Abstract
AIM: To screen lymph nodes metastasis associated long noncoding RNAs (lncRNAs) in colorectal cancer through microarray analysis.
METHODS: Metastatic lymph node (MLN), normal lymph node (NLN) and tumor tissues of 3 colorectal cancer (CRC) patients were collected during the operation and validated by pathological examinations. RNAs were extracted from MLN, NLN, and cancer tissues separately. RNA quantity and quality were measured with a NanoDrop ND-1000 spectrophotometer and RNA integrity was assessed by standard denaturing agarose electrophoresis. Agilent Feature Extraction Software (Version 11.0.1.1) was used to analyze acquired array images. Four differently expressed lncRNAs were confirmed by quantitative real-time polymerase chain reaction (qRT-PCR) in 26 subsets of MLN, NLN, and tumor tissues.
RESULTS: Of 33045 lncRNAs, 1133 were differentially expressed in MLN compared with NLN, of which 260 were up-regulated and 873 down-regulated (≥ 2 fold-change). Five hundred and forty-five lncRNAs were differentially expressed in MLN compared with tumor tissues, of which 460 were up-regulated and 85 down-regulated (≥ 2 fold-change). Compared with NLN and cancer tissues, 14 lncRNAs were specifically up-regulated and 5 specifically down-regulated in MLN. AK307796, ENST00000425785, and AK021444 were confirmed to be specifically up-regulated in MLN and ENST00000465846 specifically down-regulated in MLN by qRT-PCR in 26 CRC patients.
CONCLUSION: The specifically expressed lncRNAs in MLN may exert a partial or key role in the progress of lymph nodes metastasis of CRC.
Keywords: Long noncoding RNAs, Colorectal cancer, Lymph nodes metastasis, Quantitative real-time polymerase chain reaction, MicroRNA
Core tip: Long noncoding RNAs (lncRNAs) have been reported to be aberrantly expressed in a variety of human cancers. However, no data are available regarding their functions in the lymph nodes metastasis of colorectal cancer (CRC). Our study is the first study to focus on lymph nodes metastasis associated lncRNAs in CRC by microarray. Obvious changes of lncRNAs expression profiles were observed in metastatic lymph node, normal lymph node, and tumor tissues of CRC. These changes of lncRNAs may serve as new diagnostic biomarkers and therapeutic targets for lymph node metastasis of CRC.
INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. According to the result of IARC (International Agency for Research on Cancer), approximately 1.23 million CRC patients were diagnosed yearly worldwide and 0.6 million CRC patients died in 2008[1]. As we all know, the prime cause of death among CRC patients is metastasis such as blood metastasis and lymph node metastasis. Among various metastatic pathways, lymph node metastasis is the most frequent pathway, which plays an important role in affecting the prognosis of CRC patients. Clinically, approximately 50% of patients with lymph node metastasis of CRC will experience disease recurrence[2-4]. Lymph node metastasis is also one of the critical clinical markers to judge the tumor stage and to make specific therapeutic schedule in CRC patients. Thus, how to detect the lymph node metastasis at early stage and how to further study molecular mechanisms are essential for diagnosis and therapy of CRC.
Long noncoding RNAs (lncRNAs), longer than 200 nucleotides (nt) in length, are a kind of RNAs that do not encode proteins. LncRNAs, transcribed by RNA polymerase II (RNA pol II), were ever thought to be “transcriptional noise” without biomedical functions. However, lncRNAs have been validated to have comprehensive functions in biological processes such as inactivating X chromosome, regulating DNA metabolism, and activating transcription by recently published studies[5-7]. Increasing evidence indicated that lncRNAs play important roles in many human diseases, including various types of cancer[8]. LncRNAs were reported to be abnormally expressed in various cancers and were associated with tumor cell proliferation, growth, apoptosis, invasion, and metastasis[9-14]. When it comes to CRC, several lncRNAs have been reported to be oncogenic factors by inhibiting apoptosis and promoting cell proliferation and so on[15,16]; while several other lncRNAs have been reported to be tumor suppressive factors by inhibiting cell growth[17]. However, the roles lncRNAs play in the progress of lymph node metastasis of CRC remain unknown.
In this study, we profiled the lncRNA expression in metastatic lymph node (MLN), normal lymph node (NLN), and tumor tissues from 3 CRC patients by using Human LncRNA Array. Differentially expressed lncRNAs were identified by comparing 3 different tissues with each other. Meanwhile, we selected specifically expressed lncRNAs in MLN by comparing MLN with NLN and tumor tissues out of these specifically expressed lncRNAs in MLN, and 4 were evaluated by qRT-PCR in 26 additional subsets of MLN, NLN and tumor tissues. Our findings indicated that lncRNAs may play a significant role in the process of lymph node metastasis of CRC.
MATERIALS AND METHODS
Patient samples
Twenty-six CRC patients who were surgically treated at Second Affiliated Hospital of Nanjing Medical University from June 2011 to June 2012 were included in our study. All patients recruited in this study received neither chemotherapy nor radiotherapy before the surgery. Written informed consent was obtained from all patients and permission for this study was obtained from the ethics committee of Second Affiliated Hospital of Nanjing Medical University. A set of lymph node and tumor tissues were collected during the operation and stained with HE by two experienced pathologists. Lymph nodes were divided into MLN and NLN according to the HE staining results. All samples were frozen in liquid nitrogen and stored at -80 °C until further analysis. Out of 87 samples, 9 (3 MLN, 3 NLN and, 3 tumor tissues) from 3 patients were used for microarray analysis of lncRNAs and the others were used for an extra evaluation.
RNA extraction
Total RNAs were extracted from 26 snap frozen subsets of MLN, NLN, and tumor tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. RNA quantity and quality were measured with a NanoDrop ND-1000 spectrometer and RNA integrity was assessed by standard denaturing agarosegel electrophoresis.
RNA labeling, array hybridization and data analysis
Sample labeling and array hybridization were performed according to the Agilent one-color microarray-based gene expression analysis protocol (Agilent Technology) with minor modifications. Briefly, mRNA was purified from total RNA after removal of rRNA (mRNA-ONLY™ eukaryotic mRNA isolation kit, Epicentre). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3’ bias utilizing a random priming method. The labeled cRNAs were purified by RNeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured with a NanoDrop ND-1000 spectrometer. Each labeled cRNA (1 μg) was fragmented by adding 5 μL 10 × Blocking Agent and 1 μL of 25 × Fragmentation Buffer, then the mixture was heated at 60 °C for 30 min, and finally 25 μL 2 × GE Hybridization Buffer was added to dilute the labeled cRNA. Fifty microliters of hybridization solution was dispensed into the gasket slide and assembled to the lncRNA expression microarray slide. The slides were incubated for 17 h at 65 °C in an Agilent hybridization oven. The hybridized arrays were washed, fixed and scanned using an Agilent DNA microarray scanner (part number G2505C).
