Abstract
Aberrant expression of cyclin D1, frequently observed in human malignant disorders, has been linked to the control of G1→S cell cycle phase transition and development and progression in carcinogenesis. Cyclin D1 level changes are partially controlled by GSK-3β-dependent phosphorylation at threonine-286 (Thr286), which targets cyclin D1 for ubiquitination and proteolytic degradation. In our continuing studies on the mechanism of prostate cancer prevention by resveratrol, focusing on the role of its recently discovered target protein, quinone reductase 2 (NQO2), we generated NQO2 knockdown CWR22Rv1 using short hairpin RNA (shRNA)-mediated gene silencing approach. We found that, compared with cells expressing NQO2 (shRNA08), NQO2 knockdown cells (shRNA25) displayed slower proliferation and G1 phase cell accumulation. Immunoblot analyses revealed a significant decrease in phosphorylation of retinoblastoma Rb and cyclin D1 in shRNA25 compared with shRNA08. Moreover, shRNA25 cells showed a 37% decrease in chymotrypsin-like proteasome activity. An increase in AKT activity was also observed in shRNA25, supported by a ∼1.5-fold elevation in phosphorylation and ∼50% reduction/deactivation of GSK-3α/β at Ser21/9, which were accompanied by a decrease in phosphorylation of cyclin D1 at T286. NQO2 knockdown cells also showed attenuation of resveratrol-induced downregulation of cyclin D1. Our results indicate a hitherto unreported role of NQO2 in the control of AKT/GSK-3β/cyclin D1 and highlight the involvement of NQO2 in degradation of cyclin D1, as part of mechanism of chemoprevention by resveratrol.
Introduction
Quinone reductase 2 (NQO2) is a FAD (flavin adenine dinucleotide)-containing metallo-oxidoreductase discovered in 1961 that has been classified as a phase II detoxification enzyme due to its sequences homology with quinone reductase 1 (NQO1) (1,2). Both NQO1 and NQO2 are cytosolic flavoproteins that catalyze metabolism of quinones; NQO1 utilizes nicotinamide adenine dinucleotide and nicotinamide adenine dinucleotide phosphate as electron donors, whereas NQO2 uses N-ribosyl dihydronicotinamide as the electron donor for two- or four-electron reduction of quinones to prevent the formation of potentially harmful semiquinones. Since NQO2 utilizes non-naturally occurring co-substrate N-ribosyl dihydronicotinamide and because the exact identity and nature of quinone substrates for NQO2 remain equivocal, it is debatable whether NQO2 actually functions as a detoxification enzyme in vivo. Recent studies demonstrate that quinone reductases (NQOs) act as a novel redox switch of 20S proteasome in yeast (3) and are also involved in the control of 20S proteasome-mediated degradation of p53/p63 based on studies using cultured keratinocytes from rodents (4,5). Furthermore, NQO1 reportedly blocks 20S proteasomal degradation of tumor suppressor p33ING1b in human melanoma cells (6). These observations suggest that NQOs may participate in the control of stability of proteins via the 20S proteasome (4,7). Conceivably, NQO2 may also be involved in the turnover of proteins playing regulatory roles in various cellular processes including proliferation, cell-cycle arrest, apoptosis and DNA repair.
Overexpression of cyclin D1 is among the most commonly observed alterations in human malignant disorders, linked to their development and progression. Genetic mutations or aberrant control of cyclin D1 expression contribute to increased levels of cyclin D1 and accompanying dysregulation of the G1→S transition in carcinogenesis. Proteasome-dependent degradation of cyclin D1 is triggered by GSK-3β-mediated phosphorylation at threonine 286 (Thr286), which targets cyclin D1 for ubiquitination and proteolytic destruction (8,9). The activities of cyclin-dependent protein kinases (CDKs) 4/6 are regulated by the abundance of D-type cyclins; cyclin D/CDK4/6 complex in turn controls the phosphorylation of Rb and release of E2F transcription factor as key regulatory steps underlying the G1→S traverse and consequently uncontrolled proliferation. Evidently, therefore, GSK-3β-mediated cyclin D1 phosphorylation and turnover play an important role in controlling Rb-mediated G1→S phase transition (9–11).
The chemopreventive activities of resveratrol were first reported on skin and breast carcinomas by Jang et al. (12). Subsequently, resveratrol-mediated anti-prostrate cancer (CaP) properties were uncovered by us and others (13–19). Despite evidence demonstrating that resveratrol elicits a wide range of biological functions with multi-system benefits including chemoprevention, cardioprotection, neuroprotection, renal protection and suppression of aging, diabetes, oxidative stress and inflammation (20–25), the mechanism of action of resveratrol remains incomplete. Using resveratrol affinity column combined with matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS), we recently identified NQO2 as a distinct high affinity resveratrol-targeting protein (26), raising the question whether NQO2 might contribute to the resveratrol-elicited growth and gene control in CaP cells. In the present study, CWR22Rv1 CaP cells expressing NQO2 and containing knockdown NQO2 were used to examine the role of NQO2 in the control of proliferation and cell cycle phase transition. In addition, involvement of NQO2 in the regulation of cyclin D1 turnover via GSK-3β-mediated cyclin D1 phosphorylation and degradation was also investigated. We show that NQO2 plays a role in resveratrol-induced anti-CaP activity by targeting GSK-3β/cyclin D1-mediated cell cycle control via a posttranslational regulatory mechanism coupled to activity of the proteasomes.
Materials and methods
Reagents
Resveratrol was purchased from LKT laboratories (St Paul, MN). The primary antibodies for Rb, cyclin D1, cdk4, cdk6, NQO2, NQO1, GSK-3β, actin and secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The antibody against phosphorylated Rb (Ser780) and Rb (Ser807/811) was from Biosource International (Camarillo, CA). The antibody for AKT, p-cyclin D1 (Thr286) and AKT kinase assay kit were from Cell Signaling Technology (Beverly, MA). Fetal bovine serum, RPMI-1640, penicillin and streptomycin were purchased from Cellgro (Herndon, VA). All other chemicals and solvents used were of analytical grade.
