Summary
Objective
To improve the transparency of clinical trial generalizability and to illustrate the method using Type 2 diabetes as an example.
Methods
Our data included 1,761 diabetes clinical trials and the electronic health records (EHR) of 26,120 patients with Type 2 diabetes who visited Columbia University Medical Center of New-York Presbyterian Hospital. The two populations were compared using the Generalizability Index for Study Traits (GIST) on the earliest diagnosis age and the mean hemoglobin A1c (HbA1c) values.
Results
Greater than 70% of Type 2 diabetes studies allow patients with HbA1c measures between 7 and 10.5, but less than 40% of studies allow HbA1c<7 and fewer than 45% of studies allow HbA1c>10.5. In the real-world population, only 38% of patients had HbA1c between 7 and 10.5, with 12% having values above the range and 52% having HbA1c<7. The GIST for HbA1c was 0.51. Most studies adopted broad age value ranges, with the most common restrictions excluding patients >80 or <18 years. Most of the real-world population fell within this range, but 2% of patients were <18 at time of first diagnosis and 8% were >80. The GIST for age was 0.75.
Conclusions
We contribute a scalable method to profile and compare aggregated clinical trial target populations with EHR patient populations. We demonstrate that Type 2 diabetes studies are more generalizable with regard to age than they are with regard to HbA1c. We found that the generalizability of age increased from Phase 1 to Phase 3 while the generalizability of HbA1c decreased during those same phases. This method can generalize to other medical conditions and other continuous or binary variables. We envision the potential use of EHR data for examining the generalizability of clinical trials and for defining population-representative clinical trial eligibility criteria.
Keywords: Clinical trials, selection bias, comparative effectiveness research, electronic health records, clinical research informatics, meta-analysis
Introduction
Clinical trials are the gold standard for generating new high-quality medical evidence. Many clinical trials are designed to emphasize internal validity, and some decisions may compromise external validity. When a clinical trial has limited generalizability, the study results can be difficult to translate to the real-world population that would otherwise be the users of the study information. In fact, this common problem is a concern of both the public and the clinical research community [1, 2], and has significantly impaired the cost-benefit ratio of many clinical trials [3]. Moreover, disease-specific evidence accumulated from multiple clinical trials can greatly influence clinical decisions of care providers [4-7], but little is known about how the populations of multiple trials collectively represent the real-world population with the condition despite the fact that such knowledge is important for patient-centered outcomes research (PCOR) [8].
Each clinical trial has three populations: the real-world patient population, the target population of this study, and the study population or study sample. The real-world patient population represents the set of individuals to whom the results of the trial might be applied. The target population represents a subset of the real-world population who can be enrolled in the study according to ethical or other concerns and must be defined in advance with unambiguous inclusion and exclusion eligibility criteria. In contrast, the study population defines the type of individuals who are enrolled in clinical trials. Information about the study population is often only available after the trial is complete and is summarized in the publications for that trial. Ideally, the study population should represent the target population. However, in reality, the study population often fails to represent the target population for many reasons, such as recruitment problems [9, 10].
Many have reported the compromised generalizability of study population for clinical trials in various disease domains, such as inflammatory bowel disease [11], rheumatoid arthritis [12], heart failure [10], Alzheimer’s disease [13], and dementia [9], but these prior studies on the generalizability of study samples to the patient population largely relied on manual comparisons between the study participants for a few or a dozen of clinical trials and the patient characteristics of convenient patient samples so that they do not scale to include thousands of trials. Moreover, none has addressed corresponding differences between the target populations of clinical trials and the patient population. There is a paucity of scalable methods that systematically and efficiently assess the differences between target populations for clinical trials and the pertinent patient populations for any disease topic of interest.