Agilent feature extraction software (version 11.0.1.1) was used to analyze the array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). A normalized value is a relative number that comes from the ratio of the raw data value of the listed probe to that of the control. After quantile normalization of the raw data, lncRNAs and mRNAs with 9 samples that have flags in present or marginal were chosen for further data analysis. Differentially expressed lncRNAs and mRNAs with statistical significance were identified through Volcano Plot filtering and fold change filtering. Hierarchical clustering was performed using the Agilent GeneSpring GX software (Version 11.5.1). GO analysis and pathway analysis were performed using the standard enrichment computation method. The microarray work was performed by KangChen Bio-tech Shanghai, China.
Statistical analysis
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to verify differential expression of 4 lncRNAs that were detected to be specifically expressed in MLN by the lncRNA expression microarray. The first strand cDNA was synthesized using SuperScript™ III Reverse Transcriptase (Invitrogen), RNase Inhibitor (Epicentre), and 1.25 mmol/L dNTPs Mix. Each qRT-PCR reaction (in 10 μL) contained 2 × Super Array PCR master mix 5 μL, 10 μmol/L PCR forward primer 0.5 μL, 10 μmol/L PCR reverse primer 0.5 μL, diluted first strand cDNA synthesis reaction 2 μL, and ddH2O 2 μL. The cycling conditions consisted of an initial, single cycle of 10 min at 95 °C, followed by 40 cycles of 10 s at 95 °C, 60 s at 60 °C, and 15 s at 95 °C. For each sample, we performed qRT-PCR for target genes and a housekeeping gene. A standard curve was constructed using serial 10-fold dilutions (from 1 to 106) of the PCR products to quantify the results. According to the standard curve, the gene concentration of each sample is generated directly using Rotor-Gene Real-Time Analysis Software 6.0. For each sample, the relative amount of the target gene is determined by calculating the ratio between the concentration of the target gene and that of the housekeeping gene. For each target gene, we took the relative amount of the control sample as 1, and then the relative amount of the other samples as n fold of the control sample. The lncRNA expression differences between two groups were analyzed using Student’s t test. P < 0.05 was considered statistically significant.
RESULTS
Patients and array information
Clinical and pathological information of 3 patients were used for microarray analysis. Briefly, more than 15 lymph nodes were harvested and at least one was confirmed to be metastatic by HE staining in each patient (Figure 1).
Figure 1.
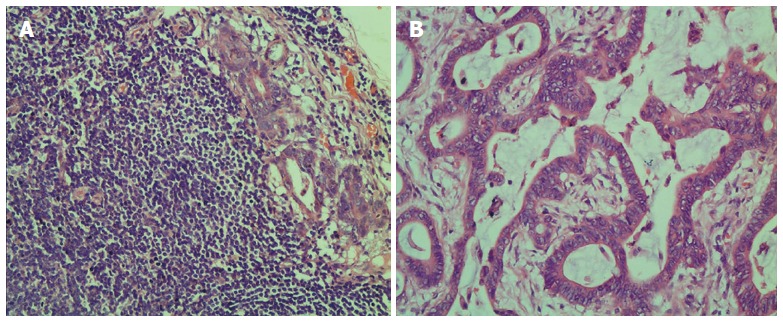
Metastatic lymph node and tumor tissues of colorectal cancer revealed by HE staining. A: Metastatic lymph node; B: Tumor tissue.
Arraystar human lncRNA microarray V2.0 is designed for the global profiling of human lncRNAs and protein-coding transcripts. A total of 33045 lncRNAs and 30215 coding transcripts can be detected by this second-generation lncRNAs microarray. The lncRNAs are carefully collected from the most authoritative databases such as RefSeq_NR, UCSC_Known Genes, Ensembl, H-invDB, UCR, lincRNA, and the related literature. Each transcript is represented by a specific exon or splice junction probe which can identify individual transcript accurately. Positive probes for housekeeping genes and negative probes are also printed onto the array for hybridization quality control.
LncRNA expression profile in MLN, NLN and tumor tissues
After quantile normalization of the raw data, lncRNAs with 9 samples that have flags in present or marginal were chosen for differentially expressed lncRNA screening. Out of 33045 lncRNAs, 20705 were in present or marginal in all 9 samples.
Quality assessment of lncRNA data after filtering
The box-plot is a convenient way to quickly visualize the distributions of a dataset. It is commonly used for comparing the distributions of the intensities from all samples. After normalization, the distributions of expression values for the samples are nearly the same, indicating that it is suitable for further analysis. The scatter-plot is a visualization method used for assessing the lncRNA expression variation (or reproducibility) between the two compared samples or two compared sample groups.
Screening of differentially expressed lncRNAs
Initially, we performed a volcano plot filtering between two groups to identify differentially expressed lncRNAs with statistical significance (fold change ≥ 2.0, P ≤ 0.05). From the data of microarray, variations of lncRNA and mRNA expression were shown in Tables 1 and 2. Briefly, compared with the NLN group, 873 probe sets representing 633 lncRNAs were down-regulated, while 260 probe sets representing 197 lncRNAs were up-regulated in the MLN group. Compared with the tumor tissue group, 85 probe sets representing 53 lncRNAs were down-regulated, while 460 probe sets representing 337 lncRNAs were up-regulated in the MLN group. Compared with the tumor tissue group, 623 probe sets representing 471 lncRNAs were down-regulated, while 2038 probe sets representing 1396 lncRNAs were up-regulated in the NLN group. Then, we performed a fold change filtering between every two samples in each patient to identify differentially expressed lncRNAs (fold change ≥ 2.0). The number of up-regulated and down-regulated lncRNAs varied in different patients compared with that of different samples. Interestingly, we found that the number of differently expressed lncRNAs between NLN and tumor tissues was much greater than those of differently expressed lncRNAs between MLN and NLN, and between MLN and tumor tissues (Figure 2), which indicated that NLN was quite different from tumor tissue.
Table 1.