Generation and selection of CWR22Rv1 cells containing stably expressed shRNA-mediated NQO2 knockdown
Human CWR22Rv1 cells were obtained from the American Tissue Culture Collection (Manassas, VA) and maintained in RPMI-1640 supplemented with penicillin, streptomycin and 10% heat-inactivated fetal bovine serum as described (27–29). Stably expressed short hairpin RNA (shRNA)-mediated NQO2-knockdown CWR22Rv1 cells were established using the following procedure. Cells were first seeded at a density of 2 × 105 cells/ml in six-well plates; following an overnight incubation, cells were transfected with control or NQO2-targeted shRNA using Lipofectamine 2000 transfection reagent and protocol provided by the manufacturer (Invitrogen, Carlsbad, CA). Control shRNA in pGFP-V-RS tGFP plasmid (TR30008, marked as shRNA08) and HuSH 29mer NQO2-targeting shRNA in pGFP-V-RS plasmid (GI344425, marked as shRNA25) were obtained from Origene Technologies (Rockville, MD) and used in this study. At 48 h post transfection, cells selection was initiated with 0.25 μg/ml puromycin (Research Products International Corp., Mt Prospect, IL) for four passages, by serially diluting and seeding cells at sequential passages at a progressively reduced density of 30, 3 and 0.3 cells per well in 96-well plates. Single colonies were picked and transferred into six-well plate for propagation and expansion. GFP-encoded plasmids containing either control shRNA08 or NQO2-targeting shRNA25 were stably expressed and cells were analyzed by fluorescent microscopy. A Zeiss microscope equipped with Axiovert 200 Imaging System (Carl Zeiss MicroImaging, Jena, Germany) was used to capture cell images at ×10 magnification. Confirmation of NQO2 knockdown by shRNA was based on results of immunoblot analysis. Using this approach, stable control and NQO2-knockdown CWR22Rv1 cell lines were established and used for experiments. They were designated as shRNA08 and shRNA25, respectively.
Preparation of whole-cell extracts and western blot analysis
Cells were collected and lysed by incubation on ice for 30 min in cold immunoprecipitation (RIPA) buffer, which contained 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1% Triton X-100, 1% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1 mM dithiothreitol and 10 μl/ml protease inhibitor cocktail from Sigma–Aldrich Corp. (St Louis, MO). The extracts were centrifuged and the clear supernatants were stored in aliquots at –80°C for further analysis. Protein concentrations of cell lysates were determined by coomassie protein assay kit (Pierce, Rockford, IL) using bovine serum albumin as standard. Aliquots of lysates (10 μg of protein) were resolved by 10% SDS–polyacrylamide gel electrophoresis followed by western blot analysis. The blots were first incubated with specific primary antibodies followed by secondary antibodies. Specific immunoreactive bands were identified and detected by enhanced chemiluminescence using protocol provided by the manufacturer (Kirkegaard & Perry Laboratories, Gaithersburg, MD). The expression of actin was monitored in parallel as loading control. The intensity of specific immunoreactive bands was quantified by densitometry and expressed as a ratio relative to the expression of actin (30,31).
NQO2 enzyme activity assay
The enzyme activity was determined spectrophotometrically at 25°C in pH 8.5 buffer (50 mM Tris, 140 mM NaCl, 0.1% Tween 20 and 1 mM dithiothreitol) using menadione as the substrate and 1, 4-dihydro-N-methyl-nicotinamide as the cosubstrate. The reaction was initiated by addition of 15–20 μg of cell lysate, and the catalysis was monitored at a wavelength of 360 nm using a BioMateTM 3 spectrophotometer (Thermo Electron Corp., Madison, WI).
Effects of NQO2 knockdown on CWR22Rv1 cell proliferation
Cell proliferation was assayed using a commercially available non-radioactive CellTiter 96® AQueous assay kit as outlined by the manufacturer (Promega Corp., Madison, WI). Briefly, the cells seed into 96-well plates (1 × 105 cells/ml, 0.1 ml per well) were maintained in culture for different times as indicated. Immediately prior to cell harvest, 20 μl of solution containing MTS [(3- 4,5-dimethylthiazol-2-yl)- 5- (3-carboxymethoxyphenyl) -2 -(4-sulfophenyl)-2H-tetrazolium] was added into each well and incubation was continued for 3 h at 37°C, to allow bioreduction of MTS to a formazan product whose absorbance was measured at 570 nm using a Tecan Sunrise microplate reader (Tecan U.S., Research Triangle Park, NC). Data from treated cells were normalized against the control cells and presented as a ratio against control values obtained at time 0.
Effects of resveratrol on proliferation of CWR22Rv1 cells containing stably expressed shRNA-mediated NQO2 knockdown
Control shRNA08 (NQO2 expressing) and shRNA25 (NQO2 knockdown) cells were established in the background of pGFP-RS shRNA expression vector, which allows constitutive expression of tGFP using the pCMV promoter. Therefore, fluorescent GFP was used to monitor proliferation in both cell lines, in response to treatment by resveratrol. Cells were seeded in 96-well plates at a density of 2 × 105 cells/ml in 200 μl of 10% fetal bovine serum/RPMI-1640 and incubated overnight before treatment with increasing doses of resveratrol (0, 6.25, 12.5, 25 and 50 μM) for 72 h. Following cell harvest, the fluorescence of control and treated cells was measured using a Synergy HT multi-mode microplate reader (BioTek Instruments, Winooski, VT). Data from treated cells were normalized against the control cells and cell proliferation was presented as ratio against control.
Cell cycle analysis
Both shRNA08 and shRNA25 cells were treated with 0 and 50 μM of resveratrol for 72 h, washed with phosphate-buffered saline and incubated with 8.0 μg/ml propidium iodide staining solution (Molecular Probes, Eugene, OR). Cell cycle phase distribution was assayed by flow cytometry (28,32,33). MultiCycle software from Phoenix Flow Systems (San Diego, CA) was used to deconvolute the cellular DNA histograms and their quantitative conversion into percentage of cells in the G1, S and G2 + M phases of the cell cycle.
The proteasome-GloTM chymotrypsin-like cell-based assay
The proteasome activity was measured by the Proteasome-GloTM chymotrypsin-like cell-based assay according to instructions provided by the manufacturer (Promega Corp.). Luciferin detection reagent and proteasome-GloTM substrate was mixed in proteasome-GloTM cell-based buffer and incubated at room temperature for 30 min before added to samples at equal volume. After a 10 min reaction, the luminescence was determined using a Turner Biosystems luminometer (Turner Biosystems, Sunnyvale, CA).
In vitro AKT kinase assay
Lysates prepared from control shRNA08 and NQO2 knockdown shRNA25 cells were used in AKT kinase assay according to instructions provided by the manufacturer (Cell Signaling Technology). Cell lysates (150 μg) were incubated overnight at 4°C with 20 μl of immobilized AKT antibody bead slurry. GSK-3 fusion protein (1 μg), used as substrate for the kinase reaction, was incubated with immobilized AKT for 30 min at 30°C and the reaction was terminated by adding 2 × SDS sample buffer. Samples were separated on SDS–polyacrylamide gel electrophoresis and immunoblotted with phospho-GSK3α/β (Ser21/9) antibody followed by detection using the LumiGLO Substrate (Cell Signaling Technology).