We face an opportunity to address this need. Lately, the promise of data-driven decision-making has been recognized broadly, especially in the past 2–3 years. Specifically, in biomedicine, the recent burgeoning adoption of the electronic health record (EHR) has made it practical to prescreen potentially eligible patients for clinical trials [14-16] and practical to electronically assess the feasibility of meeting recruitment targets. Notable related efforts include EHR4CR (http://www.ehr4cr.eu) and TRANSFoRm (http://www.transformproject.eu), and i2b2 (https://www.i2b2.org). EHRs are also useful for studying distributions of disease indicators [17, 18], such as hemoglobin A1c (HbA1c) and serum glucose, in both inpatient and outpatient populations. Meanwhile, the mandatory public registry for clinical trials, ClinicalTrials.gov [19], provides rich information from more than 160,000 clinical trials investigating thousands of diseases, facilitating systematic analysis of the distributions of the characteristics of clinical trial target populations, as reflected in recruitment eligibility criteria, which can be downloaded, parsed, and aggregated [20-28].
Therefore, we hypothesize that ClinicalTrials.gov and EHRs together offer an opportunity for using electronic data to compare the aggregated target populations in clinical trials with the real-world population captured by EHRs. Such comparisons may inform policy makers or clinical trial sponsors which patient subgroups are understudied and therefore should be considered for future clinical trial interventions, or, conversely, which patient subgroups are overly studied so that the “return of investment” of future studies would be marginal. The comparisons may also identify the gaps between existing medical evidence and the needs of the patient population to guide clinical investigators to design population-representative clinical trial eligibility criteria and select target populations that are both feasible to recruit and also meaningful clinically. A collaboratory called PACeR (Partnership to Advance Clinical Electronic Research) is studying this area [29].
This paper contributes an original method for aggregating clinical trial eligibility criteria across clinical trials of the same medical condition and comparing their distribution with the distribution of the EHR patient population with the same disease. We illustrated this type of data-driven approach for clinical trial generalizability analysis using Type 2 diabetes as an example. We aggregated two representative eligibility variables, age and HbA1c, from the eligibility criteria of 1,761 Type 2 diabetes trials, and compared them with the corresponding measures for the Type 2 diabetes patients, both inpatients and outpatients, in our EHR at Columbia University Medical Center. This study investigated the feasibility of using electronic data to assess how the aggregated target populations of Type 2 diabetes trials represent a diabetes patient population. On this basis, we discuss the implications of our results and future work.
Methods
1. Dataset Preparation
Our dataset preparation includes the following steps:
identification of Type 2 diabetes patients using EHR data;
selection of quantifiable measures to characterize and profile Type 2 diabetes patients in trials and in the EHR; and
extraction of quantifiable measures from ClinicalTrials.gov and EHR.
Step 1.1 Type 2 Diabetes Patients Identification in EHR
There are multiple methods for phenotyping or cohort identification for Type 2 diabetes using EHRs, including notable efforts ongoing at the eMERGE consortium [30-33]. Given the significant data quality problems in EHR data [34], such as information incompleteness [35], inconsistency, and inaccuracy, both structured and unstructured clinical data, including diagnosis codes, clinical notes, medication orders, and laboratory test results, should be used together to identify Type 2 diabetes patients [32]. However, implementation of such a comprehensive approach entails high costs of developing or adapting natural language processing (NLP) methods for various types of clinical notes of varying local formats due to the frequent lack of portability of NLP algorithms [36], and substantial efforts needed to reconcile inconsistent information from structured and unstructured clinical data [37, 38]. Despite the controversy around the Positive-Predictive-Value (PPV) of using administrative coding for Type 2 diabetes [39, 40] for cohort identification, investigators have found ICD-9 diagnosis codes (‘250.XX’) to be an acceptable cost-effective method to identify Type 2 diabetes patients from EHRs in several previous studies [30, 31, 33] with a PPV up to 91% [40]. Building on the success of these studies, this study used only structured data to identify Type 2 diabetes patients who had the following characteristics:
ICD-9 diagnosis codes for Type 2 diabetes on two or more occasions;
no ICD-9 diagnosis codes for Type 1 diabetes; and
HbA1c measurements on one or more occasions regardless of their temporal relationships to diagnosis times.
In addition, we used a list of published Type 2 diabetes medication names [41, 42] to query the medication order records for these patients.
