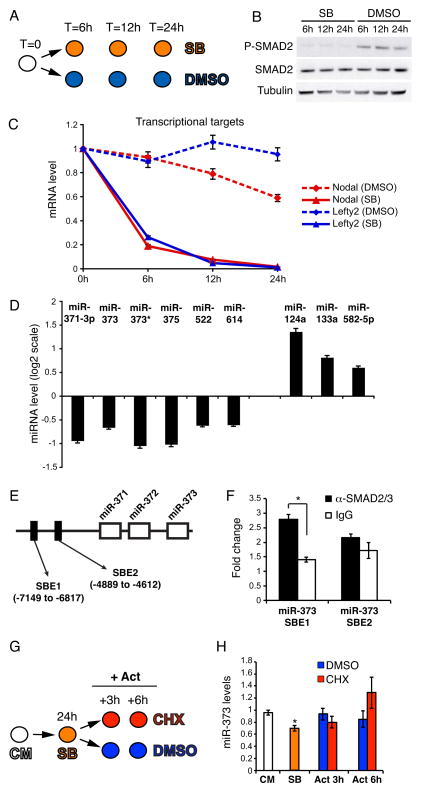
MEF-Conditioned Medium (CM) contains several ligands of the TGFβ family that are necessary to maintain hESC pluripotency [3]. To identify TGFβ-regulated miRNAs, we specifically blocked signaling through the SMAD2/3 branch of the TGFβ pathway by treating hESCs with the small inhibitory compound SB431542 (SB) [20] for 6, 12, and 24 hours (Fig. 1A). The effective block on SMAD2/3 was evidenced by strong reduction of SMAD2 C-terminal phosphorylation (Fig. 1B) and significant downregulation of two direct Smad2/3 transcriptional targets, Nodal and Lefty2 (Fig. 1C). Through miRNA profiling analysis we found 9 miRNAs showing a significant up- or down-regulation at 24 hours (Fig. 1D, Fig. S1 and Table S1). Out of these, we decided to specifically focus on mir-373 and miR-373*, which belong to a hESC-specific miRNA cluster [14]. ChIP analysis suggested that this regulation might be direct, as we identified one SMAD2/3-bound site upstream of the miR-373 cluster (Fig. 1E,F). To test whether regulation was indeed direct, we performed an induction experiment in absence of new protein synthesis. As MEF-CM contains ligands of the TGFβ family [21], we first blocked the pathway by SB and then induced the signaling by ActivinA in presence of the protein synthesis inhibitor cycloheximide (CHX) (Fig. 1G). Regardless of the presence of CHX we observed an increase of miR-373 up to its levels in CM, demonstrating direct regulation (Fig. 1H). Activation of the Nodal/Activin signaling in non MEF-CM did not increase the levels of miR-373 (Fig S2), suggesting that other signaling cues contribute to miR-373 activation under pluripotency conditions. Taken together, our results suggest that the SMAD2/3 branch of the TGFβ pathway is necessary for proper levels of miR-373 expression, whereas the related miR-302 is not changed in the same conditions (Fig. 1D, Table S1 and data not shown).
Figure 1. Inhibition of TGFβ signaling in hESCs results in deregulated miRNA expression.
(A) RUES2 cells were cultured under pluripotency conditions in the presence of either 10μM SB431542 (SB) or DMSO, and were harvested at the indicated time points. (B) Western blot showing that SB treatment leads to complete inhibition of C-terminal Smad2/3 phosphorylation (P-SMAD2), without affecting total Smad2/3 protein (SMAD2) levels. β-Tubulin was used as a loading control. (C) Real-Time qPCR analysis of Nodal and Lefty2 reveals a clear inhibition of the pathway. Data points represent average from triplicates. (D) Illumina miRNA microarray analysis of SB-treated RUES2 cells; shown here are the miRNAs whose expression was significantly (p < 0.01; FDR < 0.25) up- or down-regulated after 24 hours of SB treatment. (E) Schematic representation, not in scale, of the miR-371/372/373 locus on chromosome 19. The position of two putative SMAD2/3 binding elements (SBE) [32] is depicted. The numbers are relative to the first nucleotide of pre-miR-371. (F) Real-Time qPCR analysis of the chromatin immunoprecipitated with the indicated antibodies. Fold change is relative to a negative control genomic region on chromosome 4. Data points represent average from triplicates (*p<0.01). (G) RUES2 cells were cultured in the presence of 10μM SB431542 (SB) for 24 hours, then the medium was changed and the cells were cultured for additional 3 or 6 hours in CM containing 50ng/ml ActivinA in absence (DMSO) or presence of 100μg/ml of the protein synthesis cycloheximide (CHX). Cells were harvested at the indicated time points. CM: control untreated cells. (H) Real-Time qPCR analysis of miR-373 expression. Data points represent average from triplicates. * indicates statistically significant difference from the SB sample (p<0.01).