Reverse transcription–PCR analysis and determination of gene-specific messenger RNA expression
Total RNA was extracted from shRNA08 and shRNA25 cells using the TRIzol® reagent (Invitrogen). Isolated RNA (0.5μg) was reverse transcribed using one-step reverse transcription–PCR kit (Promega Corp.). The PCR primer sequences were: cyclin D1, forward 5′-CCC TCG GTG TCC TAC TTC AA-3′, reverse 5′-GTG TTC AAT GAA ATC GTG CG-3′; GAPDH, forward 5′-CCA CCC ATG GCA AAT TCC ATG GCA-3′, reverse 5′-TCT AGA CGG CAG GTC AGG TCC ACC-3′. The expression of GAPDH was used as a control for normalization of messenger RNA expression results. PCR condition for cyclin D1 expression was: denaturation, 94°C, 5 min, followed by 30 cycles of denaturation at 94°C, each 1 min, 1 min annealing at 60°C and 1 min extension at 72°C. PCR condition for GAPDH expression was: denaturation at 95°C, 5 min, followed by 28 cycles of denaturation at 95°C, each 30 s, 30 s annealing at 60°C and 30 s extension at 72°C.
Results
Generation of stable NQO2-knockdown CWR22Rv1 cell lines
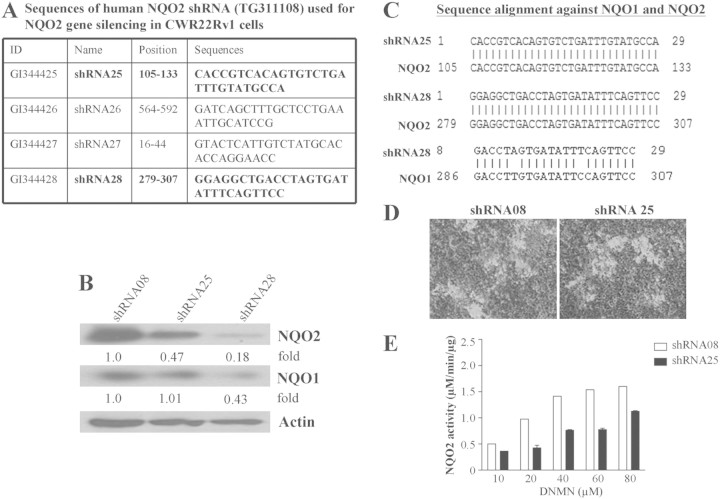
Development of a cell model represents a reasonable approach to study the role of NQO2 in resveratrol-mediated bioactivities including chemoprevention against CaP. To this end, we generated CWR22Rv1 cells harboring stable NQO2 knockdown using NQO2-specific shRNA designed on the basis of NQO2-specific shRNA sequences (shRNA25, shRNA26, shRNA27 and shRNA28) shown in Figure 1A, and the efficacy of NQO2 knockdown was confirmed by immunoblot analysis. While shRNA26 and shRNA27 did not result in knockdown of NQO2 (data not shown), in cells transfected with shRNA25, a ∼53% decrease in NQO2 expression without changes in NQO1 was observed, as compared with cells similarly transfected with control shRNA08 (Figure 1B). Surprisingly, cells transfected with NQO2-specific shRNA28 were accompanied by ∼82% and ∼57% loss of levels of both NQO2 and NQO1 relative to cells transfected with control shRNA08 (Figure 1B). The level of actin was unchanged suggesting specificity of the NQO2 knockdown (Figure 1B). Since transfection by shRNA28 resulted in knockdown of NQO1 and NQO2 expression, we aligned the nucleotide sequence of shRNA 25 and 28 against NQO1 and NQO2. Figure 1C showed that the shRNA25 nucleotide sequence was perfectly aligned with NQO2, whereas nucleotides of shRNA28 showed complementarity to both NQO1 and NQO2. Given that stable clones generated also contained the pGFP-RS shRNA expression vector, evident by constitutive expression of tGFP under control of the pCMV promoter, GFP fluorescence was used to monitor selection and identification of positive clones (Figure 1D). Whether suppression of NQO2 expression by shRNA also affected its enzyme activity was investigated in control (shRNA08) and NQO2 knockdown (shRNA25) cells. As expected, knockdown of NQO2 resulted in ∼50% reduction of its enzyme activity (Figure 1E).
Fig. 1.
Generation of NQO2 shRNA CWR22Rv1 cell lines. (A) Sequences of human NQO2 shRNA used for silencing NQO2 in CWR22Rv1 cells. (B) Western blot analysis performed to verify the efficiency of NQO2 knockdown and expression of functionally redundant NQO1 in control shRNA08 and NQO2-knockdown shRNA25 and shRNA28 CWR22Rv1 cells. The blots were stripped and reprobed with actin, which served as a loading control. The intensity of the specific immunoreactive bands was quantified by densitometry and expressed as fold differences against actin. (C) Sequence alignment of human NQO2 shRNA25 and shRNA28 sequences against human NQO1 and NQO2 complementary DNA. (D) Fluorescent images of positive control and stable NQO2-knockdown CWR22Rv1 cells. Stable clone contained the pGFP-RS shRNA expression vector, which constitutively expressed tGFP driven by the pCMV promoter. tGFP-expressing cells (light areas) carried either transfected vector plasmid or shRNA. Expression of GFP fluorescence was used to monitor success and efficacy of NQO2 knockdown and selection of positive clone. (E) NQO2 enzymatic activity in control shRNA08 and NQO2-knockdown shRNA25 cells, as assayed using methods detailed in Materials and methods.
Effects of NQO2 knockdown on proliferation and cell cycle distribution
Using the stable control shRNA08 and NQO2-knockdown shRNA25 clones as models, the effects of NQO2 knockdown on CWR22Rv1 cell proliferation were studied. Knockdown of NQO2 by shRNA25 inhibited growth relative to controls transfected with shRNA08. Compared with time 0, suppression of cell proliferation in shRNA25 cells was evident at 24 h (∼17% inhibition; Figure 2A), which increased to ∼47% at 48 h (Figure 2A). To further test NQO2 knockdown-induced growth suppressive effects, changes of cell cycle phase distribution were determined by flow cytometry. The percentage of cells in G1, S and G2 + M phases was quantified and presented in Figure 2B. NQO2 knockdown caused a decrease in S phase cell population at 24 h (32.7% in control versus 22.7% in NQO2 knockdown cells), accompanied by a concomitant accumulation in the G1 phase cell population (43.3% in control versus 52.3% in NQO2 knockdown cells), suggesting that NQO2 knockdown resulted in an altered G1→S transition. Because phosphorylation of Rb plays a pivotal role in the control of cell progression through the G1→S checkpoint, we tested whether NQO2 contributed to control of Rb status by measuring changes in pRb/Rb in NQO2 expressing (shRNA08) and knockdown (shRNA25) cells. Western blot analysis showed a significant reduction in hyperphosphorylated Rb at Ser-780 and Ser-807/811, respectively, in NQO2-knockdown shRNA25 cells compared with control shRNA08 cells at 72 h (Figure 2C). Since phosphorylation of Rb is under the control of cdk/cyclin complex, the expression of cyclins D1 and cdk4/6 were analyzed. Results in Figure 2D showed that knockdown of NQO2 caused less pronounced effects on cyclin D1/cdk4 or cyclin D1/cdk6 complexes; whether and how NQO2 status might affect the phosphorylation of cyclin D1 remains unknown. Since cyclin D1 degradation/stability is controlled by phosphorylation at Thr286 in concert with changes in cdk4/6 activity, we analyzed changes in cyclin D1-Thr286 in control shRNA08 and NQO2-knockdown shRNA25 cells. Western blot analysis demonstrated that the expression of p-cyclin D1 (Thr286) was markedly downregulated in NQO2-knockdown shRNA25 cells; in stark contrast to NQO2-expressing shRNA08 cells where no change on cyclin D1 phosphorylation at Thr286 occurred (Figure 2D). These results suggest that knockdown of NQO2 abrogates Rb activation and cyclin D1 degradation via a reduction in phosphorylation activity/capacity; albeit, because both changes in both molecular events did not occur until cells were maintained in culture for 48 and 72 h, they are unlikely to directly contribute to growth suppression or cell cycle arrest observed in NQO2-knockdown shRNA25 cells at 24 h. The observed changes, however, proffer the possibility that NQO2 plays a role in phosphorylation-dependent protein modification reactions and processes.