Summary of data from long noncoding RNA microarray for three pairs of metastatic lymph nodes, normal lymph nodes and tumor tissue of colorectal cancer
| Sample ID | lncRNA |
|||||
| Fold change ≤ 4 | Fold change > 4 | Total | Changes lncRNA | |||
| R1 | MLN vs NLN | Up-regulation | 704 | 161 | 865 | 2087 |
| Down-regulation | 1034 | 188 | 1222 | |||
| MLN vs tumor | Up-regulation | 2864 | 1720 | 4584 | 9052 | |
| Down-regulation | 2421 | 2047 | 4468 | |||
| NLN vs tumor | Up-regulation | 2915 | 1999 | 4914 | 8895 | |
| Down-regulation | 2303 | 1678 | 3981 | |||
| R2 | MLN vs NLN | Up-regulation | 1223 | 354 | 1577 | 3923 |
| Down-regulation | 1934 | 412 | 2346 | |||
| MLN vs tumor | Up-regulation | 488 | 105 | 593 | 1109 | |
| Down-regulation | 457 | 59 | 516 | |||
| NLN vs tumor | Up-regulation | 2123 | 763 | 2886 | 4514 | |
| Down-regulation | 1231 | 397 | 1628 | |||
| R3 | MLN vs NLN | Up-regulation | 1557 | 399 | 1956 | 3707 |
| Down-regulation | 1441 | 310 | 1751 | |||
| MLM vs tumor | Up-regulation | 915 | 114 | 1029 | 1572 | |
| Down-regulation | 482 | 61 | 543 | |||
| NLN vs tumor | Up-regulation | 1481 | 597 | 2078 | 3619 | |
| Down-regulation | 1102 | 439 | 1541 | |||
MLN: Metastatic lymph nodes; NLN: Normal lymph nodes; Tumor: Tumor tissue of colorectal cancer. lncRNA: Long noncoding RNA.
Table 2.
Summary of data from message RNA microarray for three pairs of metastatic lymph nodes, normal lymph nodes and tumor tissue of colorectal cancer
| Sample | Message RNA |
|||||
| ID | Fold change ≤ 4 | Fold change > 4 | Total | Changes lncRNA | ||
| R1 | Positive vs negative | Up-regulation | 1421 | 310 | 1731 | 2818 |
| Down-regulation | 910 | 177 | 1087 | |||
| Positive vs tumor | Up-regulation | 4028 | 2762 | 6790 | 10525 | |
| Down-regulation | 2112 | 1623 | 3735 | |||
| Negative vs tumor | Up-regulation | 3848 | 2542 | 6390 | 9875 | |
| Down-regulation | 2088 | 1397 | 3485 | |||
| R2 | Positive vs negative | Up-regulation | 1354 | 544 | 1898 | 4354 |
| Down-regulation | 1960 | 496 | 2456 | |||
| Positive vs tumor | Up-regulation | 571 | 129 | 700 | 1534 | |
| Down-regulation | 711 | 123 | 834 | |||
| Negative vs tumor | Up-regulation | 1806 | 858 | 2664 | 5103 | |
| Down-regulation | 1692 | 747 | 2439 | |||
| R3 | Positive vs negative | Up-regulation | 1706 | 612 | 2318 | 4317 |
| Down-regulation | 1562 | 437 | 1999 | |||
| Positive vs tumor | Up-regulation | 865 | 142 | 1007 | 1855 | |
| Down-regulation | 731 | 117 | 848 | |||
| Negative vs tumor | Up-regulation | 1328 | 756 | 2084 | 4569 | |
| Down-regulation | 1681 | 804 | 2485 | |||
MLN: Metastatic lymph nodes; NLN: Normal lymph nodes; Tumor: Tumor tissue of colorectal cancer.
Figure 2.
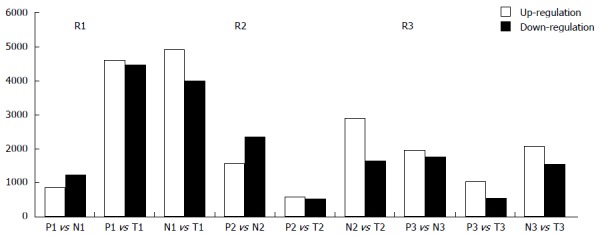
Counts of up-regulated and down-regulated long noncoding RNAs. Up-regulated and down-regulated long noncoding RNAs (lncRNAs) varied in different patients when comparing different samples: many lncRNAs were determined to be significantly up-regulated or down-regulated in metastatic lymph nodes (MLN), normal lymph node (NLN) and tumor tissues of colorectal cancer (CRC) in three patients by microarray analysis. The number of differently expressed lncRNAs between NLN and tumor tissue was much larger than those of differently expressed lncRNAs both between MLN and NLN and between MLN and tumor tissue. M: Metastatic lymph nodes; N: Normal lymph nodes; T: Tumor tissue of colorectal cancer.
Next, we collected the specifically expressed lncRNAs in MLN by comparing MLN with NLN and tumor tissues, respectively. Only 9 probe sets representing 5 lncRNAs were specifically down-regulated and 19 probe sets representing 14 lncRNAs specifically up-regulated in the MLN group compared with the NLN group and tumor tissue group (fold change ≥ 2.0, P ≤ 0.05). Of these, AK021444 (absolute fold change MLN/NLN = 7.763431269) was the most significantly up-regulated one, while BC042589 (MLN/NLN = 4.323467267) was the most significantly down-regulated one in the MLN group (Table 3). In each patient, there were 548, 193, and 134 specifically down-regulated probe sets (representing 424, 144, 113 lncRNAs), and 862, 135, and 693 specifically up-regulated ones (representing 593, 111, 522 lncRNAs) in MLN, NLN and tumor tissues, respectively.
Table 3.
Specifically expressed long noncoding RNAs in metastatic lymph nodes by comparing metastatic lymph nodes both with normal lymph nodes and tumor tissue of colorectal cancer
| Up-regulated in MLN | Down-regulated in MLN | ||
| LncRNA |
Log2 fold change (M > N and M > T) |
LncRNA |
Log2 fold change (M < N and M < T) |
| uc004aej.2 | 2.466393114 | BC042589 | -4.323467267 |
| ENST00000393311 | 2.086153236 | ENST00000429729 | -2.720521004 |
| ENST00000447552 | 2.795028512 | uc002ztx.3 | -2.196481477 |
| AK025180 | 2.885255406 | ENST00000418346 | -2.260120093 |
| AW449673 | 3.034259412 | ENST00000465846 | -3.200848076 |
| ENST00000450572 | 2.06748042 | ||
| ENST00000483126 | 2.357461429 | ||
| ENST00000425785 | 2.055384542 | ||
| CR599788 | 2.670107882 | ||
| AK097728 | 2.453511443 | ||
| uc002jsc.1 | 2.854856902 | ||
| BC036914 | 6.696784453 | ||
| AK021444 | 7.763431269 | ||
| AK307796 | 3.345445171 | ||
Metastatic lymph nodes (MLN) samples vs normal lymph node samples and MLN samples vs colorectal cancer tumor tissue, false discovery rate, 0.1%, P = 0.01. M: Metastatic lymph nodes; N: Normal lymph node; T: Tumor tissue of colorectal cancer; LncRNA: Long noncoding RNA.