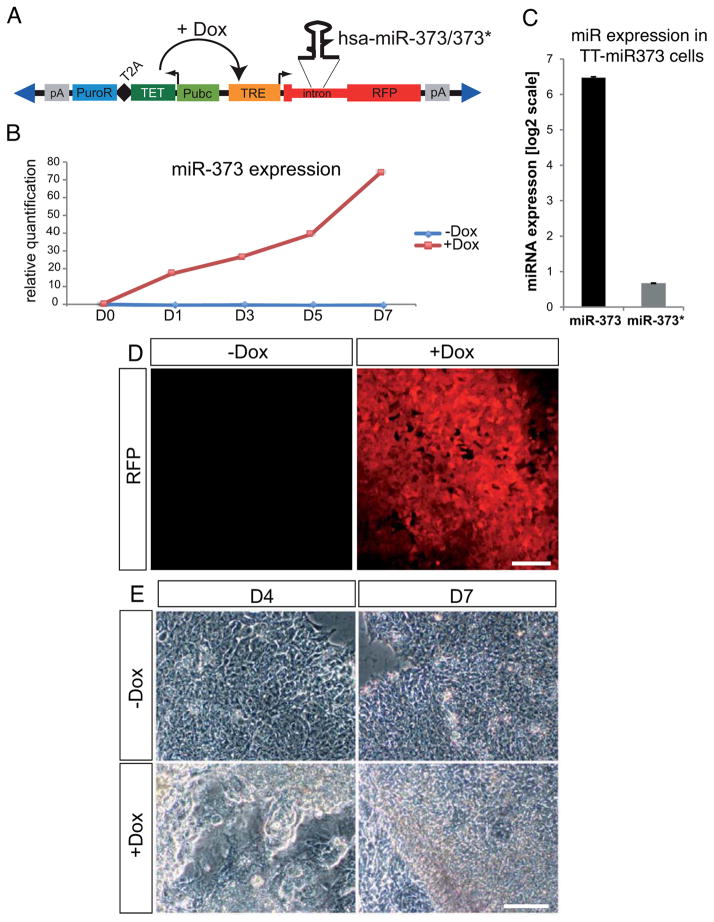
To address the role of miR-373 in hESC pluripotency, we generated a stable and clonal transgenic hESC line in which mir-373 ectopic expression can be induced by doxycycline treatment (hereafter referred to as TT-mir-373 line) (Fig. 2A). Upon doxycycline addition, expression of miR-373, but not miR-373*, was increased (Fig. 2B,C). The induction efficiency was monitored by RFP fluorescence in live cells (Fig. 2D).
Figure 2. mir-373 overexpression in hESCs using ePiggyBac-mediated transgenesis.
(A) Schematic representation of the ePiggyBac(ePB)-based inducible system used for mir-373 overexpression. RFP: Red Fluorescent Protein gene; pA: polyadenylation signal; PuroR: puromycin resistance gene; T2A: self-cleavage peptide; TET: TET transactivator protein gene; Pubc: human Ubiquitin C constitutive promoter; TRE: TET responsive element; Dox: doxycycline. Blue triangles represent terminal repeats of the transposon. (B) Real-Time miRNA qPCR analysis showing that doxycycline addition induces miR-373 expression compared to control (−dox) cells. Data points are presented as relative quantification to the values at D0. (C) Real-Time miRNA qPCR analysis on TT-mir373 induced (+Dox) cells for five days reveals that miR-373 is the predominantly expressed miRNA from the mir-373 hairpin. Results represent fold-change of expression (at a log2 scale) compared to wild-type RUES2 cells. (D) RFP expression in live TT-mir373 cells five days after addition of either 1μg/ml doxycycline (+Dox) or an equivalent volume of vehicle (H2O) (−Dox). (E) Representative bright-field images from induced (+Dox) or uninduced (−Dox) TT-mir373 cells after four (D4) or seven (D7) days of treatment. Scale bars: 100μm.