Fig. 2.
Effects of NQO2 knockdown on control of cell proliferation and cell cycle phase distribution. (A) Cell growth was monitored in control shRNA08 and NQO2-knockdown shRNA25 cells for up to 72 h; the cell numbers were determined at 24, 48 and 72 h by MTS assay as described in Materials and methods. The proliferation of cells carrying shRNA08 or shRNA25 was measured, normalized and expressed as the ratio against time 0 (set as 1). (B) Cell cycle analysis was performed in control shRNA08 and NQO2-knockdown shRNA25 cells at 72 h and changes in cell cycle distribution were analyzed by flow cytometry, and the percentage of cells in G1, S and G2M phases were calculated. (C) Western blot analysis of Rb, pRb (Ser780) and pRb (Ser 807/811) expression in control shRNA08 and NQO2-knockdown shRNA25 cells maintained in culture for 24, 48 and 72 h. (D) The total expression of phosphorylated cyclin D1 (Thr286), cyclins D1, cdk4 and cdk6 in shRNA08 and shRNA25 cells at 24, 48 and 72 h as determined by western blot analysis. The intensity of the specific immunoreactive bands was quantified by densitometry and expressed as a fold difference against actin (loading control).
Effect of NQO2 knockdown on cyclin D1 turnover
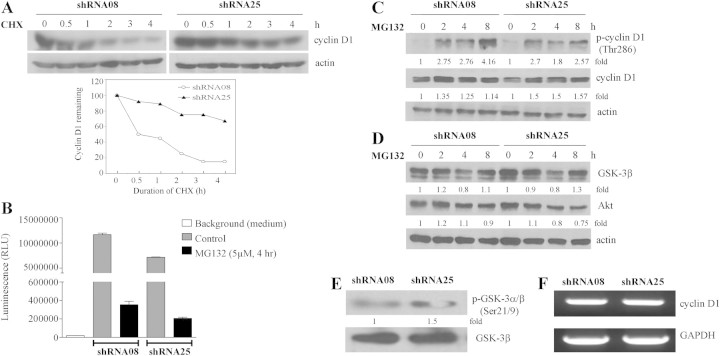
That NQO2 might contribute to control of cyclin D1 stability was further tested using control shRNA08 and NQO2-knockdown shRNA25 cells treated with cycloheximide. Western blot analysis revealed that cyclin D1 was more stable in NQO2 knockdown cells relative to control cells. As shown in Figure 3A, the half-life of cyclin D1 protein increased from ∼0.5 h in control cells to >4 h in NQO2 knockdown cells. This substantial prolongation of half-life of cyclin D1 in cells harboring partial silencing of NQO2 expression prompted us to test whether NQO2 status might affect proteasome activity. Using the chymotrypsin-like proteasome activity assay, we found a 37% decrease in proteasome activity in NQO2-knockdown shRNA25 cells compared with control shRNA08 cells (Figure 3B).
Fig. 3.
Analysis of stability of cyclin D1 protein in relation to NQO2 knockdown. (A) Effect of cycloheximide on turnover/degradation of cyclin D1 in shRNA08 or shRNA25. Cells were treated with 30 μg/ml cycloheximide to inhibit new protein synthesis for the times indicated and changes in cyclin D1 were analyzed by western blot analysis, with actin expression used as loading control. Cyclin D1 expression was quantified by densitometric analysis and the expression was presented as percentage (%) remaining relative to time 0. (B) Effect of knockdown NQO2 on proteasome activity. Both shRNA08 and shRNA25 cells were treated with or without the addition of proteasome inhibitor MG132 (5 μM) for 4 h. Changes in proteasome activity were measured using chymotrypsin-like proteasome activity assay kit obtained from Promega. (C) Effect of proteasome inhibitor MG132 on degradation of phosphorylated cyclin D1 (Thr286) and cyclin D1 in shRNA08 versus shRNA25. Cells were treated with 5 μM MG132 for 0, 2, 4 and 8 h and changes in protein expression were analyzed by western blot analysis, with actin used as loading control. (D) Effect of proteasome inhibitor MG132 on degradation of GSk-3β and AKT in shRNA08 versus shRNA25. Cells were treated with 5 μM MG132 for 0, 2, 4 and 8 h and changes in GSk-3β and AKT expression were analyzed by western blot analysis, using actin as loading control. (E) AKT kinase activity assay was performed and changes of phosphor-GSK-3α/β (Ser21/9) expression in shRNA08 and shRNA25 cells at 72 h were analyzed by western blot analysis. (F) Reverse transcription–PCR was performed to assess cyclin D1 messenger RNA changes in shRNA08 and shRNA25 cells at 72 h. The expression of GAPDH was used as internal control.
Proteasome activity in both normal shRNA08 and NQO2-knockdown shRNA25 cells was effectively inhibited by proteasome inhibitor MG132 (Figure 3B). Accordingly, the possible involvement of proteasomes in mediating cyclin D1 protein stabilization in control shRNA08 and NQO2-knockdown shRNA25 cells was further tested by exposing cultured cells to MG132 and following cyclin D1 loss by immunoblot analysis. Treatment by MG132 effectively blocked degradation of both p-cyclin D1 (Thr286) and cyclin D1 in control shRNA08 and NQO2-knockdown shRNA25 cells (Figure 3C). Furthermore, relatively less p-cyclin D1 (Thr286) was present in NQO2-knockdown shRNA25 cells compared with control shRNA08 cells (Figure 3C), thereby providing additional evidence that stability of cyclin D1 in NQO2 expressing cells is attributed to posttranslational degradative mechanisms. Conceivably, knockdown of NQO2 impinges on control of cyclin D1 protein stabilization by attenuating cyclin D1 degradation in part and in concert with the abrogation of cyclin D1 phosphorylation at Thr286 (Figure 2E).
