Abstract
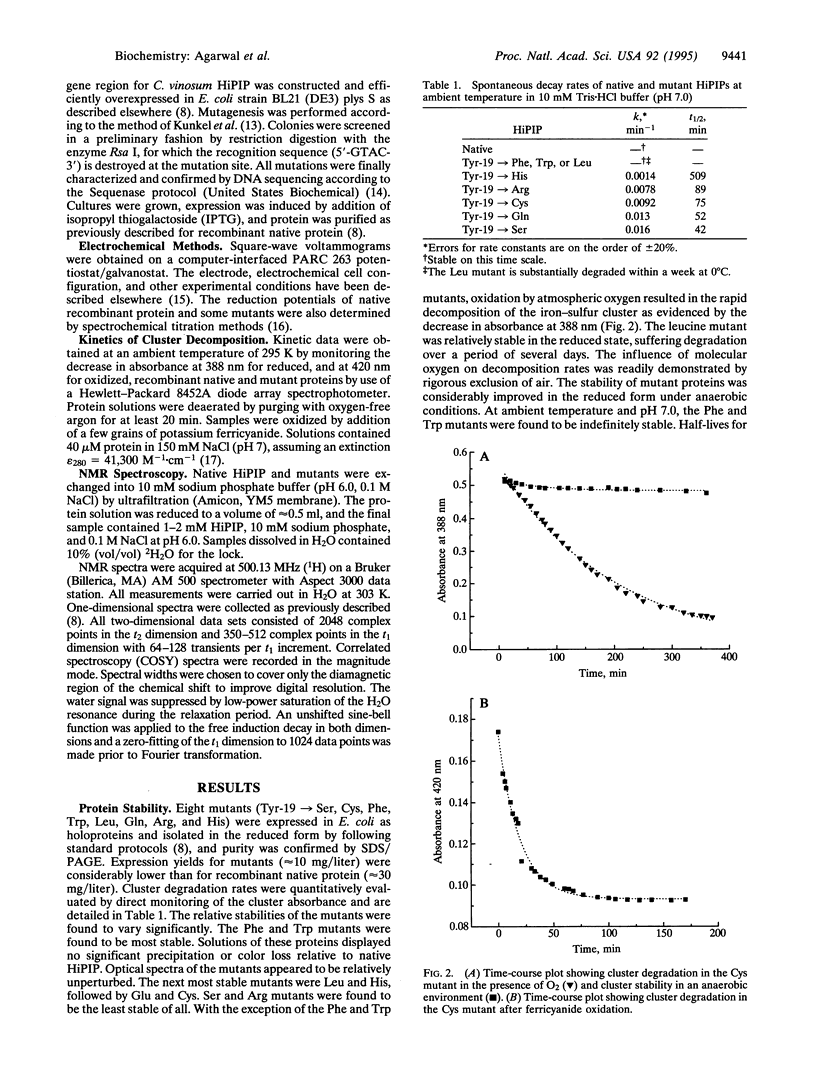
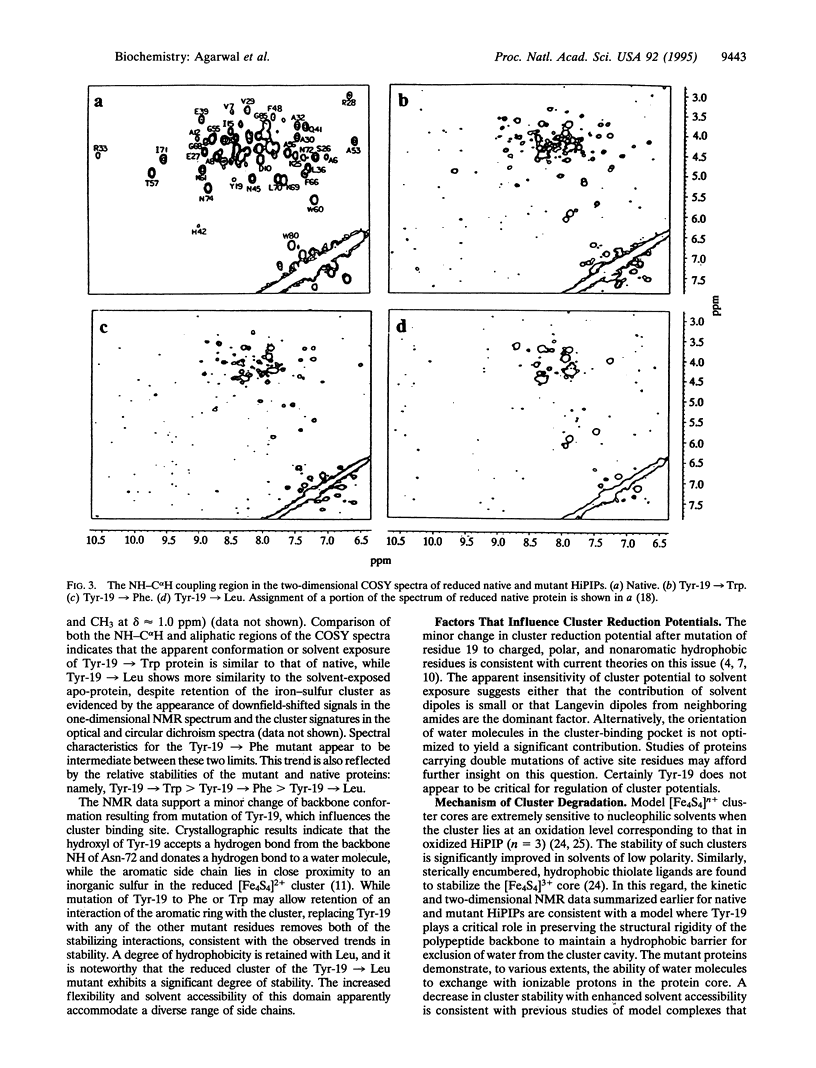
The functional role of residue Tyr-19 of Chromatium vinosum HiPIP has been evaluated by site-directed mutagenesis experiments. The stability of the [Fe4S4] cluster prosthetic center is sensitive to side-chain replacements. Polar residues result in significant instability, while nonpolar residues (especially with aromatic side chains) maintain cluster stability. Two-dimensional NMR data of native and mutant HiPIPs are consistent with a model where Tyr-19 serves to preserve the structural rigidity of the polypeptide backbone, thereby maintaining a hydrophobic barrier for exclusion of water from the cluster cavity. Solvent accessibility results in more facile oxidation of the cluster by atmospheric oxygen, with subsequent rapid hydrolysis of the [Fe4S4]3+ core.
Full text
PDF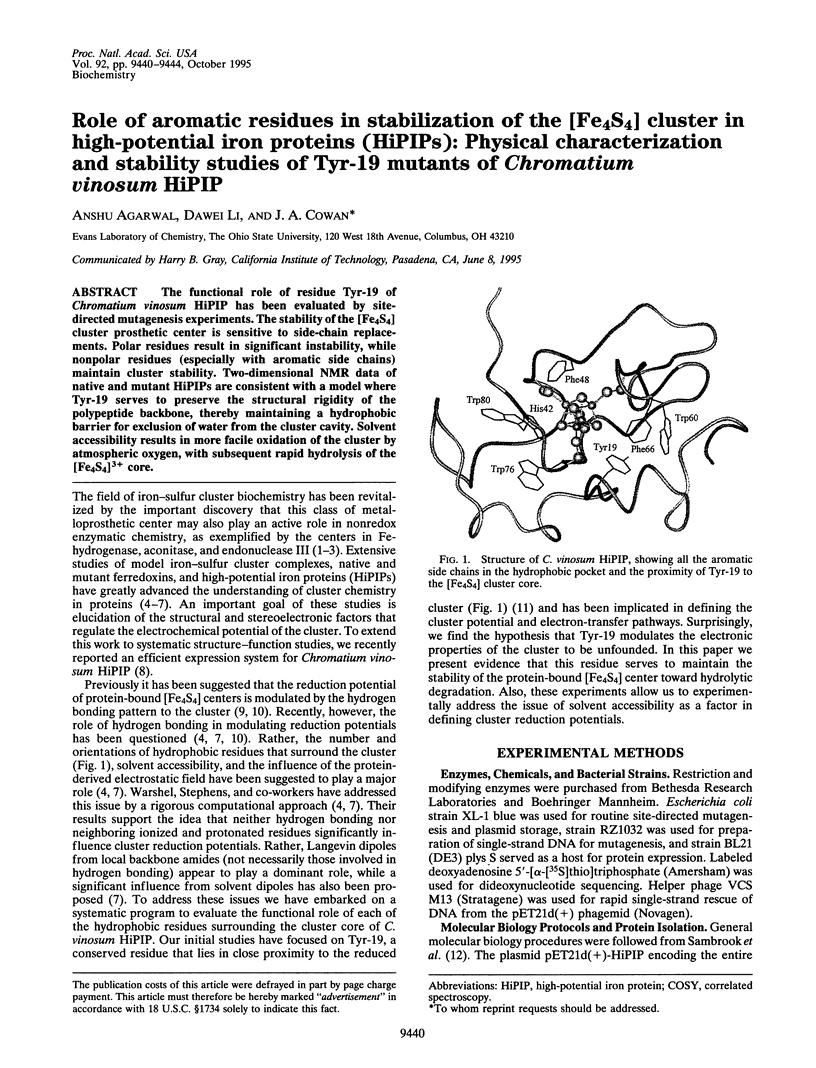


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman E., Watenpaugh K. D., Jensen L. H. NH---S hydrogen bonds in Peptococcus aerogenes ferredoxin, Clostridium pasteurianum rubredoxin, and Chromatium high potential iron protein. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4854–4858. doi: 10.1073/pnas.72.12.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Tan J., Eren M., Tevelev A., Lui S. M., Cowan J. A. Synthesis, cloning and expression of a synthetic gene for high potential iron protein from Chromatium vinosum. Biochem Biophys Res Commun. 1993 Dec 30;197(3):1357–1362. doi: 10.1006/bbrc.1993.2626. [DOI] [PubMed] [Google Scholar]
- Bartsch R. G. Purification of (4Fe-4S)1--2--ferredoxins (high-potential iron--sulfur proteins) from bacteria. Methods Enzymol. 1978;53:329–340. doi: 10.1016/s0076-6879(78)53038-3. [DOI] [PubMed] [Google Scholar]