Additionally, we determined whether lncRNAs changed gradually from tumor tissue to MLN and then from MLN to NLN. We summarized the gradually increased and decreased lncRNAs from tumor tissue to MLN then to NLN. These lncRNAs may also play an important role in facilitating the CRC tumor cell transfer from the primary tumor to lymph nodes.
As reported, lncRNAs exerted important roles in gene expression, especially their nearby coding genes. The possible relationships of lncRNAs with their nearby coding genes included natural antisense[18-20], exon sense-overlapping[21], intron sense-overlapping, intronic antisense[22,23], bidirectional[24-26], and intergenic[27]. We analyzed the relationship between lncRNAs and their nearby coding genes and each lncRNA was annotated by their associated genes and proteins.
Heat map and hierarchical clustering
Hierarchical clustering is one of the most widely used clustering methods for analyzing lncRNA expression data. Cluster analysis arranges samples into groups based on their expression levels, which allows us to hypothesize the relationships among samples. Here, hierarchical clustering was performed based on “differentially expressed lncRNAs”. The result of hierarchical clustering showed distinguishable lncRNAs expression profiles among samples which were divided into two main subgroups, signifying two different tissues.
LncRNA classification and subgroup analysis
Some specific classes of lncRNAs, such as enhancer lncRNAs, rinn lincRNAs, HOX cluster, lincRNAs near coding genes, and enhancer lncRNAs near coding genes, have been reported to be involved in the development of many diseases, especially cancers[28-31]. Thus, we analyzed the expression levels of these lncRNAs in 9 samples. LncRNAs with enhancer-like function (LncRNA-a) are identified using GENCODE annotation of the human genes[28]. Depletion of these lncRNAs led to decreased expression of the neighboring protein-coding genes, including the master regulator of hematopoiesis, SCL (also called TAL1), Snai1, and Snai2. LncRNAs were demonstrated to be necessary for the activation of gene expression[29]. Nine hundred and one enhancer lncRNAs were identified to be expressed in samples by microarray. Rinn et al[31] characterized the transcriptional landscape of the four human Hox loci and identified a total of 407 discrete transcribed regions in the four human Hox loci. Transcription of these lncRNAs may demarcate chromosomal domains of gene silencing at a distance, which contains profiling data of all probes in the four human Hox loci.
Overview of mRNA profiles
17159 mRNAs out of 30215 coding transcripts could be detected in 9 samples. Generally, hundreds of mRNAs were statistically aberrantly expressed among the 3 different kinds of tissues. GO and pathway analyses showed that the different expressed mRNAs might be involved in T/B cell receptor signaling pathway and primary immunodeficiency, which indicated that lymph node metastasis of CRC was closely associated with deficiency of immune cells.
Verifying the microarray results using qRT-PCR
To verify the result of lncRNA array, we selected 4 specifically expressed lncRNAs in MLN to confirm their expression levels by qRT-PCR in 26 CRC patients with lymph node metastasis (Table 4). The data were in agreement with the results of the microarray analysis. Out of these 4 lncRNA, AK021444, ENST00000425785, and AK307796, 3 specifically up-regulated lncRNAs in MLN, were proved to be highly expressed in MLN while ENST00000465846 to be down-regulated in MLN. Interestingly, the fold changes of all of them were smaller than that of the microarray results, indicating that the microarray result may exaggerate the difference (Figure 3).
Table 4.
Verification of expression levels of differentially expressed long noncoding RNAs in metastatic lymph nodes by quantitative real-time polymerase chain reaction in 26 colorectal cancer patients with lymph node metastasis
|
AK307796/GAPDH |
AK025180/GAPDH |
AK021444/GAPDH |
ENST00000465846/GAPDH |
|||||||||
| MLN | NLN | Tumor | MLN | NLN | Tumor | MLN | NLN | Tumor | MLN | NLN | Tumor | |
| R1 | 0.08760 | 0.03420 | 0.0358 | 0.0790 | 0.03480 | 0.03190 | 0.01370 | 0.00453 | 0.00735 | 0.02700 | 0.0347 | 0.06610 |
| R2 | 0.06670 | 0.04090 | 0.0432 | 0.1340 | 0.07770 | 0.03870 | 0.00978 | 0.00225 | 0.00560 | 0.02120 | 0.0421 | 0.06440 |
| R3 | 0.10417 | 0.04530 | 0.0524 | 0.0940 | 0.05300 | 0.04500 | 0.00762 | 0.00365 | 0.00435 | 0.02087 | 0.0334 | 0.05210 |
| R4 | 0.04708 | 0.03990 | 0.0526 | 0.0800 | 0.06020 | 0.04938 | 0.01060 | 0.00415 | 0.00541 | 0.02900 | 0.0297 | 0.05370 |
| R5 | 0.07200 | 0.03230 | 0.0608 | 0.0906 | 0.06860 | 0.05450 | 0.01030 | 0.00324 | 0.00608 | 0.02910 | 0.0390 | 0.06820 |
| R6 | 0.07950 | 0.03850 | 0.0635 | 0.1310 | 0.06070 | 0.05690 | 0.01160 | 0.00363 | 0.00764 | 0.02015 | 0.0294 | 0.05900 |
| R7 | 0.09710 | 0.03310 | 0.0401 | 0.0870 | 0.04520 | 0.03580 | 0.00813 | 0.00268 | 0.00455 | 0.02180 | 0.0303 | 0.05310 |
| R8 | 0.05620 | 0.04024 | 0.0614 | 0.1110 | 0.07154 | 0.05544 | 0.00798 | 0.00427 | 0.00706 | 0.02680 | 0.0379 | 0.06635 |
| R9 | 0.07110 | 0.03430 | 0.0414 | 0.0892 | 0.04750 | 0.03740 | 0.01320 | 0.00289 | 0.00495 | 0.02230 | 0.0315 | 0.06427 |
| R10 | 0.06120 | 0.03910 | 0.0591 | 0.0923 | 0.04680 | 0.05233 | 0.00834 | 0.00308 | 0.00751 | 0.02520 | 0.0369 | 0.05480 |
| R11 | 0.08980 | 0.04270 | 0.0398 | 0.1200 | 0.07313 | 0.03980 | 0.01340 | 0.00448 | 0.00508 | 0.02390 | 0.0325 | 0.06177 |
