Abstract
High water use efficiency (WUE) can be achieved by coordination of biomass accumulation and water consumption. WUE is physiologically and genetically linked to carbon isotope discrimination (CID) in leaves of plants. A population of 148 recombinant inbred lines (RILs) of sunflower derived from a cross between XRQ and PSC8 lines was studied to identify quantitative trait loci (QTL) controlling WUE and CID, and to compare QTL associated with these traits in different drought scenarios. We conducted greenhouse experiments in 2011 and 2012 by using 100 balances which provided a daily measurement of water transpired, and we determined WUE, CID, biomass and cumulative water transpired by plants. Wide phenotypic variability, significant genotypic effects, and significant negative correlations between WUE and CID were observed in both experiments. A total of nine QTL controlling WUE and eight controlling CID were identified across the two experiments. A QTL for phenotypic response controlling WUE and CID was also significantly identified. The QTL for WUE were specific to the drought scenarios, whereas the QTL for CID were independent of the drought scenarios and could be found in all the experiments. Our results showed that the stable genomic regions controlling CID were located on the linkage groups 06 and 13 (LG06 and LG13). Three QTL for CID were co-localized with the QTL for WUE, biomass and cumulative water transpired. We found that CID and WUE are highly correlated and have common genetic control. Interestingly, the genetic control of these traits showed an interaction with the environment (between the two drought scenarios and control conditions). Our results open a way for breeding higher WUE by using CID and marker-assisted approaches and therefore help to maintain the stability of sunflower crop production.
Introduction
Water use efficiency (WUE) as a breeding target can be defined as the ratio of biomass production to water consumption. Breeding for WUE and drought-resistant crop varieties has been a critical area of agricultural research worldwide [1]–[3]. Substantial efforts have been devoted to identifying and selecting for morphological and physiological traits that increase WUE and yield under rain-fed conditions [2], [4]–[5]. In field conditions, water consumption is usually difficult to determine. Nevertheless, WUE can be represented by measuring leaf carbon isotope discrimination (CID) [6]–[7]. Because the CID has been demonstrated to be a simple but reliable measure of WUE, the negative correlation between them has been used as an indirect method in selection to improve WUE [8]–[10]. The principle mechanisms underlying the variation of CID act through variation in the intercellular CO2 concentration (ci) maintained in leaves [6]. The value of ci is determined through the coordinated regulation of carboxylation capacity (photosynthesis) and stomatal control of leaf diffusive conductance (transpiration regulation) [6]–[7].
Genetic variation underlying quantitative traits, such as WUE and CID, which are generally under considerable environmental influence, is governed by quantitative trait loci (QTL) [11]–[14]. QTL mapping provides a starting point in breeding programs [15]–[16] and for cloning of the causal mutations by fine mapping.
QTL mapping of WUE is rarely reported. Four QTL associated with WUE have been identified in soybean [17]. The inheritance of WUE has been studied using simple sequence repeat (SSR) markers in alfalfa [18]. In contrast, QTL mapping of CID has been reported by numerous authors. The first QTL identified for CID was reported by Martin and Nienhuis [19]. These authors identified four QTL associated with CID in tomato. Since that time, QTL for CID have been identified across a wide range of species, for example in cotton [20], rice [21], barley [22], Arabidopsis [23], and in wheat [24]. However, to our knowledge, QTL of WUE and CID in sunflower have never been reported.
Most of the work identifying QTL of WUE and CID has been done in well-watered conditions, with only one study in a drought situation. There is no report on the QTL identification of WUE and CID of crops subjected to different scenarios of water deficit establishment.
The objectives of the present study are to identify QTL controlling WUE and CID in a population of RILs of sunflower, and to compare QTL associated with these traits in a dual drought scenario: (i) a progressively water-stressed establishment and (ii) a stable water deficit treatment. We are interested in providing new insights into the genetic architecture of WUE and CID, and in contributing to the potential of sunflower breeding by improved WUE.
Materials and Methods
Plant materials
A population of 150 recombinant inbred lines (RILs) was used in two experiments. A population of these RILs was named INEDI and was obtained by single seed descent (self-pollination to at least F8) from a cross between XRQ and PSC8 [25].
Experiments and trait measurements
Two experiments were conducted in spring 2011 (Exp. 2011) and in spring 2012 (Exp. 2012) under quite similar weather conditions. Plants were grown in a greenhouse at the INRA Auzeville station, Toulouse, France (43°31′46,94″ N; 1°29′59,71″ E). Greenhouse air temperature was set at 25/18±2°C (day/night) and relative humidity was about 55–75±5%.
Three seeds per genotype were sown in a pot (volume: 2 liters) at the beginning of the experiments. The pots contained a mixture of 50% soil (collected from the field), 30% organic matter and 20% sand. These pots were arranged on 100 balances (maximum capacity 30 kg, precision 2 g, model SXS, GRAM, Spain), with six pots per balance (total pot number in greenhouse was 600). Each pot was then covered with a 3 mm layer of polystyrene sheet with a hole in the middle to allow normal plant growth, thus reducing the evaporation of water from the soil surface. Throughout the experiments, the amounts of water in the pots were determined by weighing the pots every day. This weighing recorded the amount of daily water loss, corresponding to the daily transpiration of the plants. For each pot, at the end of the experiment, cumulative daily transpiration was called CWT (the cumulative water transpired). Biomass was separated into leaves and stems at harvest. Total dry aerial biomass (BM) was obtained after drying at 80°C for 48 h. WUE was determined at the end of the experiment, defined as the ratio of BM to CWT. In addition, a dual drought scenario strategy for the two experiments (explained in detail below) was studied.
Experiment conducted in 2011: scenario of progressive water stress
A randomized complete block design with three replicates was used for the progressive water stress treatments (three replicates×150 genotypes = 450 plants; called WS). There was another replicate (150 plants) that was considered as a well-watered treatment, called WW.
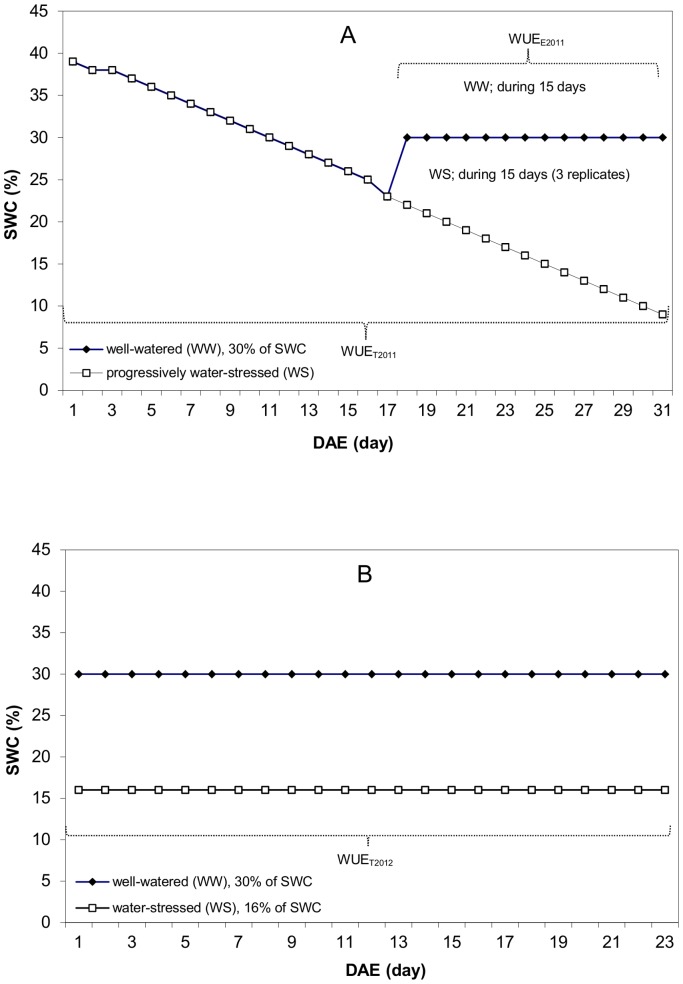
At 1 day after emergence (DAE), 17 days after sowing (DAS), all 600 pots were watered to field capacity, corresponding to 39.5% of soil water content (SWC). These 600 pots (WW and WS) were kept without irrigation until 17 DAE (Fig. 1A). In these conditions, stomatal conductance of the plant was still not affected. We calculated that stomatal conductance started to decrease at an average SWC of about 21% (unpublished data).
Figure 1. Principles of the water treatments used in this study.
(A) In experiment 2011, three replicates (each of 150 plants) were subjected to progressive water-stress by water withholding from 1 to 31 DAE. In this experiment a control replicate (150 plants) was watered to maintain non-stressful conditions (SWC = 30%). (B) In Experiment 2012, two replicates (each of 150 plants) were maintained at in stressful conditions SWC = 16% from 1 to 23 DAE whereas two other replicates (each of 150 plants) were irrigated to maintain non-stressful conditions (SWC = 30%). DAE: day after emergence.
Starting at 17 DAE, when genotypes reached around 23% of SWC, we irrigated the WW treatment to 30% of SWC and we maintained this SWC by daily irrigation. The WS treatment was kept without irrigation until harvest (during 15 days).