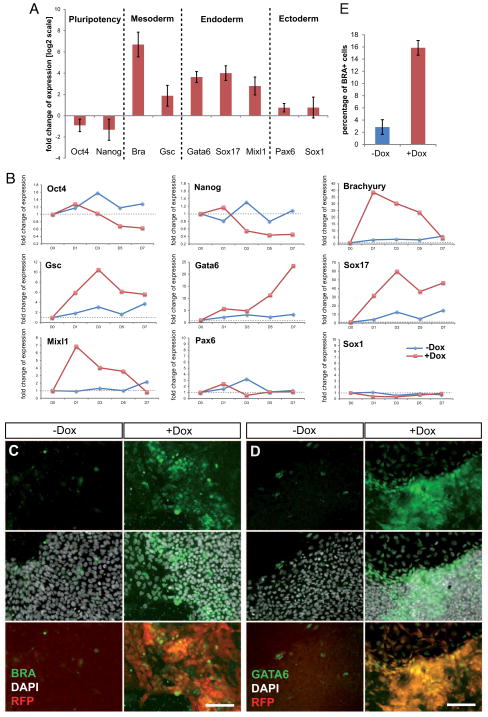
We then examined the effects of miR-373 overexpression on hESC growth. We observed the first signs of morphological differentiation as early as day 3 of induction (Fig. S3), with flattening out and detaching of cells from the edges of the colonies (Fig. S3B/B′). Subsequently, most colonies lost their typical monolayer appearance and formed three-dimensional structures. Also, a loss-of-contact inhibition was observed at the borders of some colonies (Fig. 2E and Fig. S3). At the molecular level we found that after 5 days of induction TT-mir-373 cells showed a slight reduction in the levels of pluripotency markers, whereas mesoderm and endoderm markers were strongly upregulated (Fig. 3A). Ectodermal markers do not seem to be significantly affected at this time. A detailed time course gene expression analysis revealed that early mesodermal and endodermal markers Bra and Mixl1 showed a very strong early upregulation at D1 and then gradually decreased; Gsc showed a similar pattern of upregulation, yet its expression remained strong at D7. Endodermal marker Gata6 showed a gradual increase in its expression levels until it reached a ~25-fold increase at D7, and Sox17 showed very strong upregulation, reaching a 60-fold peak at D3 and then remaining robust all throughout the time course of analysis (Fig. 3B). This pattern of expression is consistent with an initial transient induction of factors (Bra and Mixl1) known to be involved in gastrulation and subsequently decreasing during embryogenesis, whereas transcripts that also play a role during later stages (Gsc, Gata6 and Sox17) are maintained at high levels until the end of our analysis.
Figure 3. Overexpression of miR-373 in hESCs leads to mesendodermal differentiation.
(A) Real-Time qPCR analysis of gene expression was performed on induced (+Dox) and uninduced (−Dox) TT-miR-373 cells at day 5. Data are shown here as fold-change of expression (on a log2 scale) in +Dox compared to −Dox cells, and represent average values from three biological replicates ±s.dev. (B) Real-Time qPCR analysis of gene expression over the course of seven days of mir-373 overexpression (D0 to D7). Data are represented as relative quantification to D0. Note different scales on y axes for each gene. (C, D) TT-miR-373 cells were left untreated (−Dox) or induced with doxycycline (+Dox) for five (for BRA) or seven (for GATA6) days and stained with an anti-BRA (C) or anti-GATA6 (D) antibody. DAPI was used for nuclear staining; reporter RFP was also visualized. Scale bars: 100μm. (E) Quantification of BRA+ TT-miR-373 cells after 5 days of mir-373 overexpression.
At the single cell level, numerous BRA-positive cells were present in Dox-induced TT-mir-373 colonies (Fig. 3C and Fig. S4). After five days of miR-373 overexpression, ~16% of +Dox cells were positive for BRA, when only ~2% of −Dox cells were positive for this marker (Fig. 3E). GATA6 or SOX17-positive cells were slightly differently dispersed, appearing grouped in clusters either close to the borders or at the center of the colonies (Fig. S5). Despite miR-373* is only minimally increased in induced cells, compared to the strong upregulation of miR-373 (Fig. 2C), we cannot exclude its possible contribution to the observed effects.
Taken together, these findings show that miR-373 overexpression causes hESCs to exit from pluripotency and engage in mesendodermal differentiation. Conversely, inhibition of miR-373 did not exert any significant effect on pluripotency or lineage markers (Fig. S6). This may be due to the presence in hESCs of high levels of related miRNAs, such as miR-302 [11].