Step 1.2 Quantifiable Measure Selection for Characterizing Type 2 Diabetics
We compared the trial target population to the EHR patient population for Type 2 diabetes by HbA1c and age distributions. Frequent eligibility criteria for Type 2 diabetes trials include serum creatinine, glucose, HbA1c, and body mass index. For this feasibility study that intends to generalize to any disease variable in the future, we chose HbA1c to characterize Type 2 diabetes patients because it is commonly available in clinical trial eligibility criteria [21] and EHR. The count of HbA1c measures in a diabetes patient’s EHR ranges from 1 to 172 (►Supplement Figure 1). We chose age as a demographic criterion to complement the HbA1c evaluation.
Fig. 1.
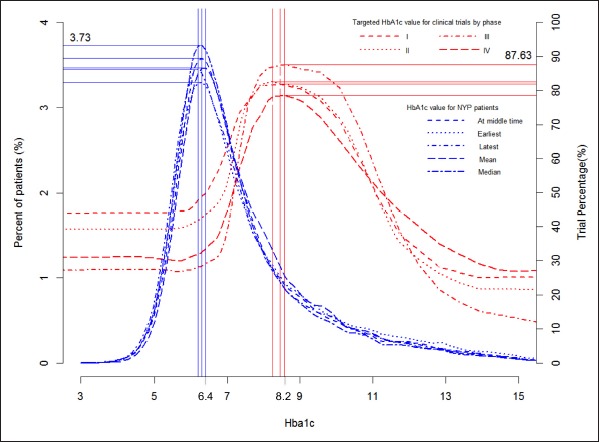
For each HbA1c value on X, the red lines indicate the percentage of trials of different phases whose eligibility criteria include that value and the blue lines indicate the percentage of patients whose earliest, latest, mean, median, or middle- collection-time HbA1c was that value.
Step 1.3 Data Extraction from ClinicalTrials.gov and EHR
Two datasets were needed for the proposed analysis. The trial summary dataset included the eligibility criteria for age and HbA1c for Type 2 diabetes trials from ClinicalTrials.gov for profiling the aggregated target populations. The real-world population summary dataset included our EHR data for date of birth and all HbA1c values with timestamps for the Type 2 diabetes patients sampled using the aforementioned method.
To develop the trial summary dataset, in March 2012, we searched ClinicalTrials.gov using the keyword “diabetes” in the “condition” field of the online trial search form and identified 5,652 Type 2 diabetes mellitus trials (T1). Among these trials, we identified a subset of 4,765 trials with a known status (i.e., closed or open) and valid age criteria with specific value ranges (T2). Within T2, we identified 1,761 trials with known status and HbA1c criteria with specific value ranges (T3). We included T2 for analyzing age distribution and T3 for analyzing HbA1c distribution. We downloaded the following structured information for each of the 1,761 trials: ClinicalTrials.gov registry number (NCT ID), title, recruitment status, conditions, interventions, sponsors, gender, age groups, phases, start date, sponsor, inclusion criteria, and exclusion criteria.
Clinical trials for Type 2 diabetes focus on patients from different age groups or with varying HbA1c values. For example, trial NCT00174681 requires HbA1c between 6% and 8%, whereas trial NCT01341067 requires HbA1c between 7% and 17%. One author (Zhang) extracted the value ranges for age from our trial set T2 (N = 4,765) and the value ranges for HbA1c from our trial set T3 (N = 1,761) respectively through manual review of their free-text eligibility criteria. In addition, in most of trials, eligibility criteria are expressed as separate inclusion criteria and exclusion criteria. We transformed all exclusion criteria for age and HbA1c into inclusion criteria. For example, if a trial excluded “patients with age >75 years old”, we manually replaced this exclusion criterion with its equivalent inclusion criterion “patients with age ≤75 years old”.