Studies have shown that GSK-3β plays a role in phosphorylation of cyclin D1 and control of its stability; moreover, the activity of GSK-3β can be effectively inhibited by upstream kinase (9). Therefore, we next investigated whether the differential expression of cyclin D1 phosphorylation in normal and NQO2 knockdown cells may be due to the changes in proteasome activity. Specifically, the possibility that proteasomes are involved in modulating AKT and GSK-3β protein stabilization and contributing to the effects on cyclin D1 was tested in control shRNA08 and NQO2-knockdown shRNA25 cells exposed to MG132. We found that the status of NQO2 had little effect on GSK-3β and AKT protein stability (Figure 3D). To additionally investigate whether NQO2 is involved in AKT-mediated GSK-3β control, we measured AKT activity in control shRNA08 and NQO2-knockdown shRNA25 cells using an in vitro AKT kinase assay kit (see Materials and methods). Compared with control cells, NQO2 knockdown caused an estimated 1.5-fold increase in phosphorylation of GSK-3α/β at Ser21/9 (Figure 3E), indicating that NQO2 knockdown may be accompanied by an increase in AKT activity concomitant with deactivation of GSK-3 that consequentially contributes to the stabilization of cyclin D1 in NQO2-knockdown shRNA25 cells. Notably, these results are consistent with and lend support to the observed inhibition of cyclin D1 phosphorylation in NQO2-knockdown shRNA25 cells (Figure 2E).
Next, we tested whether NQO2 also affected cyclin D1 expression at the transcriptional level by using reverse transcription–PCR to assess changes in cyclin D1 messenger RNA levels. No difference in cyclin D1 messenger RNA expression was observed between control and NQO2 knockdown cells (Figure 3F).
Role of NQO2 in CWR22Rv1 growth control in response to resveratrol
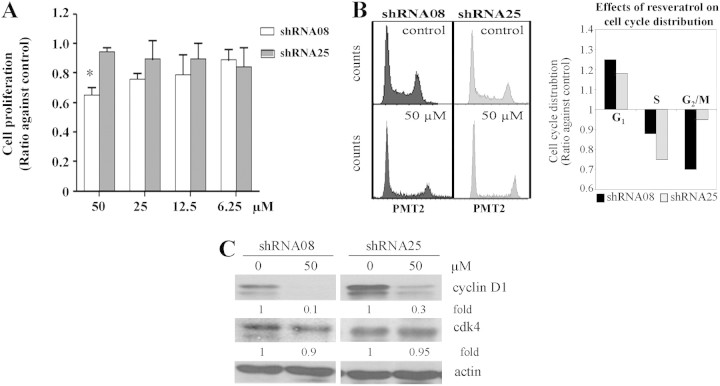
To assess the involvement of NQO2 in resveratrol-induced growth suppression, control shRNA08 and NQO2-knockdown shRNA25 cells were exposed to increasing dose of resveratrol. Concentration-dependent inhibition of proliferation by resveratrol occurred in control shRNA08 cells but not in NQO2-knockdown shRNA25 cells (Figure 4A), supporting the premise that NQO2 plays a role in resveratrol-induced growth inhibition.
Fig. 4.
Role of NQO2 on CWR22Rv1 growth control and cell cycle phase distribution by resveratrol. (A) Effect of resveratrol on cell growth in shRNA08 and shRNA25 cells. Cells were treated with increasing doses of resveratrol (0, 6.25, 12.5, 25 and 50 μM) for 72 h and effects on cell proliferation were analyzed. The intensity of fluorescence was measured and normalized against control and expressed as the ratio against control. (B) Effects of resveratrol on the changes of cell cycle phase distribution in shRNA08 and shRNA25 cells. Cells were treated with resveratrol (0 and 50 μM) for 72 h and the effect on cell cycle distribution was analyzed by flow cytometry. The calculated percentages of cells in G1, S and G2M phases were represented as histograms. (C) The protein expression levels of cyclins D1 and cdk4 in shRNA08 and shRNA25 cells were determined by western blot analysis. The intensity of the specific immunoreactive bands was quantified by densitometry and expressed as a fold difference against actin (loading control).
To further investigate whether NQO2 actively controls resveratrol-induced cell cycle change, flow cytometric analysis was performed. A more pronounced G1 accumulation accompanied by G2M blockade was found in control cells, as opposed to depletion of S phase in NQO2 knockdown cells, at 72 h exposure to 50 μM resveratrol (Figure 4B). Modulation of sensitivity to resveratrol-induced cell cycle changes by NQO2 knockdown was also evident in analysis of expression of cell cycle regulatory proteins cyclin D1/cdk4 in resveratrol-treated control and NQO2 knockdown cells. Results in Figure 4C show that treatment by resveratrol caused a comparably less pronounced reduction in cyclin D1 and cdk4 expression in NQO2-knockdown shRNA25 cells, relative to NQO2-expressing shRNA08 cells.
Role of NQO2 on cyclin D1 control in resveratrol-treated cells
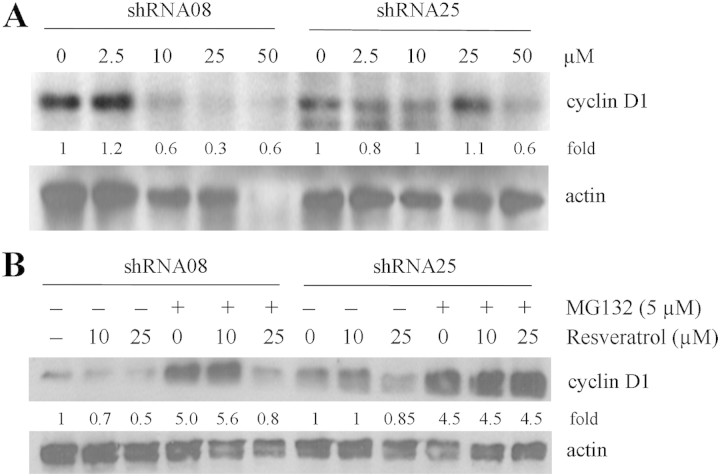
To obtain additional information on whether NQO2 contributes to resveratrol-induced growth control via modulation of cyclin D1 turnover, western blot analyses were performed demonstrating that cyclin D1 protein was more stable in NQO2 knockdown cells compared with control cells. For example, exposure to 10 μM resveratrol sufficed to downregulate cyclin D1 protein expression in control shRNA08, whereas 50 μM resveratrol was required to cause a comparable decrease in cyclin D1 levels in NQO2-knockdown shRNA25 cells (Figure 5A). These results indicate that NQO2 at least in part contributes to resveratrol-induced downregulation of cyclin D1 expression in concordance with the inhibition of cell growth; it may be surmised that at low doses of resveratrol (2.5–10 μM), NQO2 plays an integral role in mediating cyclin D1 expression/stability by resveratrol, whereas at >25 μM resveratrol, control of cyclin D1 occurs independent of the presence of NQO2.