Two determinations of WUE were made. The first was the total water use efficiency, WUET2011, calculated by dividing the BM by the CWT31d. CWT31d is the cumulative water transpired during 31 days (from 1 to 31 DAE). The second calculation of WUE was made during the period when the two treatments differed in their soil water content (WW and WS), from 17 to 31 DAE, and called WUEE2011 (water use efficiency “estimation”). WUEE2011 was calculated by dividing the “estimated biomass” (BME), by the CWT15d, calculated from 17 to 31 DAE. BME = BM – BM17, where BM17 is the biomass estimated at 17 DAE. In addition, the BM17 was calculated as follows: BM17 = (LA17/LA31)×BM, where LA17 and LA31 are the leaf areas measured on 17 and 31 DAE, respectively.
Experiment conducted in 2012: scenario of stable SWC
A randomized complete block design with two treatments and two replicates was performed (300 pots per treatment). Treatments consisted of two levels of stable SWC which was imposed: well-watered (30% of SWC, namely WW) and water-stressed (16% of SWC, namely WS) (Fig. 1B).
At 1 DAE (19 DAS), stable water contents corresponding to 30% of SWC (WW) and 16% of SWC (WS) were maintained for 23 days (Fig. 1B). WUE was calculated by dividing the BM by the CWT23d (WUET2012), where CWT23d is the cumulative water transpired during 23 days (from 1 to 23 DAE).
Determination of carbon isotope discrimination (CID)
Carbon isotope composition (δ) was calculated relative to the international Pee Dee Belemnite (PDB) standard [26]: δplant = (Rsa – Rsd)/Rsd×1000 [‰] where Rsa and Rsd are the 13C:12C ratios of the sample and the standard, respectively [27]. Carbon isotope discrimination (CID), a factor related to isotope fractionation by the photosynthetic process relative to the source carbon was then estimated as CID = (δair – δplant)/(1+ δplant/1000) where δair is the 13C composition of atmospheric CO2, which is assumed to be −8.0‰ [26]. Before calculating CID, oven-dried leaves of each plant were ground into a homogenous fine powder and 2–3 mg subsamples were weighed and placed into tin capsules (Elemental Microanalysis, UK) to be analyzed using a continuous flow Isotope Ratio Mass Spectrometer (Sercon Ltd., Cheshire, UK) at UC Davis Stable Isotope Facility (California, USA).
Genetic map construction
A set of 9832 SNPs were used to produce an Infinium HD iSelect BeadChip (Infinium). These SNPs were selected from either genomic re-sequencing or transcriptomic experiments. The gDNA from the INEDI RILs population obtained from the cross between XRQ and PSC8 lines (210 samples) were genotyped with the Infinium array. All genotyping experiments were performed by Integragen (IntegraGen SA, Genopole Campus 1 - Genavenir 8, 5 rue Henri Desbruères, 91000 Evry, France) and the genotypic data were obtained with the Genome Studio software (Illumina) with automatic and manual calling. From the 9832 SNPs, 7094 were technically functional with more than 200 samples having a genotyping data. From this set of 7094 markers, 2576 were polymorphic between XRQ and PSC8 and 2164 did not show distortion of segregation in the population. We used CarthaGène v1.3 [34] to build the genetic maps. We added the genotypic data of markers from a consensus map [35] to the set of the 2164 SNPs to assign them to the appropriate LG to the group 0.3 8 in CarthaGène. They were ordered using the lkh 1 -1 function in CarthaGène for each group. The genetic map consisted of 2610 markers located on the 17 LG for a total genetic distance of 1863.1 cM and grouped on 999 different loci. All data will be available through the www.heliagene.org portal.
Statistical and QTL analysis
The data were first tested for normal distribution with the Kolmogorov-Smirnov test. These data were subjected to analysis of variance (ANOVA) and phenotypic correlation analysis (Pearson’s correlation) using the software of statistical package PASW statistics 18 (IBM, New York, USA). Means were compared using a Student-Newman-Keuls (SNK) test (P<0.05). The broad sense heritability (h2) was then computed from the estimates of genetic (σ2g) and residual (σ2e) variances derived from the expected mean squares of the analyses of variance as h2 = σ2g/(σ2g+ σ2e/r), where r was the number of replicates.
QTL identification was performed using MCQTL, software for QTL analysis (http://carlit.toulouse.inra.fr/MCQTL/). The MCQTL software package can be used to perform QTL mapping in a multi-cross design. It allows the analysis of the usual populations derived from inbred lines [28]. MCQTL package is comprised of three software applications. The first component, TranslateData reads data from MAPMAKER [29] like files. The second component, ProbaPop computes QTL genotype probabilities given marker information at each chromosome location for each family and stores them in XML formatted files. The last component, Multipop builds the pooled model using the genotype probabilities, computes Fisher tests and estimates the model parameters [28]. The statistical significance of QTLs was assessed using the MCQTL test, which is equal to –log(P-value (F-test)), as described in the MCQTL user guide.
Significant thresholds (P<0.05) for QTL detection were calculated for each dataset using 1000 permutations [30] and a genome-wide error rate of 0.01 (Type I error). The corresponding type I error rate at the whole-genome level was calculated as a function of the overall number of markers in the map and the number of markers in each linkage group [31]. In our analysis, the threshold for the Fisher test (–log(P-value (F-test))) was 3.69 for both experiments. This threshold was an average of several thresholds of the traits at a significance level of 5% and was determined after 1000 permutations.
In each experiment, the QTL detection was also performed to identify QTL for the phenotypic response (called “response QTL”), calculated as the difference between two different water treatments (WW and WS). This allowed us to detect chromosome regions having quantitative effects on traits, depending on the environment [32]–[33].
Results
Genotypic variability and phenotypic correlation between water use efficiency (WUE) and carbon isotope discrimination (CID)
In general, a normal distribution was observed for WUE and CID traits across the two experiments and water treatments, except for WUET2012 and CID in Exp. 2012 at WW conditions, the distributions deviate from normality according to the Kolmogorov-Smirnov test (Fig. 2 and 3). As normalizing data through transformation may misrepresent differences among individuals by pulling skewed tails toward the center of the distribution [30], all phenotypic analyses were performed on untransformed data.
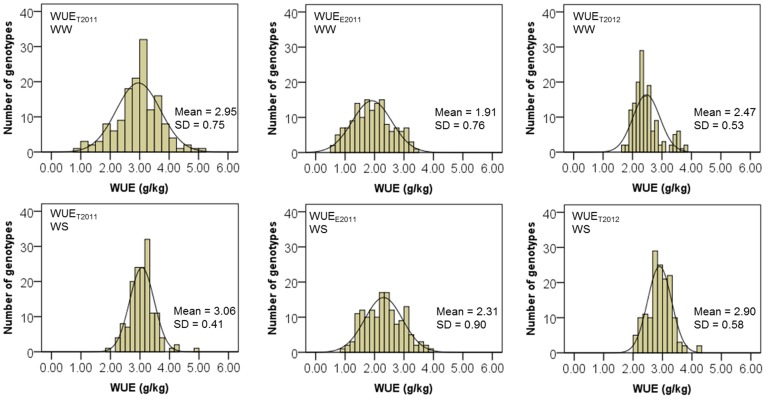
Figure 2. Frequency distribution for water use efficiency (WUE) in Exp. 2011 and 2012 of 150 recombinant inbred lines (RILs).
WUET2011: total water use efficiency “total” in Exp. 2011; WUEE2011: water use efficiency “estimation” in Exp. 2011; WUET2012: water use efficiency “total” in Exp. 2012. WW: well-watered; WS: water-stressed. For WUET2011 and WUEE2011 at WW, data represent 150 RILs (n = 150); for WUET2011 and WUEE2011 at WS, data represent mean of three replicates of 150 RILs (n = 150); for WUET2012 at WW and WS, data represent mean of two replicates of 150 RILs (n = 150). SD: standard deviation.
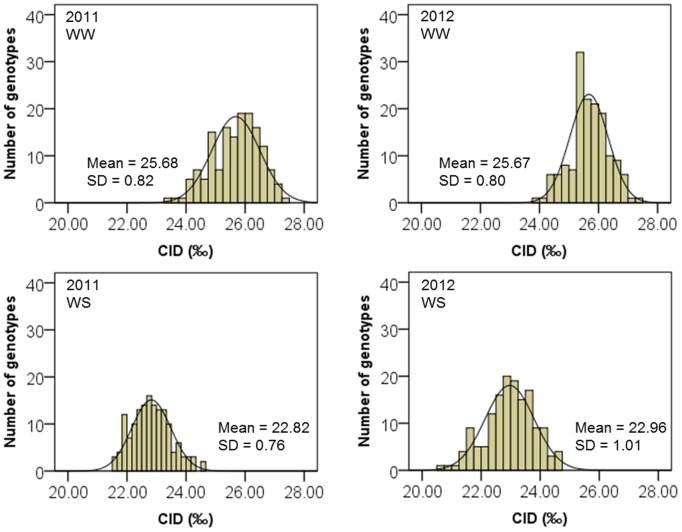
Figure 3. Frequency distribution for carbon isotope discrimination (CID) in Exp. 2011 and 2012 of 150 recombinant inbred lines (RILs).
WW: well-watered; WS: water-stressed. For CID in Exp. 2011 at WW, data represent 150 RILs (n = 150); for CID in Exp. 2012 at WS, data represent mean of three replicates of 150 RILs (n = 150); for CID in 2012 at WW and WS, data represent mean of two replicates of 150 RILs (n = 150). SD: standard deviation.