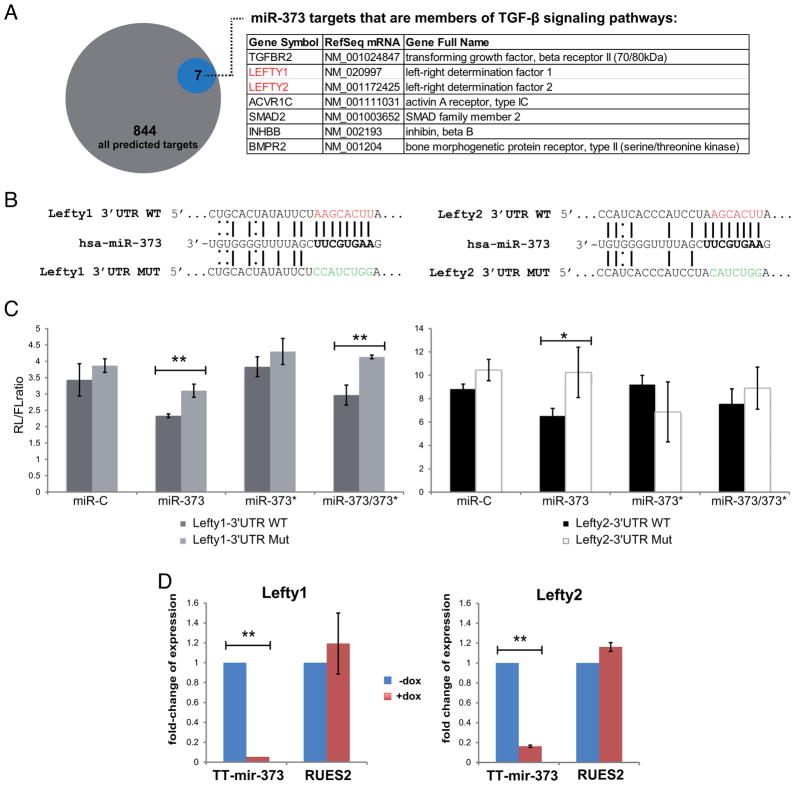
To study the targets of miR-373, we compared 844 predicted miR-373 targets (TargetScan [22]) with a list of 2042 genes that are expressed in RUES2 cells (Ismailoglu and Brivanlou, personal communication), and found 27 to be common (Fig S7). Among them we found Lefty1 and Lefty2, known inhibitors of Nodal ligands providing cue signals for the Smad2/3 branch of the TGFβ pathway. We tested whether the two human Lefty genes are bona fide targets of miR-373 using a standard luciferase-based reporter assay in HeLa cells. A miR-373 mimic caused repression of Lefty1- and Lefty2- WT 3′UTR, which was significantly relieved when their seeds were mutated (Fig. 4C). miR-373* mimic had no significant effect on either Lefty1 or Lefty2. Accordingly, miR-373 induction in hESCs led to a significant reduction in the levels of both Lefty mRNAs (Fig. 4D). Taken together, these findings suggest that miR-373, but not miR-373*, targets Lefty1 and Lefty2.
Figure 4. miR-373, but not miR-373*, targets Lefty1 and Lefty2.
(A) TargetScan predicts that miR-373 targets 844 3′UTR sequences. Members of the TGF-β signaling family are indicated in the table. (B) Schematic representation of the binding of miR-373 on either the wild-type (WT) or mutated (MUT) Lefty1 or Lefty2 3′UTRs; WT seeds are marked in red, MUT seeds in green, Watson-Crick base pairing with a straight line and U-G wobbles with a dotted line. (C) Luciferase assay in HeLa cells using Lefty1 and Lefty2 3′UTR Renilla(RL)-reporter constructs; Firefly (FL)-luciferase was used as a transfection control. Data points represent the RL/FL ratio and are average values from triplicates. Error bars represent standard deviations. *p<0.05, **p<0.01. (D) Real-Time qPCR analysis of Lefty1 and Lefty2 expression after 5 days of doxycycline treatment in TT-miR-373 and RUES2 cells. Data points are presented as fold-change of expression relative to uninduced (−Dox, blue bars) cells, ±s.dev.