Fig. 5.
Effects of NQO2 on resveratrol-mediated cyclin D1 control. (A) Western blot analysis of cyclin D1 protein expression in control shRNA08 and NQO2-knockdown shRNA25 cells following treatment by increasing doses of resveratrol (0, 2.5, 10, 25 and 50 μM) for 72 h. The blots were also probed for actin, which served as loading control. (B) Effect of proteasome inhibitor MG132 on resveratrol-induced cyclin D1 degradation in shRNA08 versus shRNA25. Cells were treated 0, 10 or 25 μM of resveratrol with or without 5 μM MG132 for 4 h and changes in cyclin D1 expression was analyzed by western blot analysis, with actin level used as loading control.
The possibility that proteasomes are involved in resveratrol-mediated cyclin D1 protein degradation was further tested by culturing both cell lines differing in NQO2 status with addition of resveratrol alone or in combination with proteasome inhibitor MG132. Changes of immunoreactive cyclin D1 levels definitively showed that MG132 effectively blocked resveratrol-induced degradation of cyclin D1 in NQO2-knockdown shRNA25 cells (Figure 5B). In shRNA08, however, MG132 only blocked cyclin D1 degradation at 10 μM but not at 25 μM resveratrol-treated cells (Figure 5B). These results fortify the notion that NQO2 participates in proteasome-mediated degradation of cyclin D1 at low dose of resveratrol (≤10 μM); in contrast, at ≥25 μM resveratrol, mechanisms separate from the proteasomes or NQO2 may be critically involved in mediating the turnover of cyclin D1.
Discussion
Resveratrol exerts significant chemopreventive and therapeutic effects in various cancers; however, the mechanisms contributing to its anti-tumorigenic activities are only partially understood. Previously, we have identified NQO2 as a novel resveratrol-targeting protein; details by which NQO2 contributes to resveratrol-mediated anti-CaP activity were investigated in this study. Knockdown of NQO2 in CWR22Rv1 cells resulted in growth suppression, G1→S arrest as well as downregulation of Rb and cyclin D1 phosphorylation (Figure 2) suggesting that NQO2 plays a role in Rb/cyclin D1-mediated growth and cell cycle control. Overexpression of cyclin D1 is commonly observed in different human cancers and whether NQO2 controls cyclin D1 stability and turnover has not been previously reported. Using knockdown NQO2 cells, we found that cyclin D1 turnover is significantly attenuated in NQO2-knockdown shRNA25 cells. Moreover, NQO2-mediated cyclin D1 degradation can be effectively blocked by proteasome inhibitor MG-132 in NQO2 expression shRNA08 cells (Figure 3B), providing support that NQO2 has an integral role in proteasome-mediated cyclin D1 degradation, and also is an important mediator of resveratrol-mediated growth control via cyclin D1. In previous studies, resveratrol has been reported to effectively downregulate cyclin D1 in various cancer types (34–36). Herein, we demonstrate for the first time that NQO2 participates in resveratrol-induced changes of cyclin D1 level via control of its stability and turnover; knockdown of NQO2 attenuates downregulation of cyclin D1 and similarly abrogates resveratrol-induced growth suppression (Figures 4 and 5).
It is noteworthy that the newly revealed role of NQO2 differs significantly from its traditional biochemical assignment as a phase II detoxification enzyme for protection of cells against free radicals and toxic oxygen metabolites. This may not be all surprising since unlike the more comprehensively studied phase II enzyme NQO1, which utilizes nicotinamide adenine dinucleotide/nicotinamide adenine dinucleotide phosphate to carry out the detoxification reaction, NQO2 has always been considered enigmatic as it exclusively uses the unusual, non-naturally occurring co-substrate N-ribosyl dihydronicotinamide to reduce harmful semiquinones—in part the basis for the ongoing debate as to the ‘real’ physiological and biological function of NQO2. Indeed, yet to be discovered the role of NQO2 is supported by recent demonstration that NQOs (NQO1 and NQO2) impart stability and protection of tumor suppressor p53 against degradation catalyzed by 20S proteasomes (4). In the current study, we showed that knockdown of NQO2 is accompanied by hyperphosphorylation and deactivation of GSK-3α/β at Ser21/9, which in turn impinges on cyclin D1 phosphorylation and blocks its degradation by proteasomes. Whether NQO2 facilitates GSK-3β-mediated cyclin D1 proteolysis by direct binding with 20S proteasomes to effect inhibition of proteasomal-mediated protein degradation, by targeted posttranslational modification of cyclin D1, or alternatively by interacting with AKT to suppress its phosphorylation-dependent activation to in turn interfere with phosphorylation of GSK-3β remain to be elucidated. A hypothetic model is shown in Figure 6, depicting these possibilities.
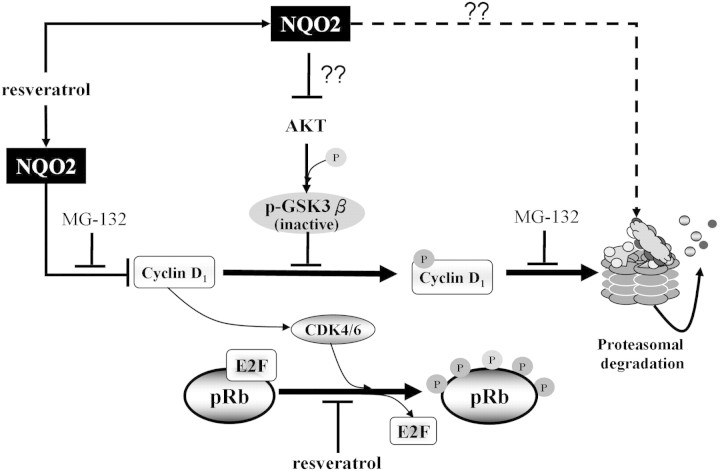
Fig. 6.
A proposed mechanism on the role of NQO2 in regulation of stability of cyclin D1 and functioning as a mediator of resveratrol-induced suppression of growth and cell cycle control. Our hypothesis is that NQO2 regulates GSK-3β-mediated cyclin D1 degradation by modulating proteasome activity and by interacting with AKT and/or inhibiting its phosphorylation-dependent activation; in turn, GSK-3β phosphorylation by AKT is reduced, resulting in a more active GSK-3β capable of phosphorylating cyclin D1 for degradation by proteasomes. This sequence of events is effectively attenuated in NQO2 knockdown cells or when cells are exposed to resveratrol, which acts by binding and sequestration of NQO2, rendering it inaccessible or incapable of interaction with AKT.