| R12 | 0.06250 | 0.04133 | 0.0605 | 0.0827 | 0.07010 | 0.03670 | 0.00780 | 0.00397 | 0.00562 | 0.02638 | 0.0355 | 0.05610 |
| R13 | 0.10250 | 0.03570 | 0.0423 | 0.1050 | 0.04990 | 0.05060 | 0.01270 | 0.00319 | 0.00683 | 0.02310 | 0.0339 | 0.05690 |
| R14 | 0.05850 | 0.03592 | 0.0537 | 0.1070 | 0.05150 | 0.05470 | 0.01310 | 0.00408 | 0.00697 | 0.02556 | 0.0389 | 0.06105 |
| R15 | 0.04981 | 0.03490 | 0.0449 | 0.0983 | 0.06360 | 0.04074 | 0.00883 | 0.00313 | 0.00719 | 0.02497 | 0.0331 | 0.05940 |
| R16 | 0.09623 | 0.03680 | 0.0630 | 0.0962 | 0.05080 | 0.04980 | 0.00847 | 0.00375 | 0.00463 | 0.02350 | 0.0383 | 0.06370 |
| R17 | 0.08120 | 0.04080 | 0.0507 | 0.1030 | 0.06750 | 0.04230 | 0.01210 | 0.00335 | 0.00652 | 0.02090 | 0.0305 | 0.06010 |
| R18 | 0.07800 | 0.03623 | 0.0627 | 0.0839 | 0.04820 | 0.04850 | 0.01240 | 0.00367 | 0.00478 | 0.02860 | 0.0365 | 0.06505 |
| R19 | 0.09910 | 0.03800 | 0.0462 | 0.1140 | 0.07084 | 0.04472 | 0.00932 | 0.00349 | 0.00531 | 0.02050 | 0.0311 | 0.05750 |
| R20 | 0.08452 | 0.03524 | 0.0566 | 0.0998 | 0.05473 | 0.04634 | 0.01160 | 0.00381 | 0.00736 | 0.02475 | 0.0363 | 0.06810 |
| R21 | 0.09110 | 0.04210 | 0.0437 | 0.0815 | 0.06680 | 0.04360 | 0.00956 | 0.00341 | 0.00759 | 0.02410 | 0.0318 | 0.05530 |
| R22 | 0.06780 | 0.03794 | 0.0521 | 0.1160 | 0.06154 | 0.04740 | 0.00868 | 0.00403 | 0.00592 | 0.02600 | 0.0391 | 0.06287 |
| R23 | 0.09380 | 0.04392 | 0.0475 | 0.1210 | 0.05823 | 0.04580 | 0.01250 | 0.00275 | 0.00578 | 0.02430 | 0.0329 | 0.05450 |
| R24 | 0.06781 | 0.03900 | 0.0553 | 0.0881 | 0.06014 | 0.05143 | 0.00907 | 0.00441 | 0.00622 | 0.02730 | 0.0376 | 0.06590 |
| R25 | 0.07430 | 0.04180 | 0.0491 | 0.1150 | 0.05680 | 0.04150 | 0.01190 | 0.00295 | 0.00517 | 0.02279 | 0.0309 | 0.05830 |
| R26 | 0.05320 | 0.03753 | 0.0579 | 0.1020 | 0.06840 | 0.05630 | 0.01290 | 0.00421 | 0.00636 | 0.02813 | 0.0385 | 0.06668 |
MLN: Metastatic lymph nodes; NLN: Normal lymph nodes; Tumor: Tumor tissue of colorectal cancer; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
Figure 3.
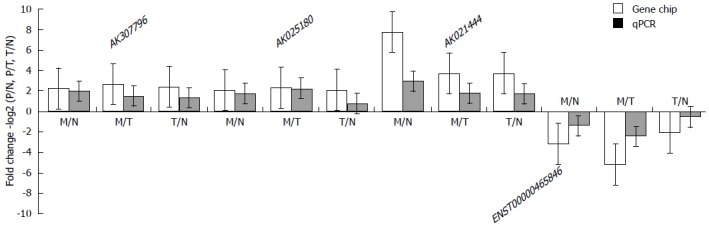
Comparison between microarray data and quantitative real-time polymerase chain reaction results. AK021444, ENST00000425785, AK307796 and ENST00000465846 were determined to be differentially expressed by microarray in 3 colorectal cancer patients, and this result was validated by quantitative polymerase chain reaction result (qPCR). AK021444, ENST00000425785, and AK307796, 3 specifically up-regulated lncRNAs in metastatic lymph nodes (MLN), were confirmed to be highly expressed in MLN, while ENST00000465846 was confirmed to be down-regulated in MLN. The validation results for the four long noncoding RNAs (lncRNAs) indicated that the microarray data correlated well with the qPCR results. M: Metastatic lymph nodes; N: Normal lymph nodes; T: Tumor tissue of colorectal cancer.
DISCUSSION
Recent advances in genome analysis, including massively parallel sequencing and microarray, have shown that a much greater portion of the human genome is pervasively transcribed into RNA than previously recognized. Moreover, much of the evidence emerging in recent years has highlighted the biological and pathological importance of RNAs that lack protein-coding potential; these are collectively referred to as noncoding RNAs (ncRNAs)[32]. MiRNAs are a kind of well-defined ncRNAs that play critical roles in many diseases including cancer. In contrast, functions of a majority of lncRNAs remain unknown, but recent studies have begun to shed light on the critical roles played by lncRNAs in a variety of cellular processes such as tumorigenesis and malignancy transformation in various types of cancer[9,33-35]. However, the potential use of lncRNA expression profiling to identify lymph node metastasis of CRC has not been systematically explored. To uncover the potential role lncRNAs play in lymph node metastasis of CRC, we investigated the lncRNA expression signatures in MLN, NLN, and tumor tissues of CRC.
Arraystar Human LncRNA Microarray V2.0 is one of the most commonly used commercial microarrays in human cancer profiling[35,36]. LncRNAs were collected from databases that were very comprehensive and authoritative. It allows the analysis of mRNA and lncRNA expression at the same time and is easier to follow than transcript sequencing analysis.
In this study, to be comprehensive, we selected 3 CRC patients including 1 right colon cancer patient, 1 left colon cancer patient, and 1 rectal cancer patient. The results indicated that rectal cancer and colon cancer had a relatively greater diversity in lncRNA expression. For instance, 9051 lncRNAs altered between MLN and tumor tissues in the rectal patient while only 1109 and 1572 lncRNAs altered in the two colon cancer patients, respectively. The lncRNA expression features may be greatly different between colon cancer and rectal cancer, but more information was needed to confirm this disparity. In each patient, to reduce the histological difference, we compared the MLN with NLN and tumor tissues, respectively. The numbers of differentially expressed lncRNAs were 389 between MLN and tumor tissue, 829 between MLN and NLN, and 1867 between NLN and tumor tissue. Our results indicated that tumor tissue and lymph nodes are two quite different kinds of tissues. Thus, it was reasonable to compare MLN both with NLN and tumor tissue to identify the specifically altered lncRNAs in MLN.