- Bruice T. C., Maskiewicz R., Job R. The Acid-Base Properties, Hydrolytic Mechanism, and Susceptibility to O(2) Oxidation of Fe(4)S(4)(SR)(4) Clusters. Proc Natl Acad Sci U S A. 1975 Jan;72(1):231–234. doi: 10.1073/pnas.72.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. W., Jr, Kraut J., Freer S. T., Alden R. A. Comparison of oxidation-reduction site geometries in oxidized and reduced Chromatium high potential iron protein and oxidized Peptococcus aerogenes ferredoxin. J Biol Chem. 1974 Oct 10;249(19):6339–6346. [PubMed] [Google Scholar]
- Cunningham R. P., Asahara H., Bank J. F., Scholes C. P., Salerno J. C., Surerus K., Münck E., McCracken J., Peisach J., Emptage M. H. Endonuclease III is an iron-sulfur protein. Biochemistry. 1989 May 16;28(10):4450–4455. doi: 10.1021/bi00436a049. [DOI] [PubMed] [Google Scholar]
- Dutton P. L. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- Emptage M. H., Kent T. A., Kennedy M. C., Beinert H., Münck E. Mössbauer and EPR studies of activated aconitase: development of a localized valence state at a subsite of the [4Fe-4S] cluster on binding of citrate. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4674–4678. doi: 10.1073/pnas.80.15.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J., Albrand J. P., Moulis J. M., Wemmer D. E. Sequence-specific assignments of the 1H nuclear magnetic resonance spectra of reduced high-potential ferredoxin (HiPIP) from Chromatium vinosum. Biochemistry. 1992 Jun 23;31(24):5632–5639. doi: 10.1021/bi00139a029. [DOI] [PubMed] [Google Scholar]