Higher mean values for WUE for WS (2.31 to 3.06 g.kg−1) than for WW (1.91 to 2.95 g.kg−1) (Table S1 and S2) were observed in each experiment. In contrast, higher mean values for CID for WW than for WS were also observed in each experiment. In addition, a similar range of WUE and CID values was observed in both experiments for both WW and WS (for WUE in Exp. 2011 was represented by the WUEE2011). In addition, significant genotypic effects were detected for all traits in Exp. 2011 (Table S1), and significant genotypic and SWC effects were detected for all traits in Exp. 2012 (Table 1).
Table 1. Heritability (h 2) and mean square (MS) of analysis of variance (ANOVA) for water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) for 150 recombinant inbred lines (RILs), two stable soil water contents (SWC) and two replicates in Exp. 2012 (n = 600).
| Trait | h2 | MS | ||
| Genotype | Soil water content | Genotype×soil water content | ||
| WUET2012 | 0.26 | 0.50*** | 28*** | 0.25ns |
| CID | 0.41 | 1.68*** | 1100*** | 0.53ns |
| BM | 0.36 | 0.51*** | 180*** | 0.29** |
| CWT23d | 0.36 | 40862*** | 31746440*** | 25565*** |
**Significant at P<0.01,
***significant at P<0.001.
Not significant.
The heritabilities of CID were usually higher than those of WUE in both experiments (CID with WUET2011 or WUET2012), except that the heritability of WUEE2011 was higher than that of CID ( and 1).
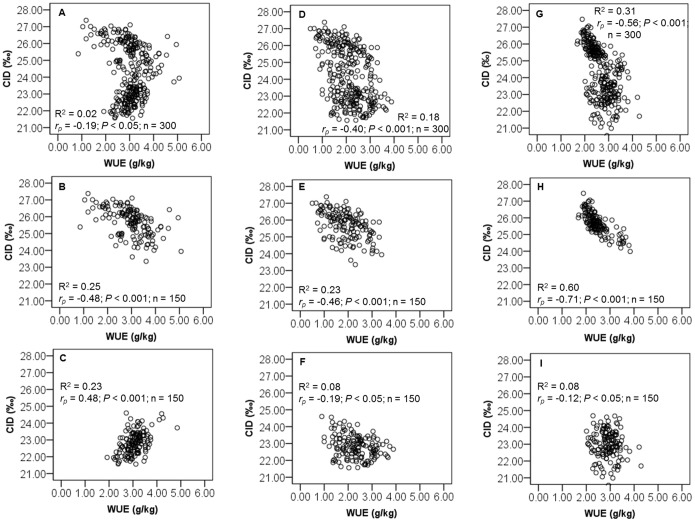
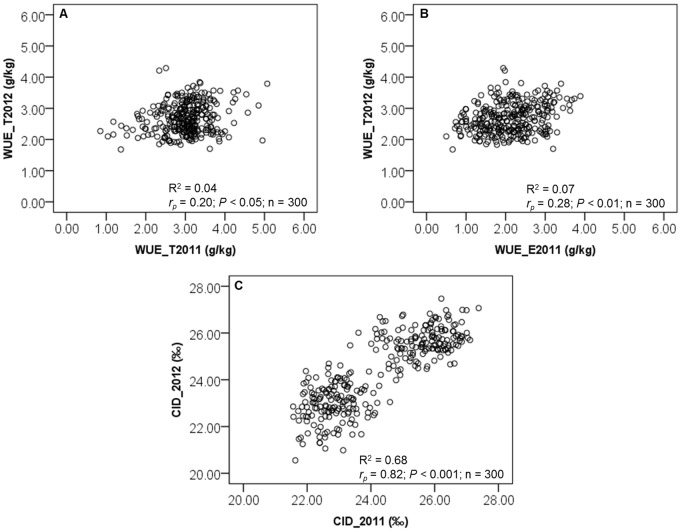
Significant negative correlations were observed between WUE and CID in both experiments (rp = −0.197, P<0.05; rp = −0.409, P<0.001; rp = −0.565, P<0.001 for the correlations of WUET2011, WUEE2011, WUET2012 with the CID, respectively; Fig. 4, Table S3, S4 and S5). However, when we determined the correlation between WUE and CID for each treatment, we observed a positive correlation between the WUET2011 and CID in Exp. 2011 for WS (Fig. 4 and Table S4). In addition, a significant phenotypic correlation was observed between Exp. 2011 and 2012 for both WUE and CID (Fig. 5).
Figure 4. Relationship between water use efficiency (WUE) and carbon isotope discrimination (CID) of 150 recombinant inbred lines (RILs) in Exp. 2011 and Exp. 2012.
Relationship between (A) WUET2011 and CID in Exp. 2011, (B) WUET2011 and CID at WW in Exp. 2011, (C) WUET2011 and CID at WS in Exp. 2011, (D) WUEE2011 and CID in Exp. 2011, (E) WUEE2011 and CID at WW in Exp. 2011, (F) WUEE2011 and CID at WS in Exp. 2011, (G) WUET2012 and CID in Exp. 2012, (H) WUET2012 and CID at WW in Exp. 2012; (I) WUET2012 and CID at WS in Exp. 2012. Phenotypic correlation (rp) value is provided in each graph.
Figure 5. Relationship between (A, B) WUE and (B) CID values for 150 recombinant inbred lines (RILs) determined in two separate experiments (Exp. 2011 and 2012).
For each trait and experiment, mean of well-watered (WW) and water-stressed (WS) plants were grouped together (n = 300). Phenotypic correlation (rp) value is provided in each graph.
QTL identified for water use efficiency (WUE)
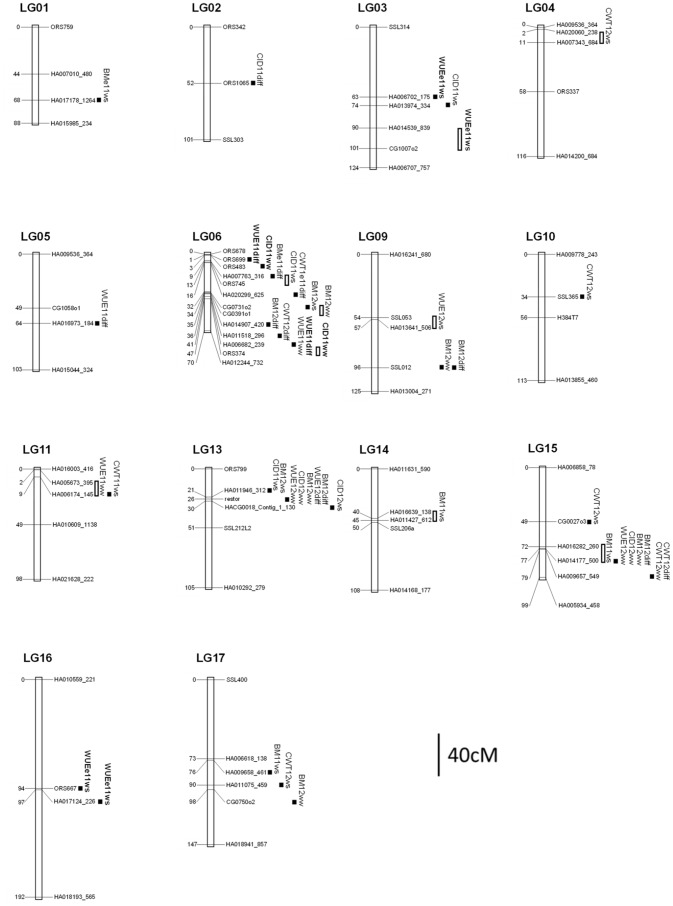
In Exp. 2011, two QTL for WUET2011 were detected for WW and four QTL for WUEE2011 were detected for WS (Table 2). For WW, the QTL were located on LG06 and LG11 with the highest likelihood odds ratio (LOD) value at 3 cM (QTL of WUE11ww.11.1) (Fig. S1). The marker for the QTL of WUE11ww.11.1 was identified between the markers of HA005673_395 and HA006174_145 (Fig. 6). For WS, the QTL were located on chromosomes LG03 and LG16 (two QTL for each chromosome) with the highest LOD value at 6 cM, the QTL of WUEe11ws.16.2, and the marker of this QTL was HA017124_226. A “response QTL” for WUE (WUE11diff.06.2) was collocated with QTL of WUE11ww.06.1. In addition, two other “response QTL” were found on LG05 and LG06. The additive effects of the WUE11ww.06.1 and WUE11ww.11.1 were −0.14 and 0.11 while the additive effects of the WUEe11ws.03.1, WUEe11ws.03.2, WUEe11ws.16.1, and WUEe11ws.16.2 were −0.13, 0.13, 0.38 and −0.44, respectively.