The control of cyclin D1 by NQO2 observed in the present studies may be germane to the previously reported role of NQO2 in protection of degradation of tumor suppressor p53 by 20S proteasomes (4). As noted by Gong et al. (4), both NQO2 and NQO1 confer stability on p53 by preventing 20S proteasomal activity in human hepatoma Hep-G2 cells and keratinocytes obtained from tumors in wild-type, NQO2 null and NQO1-null mice (4). Thus, it is of interest to further analyze control of p53 stability in normal shRNA08 and NQO2-knockdown shRNA25 cells as underscored by the following considerations. First, shRNA28 cells (Figure 1B), uniquely showing both NQO1 and NQO2 knockdowns, might be expected to have least abundantly expressed p53 compared with shRNA08 and shRNA25; this possibility is supported by results of our preliminary analysis (results not shown). Second, compared with wild-type p53, mutant p53 has been shown to display increased binding to NQO1 resulting in its resistance to ubiquitin-independent degradation (37); this observation is of particular relevance to CWR22Rv1 cells, known to contain mutated p53 (38). Finally, as previously suggested, protection of p53 from 20S proteasome-mediated degradation may require its separate but simultaneous interaction with NQO1 and NQO2 (4), suggesting that the cellular level of NQO1 relative to NQO2 may be critically important in modulating the stability of p53 and determining its expressed levels. Accordingly, as applied to CWR22Rv1 cells, p53 may display both increased and decreased expression as reflection of its dynamic and temporal stability and turnover changes, perhaps resulting from redundancy in and/or preferred interaction with NQO1, relative to NQO2. These possibilities and details of the control of p53 stability in normal shRNA08 and NQO2-knockdown shRNA25 cells are currently under investigation in our laboratory. Since tumor suppressor gene p53 plays an important function in protection of genomic stability, modulation of ubiquitin-independent p53 degradation by NQO2, alone and in partnership with NQO1, offers the intriguing hypothesis that NQOs play a critical yet to be defined role in control of chromosomal integrity and development of genetic variations in normal and diseased cells.
Our results support the interpretation that deletion of NQO2 prevents cyclin D1 phosphorylation and degradation, secondary to inhibition of GSK-3β activity via activation of AKT. The molecular details on how deletion of NQO2 results in AKT activation remain to be investigated and elucidated, especially since it was reported that activation of several cell survival kinases including AKT by exposure to tumor necrosis factor was effectively abrogated in NQO2-knockdown keratinocytes (39). In unpublished results, we found that NQO2 and AKT coeluted from resveratrol affinity column, suggesting NQO2 may interact with AKT thereby affecting AKT signaling. Moreover, since AKT is considered critical for cancer cell survival, its ability to interact with NQO2 raises the possibility that NQO2 plays a novel role as a cancer risk modulator, with an integral participation in cancer prevention by resveratrol.
When traditional role of NQO2 as a detoxification enzyme is considered in combination with current findings showing that cyclin D1 degradation is regulated by NQO2, it may be surmised that NQO2 contributes to the cancer prevention by direct and indirect mechanisms: (i) NQO2 as a phase II enzyme affords protection against carcinogenicity, mutagenicity and other toxicities caused by carcinogenic xenobiotics; (ii) NQO2 mediates resveratrol-elicited control of cell proliferation via regulation of stability of cyclin D1; (iii) NQO2 as a distinct resveratrol-targeting protein for resveratrol may also serve as a on/off molecular switch for resveratrol to in turn function in its capacity as a phytochemical enzyme activator or inhibitor. Studies to test the added roles of NQO2, in chemoprevention and anti-CaP activities of resveratrol, are underway in our laboratory.
Funding
This research was supported in part by the Intramural Sponsored Research Program of New York Medical College to T.-C.H. (49-432-1 and 49-466-1).
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CaP
prostrate cancer
- CDK
cyclin-dependent protein kinase
- NQO
quinone reductase
- SDS
sodium dodecyl sulfate
- shRNA
short hairpin RNA
References
- 1.Long DJ, II, et al. NRH:quinone oxidoreductase2 (NQO2) Chem. Biol. Interact. 2000;129:99–112. doi: 10.1016/s0009-2797(00)00200-3. [DOI] [PubMed] [Google Scholar]
- 2.Chen S, et al. Structure-function studies of DT-diaphorase (NQO1) and NRH: quinone oxidoreductase (NQO2) Free Radic. Biol. Med. 2000;29:276–284. doi: 10.1016/s0891-5849(00)00308-7. [DOI] [PubMed] [Google Scholar]
- 3.Sollner S, et al. Quinone reductase acts as a redox switch of the 20S yeast proteasome. EMBO Rep. 2009;10:65–70. doi: 10.1038/embor.2008.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong X, et al. NRH:quinone oxidoreductase 2 and NAD(P)H: quinone oxidoreductase 1 protect tumor suppressor p53 against 20s proteasomal degradation leading to stabilization and activation of p53. Cancer Res. 2007;67:5380–5388. doi: 10.1158/0008-5472.CAN-07-0323. [DOI] [PubMed] [Google Scholar]
- 5.Patrick BA, et al. Disruption of NAD(P)H:quinone oxidoreductase 1 gene in mice leads to 20S proteasomal degradation of p63 resulting in thinning of epithelium and chemical-induced skin cancer. Oncogene. 2011;30:1098–1107. doi: 10.1038/onc.2010.491. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Garate M, et al. NAD(P)H quinone oxidoreductase 1 inhibits the proteasomal degradation of the tumour suppressor p33(ING1b) EMBO Rep. 2008;9:576–581. doi: 10.1038/embor.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sollner S, et al. New roles of flavoproteins in molecular cell biology: an unexpected role for quinone reductases as regulators of proteasomal degradation. FEBS J. 2009;276:4313–4324. doi: 10.1111/j.1742-4658.2009.07143.x. [DOI] [PubMed] [Google Scholar]
- 8.Diehl JA, et al. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 9.Diehl JA, et al. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alt JR, et al. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukas J, et al. Convergence of mitogenic signalling cascades from diverse classes of receptors at the cyclin D-cyclin-dependent kinase-pRb-controlled G1 checkpoint. Mol. Cell. Biol. 1996;16:6917–6925. doi: 10.1128/mcb.16.12.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang M, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh TC, et al. Differential effects on growth, cell cycle arrest, and induction of apoptosis by resveratrol in human prostate cancer cell lines. Exp. Cell Res. 1999;249:109–115. doi: 10.1006/excr.1999.4471. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh TC, et al. Grape-derived chemopreventive agent resveratrol decreases prostate-specific antigen (PSA) expression in LNCaP cells by an androgen receptor (AR)-independent mechanism. Anticancer Res. 2000;20:225–228. [PubMed] [Google Scholar]