Conclusions
Even though miRNAs have been shown to participate in the control of crucial hESC properties [8, 10], little is known about their upstream regulation. Here, we describe that some miRNAs are downstream targets of Activin/Nodal signaling. In particular, the levels of miR-373/373* decrease in response to Smad2/3 inhibition. Our analysis was carried out within 24 hours after the SB treatment; at this time, direct transcriptional targets of Smad2/3, such as Nodal and Lefty genes, are already downregulated, while indirect targets, such as the POU5F1 gene, are not [21]. Together with our ChIP analysis and the evidence that Activin-mediated induction of miR-373 is independent from protein synthesis, this suggests that miR-373/373* is likely to be a direct target of Smad2/3, although we cannot exclude that other signaling cues, such as the Wnt pathway [23], participate to the regulation of miR-373/373* transcription.
Among miRNAs belonging to the miR-302 family in mammals, the miR-302/367 cluster is highly conserved in mice and humans, and is directly under the control of the core TFs Nanog, Sox2 and Oct4 [9, 24, 25]. Conversely, the human miR-371/372/373 cluster shows poor similarity with the mouse homolog (the miR-290 cluster) and although its regulation in the context of human cancer cells is beginning to be studied [23, 26, 27], to our knowledge, nothing is currently known about its regulation in hESCs. Recent studies identified miRNAs under the control of SMAD2/3 in mouse adult and embryonic stem cells [28, 29]. Interestingly, the subset of miRNAs regulated by this branch of the TGFβ pathway is different in mouse and human ESCs, with the remarkable exception of the miR-290/miR-371/372/373 cluster, whose regulation seems conserved.
In our previous work [11] we have focused on the role of miR-302 during hESC differentiation, as a model for human embryonic development. Conversely, in the present paper we investigated the role of miR-373 under pluripotency conditions, before induction of differentiation. We found here that miR-302 levels are not affected by Nodal/Activin signaling inhibition, while miR-373 is under the direct control of SMAD2/3. Therefore, despite belonging to the same family, the two miRNAs may be not totally redundant in hESCs. Human ESCs express prevalently miR-302/367, while mouse ESCs (mESCs) are dominated by miR-290 members. Interestingly, the conversion from a naïve to a primed state in mESCs correlates with a switch between the miR-302/367 and the miR-290 clusters in terms of miRNA abundance [30]. The primed state of mESCs corresponds to mouse epiblast stem cells (EpiSCs), which are more similar to hESCs than mESCs [31]. Accordingly, miR-373 in hESCs is less abundant than miR-302. Our present and previous work [11] shows that members of both clusters converge on a common target, Lefty. However, while upregulation of miR-302 had no consequences on undifferentiated hESCs [11], increased levels of miR-373 led to differentiation of hESCs towards mesendoderm. Since the induction of mesendoderm upon miR-373 overexpression could be only marginally rescued by a recombinant Lefty protein (Fig S8), other targets probably contribute to this biological effect.
In conclusion, a specific amount of miR-373 specified by a discrete level of Activin/Nodal signaling must be maintained by the cell in order to prevent differentiation. According to our model, such specific Nodal/Activin signaling threshold activates both miR-373 and the core TF circuitry that, in its turn, promotes transcription of the miR-302/367 cluster. Moreover, both Nodal and Lefty genes are positively regulated by Smad2/3, thus establishing positive and negative feedback interactions. In this view, a critical role played by miR-373 and miR-302 in the TGFβ pathway might be the maintenance of a balance between agonists, such as Nodal, and antagonists, such as Lefty.
Supplementary Material
Highlights.
A subset of miRNAs is controlled by the TGFβ pathway (Nodal/Activin branch) in hESCs
One of them is the miR-302 family member, miR-373
Overexpression of miR-373 leads to differentiation along the mesendodermal lineage
The Nodal inhibitor, Lefty, is a direct target of miR-373
Acknowledgments
Grant acknowledgment: A.H.B. was supported for this work by NIH grant #5R01GM097615
We are grateful to Dr. Mayte Suarez-Farinas for help with the bioinformatics analysis of the miRNA profiling results. We thank Dena Sweeney for technical assistance, Dr. Aryeh Warmflash for quantitative analysis of differentiation, Dr. Gist Croft for critical discussions and all the members of the AHB laboratory for helpful discussion. This work was supported by an NIH grant (#5R01GM097615) to AHB.