Although hundreds of lncRNAs were deregulated when comparing the MLN group with the NLN group and tumor tissue group, respectively, only 14 lncRNAs were specifically up-regulated in MLN (both compared with NLN and tumor tissue): uc004aej.2, ENST00000393311, ENST00000447552, AK025180, AW449673, ENST00000450572, ENST00000483126, ENST00000425785, CR599788, AK097728, uc002jsc.1, BC036914, AK021444, and AK307796; and only 5 were specifically down-regulated in MLN: BC042589, ENST00000429729, uc002ztx.3, ENST00000465846, ENST00000418346. These lncRNAs were specifically expressed in MLN, indicating that their potential important role in facilitating the occurrence of lymph node metastasis of CRC. Unfortunately, after careful retrieval, no additional information of direct functions of these lncRNAs was available except the papers that first described their discoveries[37,38]. Further research is urgently needed to clarify the functions and the underlying mechanism of these lncRNAs in the process of lymph node metastasis of CRC.
Generally, tumor cells transfer from the primary tumor to adjacent lymph nodes and finally reach distant lymph nodes. To determine whether certain lncRNAs gradually change during lymph node metastasis, we identified those lncRNAs that gradually up-regulated and down-regulated from tumor tissue to MLN then from MLN to NLN. Four and 66 lncRNAs were identified to be gradually up-regulated and down-regulated from tumor tissue to MLN and NLN, respectively. Unfortunately, as identified above, functions of these lncRNAs were also unclear. We supposed that these gradually changed lncRNAs may also play partial roles in the progress of lymph node metastasis. But more evidence should be collected to confirm this hypothesis.
To validate the consistency of the gene chip, we selected 4 specifically expressed lncRNAs (AK021444, ENST00000425785, AK307796, and ENST00000465846) to evaluate their expression in 26 CRC patients with lymph node metastasis. The results of qRT-PCR were consistent well with the chip results. AK021444 is a 1611 bp lncRNA collected from ncRNA Expression Database (http://jsm-research.imb.uq.edu.au/nred/cgi-bin/ncrnadb.pl). It is located on chromosome 13 and associated with gene (nearby gene) POSTN (periostin). All of 4 isoforms of periostin protein could be detected to be associated with AK021444 in our samples. POSTN was reported to be up-regulated in various cancers including non-small cell lung cancer, pancreatic cancer, breast cancer, and CRC[39-42]. Furthermore, Bao et al[41] reported that POSTN in those hepatic metastases was 2.6-fold of that in the matched colon primary tumors. POSTN was also reported to be overexpressed in serum of CRC patients and the overexpression of POSTN was correlated with distant metastasis of CRC[43]. In addition, POSTN was reported to be correlated with lymph node metastasis of many other malignant neoplasms[43,44]. In our study, AK021444 was significantly up-regulated in MLN compared with the primary tumor, indicating that it is in accordance with POSTN expression. However, whether AK021444 can regulate lymph node metastasis of CRC and what the underlying molecular mechanism is need further study.
ENST00000425785, located on chromosome 1, was a 1258 bp lncRNA transcribed from the antisense strand of the DPYD/DPD (dihydropyrimidine dehydrogenase) gene. DPYD/DPD played a key role in the pharmacokinetics of 5-FU and was considerably involved in the antitumour efficacy of 5-FU[45-48]. DPD was reported to be up-regulated in tumour tissues and associated with poor prognosis of CRC in various studies[49-52]. The increase of intra-tumoral DPD level was reported to be one of the mechanisms of drug resistance[50-53]. Shirota et al[53] considered that the mRNA level of DPDY reflected tumor progression in CRC and a high level of DPDY was associated with lymph node metastasis of CRC. However, the mechanism by which ENST00000425785 interacts with DPYD/DPD and the biological functions of ENST00000425785 need further study.
AK307796 was another specifically up-regulated lncRNA in MLN. AK307796 was located on chromosome 6q, and the annotated nearby coding gene was ARHGAP18. ARHGAP18 was thought to be specifically expressed in the epididymis[54]. However, a recently published study defined ARHGAP18 as one of the crucial factors for the regulation of RhoA for the control of cell shape, spreading, and migration. Knockdown of ARHGAP18 inhibited cell migration while overexpression of ARHGAP18 promoted cell migration[55]. In accordance with our results, overexpression of AK307796 might promote the CRC cancer cell migration to lymph nodes. ENST00000465846 was a 419 bp lncRNA transcribed from chromosome 17. The annotated nearby coding gene was HOXB3-6. HOX genes control diverse cellular processes by regulating the expression of many downstream target genes. Thus, it is typical for an individual homeoprotein to confer pleiotropic effects on cell behavior, including alterations in proliferation, survival, migration, and invasion[56-60]. Genes of the HOX family are aberrantly expressed in various cancers such as leukemia, melanoma, and breast, ovarian, cervical, esophageal, prostate, and colorectal cancers[61-68]. However, the relationship of HOXB with ENST00000465846 remains unclear.
Our findings demonstrated differential lncRNA expression patterns between MLN, NLN, and tumor tissues of CRC, which can have potential implications in diagnosis and treatment of lymph node metastasis of CRC. Clinically, tumor markers of lymph node metastasis of CRC were rare. Even several markers were reported, their specificity and sensitivity were also limited. New molecular markers predicting lymph node metastasis of CRC are urgently needed. It has been reported that lncRNAs demonstrated higher specificity than that of protein-coding mRNAs[12,69,70], and had the advantages of being detectable in the blood[71] and urine[69,70] of cancer patients by conventional PCR methods. The four identified distinctive lncRNAs in MLN in the present study may potentially aid the development of specific diagnostic markers and novel therapeutic markers. However, it was too early for us to explore the 4 lncRNAs to be the possible clinical biomarkers for CRC just based on the present data.
As far as we know, this is the first study to describe the expression profiles of human lncRNAs in MLN, NLN, and tumor tissues of CRC by microarray assay. In this study, we have identified hundreds of differentially expressed lncRNAs among these 3 different tissues. Thirteen lncRNAs and 5 lncRNAs were identified to be specifically up-regulated and down-regulated in MLN, respectively. AK021444, ENST00000425785, AK307796, and ENST00000465846 were validated to be specifically expressed in MLN of CRC. Our findings suggest that lncRNAs may play important roles in lymph node metastasis of CRC. The clinical implications of our findings include the potential use of lncRNAs as molecular diagnosis markers and therapeutic targets. However, this is just a pilot study, and further studies are needed to expand the sample size for clinical research, and determine whether these lncRNAs can serve as new diagnostic biomarkers and therapeutic targets for lymph node metastasis of CRC.