Table 2. Significant quantitative trait loci (QTL) detected for water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) under under well-watered and progressive water-stressed treatments in Exp. 2011.
| Trait | Treatment | Chromosome | QTL name | QTL position (cM) | Inferior position (cM) | Superior position (cM) | R2a (%) | R2 globalb (%) | Additive effect c |
| WUET2011 | WW | LG06 | WUE11ww.06.1 | 41.1 | 0 | 69.5 | 7 | 13 | −0.14 |
| WW | LG11 | WUE11ww.11.1 | 7.8 | 0 | 17.4 | 9 | 13 | 0.11 | |
| CID | WW | LG06 | CID11ww.06.1 | 3 | 0.45 | 14.4 | 12 | 17 | −0.15 |
| WW | LG06 | CID11ww.06.2 | 47.3 | 23.6 | 60.1 | 8 | 17 | 0.12 | |
| WUEE2011 | WS | LG03 | WUEe11ws.03.1 | 63.2 | 53.8 | 95.7 | 7 | 21 | −0.13 |
| WS | LG03 | WUEe11ws.03.2 | 97.7 | 63.9 | 124 | 5 | 21 | 0.13 | |
| WS | LG16 | WUEe11ws.16.1 | 94.1 | 92.7 | 96.1 | 11 | 21 | 0.38 | |
| WS | LG16 | WUEe11ws.16.2 | 97.1 | 96.1 | 99.3 | 15 | 21 | −0.44 | |
| CID | WS | LG03 | CID11ws.03.1 | 73.6 | 52 | 76.2 | 15 | 25 | −0.13 |
| WS | LG06 | CID11ws.06.1 | 11.3 | 0 | 16.6 | 9 | 25 | −0.1 | |
| WS | LG13 | CID11ws.13.1 | 21.2 | 0 | 36.5 | 10 | 25 | −0.13 | |
| BM | WS | LG14 | BM11ws.14.1 | 42.4 | 0 | 108 | 5 | 17 | 0.01 |
| WS | LG15 | BM11ws.15.1 | 76.3 | 0 | 98.9 | 7 | 17 | 0.02 | |
| WS | LG17 | BM11ws.17.1 | 76.1 | 0 | 112 | 6 | 17 | −0.02 | |
| BME | WS | LG01 | BMe11ws.01.1 | 67.8 | 46.3 | 74.9 | 9 | 9 | 0.02 |
| CWT31d | WS | LG11 | CWTe11ws.11.1 | 9.1 | 0 | 20.5 | 7 | 7 | −4.28 |
WW: well-watered, WS: progressive water-stressed.
Phenotypic variance explained by QTL effect.
Total of phenotypic variances explained by QTL effects.
Additive effect estimated as one-half the difference in homozygotes carrying either allele of parents (XRQ or PSC8). Positive values indicate that XRQ allele increases the trait value, while negative values indicate that PSC8 allele increases the trait value.
Figure 6. Genetic locations of QTL for water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) in the progressive stress experiment (2011) and the stable stress experiment (2012).
Numbers on the left of linkage groups (LG) indicate the cumulative distance in centimorgan (cM) to the first marker at the top LG. Marker names and QTL are specified to the right of LG. The same QTLs which are found in a LG are shown in bold. Not all these chromosomes contain the complete markers (each chromosome has only been provided by the markers at the top, middle and bottom of LG as well as the markers for identified QTLs). QTL confidence intervals were estimated using the two-LOD confidence region.
In Exp. 2012, two QTL for WUET2012 were detected at WW and one QTL for WUET2012 at WS (Table 3). For WW, the QTL were detected on chromosome LG13 and LG15 with the highest LOD value at 25 cM, the QTL of WUE12ww.13.1, and the markers for this QTL was restor (Fig. 6, Fig. S1). For WS, a QTL was detected on chromosome LG09 (QTL of WUE12ws.09.1) with the LOD value at 3 cM. The marker for the QTL of WUE12ws.09.1 was identified between the markers of SSL053 and HA013641_506. In addition, a “response QTL” for WUE (WUE12diff.13.1) was co-located with the QTL of WUE12ww.13.1 and CID12ww.13.1 (Table 4). The additive effects of WUE12ws.09.1, WUE12ww.13.1 and WUE12ww.15.1 were 0.20, 0.04 and −0.06, respectively.
Table 3. Significant quantitative trait loci (QTL) detected for water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) under well-watered and water-stressed treatments in Exp. 2012.
| Trait | Treatment | Chromosome | QTL name | QTL position (cM) | Inferior position (cM) | Superior position (cM) | R2a (%) | R2 globalb (%) | Additive effect c |
| WUET2012 | WW | LG13 | WUE12ww.13.1 | 26.2 | 25.14 | 26.95 | 42 | 45 | 0.2 |
| WW | LG15 | WUE12ww.15.1 | 77.1 | 13.94 | 90.73 | 6 | 45 | 0.04 | |
| CID | WW | LG13 | CID12ww.13.1 | 26.2 | 4.29 | 37.43 | 21 | 26 | 0.2 |
| WW | LG15 | CID12ww.15.1 | 77.1 | 0 | 98.90 | 6 | 26 | 0.07 | |
| BM | WW | LG06 | BM12ww.06.1 | 33.6 | 29.2 | 40.74 | 13 | 40 | 0.1 |
| WW | LG09 | BM12ww.09.1 | 95.5 | 88.25 | 114.1 | 9 | 40 | 0.08 | |
| WW | LG13 | BM12ww.13.1 | 26.2 | 24.16 | 37.81 | 2 | 40 | 0.17 | |
| WW | LG15 | BM12ww.15.1 | 77.1 | 47.47 | 82.62 | 12 | 40 | 0.1 | |
| CWT23d | WW | LG15 | CWT12ww.15.1 | 79.1 | 40.28 | 87.03 | 7 | 7 | 26.07 |
| WUET2012 | WS | LG09 | WUE12ws.09.1 | 55.5 | 33.28 | 83.25 | 9 | 9 | −0.06 |
| CID | WS | LG13 | CID12ws.13.1 | 30.8 | 0 | 62.45 | 7 | 7 | 0.14 |
| BM | WS | LG06 | BM12ws.06.1 | 31.6 | 29.09 | 35.10 | 13 | 26 | 0.02 |
| WS | LG13 | BM12ws.13.1 | 21.2 | 0 | 29.86 | 9 | 26 | 0.02 | |
| WS | LG17 | BM12ws.17.1 | 98.2 | 68.14 | 111 | 7 | 26 | 0.02 | |
| CWT23d | WS | LG04 | CWT12ws.04.1 | 6 | 0 | 21.48 | 10 | 30 | −6.17 |
| WS | LG10 | CWT12ws.10.1 | 33.6 | 0 | 112.5 | 4 | 30 | 3.23 | |
| WS | LG15 | CWT12ws.15.1 | 49.5 | 28.33 | 80.98 | 10 | 30 | 5.32 | |
| WS | LG17 | CWT12ws.17.1 | 89.8 | 74.7 | 92.29 | 12 | 30 | −6.78 |
WW: well-watered (30% of SWC), WS: water-stressed (16% of SWC).
Phenotypic variance explained by QTL effect.
Total of phenotypic variances explained by QTL effects.
Additive effect estimated as one-half the difference in homozygotes carrying either allele of parents (XRQ or PSC8). Positive values indicate that XRQ allele increases the trait value, while negative values indicate that PSC8 allele increases the trait value.
Table 4. Significant “response quantitative trait loci (QTL)” detected for water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) in Exp. 2011 and Exp. 2012.
| Trait | Experiment | Chromosome | QTL name | QTL position (cM) | Inferior position (cM) | Superior position (cM) | R2a (%) | R2 globalb (%) | Additive effectc |
| WUET2011 | 2011 | LG05 | WUE11diff.05.1 | 64 | 0 | 103 | 5 | 14 | −0.7 |
| 2011 | LG06 | WUE11diff.06.1 | 1.3 | 0 | 22.9 | 7 | 14 | −0.1 | |
| 2011 | LG06 | WUE11diff.06.2 | 41.1 | 3 | 69.5 | 6 | 14 | 0.08 | |
| CID | 2011 | LG02 | CID11diff.02.1 | 52.1 | 0 | 101 | 7 | 7 | −0.1 |
| BME | 2011 | LG06 | BMe11diff.06.1 | 9.3 | 0 | 21.6 | 8 | 8 | −0.1 |
| CWT15d | 2011 | LG06 | CWTe11diff.06.1 | 15.9 | 0 | 22.4 | 9 | 9 | −19 |
| WUET2012 | 2012 | LG13 | WUE12diff.13.1 | 26.2 | 21.9 | 41.6 | 18 | 18 | 0.14 |
| BM | 2012 | LG06 | BM12diff.06.1 | 34.7 | 28.3 | 43 | 10 | 39 | 0.07 |
| 2012 | LG09 | BM12diff.09.1 | 95.5 | 0.1 | 111 | 10 | 39 | 0.07 | |
| 2012 | LG13 | BM12diff.13.1 | 26.2 | 24.1 | 38.8 | 20 | 39 | 0.14 | |
| 2012 | LG15 | BM12diff.15.1 | 77.1 | 74.1 | 82.2 | 13 | 39 | 0.09 | |
| CWT23d | 2012 | LG06 | CWT12diff.06.1 | 35.5 | 24.9 | 43.7 | 10 | 16 | 25.5 |
| 2012 | LG15 | CWT12diff.15.1 | 79.1 | 0 | 98.9 | 6 | 16 | 19.1 |
Phenotypic variance explained by QTL effect.
Total of phenotypic variances explained by QTL effects.
Additive effect estimated as one-half the difference in homozygotes carrying either allele of parents (XRQ or PSC8). Positive values indicate that XRQ allele increases the trait value, while negative values indicate that PSC8 allele increases the trait value.