- Higuchi Y., Yasuoka N., Kakudo M., Katsube Y., Yagi T., Inokuchi H. Single crystals of hydrogenase from Desulfovibrio vulgaris Miyazaki F. J Biol Chem. 1987 Feb 25;262(6):2823–2825. [PubMed] [Google Scholar]
- Jensen G. M., Warshel A., Stephens P. J. Calculation of the redox potentials of iron-sulfur proteins: the 2-/3-couple of [Fe4S*4Cys4] clusters in Peptococcus aerogenes ferredoxin, Azotobacter vinelandii ferredoxin I, and Chromatium vinosum high-potential iron protein. Biochemistry. 1994 Sep 13;33(36):10911–10924. doi: 10.1021/bi00202a010. [DOI] [PubMed] [Google Scholar]
- Job R. C., Bruice T. C. Iron-sulfur clusters II: Kinetics of ligand exchange studied on a water-soluble Fe(4)S(4)(SR)(4) cluster. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2478–2482. doi: 10.1073/pnas.72.7.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Langen R., Jensen G. M., Jacob U., Stephens P. J., Warshel A. Protein control of iron-sulfur cluster redox potentials. J Biol Chem. 1992 Dec 25;267(36):25625–25627. [PubMed] [Google Scholar]
- Martín A. E., Burgess B. K., Stout C. D., Cash V. L., Dean D. R., Jensen G. M., Stephens P. J. Site-directed mutagenesis of Azotobacter vinelandii ferredoxin I: [Fe-S] cluster-driven protein rearrangement. Proc Natl Acad Sci U S A. 1990 Jan;87(2):598–602. doi: 10.1073/pnas.87.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskiewicz R., Bruice T. C. Dependence of the rates of dissolution of the Fe4S4 clusters of Chromatium vinosum high-potential iron protein and ferredoxin on cluster oxidation state. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5231–5234. doi: 10.1073/pnas.74.12.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskiewicz R., Bruice T. C. Kinetic study of the dissolution of Fe4S4(2-)-cluster core ions of ferredoxins and high potential iron protein. Biochemistry. 1977 Jun 28;16(13):3024–3029. doi: 10.1021/bi00632a033. [DOI] [PubMed] [Google Scholar]