- 15.Narayanan BA, et al. Differential expression of genes induced by resveratrol in LNCaP cells: P53-mediated molecular targets. Int. J. Cancer. 2003;104:204–212. doi: 10.1002/ijc.10932. [DOI] [PubMed] [Google Scholar]
- 16.Jones SB, et al. Resveratrol-induced gene expression profiles in human prostate cancer cells. Cancer Epidemiol. Biomarkers Prev. 2005;14:596–604. doi: 10.1158/1055-9965.EPI-04-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayanan NK, et al. Resveratrol-induced cell growth inhibition and apoptosis is associated with modulation of phosphoglycerate mutase B in human prostate cancer cells: two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis and mass spectrometry evaluation. Cancer Detect. Prev. 2004;28:443–452. doi: 10.1016/j.cdp.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Gao S, et al. Modulation of androgen receptor-dependent transcription by resveratrol and genistein in prostate cancer cells. Prostate. 2004;59:214–225. doi: 10.1002/pros.10375. [DOI] [PubMed] [Google Scholar]
- 19.Lin HY, et al. Resveratrol induced serine phosphorylation of p53 causes apoptosis in a mutant p53 prostate cancer cell line. J. Urol. 2002;168:748–755. [PubMed] [Google Scholar]
- 20.Ates O, et al. Neuroprotection by resveratrol against traumatic brain injury in rats. Mol. Cell. Biochem. 2007;294:137–144. doi: 10.1007/s11010-006-9253-0. [DOI] [PubMed] [Google Scholar]
- 21.Das DK, et al. Resveratrol in cardioprotection: a therapeutic promise of alternative medicine. Mol. Interv. 2006;6:36–47. doi: 10.1124/mi.6.1.7. [DOI] [PubMed] [Google Scholar]
- 22.Tikoo K, et al. Change in histone H3 phosphorylation, MAP kinase p38, SIR 2 and p53 expression by resveratrol in preventing streptozotocin induced type I diabetic nephropathy. Free Radic. Res. 2008;42:397–404. doi: 10.1080/10715760801998646. [DOI] [PubMed] [Google Scholar]
- 23.Anekonda TS. Resveratrol—a boon for treating Alzheimer's disease? Brain Res. Rev. 2006;52:316–326. doi: 10.1016/j.brainresrev.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Gatz SA, et al. Take a break—resveratrol in action on DNA. Carcinogenesis. 2008;29:321–332. doi: 10.1093/carcin/bgm276. [DOI] [PubMed] [Google Scholar]
- 25.Csaki C, et al. Regulation of inflammation signalling by resveratrol in human chondrocytes in vitro. Biochem. Pharmacol. 2008;75:677–687. doi: 10.1016/j.bcp.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, et al. Identification and purification of resveratrol targeting proteins using immobilized resveratrol affinity chromatography. Biochem. Biophys. Res. Commun. 2004;323:743–749. doi: 10.1016/j.bbrc.2004.08.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiPietrantonio AM, et al. Specific processing of poly(ADP-ribose) polymerase, accompanied by activation of caspase-3 and elevation/reduction of ceramide/hydrogen peroxide levels, during induction of apoptosis in host HL-60 cells infected by the human granulocytic ehrlichiosis (HGE) agent. IUBMB Life. 2000;49:49–55. doi: 10.1080/713803590. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh TC, et al. Effects of extracts of Coriolus versicolor (I'm-Yunity) on cell-cycle progression and expression of interleukins-1 beta,-6, and -8 in promyelocytic HL-60 leukemic cells and mitogenically stimulated and nonstimulated human lymphocytes. J. Altern. Complement. Med. 2002;8:591–602. doi: 10.1089/107555302320825101. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh TC, et al. Induction of cell cycle changes and modulation of apoptogenic/anti-apoptotic and extracellular signaling regulatory protein expression by water extracts of I'm-Yunity (PSP) BMC Complement. Altern. Med. 2006;6:30. doi: 10.1186/1472-6882-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh TC. Uptake of resveratrol and role of resveratrol-targeting protein, quinone reductase 2, in normally cultured human prostate cells. Asian J. Androl. 2009;11:653–661. doi: 10.1038/aja.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh TC. Antiproliferative effects of resveratrol and the mediating role of resveratrol targeting protein NQO2 in androgen receptor-positive, hormone-non-responsive CWR22Rv1 cells. Anticancer Res. 2009;29:3011–3017. [PubMed] [Google Scholar]
- 32.DiPietrantonio AM, et al. Regulation of G1/S transition and induction of apoptosis in HL-60 leukemia cells by fenretinide (4HPR) Int. J. Cancer. 1998;78:53–61. doi: 10.1002/(sici)1097-0215(19980925)78:1<53::aid-ijc10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 33.Darzynkiewicz Z, et al. Flow cytometry in analysis of cell cycle and apoptosis. Semin. Hematol. 2001;38:179–193. doi: 10.1016/s0037-1963(01)90051-4. [DOI] [PubMed] [Google Scholar]
- 34.Ahmad N, et al. Resveratrol causes WAF-1/p21-mediated G(1)-phase arrest of cell cycle and induction of apoptosis in human epidermoid carcinoma A431 cells. Clin. Cancer Res. 2001;7:1466–1473. [PubMed] [Google Scholar]
- 35.Benitez DA, et al. Mechanisms involved in resveratrol-induced apoptosis and cell cycle arrest in prostate cancer-derived cell lines. J. Androl. 2007;28:282–293. doi: 10.2164/jandrol.106.000968. [DOI] [PubMed] [Google Scholar]
- 36.Aggarwal BB, et al. Role of resveratrol in prevention and therapy of cancer: preclinical and clinical studies. Anticancer Res. 2004;24:2783–2840. [PubMed] [Google Scholar]
- 37.Asher G, et al. P53 hot-spot mutants are resistant to ubiquitin-independent degradation by increased binding to NAD(P)H:quinone oxidoreductase 1. Proc. Natl Acad. Sci. USA. 2003;100:15065–15070. doi: 10.1073/pnas.2436329100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devlin HL, et al. Impairment of the DNA repair and growth arrest pathways by p53R2 silencing enhances DNA damage-induced apoptosis in a p53-dependent manner in prostate cancer cells. Mol. Cancer Res. 2008;6:808–818. doi: 10.1158/1541-7786.MCR-07-2027. [DOI] [PubMed] [Google Scholar]
- 39.Ahn KS, et al. Deficiency of NRH:quinone oxidoreductase 2 differentially regulates TNF signaling in keratinocytes: up-regulation of apoptosis correlates with down-regulation of cell survival kinases. Cancer Res. 2007;67:10004–10011. doi: 10.1158/0008-5472.CAN-07-2213. [DOI] [PubMed] [Google Scholar]