COMMENTS
Background
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. The prime cause of death among CRC patients is metastasis such as blood metastasis and lymph node metastasis. Lymph node metastasis plays an important role in affecting the prognosis of CRC patients. Thus, how to detect lymph node metastasis at early stage and how to further study molecular mechanisms are essential for diagnosis and therapy of CRC. Long noncoding RNAs (lncRNAs) have been validated to have comprehensive functions in biological processes. When it comes to CRC, several lncRNAs have been reported to be oncogenic factors by inhibiting apoptosis and promoting cell proliferation and so on. However, the roles lncRNAs play in the progress of lymph node metastasis of CRC remain unknown. The authors findings indicated that lncRNAs may play a significant role in the process of lymph node metastasis of CRC.
Research frontiers
LncRNAs have been validated to have comprehensive functions in biological processes such as inactivating X chromosome, regulating DNA metabolism, and activating transcription by recently published studies. Increasing evidence indicated that lncRNAs play important roles in many human diseases, including various types of cancer. LncRNAs were reported to be abnormally expressed in various cancers and were associated with tumor cell proliferation, growth, apoptosis, invasion, and metastasis.
Innovations and breakthroughs
In this study, authors have identified hundreds of differently expressed lncRNAs among the 3 different tissues. Thirteen lncRNAs and 5 lncRNAs were identified to be specifically up-regulated and down-regulated in metastatic lymph node (MLN), respectively. AK021444, ENST00000425785, AK307796, and ENST00000465846 were validated to be specifically expressed in MLN of CRC.
Applications
The findings suggest that lncRNAs may play important roles in lymph node metastasis of CRC. The clinical implications of our findings include the potential use of lncRNAs as molecular diagnosis markers and therapeutic targets.
minology
LncRNAs, longer than 200 nucleotides (nt) in length, are a kind of RNAs that do not encode proteins.
Peer review
LncRNAs was recently found to be frequently dysregulated in various diseases. Although previous data suggested that lncRNAs are likely to have biological effects, this possibility needs to be confirmed by specific hypothesis-driven studies. Understanding the precise molecular mechanisms by which lncRNAs function in various diseases will be critical for exploring new potential strategies for early diagnosis and therapy.
Footnotes
Supported by Natural Science Foundation Project of Jiangsu Province, No. BK2012872; and the Science and Technology Projects, Health Department of Jiangsu Province, No. H201207
P- Reviewers: De Re V, Mihaila RG S- Editor: Gou SX L- Editor: Wang TQ E- Editor: Wang CH
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. GLOBOCAN 2008 v2.0, Cancer incidence and mortality worldwide: IARC CancerBase No. 10 [Internet] Lyon: International Agency for Research on Cancer; 2010. [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Nicastri DG, Doucette JT, Godfrey TE, Hughes SJ. Is occult lymph node disease in colorectal cancer patients clinically significant? A review of the relevant literature. J Mol Diagn. 2007;9:563–571. doi: 10.2353/jmoldx.2007.070032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iddings D, Ahmad A, Elashoff D, Bilchik A. The prognostic effect of micrometastases in previously staged lymph node negative (N0) colorectal carcinoma: a meta-analysis. Ann Surg Oncol. 2006;13:1386–1392. doi: 10.1245/s10434-006-9120-y. [DOI] [PubMed] [Google Scholar]
- 5.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 11.Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 12.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 14.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 16.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 17.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 19.Okada Y, Tashiro C, Numata K, Watanabe K, Nakaoka H, Yamamoto N, Okubo K, Ikeda R, Saito R, Kanai A, et al. Comparative expression analysis uncovers novel features of endogenous antisense transcription. Hum Mol Genet. 2008;17:1631–1640. doi: 10.1093/hmg/ddn051. [DOI] [PubMed] [Google Scholar]
- 20.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 22.Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, El-Jundi TA, da Silva AM, Reis EM, Verjovski-Almeida S. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AV, Almeida GT, Egidio CM, Paquola AC, Machado AA, Festa F, et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–6692. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- 24.Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P. Replication and transcription: shaping the landscape of the genome. Nat Rev Genet. 2005;6:669–677. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- 25.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 26.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7 Suppl 1:S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 33.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 36.Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, Xia D, Yang Y, Deng K, Xiao H, et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 39.Soltermann A, Tischler V, Arbogast S, Braun J, Probst-Hensch N, Weder W, Moch H, Kristiansen G. Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin Cancer Res. 2008;14:7430–7437. doi: 10.1158/1078-0432.CCR-08-0935. [DOI] [PubMed] [Google Scholar]
- 40.Baril P, Gangeswaran R, Mahon PC, Caulee K, Kocher HM, Harada T, Zhu M, Kalthoff H, Crnogorac-Jurcevic T, Lemoine NR. Periostin promotes invasiveness and resistance of pancreatic cancer cells to hypoxia-induced cell death: role of the beta4 integrin and the PI3k pathway. Oncogene. 2007;26:2082–2094. doi: 10.1038/sj.onc.1210009. [DOI] [PubMed] [Google Scholar]
- 41.Bao S, Ouyang G, Bai X, Huang Z, Ma C, Liu M, Shao R, Anderson RM, Rich JN, Wang XF. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell. 2004;5:329–339. doi: 10.1016/s1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- 42.Shao R, Bao S, Bai X, Blanchette C, Anderson RM, Dang T, Gishizky ML, Marks JR, Wang XF. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol. 2004;24:3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben QW, Zhao Z, Ge SF, Zhou J, Yuan F, Yuan YZ. Circulating levels of periostin may help identify patients with more aggressive colorectal cancer. Int J Oncol. 2009;34:821–828. doi: 10.3892/ijo_00000208. [DOI] [PubMed] [Google Scholar]