- Moulis J. M., Scherrer N., Gagnon J., Forest E., Petillot Y., Garcia D. Primary structure of Chromatium tepidum high-potential iron-sulfur protein in relation to thermal denaturation. Arch Biochem Biophys. 1993 Aug 15;305(1):186–192. doi: 10.1006/abbi.1993.1409. [DOI] [PubMed] [Google Scholar]
- Peisach J., Orme-Johnson N. R., Mims W. B., Orme-Johnson W. H. Linear electric field effect and nuclear modulation studies of ferredoxins and high potential iron-sulfur proteins. J Biol Chem. 1977 Aug 25;252(16):5643–5650. [PubMed] [Google Scholar]
- Rayment I., Wesenberg G., Meyer T. E., Cusanovich M. A., Holden H. M. Three-dimensional structure of the high-potential iron-sulfur protein isolated from the purple phototrophic bacterium Rhodocyclus tenuis determined and refined at 1.5 A resolution. J Mol Biol. 1992 Nov 20;228(2):672–686. doi: 10.1016/0022-2836(92)90849-f. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoesz J. D., Redfield A. G. Cross relaxation and spin diffusion effects on the proton NMR of biopolymers in H2O. Solvent saturation and chemical exchange in superoxide dismutase. FEBS Lett. 1978 Jul 15;91(2):320–324. doi: 10.1016/0014-5793(78)81201-0. [DOI] [PubMed] [Google Scholar]
- Tedro S. M., Meyer T. E., Bartsch R. G., Kamen M. D. Primary structures of high potential, four-iron-sulfur ferredoxins from the pruple sulfur photosynthetic bacteria, Thiocapsa roseopersicina and chromatium gracile. J Biol Chem. 1981 Jan;256(2):731–735. [PubMed] [Google Scholar]