QTL identified for carbon isotope discrimination (CID)
In Exp. 2011, two QTL for CID were detected at WW and three QTL for CID were detected at WS (Table 2). For WW, the QTL were located on the same chromosomes of LG06 with the highest LOD value at 4.5 cM, QTL of CID11ww.06.1, and the marker of this QTL was ORS483 (Fig. 6, Fig. S2). For WS, the QTL were identified on chromosomes LG03, LG06 and LG13 with the highest LOD value at 5.5 cM, the QTL of CID11ws.03.1, and the marker of this QTL was HA013974_334. Besides, there was one “response QTL” detected for CID on chromosome LG02 (CID11diff.02.1) (Table 4). The additive effects were −0.15 and 0.12 (for QTL of CID11ww.06.1 and CID11ww.06.2) while the additive effects were −0.13, −0.10, −0.13 (for the QTL of CID11ws.03.1, CID11ws.06.1 and CID11ws.13.1) (Table 2).
In Exp. 2012, two QTL for CID were detected at WW and one QTL for CID at WS (Table 3). For WW, the QTL were found on chromosomes LG13 and LG15 with the highest LOD value of 8.5 cM, the QTL of CID12ww.13.1, and the marker for this QTL was restor (Fig. 6, Fig. S2). For WS, a QTL was found on chromosome LG13 with an LOD value of 2.5 cM; the QTL of CID12ws.13.1, and the marker for this QTL was HACG0018_Contig_1_130. The additive effects for CID12ww.13.1 and CID12ww.15.1 were 0.20 and 0.07, respectively. The additive effect of the QTL of CID at WS (CID12ws.13.1) was 0.14.
QTL identified for related traits: biomass (BM) and cumulative water transpired (CWT)
In Exp. 2011, three significant QTL for BM, and one QTL for each of BME and CWT31d at WS were identified (Table 2). These QTL were detected on chromosomes LG14, LG15, LG17, LG01 and LG11. There were only two “response QTL” detected for each of BME and CWT15d. These QTL were detected on the same chromosome, LG06.
In Exp. 2012, seven QTL were identified for BM under both levels of SWC. For CWT23d, five significant QTL were detected under both levels of SWC. Further, six “response QTL” for BM and CWT23d were identified on chromosomes LG06, LG09, LG13 and LG15.
Discussion
Genetic variation and relationship between WUE and CID
In our experiments, increasing drought lead to an increase in WUE and a decrease in CID. This result was previously reported by Lauteri et al. [36] in sunflower and is well known in other crops, such as durum wheat (Triticum turgidum L.) [37], rice (Oryza sativa L.) [38] and eucalyptus (Eucalyptus microtheca) [39]. In addition, a similar range of values for WUE and CID was observed in the two experiments even though their water stress patterns differed. That was likely because the population had been constructed from parents that had specific responses in non-limited and limited water availability [40]–[41]. From the phenotypic data, XRQ exhibited low WUE while PSC8 exhibited high WUE (unpublished data).
CID is highly heritable trait and its heritability is usually higher rather than WUE [7], [11]. Nevertheless, in the present study, both of CID and WUE were influenced by environmental variation because the heritability values were below 50% [24]. A previous study [42] has shown that heritabilities for CID, measured on detached sunflower leaves, were above 50% (74–96%), indicating that genetic variance for CID was dominant. However, this result was obtained for plants grown in optimal watering conditions. Consequently, CID appeared dependent on genetic and environmental control. This trait is genetically complex [43], and its expression in leaves and other plant tissues varies with the water supply. In drought conditions, Rebetzke et al. [24] reported that low soil water availability decreases stomatal conductance, which can reduce genetic variance and heritability of CID.
Our work demonstrated the clear relationship between WUE and CID in different water regimes. For each water regime and all genotypes, we observed negative correlations between WUE and CID. These results are in accordance with those of previous work in sunflower [36], [42], and with those of numerous authors working on other crops [6], [8], [44]–[47]. In one case of progressive water stress, WUET2011 and CID, were positively correlated. This was probably due to the high variability of the soil water content during the progressive drought establishment (SWC was gradually decreased). A similar result was reported on alfalfa genotypes [48]: WUE (mg of dry matter per g H2O) was positively correlated with CID for plants subjected to progressive water stress during 7 days.
In the WW treatment, the high WUE was correlated with high BM and high CWT, while for the WS treatment the high WUE was still correlated to high BM but with low CWT. If increase in WUE is associated with reduced transpiration, such genotypes are often referred to as “conductance type”. On the other hand, if increase in WUE is correlated with increased photosynthesis, such genotypes can be categorized as “capacity types” [49]–[50]. Accordingly, the sunflower genotypes in our study can be categorized as an intermediate between “conductance” and “capacity” type, unlike rice genotypes that have been categorized as “conductance type” [51]. In addition, our results were in agreement with several authors [39], [52]–[53] who have suggested that plants that use water more efficiently by producing greater biomass for a given quantity of water transpired would grow more rapidly, resulting in a positive correlation between WUE and biomass production.
QTL identified for WUE and CID
Our study is the first to identify QTL for WUE and CID in sunflower subjected to drought. In Exp. 2011, significant regions affecting WUE were identified on four different chromosomes (LG03, LG06, LG11, LG16) in two water treatments and significant regions affecting CID were identified on three different chromosomes (LG03, LG06, LG13) for the same two water treatments. From these QTL, we observed a decrease and an increase of additive effects (XRQ), indicating that genes having both negative and positive effects had been involved in the difference in WUE and CID between the parental lines [54]. In Exp. 2012, the QTL for WUE were detected on three different chromosomes in two water treatments (LG09, LG13 and LG15) and the QTL for CID were identified on two different chromosomes in these two water treatments (LG13 and LG15). All these QTL increased the values of additive effects except the QTL of WUE12ws.09.1, indicating that XRQ allele increased the traits. These findings provide an explanation for the underlying genetic basis of the transgressive variation observed in the segregating population. This is in accordance with the argument proposed by Chapman et al. [55] and Vargas et al. [56], namely that a given QTL can have positive or negative additive effects, or none at all, depending on the drought scenario.
The WUE and CID were controlled by several QTL with small genetic additive (XRQ) effects, indicating that WUE and CID were genetically complex traits [2], [57]. Reports evaluating genetic analysis for CID in other crops like soybean [58], cotton [59] and rice [54] have identified multiple QTL of smaller effect associated with the trait. However, in the present study, the QTL for WUE and CID explained 42% and 21% of the highest phenotypic variance (R2). These R2 values are higher than those found by previous authors for other crops, for example, rice [10], [54], wheat [24] and barley [60]–[61].
Expression of QTL for WUE and CID across experiments and water treatments
The locations of QTL might be affected by growth stage [54] and/or environmental change [62]–[63]. In our results, the QTL for WUET2011 and WUEE2011 were found on chromosomes LG03, LG06, LG11 and LG16 (under WW and WS), whereas the QTL for WUET2012 were found on chromosomes LG09, LG13 and LG15 (under WW and WS). These results showed that the expression of QTL for WUE differs with micro-environmental variations. This variation can be explained by the different water regimes in Exp. 2011 and Exp. 2012.
When the same mapping population is phenotyped in different environments, some QTL could be detected in one environment but not in others [63]. Collins et al. [64] noted that QTL can be categorized according to the stability of their effects across environmental conditions. A “constitutive” QTL is consistently detected across most environments, while an “adaptive” QTL is detected only in specific environmental conditions or increases in expression with the level of an environmental factor.
The QTL for CID in Exp. 2011 were detected on chromosomes LG03, LG06 and LG13 (WW and WS), whereas the QTL for CID in Exp. 2012 were detected on chromosomes LG13 and LG15 (under WW and WS). These results indicate that the expression of QTL for CID differs in the two experiments and different water regimes. Despite CID variation is influenced by stomatal conductance and photosynthetic capacity variations [7], [37], several QTL of the different water regimes have been detected on the same chromosome [65]–[67]. This was the case in our study, where the three QTL for CID of the three different water regimes were detected on the same chromosome (LG13). Therefore, the QTL for CID in this study can be considered as a “constitutive” QTL. Additionally, the constitutive QTL for CID was consistent with the result of phenotypic correlation that genotypic ranking for this trait was consistently maintained in the two experiments.
Some QTL for WUE and CID and related traits were located on the same chromosome or on a similar QTL position (co-localization). The QTL for WUET2012 for WW (WUE12ww.13.1 and WUE12ww.15.1) had a similar QTL position (26.20 and 77.10 cM) as the QTL for CID for WW (CID12ww.13.1, CID12ww.15.1). The QTL for CID (CID12ws.13.1) for WS was associated with the QTL for WUET2012 for WW (WUE12ww.13.1). This QTL was detected on chromosome LG13 (QTL position: 30.80 cM) near the QTL of CID12ww.13.1. The occurrence of QTL associated with different traits at the same locus may be explained by the fact that (i) the QTL are closely linked genetically or (ii) a single locus controls multiple traits and a gene may have pleiotropic effects [54].
We have observed a common genetic basis for WUE and CID in each experiment. Using the same mapping population under different water stress treatments helped us to characterize consistent genomic region (by QTL). Kiani et al. [68] indicated that QTL which was induced only by drought might be associated with mechanism(s) of sunflower drought response and they proposed that the QTL which can reduce trait difference between well-watered and water-stressed conditions should have an effect on drought tolerance because of their contribution to trait stability. Our study in Exp. 2011 showed that the QTL for CID on chromosome LG06 were repeatable across two different water treatments (WW and WS). In Exp. 2012, the QTL for CID on chromosome LG13 have been repeatable across two different water treatments (WW and WS).