- 44.Takanami I, Abiko T, Koizumi S. Expression of periostin in patients with non-small cell lung cancer: correlation with angiogenesis and lymphangiogenesis. Int J Biol Markers. 2008;23:182–186. doi: 10.1177/172460080802300308. [DOI] [PubMed] [Google Scholar]
- 45.Hisamitsu K, Tsujitani S, Yamaguchi K, Fukuda K, Konishi I, Kaibara N. Expression of dihydropyrimidine dehydrogenase in cancer cells but not in stromal cells predicts the efficacy of fluorouracil treatment in patients with gastric carcinoma. Anticancer Res. 2004;24:2495–2501. [PubMed] [Google Scholar]
- 46.Terashima M, Irinoda T, Fujiwara H, Nakaya T, Takagane A, Abe K, Yonezawa H, Oyama K, Inaba T, Saito K, et al. Roles of thymidylate synthase and dihydropyrimidine dehydrogenase in tumor progression and sensitivity to 5-fluorouracil in human gastric cancer. Anticancer Res. 2002;22:761–768. [PubMed] [Google Scholar]
- 47.Terashima M, Fujiwara H, Takagane A, Abe K, Irinoda T, Nakaya T, Yonezawa H, Oyama K, Saito K, Kanzaki N, et al. Prediction of sensitivity to fluoropyrimidines by metabolic and target enzyme activities in gastric cancer. Gastric Cancer. 2003;6 Suppl 1:71–81. doi: 10.1007/s10120-003-0221-z. [DOI] [PubMed] [Google Scholar]
- 48.Zhu AX, Puchalski TA, Stanton VP, Ryan DP, Clark JW, Nesbitt S, Charlat O, Kelly P, Kreconus E, Chabner BA, et al. Dihydropyrimidine dehydrogenase and thymidylate synthase polymorphisms and their association with 5-fluorouracil/leucovorin chemotherapy in colorectal cancer. Clin Colorectal Cancer. 2004;3:225–234. doi: 10.3816/CCC.2004.n.003. [DOI] [PubMed] [Google Scholar]
- 49.Diasio RB, Johnson MR. Dihydropyrimidine dehydrogenase: its role in 5-fluorouracil clinical toxicity and tumor resistance. Clin Cancer Res. 1999;5:2672–2673. [PubMed] [Google Scholar]
- 50.Jiang W, Lu Z, He Y, Diasio RB. Dihydropyrimidine dehydrogenase activity in hepatocellular carcinoma: implication in 5-fluorouracil-based chemotherapy. Clin Cancer Res. 1997;3:395–399. [PubMed] [Google Scholar]
- 51.Etienne MC, Chéradame S, Fischel JL, Formento P, Dassonville O, Renée N, Schneider M, Thyss A, Demard F, Milano G. Response to fluorouracil therapy in cancer patients: the role of tumoral dihydropyrimidine dehydrogenase activity. J Clin Oncol. 1995;13:1663–1670. doi: 10.1200/JCO.1995.13.7.1663. [DOI] [PubMed] [Google Scholar]
- 52.Beck A, Etienne MC, Chéradame S, Fischel JL, Formento P, Renée N, Milano G. A role for dihydropyrimidine dehydrogenase and thymidylate synthase in tumour sensitivity to fluorouracil. Eur J Cancer. 1994;30A:1517–1522. doi: 10.1016/0959-8049(94)00216-r. [DOI] [PubMed] [Google Scholar]
- 53.Shirota Y, Ichikawa W, Uetake H, Yamada H, Nihei Z, Sugihara K. Intratumoral dihydropyrimidine dehydrogenase messenger RNA level reflects tumor progression in human colorectal cancer. Ann Surg Oncol. 2002;9:599–603. doi: 10.1007/BF02573898. [DOI] [PubMed] [Google Scholar]
- 54.Li X, Liu Q, Liu S, Zhang J, Zhang Y. New member of the guanosine triphosphatase activating protein family in the human epididymis. Acta Biochim Biophys Sin (Shanghai) 2008;40:855–863. [PubMed] [Google Scholar]
- 55.Neisch AL, Formstecher E, Fehon RG. Conundrum, an ARHGAP18 orthologue, regulates RhoA and proliferation through interactions with Moesin. Mol Biol Cell. 2013;24:1420–1433. doi: 10.1091/mbc.E12-11-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 57.Chen H, Sukumar S. Role of homeobox genes in normal mammary gland development and breast tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:159–175. doi: 10.1023/a:1025996707117. [DOI] [PubMed] [Google Scholar]
- 58.Chen H, Sukumar S. HOX genes: emerging stars in cancer. Cancer Biol Ther. 2003;2:524–525. doi: 10.4161/cbt.2.5.525. [DOI] [PubMed] [Google Scholar]
- 59.Del Bene F, Wittbrodt J. Cell cycle control by homeobox genes in development and disease. Semin Cell Dev Biol. 2005;16:449–460. doi: 10.1016/j.semcdb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 60.Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer. 2005;41:2428–2437. doi: 10.1016/j.ejca.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 61.Chen KN, Gu ZD, Ke Y, Li JY, Shi XT, Xu GW. Expression of 11 HOX genes is deregulated in esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1044–1049. [PubMed] [Google Scholar]
- 62.Coletta RD, Jedlicka P, Gutierrez-Hartmann A, Ford HL. Transcriptional control of the cell cycle in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2004;9:39–53. doi: 10.1023/B:JOMG.0000023587.40966.f6. [DOI] [PubMed] [Google Scholar]
- 63.López R, Garrido E, Vázquez G, Piña P, Pérez C, Alvarado I, Salcedo M. A subgroup of HOX Abd-B gene is differentially expressed in cervical cancer. Int J Gynecol Cancer. 2006;16:1289–1296. doi: 10.1111/j.1525-1438.2006.00603.x. [DOI] [PubMed] [Google Scholar]
- 64.Maeda K, Hamada J, Takahashi Y, Tada M, Yamamoto Y, Sugihara T, Moriuchi T. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int J Cancer. 2005;114:436–441. doi: 10.1002/ijc.20706. [DOI] [PubMed] [Google Scholar]
- 65.Maroulakou IG, Spyropoulos DD. The study of HOX gene function in hematopoietic, breast and lung carcinogenesis. Anticancer Res. 2003;23:2101–2110. [PubMed] [Google Scholar]
- 66.Waltregny D, Alami Y, Clausse N, de Leval J, Castronovo V. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate. 2002;50:162–169. doi: 10.1002/pros.10045. [DOI] [PubMed] [Google Scholar]
- 67.De Vita G, Barba P, Odartchenko N, Givel JC, Freschi G, Bucciarelli G, Magli MC, Boncinelli E, Cillo C. Expression of homeobox-containing genes in primary and metastatic colorectal cancer. Eur J Cancer. 1993;29A:887–893. doi: 10.1016/s0959-8049(05)80432-0. [DOI] [PubMed] [Google Scholar]
- 68.Kanai M, Hamada J, Takada M, Asano T, Murakawa K, Takahashi Y, Murai T, Tada M, Miyamoto M, Kondo S, et al. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol Rep. 2010;23:843–851. [PubMed] [Google Scholar]
- 69.Hessels D, Klein Gunnewiek JM, van Oort I, Karthaus HF, van Leenders GJ, van Balken B, Kiemeney LA, Witjes JA, Schalken JA. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8–15; discussion 15-16. doi: 10.1016/s0302-2838(03)00201-x. [DOI] [PubMed] [Google Scholar]
- 70.Tinzl M, Marberger M, Horvath S, Chypre C. DD3PCA3 RNA analysis in urine--a new perspective for detecting prostate cancer. Eur Urol. 2004;46:182–186; discussion 187. doi: 10.1016/j.eururo.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 71.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]