Footnotes
Author contribution: Alessandro Rosa: concept and design, experiment design, collection, analysis and interpretation of data, manuscript writing; Marilena D. Papaioannou: experiment design, collection, analysis and interpretation of data, manuscript writing; Joanna E. Krzyspiak: experiment collection. Ali H. Brivanlou: concept and design, interpretation of data, manuscript writing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pera MF, Tam PPL. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–720. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 2.Warmflash A, Arduini BL, Brivanlou AH. The molecular circuitry underlying pluripotency in embryonic stem cells. Wiley Interdiscip Rev Syst Biol Med. 2012;4:443–456. doi: 10.1002/wsbm.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James D, Levine AJ, Besser D, Brivanlou AH. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 4.Smith JR, Vallier L, Lupo G, Alexander M, Harris WA, Pedersen RA. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev Biol. 2008;313:107–17. doi: 10.1016/j.ydbio.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MWB, Cho CH-H, Martinez A, Rugg-Gunn P, Brons G, Pedersen RA. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh AM, Reynolds D, Cliff T, Ohtsuka S, Mattheyses AL, Sun Y, Menendez L, Kulik M, Dalton S. Signaling Network Crosstalk in Human Pluripotent Cells: A Smad2/3-Regulated Switch that Controls the Balance between Self-Renewal and Differentiation. Cell Stem Cell. 2012;10:312–326. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez NJ, Gregory RI. MicroRNA Gene Regulatory Pathways in the Establishment and Maintenance of ESC Identity. Cell Stem Cell. 2010;7:31–35. doi: 10.1016/j.stem.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosa A, Brivanlou AH. microRNAs in early vertebrate development. Cell Cycle. 2009;8:3513–3520. doi: 10.4161/cc.8.21.9847. [DOI] [PubMed] [Google Scholar]
- 11.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 Family Controls Mesendodermal Fate Specification via Species-Specific Target Selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. Embo J. 2010;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 14.Suh MR, Lee Y, Kim JY, Kim S-K, Moon S-H, Lee JY, Cha K-Y, Chung HM, Yoon HS, Moon SY, Kim VN, Kim K-S. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- 16.James D, Noggle SA, Swigut T, Brivanlou AH. Contribution of human embryonic stem cells to mouse blastocysts. Dev Biol. 2006;295:90–102. doi: 10.1016/j.ydbio.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 17.Lacoste A, Berenshteyn F, Brivanlou AH. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Arduini BL, Brivanlou AH. Modulation of FOXD3 Activity in Human Embryonic Stem Cells Directs Pluripotency and Paraxial Mesoderm Fates. Stem Cells. 2012;30:2188–2198. doi: 10.1002/stem.1200. [DOI] [PubMed] [Google Scholar]
- 19.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inman GJ, Nicolás FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 21.Besser D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J Biol Chem. 2004;279:45076–45084. doi: 10.1074/jbc.M404979200. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Zhou AD, Diao LT, Xu H, Xiao Z-D, Li J-H, Zhou H, Qu L-H. β-Catenin/LEF1 transactivates the microRNA-371–373 cluster that modulates the Wnt/β-catenin-signaling pathway. Oncogene. 2012;31:2968–2978. doi: 10.1038/onc.2011.461. [DOI] [PubMed] [Google Scholar]
- 24.Barroso-delJesus A, Romero-López C, Lucena-Aguilar G, Melen GJ, Sanchez L, Ligero G, Berzal-Herranz A, Menendez P. Embryonic stem cell-specific miR302–367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairo S, Wang Y, de Reyniès A, Duroure K, Dahan J, Redon M-J, Fabre M, McClelland M, Wang XW, Croce CM, Buendia M-A. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci USA. 2010;107:20471–20476. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keklikoglou I, Zhang JD, Heckmann D, Shavinskaya A, Allgayer H, Gückel B, Fehm T, Schneeweiss A, Sahin O, Wiemann S, Tschulena U. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-kappaB and TGF-beta signaling pathways. Oncogene. 2012;31:4150–63. doi: 10.1038/onc.2011.571. [DOI] [PubMed] [Google Scholar]
- 28.Redshaw N, Camps C, Sharma V, Motallebipour M, Guzman-Ayala M, Oikonomopoulos S, Thymiakou E, Ragoussis J, Episkopou V. TGF-β/Smad2/3 signaling directly regulates several miRNAs in mouse ES cells and early embryos. PLoS ONE. 2013;8:e55186. doi: 10.1371/journal.pone.0055186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE. 2011;6:e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jouneau A, Ciaudo C, Sismeiro O, Brochard V, Jouneau L, Vandormael-Pournin S, Coppée J-Y, Zhou Q, Heard E, Antoniewski C, Cohen-Tannoudji M. Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA. 2012;18:253–264. doi: 10.1261/rna.028878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.