All these QTL which are common across different water treatments might be useful for marker-assisted selection (MAS). Identification of QTL influencing several traits could increase the efficiency of marker-assisted selection (MAS) and hasten genetic progress [69]. Ribaut et al. [70] noticed that in the design of the best-possible breeding strategy using MAS, additional traits and criteria have to be considered. For each trait of interest, some of the criteria are the number of QTL detected, the percentage of phenotypic variance that they explain, the total percentage of the genome that they represent, and their stability across different environments. Regarding these arguments, our study has shown that CID is the most interesting trait and should be useful for MAS, where three QTL overlapped on chromosome LG06 (CID for WW and WS in Exp. 2011), and three QTL across three different water treatments were co-localized on chromosome LG13 with phenotypic variance (R2) ranges from 7 to 21%. Further, these QTL and other co-localized QTL on chromosomes LG06 and LG13 were identified in the near-centromeric region (inferior to superior position explained from 0 to 60.06 cM, and from 0 to 62.45 cM for LG06 and LG13, respectively), because those chromosomes are classified as a metacentric type [71]–[72].
Co-localization of QTL for WUE and CID with related traits
In this study, we also detected QTL for the related traits BM and CWT on the same chromosome of the QTL for WUE and/or CID (for WW and WS). These were observed in Exp. 2012, where two of four QTL for BM for WW (BM12ww.13.1 and BM12ww.15.1) were detected on chromosomes LG13 and LG15, and co-located with the QTL for WUET2012 and CID for WW (WUE12ww.13.1, CID12ww.15.1). For WS, the identifications of the QTL for the related traits showed a similar trend. The QTL of BM12ws.13.1 (QTL position: 21.20 cM) was detected on chromosome LG13, as the QTL of CID11ws.13.1, CID12ww.13.1, CID12ws.13.1 and WUE12ww.13.1 have been identified. These indicated the possibility of genetic association of WUE and CID with the accumulation of biomass. Consistent with this, Kiani et al. [68] identified a QTL for total dry matter in water-stressed conditions on chromosome LG13 using another population of sunflower. Interestingly, this QTL overlapped with osmotic adjustment, grain yield, and plant height. Thereby the common genetic basis for WUE, CID, productivity and osmotic adjustment will lead to an improved understanding of drought tolerance genes. In addition, evidence of overlapping QTL of productivity and osmotic adjustment have been observed by several authors [73]–[75]. However, further study is obviously required to determine the genetic control of osmotic adjustment or hydraulic conductance and their inter-relationships with WUE and CID.
For CWT, the QTL of CWT12ww.15.1 was detected on chromosome LG15 with the QTL position at 79.10 cM near the marker at position of 77.10 cM where the QTL of WUE12ww.15.1 and CID12ww.15.1 have been identified. Not far from these positions, a QTL of CWT12ws.15.1 was also detected (QTL position: 49.5 cM). These indicated out that the cumulative water transpired in WW and WS is genetically and closely related with WUE and CID in non-limited water availability. In addition, the maintenance of biomass accumulation under stable water stress should be considered as an efficiency process between transpiration, biomass accumulation and its partitioning between non-drought and drought conditions [64]. Therefore, the increase in WUE (i.e. the amount of biomass produced per unit of transpired water) might seem to be ideal candidate mechanism for drought-prone environments.
Identifying the “response QTL” for WUE and CID
In our work, we calculated the “response QTL” to provide new insight into the genetic architecture of WUE and CID, which, unlike a “common” phenotypic trait, is rarely considered in QTL analysis. Water use traits and their response are of primary importance to plant growth and survival. Although we have a growing understanding of the genetic and molecular drivers of water use traits and WUE as well as CID, response QTL of those traits has received relatively little attention.
We detected three QTL of “response QTL” for WUE on chromosomes LG06 and LG13. From these two chromosomes we have also identified the QTL for WUET2011 and WUET2012 for WW, indicating, at least under the conditions imposed in these experiments, that response QTL was controlled by loci that determine the main trait value under a specific treatment. This was in agreement with Kliebenstein et al. [76]–[77] who evaluated the response QTL between control and methyl jasmonate (MeJa)-treated plants of Arabidopsis thaliana. They reported that significant QTL that influenced response between control and MeJa-treated plants also affected the main trait value in at least one of the two environments, which was called the “allelic sensitivity” model.
In contrast, an independent response QTL, was also observed for several traits, for example the response QTL for WUET2011 on chromosome LG15 (WUE11diff.05.1), CID on chromosome LG02 (CIDdiff11.02.1), and CWT23d on chromosome LG06 (CWT12diff.06.1). This observation was not consistent with Kliebenstein et al. [77], however, it was in agreement with an argument of Schlichting and Pigliucci [78] who suggested the “gene regulation” model must exist, and is not always controlled by loci that are expressed within at least one of the two environments.
As for the prospects for these aspects, characterization of the genes underlying QTL that control the differential WUE and CID regulation might generate a detailed understanding of the molecular and biochemical basis for water use traits in sunflower and how this alters phenotypic response in more complex environments.
Importance of high WUE or low CID for sunflower breeding: use of the identified markers for MAS
This is the first genetic quantitative analysis and QTL mapping for WUE and CID in sunflower. We investigated two drought scenarios and evaluated genetic variation of sunflower lines to identify genetic control and physiological processes that could explain genotypic differences in the response to drought stress. The present study proved that, in sunflower, selection for CID can be considered in initial screening to improve WUE. However, this merits further investigation in other populations.
Many QTL (particularly for CID) have been reported in the literature. However, very few with large effects have been adequately exploited in crop breeding programs. The majority of the favorable alleles for identified QTL are to be found in journals on library shelves rather than in crop cultivars improved by introgression or selection of these favorable QTL alleles [79]. Nevertheless, Condon et al. [80] reported the release of a new high-yielding wheat variety in droughted environments after a breeding process in which selection for low CID in non-droughted plants led to high WUE.
In conclusion, our results emphasize that the near-centromeric region of chromosomes LG06 and LG13 are a “reliable” region for MAS due to the co-localization of the QTL for CID with several QTL for WUE, BM and CWT. Indeed, the best strategy for using molecular markers should combine selection for QTL involved in the expression of CID.
This paper complements the study of Vincourt et al. [25] and Rengel et al. [81] that exploited the INEDI RIL population in analyzing genetic variation of agronomic and physiological traits, making it possible to establish strategies for a sunflower breeding program and provide a basis for identification of the molecular components of a genotype x environment interaction.
Supporting Information
Genetic maps and LOD positions showing the locations of QTLs controlling WUE identified by MCQTL.
(DOCX)
Genetic maps and LOD positions showing the locations of QTLs controlling CID identified by MCQTL.
(DOCX)
Genotypic variation of water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) for 150 recombinant inbred lines (RILs) under well-watered (WW) and progressively water-stressed (WS) treatments in Exp. 2011.
(DOCX)
Genotypic variation of water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) for 150 recombinant inbred lines (RILs) for well-watered (WW) and water-stressed (WS) in Exp. 2012.
(DOCX)
Phenotypic correlations ( rp ) between water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) of 150 recombinant inbred lines (RILs) in Exp. 2011 and Exp. 2012.
(DOCX)
Phenotypic correlations ( rp ) among water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) of 150 recombinant inbred lines (RILs) under well-watered (WW) and progressive water-stressed (WS) treatments in Exp. 2011.
(DOCX)
Phenotypic correlations ( rp ) among water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) of 150 recombinant inbred lines (RILs) under well-watered (WW) and water-stressed (WS) treatments in Exp. 2012.
(DOCX)
Acknowledgments
We wish to thank team of Laboratoire des Interactions Plantes-Microorganismes (LIPM), INRA of Toulouse for providing sunflower materials, genetic maps, and their expert help in QTL analysis. In addition, the authors sincerely thank M. Labarrere for his contribution during the experiments.
Thierry Lamaze and Philippe Grieu are supervisors of Afifuddin Latif Adiredjo.
Funding Statement
The work was supported by a French Government Scholarship (Bourse du gouvernement Français, BGF) and a co-funding by Directorate general of Higher Education, Ministry of Education and Culture, Republic of Indonesia (Beasiswa Luar Negeri, BLN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ehleringer JR, Hall AE, Farquhar GD (1993) Stable isotopes and plant carbon – water relations. San Diego: Academic Press.
- 2. Richards RA, Rebetzke GJ, Condon AG, van Herwaarden AF (2002) Breeding opportunities for increasing the efficiency of water use and crop yield in temperate cereals. Crop Sci 42: 111–121. [DOI] [PubMed] [Google Scholar]
- 3. Cattivelli L, Rizza F, Badeck FW, Mazzucotelli E, Mastrangelo AM, et al. (2008) Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crops Res 105: 1–14. [Google Scholar]
- 4. Blum A (1996) Crop responses to drought and the interpretation of adaptation. Plant Growth Regul 20: 135–148. [Google Scholar]
- 5. Richards RA (1996) Defining selection criteria to improve yield under drought. Plant Growth Regul 20: 157–166. [Google Scholar]
- 6. Farquhar GD, Richards RA (1984) Isotopic composition of plant carbon correlates with water use efficiency of wheat genotypes. Aust J Ag Res 11: 539–552. [Google Scholar]
- 7. Condon AG, Richards RA, Rebetzke GJ, Farquhar GD (2002) Improving water use efficiency and crop yield. Crop Sci 42: 122–132. [DOI] [PubMed] [Google Scholar]
- 8. Condon AG, Richards RA, Farquhar GD (1993) Relationships between carbon isotope discrimination, water-use efficiency and transpiration efficiency for dryland wheat. Aust J Ag Res 4: 1693–1711. [Google Scholar]
- 9. Xu Y, This D, Pausch RC, Vonhof WM, Coburn JR, et al. (2009) Leaf-level water use efficiency determined by carbon isotope discrimination in rice seedlings: genetic variation associated with population structure and QTL mapping. Theor Appl Genet. 118: 1065–1081. [DOI] [PubMed] [Google Scholar]
- 10. This D, Comstock J, Courtois B, Xu Y, Ahmadi N, et al. (2010) Genetic analysis of water use efficiency in rice (Oryza sativa L.) at the leaf level. Rice 3: 72–86. [Google Scholar]
- 11. Hall AE, Richards RA, Condon AG, Wright GC, Farquhar GD (1994) Carbon isotope discrimination and plant breeding. Plant Breeding Reviews 12: 81–113. [Google Scholar]
- 12. Li Z, Pinson SRM, Stansel JW, Park WD (1995) Identification of quantitative trait loci (QTLs) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor Appl Genet 91: 374–381. [DOI] [PubMed] [Google Scholar]
- 13. Juenger TE, McKay JK, Hausmann N, Keurentjes JJB, Sen S, et al. (2005) Identification and characterization of QTL underlying whole plant physiology in Arabidopsis thaliana: δ13C, stomatal conductance and transpiration efficiency. Plant Cell Environ 28: 697–708. [Google Scholar]
- 14. Austin DF, Lee M (1996) Genetic resolution and verification of quantitative trait loci for flowering and plant height with recombinant inbred lines of maize. Genome 39: 957–968. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Q (2007) Strategies for developing Green Super Rice. P Natl Acad Sci USA 104: 16402–16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen J, Chang SX, Anyia AO (2011) Gene discovery in cereals through quantitative trait loci and expression analysis in water-use efficiency measured by carbon isotope discrimination. Plant Cell Environ 34: 2009–2023. [DOI] [PubMed] [Google Scholar]
- 17. Mian MAR, Bailey MA, Ashley DA, Wells R, Carter TE, et al. (1996) Molecular markers associated with water use efficiency and leaf ash in soybean. Crop Sci 36: 1252–1257. [Google Scholar]
- 18.Julier B, Bernard K, Gibelin C, Huguet T, Lelièvre F (2010) QTL for water use efficiency in alfalfa. In: Huyghe C, ed. Sustainable Use of Genetic Diversity in Forage and Turf Breeding. The Netherlands: Springer.
- 19. Martin B, Nienhuis J (1989) Restriction fragment length polymorphisms associated with water use efficiency in tomato. Science 243: 1725–1728. [DOI] [PubMed] [Google Scholar]
- 20. Saranga Y, Menz M, Jiang C, Wright RJ, Yakir D, et al. (2001) Genomic dissection of genotype x environment interactions conferring adaptation of cotton to arid conditions. Genome Res 11: 1988–1995. [DOI] [PubMed] [Google Scholar]
- 21. Price AH, Cairns JE, Horton P, Jones HG, Griffiths H (2002) Linking drought-resistance mechanisms to drought avoidance in upland rice using a QTL approach: progress and new opportunities to integrate stomatal and mesophyll responses. J Exp Bot 53: 989–1004. [DOI] [PubMed] [Google Scholar]
- 22. Forster BP, Ellis RP, Moir J, Talam V, Sanguineti MC, et al. (2004) Genotype and phenotype associations with drought tolerance in barley tested in North Africa. Ann Appl Biol 144: 157–168. [Google Scholar]
- 23. Hausmann NJ, Juenger TE, Stowe SSK, Dawson TE, Simms EL (2005) Quantitative trait loci affecting δ 13C and response to differential water availability in Arabidopsis thaliana . Evolution 59: 81–96. [PubMed] [Google Scholar]
- 24. Rebetzke GJ, Condon AG, Farquhar GD, Appels R, Richards RA (2008) Quantitative trait loci for carbon isotope discrimination are repeatable across environments and wheat mapping populations. Theor Appl Genet 118: 123–137. [DOI] [PubMed] [Google Scholar]
- 25. Vincourt P, As-sadi F, Bordat A, Langlade NB, Gouzy J, et al. (2012) Consensus mapping of major resistance genes and independent QTL for quantitative resistance to sunflower downy mildew. Theor Appl Genet 125: 909–920. [DOI] [PubMed] [Google Scholar]
- 26. Farquhar GD, Ehleringer JR, Hubick KT (1989) Carbon isotope discrimination and photosynthesis. Ann Rev Plant Physiol Plant Mol Biol 40: 503–537. [Google Scholar]
- 27. Craig H (1957) Isotopic standards for carbon and oxygen and correction factors for mass spectrometric analysis of carbon dioxide. Geochimi Cosmochim Ac 12: 133–149. [Google Scholar]
- 28. Jourjon MF, Jasson S, Marcel J, Ngom B, Mangin B (2005) MCQTL: multi-allelic QTL mapping in multi-cross design. Bioinformatics 21: 128–130. [DOI] [PubMed] [Google Scholar]
- 29.Lincoln SE, Daly MJ, Lander ES (1993) Constructing genetic linkage maps with MAPMAKER/EXP version 3.0. A tutorial and reference manual. Technical Report, 3rd edn, Whitehead Institute for Biomedical Research.
- 30. Churchill GA, Doerge RW (1994) Emperical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brendel O, Pot D, Plomion C, Rozenberg P, Guehl JM (2002) Genetic parameters and QTL analysis of δ13C and ring width in maritime pine. Plant, Cell & Environment 25: 945–953. [Google Scholar]
- 32. Kleibenstein DJ, Figureuth A, Mitchell-Olds T (2002) Genetic architecture of plastic methyl jasmonate responses in Arabidopsis thaliana. Genetics 161: 1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ungerer MC, Halldorsdottir SS, Purugganan MA, Mackay TFC (2003) Genotype–environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana . Genetics 165: 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Givry S, Bouchez M, Chabrier P, Milan D, Schiex T (2005) CarthaGene: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21: 1703–1704. [DOI] [PubMed] [Google Scholar]
- 35. Cadic E, Coque M, Vear F, Besset GB, Pauquet J, et al. (2013) Combined linkage and association mapping of flowering time in Sunflower (Helianthus annuus L.). Theor Appl Genet 126: 1337–1356. [DOI] [PubMed] [Google Scholar]
- 36.Lauteri M, Brugnoli E, Spaccino L (1993) Carbon isotope discrimination in leaf soluble sugars and in whole-plant dry matter in Helianthus annuus L. Grown under different water conditions. In: Ehleringer JR, et al.., eds. Stable isotopes and plant carbon – water relations. San Diego. Academic Press, inc. 93–108.
- 37. Condon AG, Farquhar GD, Richards RA (1990) Genotypic variation in carbon isotope discrimination and transpiration efficiency in wheat. Leaf gas exchange and whole plant studies. Aust J Plant Physiol 17: 9–22. [Google Scholar]
- 38. Dingkuhn M, Farquhar GD, SK De D, O’Toole JC, Datta SK (1991) Discrimination of 13C among upland rices having different water use efficiencies. Aust J Agri Res 42: 1123–1131. [Google Scholar]
- 39. Li C (1999) Carbon isotope composition, water-use efficiency and biomass productivity of Eucalyptus microtheca populations under different water supplies. Plant Soil 214: 165–171. [Google Scholar]
- 40. Lauteri M, Scartazza A, Guido MC, Brugnoli E (1997) Genetic variation in photosynthetic capacity, carbon isotope discrimination and mesophyll conductance in provenances of Castanea sativa adapted to different environments. Funct Ecol 11: 675–683. [Google Scholar]
- 41. Brendel O, Thiec DL, Saintagne CS, Bodénès C, Kremer A, et al. (2008) Quantitative trait loci controlling water use efficiency and related traits in Quercus robur L. Tree Genet Genomes. 4: 263–278. [Google Scholar]
- 42. Lambrides CJ, Chapman SC, Shorter R (2004) Genetic variation for carbon isotope discrimination in sunflower: Association with transpiration efficiency and evidence for cytoplasmic inheritance. Crop Sci 44: 1642–1653. [Google Scholar]
- 43. Condon AG, Richards RA, Farquhar GD (1992) The effect of variation in soil water availability, vapor pressure deficit and nitrogen nutrition on carbon isotope discrimination in wheat. Aust J Agr Res 43: 935–947. [Google Scholar]
- 44. O’Leary MH (1988) Carbon isotopes in photosynthesis. Bioscience 38: 325–336. [Google Scholar]
- 45. Rytter RM (2005) Water use efficiency, carbon isotope discrimination and biomass production of two sugar beet varieties under well-watered and dry conditions. J. Agron Crop Sci 191: 426–438. [Google Scholar]
- 46. Misra SC, Shinde S, Geerts S, Rao VS, Monneveux P (2010) Can carbon isotope discrimination and ash content predict grain yield and water use efficiency in wheat?. Agr Water Manage 97: 57–65. [Google Scholar]
- 47. Rizza F, Ghashghaie J, Meyer S, Matteu L, Mastrangelod AM, et al. (2012) Constitutive differences in water use efficiency between two durum wheat cultivars. Field Crops Res 125: 49–60. [Google Scholar]
- 48. Erice G, Louahlia S, Irigoyen JJ, Díaz MS, Alami IT, et al. (2011) Water use efficiency, transpiration and net CO2 exchange of four alfalfa genotypes submitted to progressive drought and subsequent recovery. Environ Exp Bot 72: 123–130. [Google Scholar]
- 49.Farquhar GD, Lloyd J (1993) Carbon and oxygen isotope effects in the exchange of carbon dioxide between terrestrial plants and the atmosphere. In: Ehleringer JR, et al.., eds. Stable isotopes and plant carbon – water relations. San Diego. Academic Press, inc. 40–70.
- 50. Scheidegger Y, Saurer M, Bahn M, Seigwolf RTW (2000) Linking stable oxygen and carbon isotopes with stomatal conductance and photosynthetic capacity: A conceptual model. Oecologia 125: 350–357. [DOI] [PubMed] [Google Scholar]
- 51. Impa SM, Nadaradjan S, Boominathan P, Shashidhar G, Bindumadhava H, et al. (2005) Carbon isotope discrimination accurately reflects variability in WUE measured at a whole plant level in rice. Crop Sci 45: 2517–2522. [Google Scholar]
- 52. Martin B, Thorstenson YR (1988) Stable carbon isotope composition (δ 13C), water use efficiency and biomass productivity of Lycopersicon esculentum, Lycopersicon pennellii, and the F1 hybrid. Plant Physiol 88: 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright GC, Hubick KT, Farquhar GD, Rao RCN (1993) Genetic and environmental variation in transpiration efficiency and its correlation with carbon isotope discrimination and specific leaf area in peanut. In: Ehleringer JR, et al.., eds. Stable isotopes and plant carbon – water relations. San Diego. Academic Press, inc. 247–267.
- 54. Laza MR, Kondo M, Ideta O, Barlaan E, Imbe T (2006) Identification of quantitative trait loci for δ13C and productivity in irrigated lowland rice. Crop Sci 46: 763–773. [Google Scholar]
- 55. Chapman S, Cooper M, Podlich D, Hammer G (2003) Evaluating plant breeding strategies by simulating gene action and dryland environment effects. Agron J 95: 99–113. [Google Scholar]
- 56. Vargas MF, van Eeuwijk A, Crossa J, Ribaut JM (2006) Mapping QTLs and QTL X environment interaction for CIMMYT maize drought stress program using factorial regression and partial least squares methods. Theor Appl Genet 112: 1009–1023. [DOI] [PubMed] [Google Scholar]
- 57. Ceccarelli S, Acevedo E, Grando S (1991) Breeding for yield stability in unpredictable environments: single traits interaction between traits, and architecture of genotypes. Euphytica 56: 169–185. [Google Scholar]
- 58. Specht JE, Chase K, Macrander M, Graef GL, Chung J, et al. (2001) Soybean response to water. Crop Sci 41: 493–509. [Google Scholar]
- 59. Saranga Y, Jiang CX, Wright RJ, Yakir D, Paterson AH (2004) Genetic dissection of cotton physiological responses to arid conditions and their interrelationships with productivity. Plant Cell Environ 27: 263–277. [Google Scholar]
- 60. Diab A, Merah TB, This D, Ozturk N, Benscher D, et al. (2004) Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theor Appl Genet 109: 1417–1425. [DOI] [PubMed] [Google Scholar]
- 61. Ellis RP, Forster BP, Gordon DC, Handley LL, Keith RP, et al. (2002) Phenotype/genotype associations for yield and salt tolerance in a barley mapping population segregating for two dwarfing genes. J Exp Bot 53: 1163–76. [DOI] [PubMed] [Google Scholar]
- 62.Xu Y, Zhu L, Chen Y, Lu C, Shen L, et al.. (1997) Tagging genes for photo-thermo sensitivity in rice using RFLP and microsatellite markers. In: Plant and Animal Genome V, Plant and Animal Genome Conference Organizing Committee, San Diego, California, Poster 149.
- 63.Xu Y (2002) Global view of QTL: Rice as a model. In Kang MS, ed. Quantitative genetics, genomics and plant breeding. Wallingford, UK. CAB International, 109–134.
- 64. Collins NC, Tardieu F, Tuberosa R (2008) Quantitative trait loci and crop performance under abiotic stress: where do we stand ?. Plant Physiol 147: 469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Morgan JA, LeCain DR, McCaig TN, Quick JS (1993) Gas exchange, carbon isotope discrimination and productivity in winter wheat. Crop Sci 33: 178–186. [Google Scholar]
- 66.Reynolds MP, Delgado MI, Gutierrez RM, Larque SA (2000) Photosynthesis of wheat in a warm, irrigated environment I. Genetic diversity and crop productivity. Field Crops Res 66, 37–50.
- 67. Teulat B, Merah O, Sirault X, Borries C, Waugh R, et al. (2002) QTLs for grain carbon isotope discrimination in field-grown barley. Theor Appl Genet 106: 118–126. [DOI] [PubMed] [Google Scholar]
- 68. Kiani SP, Maury P, Nouri L, Ykhlef N, Grieu P, et al. (2009) QTL analysis of yield-related traits in sunflower under different water treatments. Plant Breeding 128: 363–373. [Google Scholar]
- 69. Upadyayula N, da Silva HS, Bohn MO, Rocheford TR (2006) Genetic and QTL analysis of maize tassel and ear inflorescence and architecture. Theor Appl Genet 112: 592–606. [DOI] [PubMed] [Google Scholar]
- 70. Ribaut JM, Jiang C, Gonzalez-de-Leon D, Edmeades GO, Hoisington DA (1997) Identification of quantitative trait loci under drought conditions in tropical maize. 2. Yield components and marker-assisted selection strategies. Theor Appl Genet 94: 887–896. [Google Scholar]
- 71. Ceccarelli M, Sarri V, Natali L, Giordani T, Cavallini A, et al. (2007) Characterization of the chromosome complement of Helianthus annuus by in situ hybridization of a tandemly repeated DNA sequence. Genome 50: 429–434. [DOI] [PubMed] [Google Scholar]
- 72. Feng J, Liu LZ, Cai X, Jan CC (2013) Toward a Molecular Cytogenetic Map for Cultivated Sunflower (Helianthus annuus L.) by Landed BAC/BIBAC Clones. G3-Genes Genome Genet 3: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Teulat B, This D, Khairallah M, Borries C, Ragot C, et al. (1998) Several QTLs involved in osmotic-adjustment trait variation in barley (Hordeum vulgare L.). Theor Appl Genet 96: 688–698. [Google Scholar]
- 74. Rachid Al-chaarani G, Gentzbittel L, Huang X, Sarrafi A (2004) Genotypic variation and identification of QTLs for agronomic traits using AFLP and SSR in recombinant inbred lines of sunflower (Helianthus annuus L). Theor Appl Genet 109: 1353–1360. [DOI] [PubMed] [Google Scholar]
- 75. Saranga Y, Jiang CX, Wright RJ, Yakir D, Paterson AH (2004) Genetic dissection of cotton physiological responses to arid conditions and their inter-relationships with productivity. Plant Cell Environ 27: 263–277. [Google Scholar]
- 76. Via SR, Gomulkiewicz R, De jong R, Scheiner SM, Schlicting CD, et al. (1995) Adaptive phenotypic plasticity. Evolution 39: 505–522. [DOI] [PubMed] [Google Scholar]
- 77. Kliebenstein DJ, Figuth A, -Olds TM (2002) Genetic Architecture of Plastic Methyl Jasmonate Responses in Arabidopsis thaliana . Genetics 161: 1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schlighting CD, Pigliucci M (1998) Phenotypic evolution: A reactionnorm perspective. Sunderland, MA: Sinauer associates.
- 79. Hao Z, Liu X, Li X, Xie C, Li M, et al. (2009) Identification of quantitative trait loci for drought tolerance at seedling stage by screening a large number of introgression lines in maize. Plant Breeding 128: 337–341. [Google Scholar]
- 80. Condon AG, Richards RA, Rebetzke J, Farquhar GD (2004) Breeding for high water-use efficiency. J Exp Bot 55: 2447–2460. [DOI] [PubMed] [Google Scholar]
- 81. Rengel D, Arribat S, Maury P, Magniette MLM, Hourlier T, et al. (2012) A Gene-Phenotype Network Based on Genetic Variability for Drought Responses Reveals Key Physiological Processes in Controlled and Natural Environments. PLoS ONE 7: e45249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic maps and LOD positions showing the locations of QTLs controlling WUE identified by MCQTL.
(DOCX)
Genetic maps and LOD positions showing the locations of QTLs controlling CID identified by MCQTL.
(DOCX)
Genotypic variation of water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) for 150 recombinant inbred lines (RILs) under well-watered (WW) and progressively water-stressed (WS) treatments in Exp. 2011.
(DOCX)
Genotypic variation of water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) for 150 recombinant inbred lines (RILs) for well-watered (WW) and water-stressed (WS) in Exp. 2012.
(DOCX)
Phenotypic correlations ( rp ) between water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) of 150 recombinant inbred lines (RILs) in Exp. 2011 and Exp. 2012.
(DOCX)
Phenotypic correlations ( rp ) among water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) of 150 recombinant inbred lines (RILs) under well-watered (WW) and progressive water-stressed (WS) treatments in Exp. 2011.
(DOCX)
Phenotypic correlations ( rp ) among water use efficiency (WUE), carbon isotope discrimination (CID), biomass (BM) and cumulative water transpired (CWT) of 150 recombinant inbred lines (RILs) under well-watered (WW) and water-stressed (WS) treatments in Exp. 2012.
(DOCX)