To develop the real-world population summary, we used the Columbia University Medical Center’s clinical data warehouse [43], which contains over 20 years of health information for about 4.5 million patients with diverse ethnicities. We used all 20 ICD-9 codes for Type 2 diabetes — 250.00, 250.02, 250.10, 250.12, 250.20, 250.22, 250.30, 250.32, 250.40, 250.42, 250.50, 250.52, 250.60, 250.62, 250.70, 250.72, 250.80, 250.82, 250.90, 250.92 to identify 116,308 patients who had at least one of these codes at least once (P1). Among P1, 63,568 patients received at least one code on at least two clinical encounters (P2), in which 45,285 had no diagnosis code for Type 1 diabetes (P3). Among P3, 26,120 patients had both date of birth and HbA1c values and formed the patient population for this study (P4). This patient population (P4) was 51% female, 63 years old on average, 45.9% White, 35.7% Hispanic or Other, 16% Black, and 1.7% Asian. We divided P4 into two groups: patients with orders for at least one of the Type 2 diabetes medications (P5, N = 19,096) and patients without orders for Type 2 diabetes medications (P6, N = 7,024).
2. Data Analysis
Our analysis also consisted of three steps. We first aggregated eligibility criteria from all the sample Type 2 diabetes trials and drew the histograms of the percentages of trials over the values for age or HbA1c. We further stratified the trials by phase, gender, and race, and drew the histograms of trials for HbA1c and age for each phase. Then we drew the histograms of the percentages of Columbia University Medical Center’s Type 2 diabetes patients over the range of values for age and HbA1c. Because some patients in our sample had more than one HbA1c value, we generated separate histograms for the earliest, latest, median, mean, and middle-measurement-time HbA1c values. On this basis, we juxtaposed the histograms for trials and patients in the same figure to contrast the distributions of trials and patients over HbA1c values and age values.
Step 2.1 Generating the histograms for Type 2 Diabetes Trials over the HbA1c and age value ranges respectively
Since we used the same method to derive a distribution function for both age and HbA1c, next we describe this process for calculating the trial distribution within variable value ranges using HbA1c as an example. This process can be illustrated with a simple scenario using the aforementioned trials NCT00174681 and NCT01341067. In this scenario, trial NCT00174681 covers value range [6%, 8%], and trial NCT01341067 covers value range [7%, 17%]. According to the method introduced below, five connected bins would be created: (-∞, 6%), [6%, 7%], (7%, 8%], (8%, 17%], and (17%, ∞+), which together cover the entire value range (-∞, ∞+). Stepwise bin divisions and results are shown in ► Table 1.
Table 1.
Stepwise bin divisions and calculation of trial counts in each bin. NCT00174681: Tulip Study: Testing the Usefulness of Lantus When Initiated Prematurely In Patients With Type 2 Diabetes. NCT01341067: Continuous Glucose Monitoring (CGM) in Subjects With Type 2 Diabetes (DexlonT2)
| Bin | Included trial(s) | Number of trials |
|---|---|---|
| (-∞, 6%) | N/A | 0 |
| [6%, 7%] | NCT00174681 | 1 |
| (7%, 8%] |
NCT00174681, NCT01341067 |
2 |
| (8%, 17%] | NCT01341067 | 1 |
| (17%, ∞+) | N/A | 0 |
We hypothesized that eligible target populations are evenly distributed within each bin so that each value within the range of a bin is assigned the same number of trials. For example, if there are N trials that recruit patients with HbA1c between 8% and 17%, there are N trial opportunities for each discrete value within this range (e.g., 9%, 10%, etc). On this basis, we drew the histograms for trials, whose X-axis represent HbA1c or age value and whose Y-axis represents the percentage of trials among the trials of each phase or among all the trials recruiting patients with that value.
Next we describe the algorithm for the above procedure. Below, [ ] means being inclusive, while () means being non-inclusive; ∞ refers to negative infinity, whereas ∞+ refers to positive infinity. Each trial Ti (I = 1, 2, … 1,761) has a value range [HbA1ci-min, HbA1ci-min]. We used the minimum lower bound, designated as HbA1ci-min, and the maximum upper bound designated as HbA1ci-min, to create two additional value ranges, negative infinity (∞, HbA1ci-min) and positive infinity (HbA1ci-min, ∞+). These two value ranges along with the existing value range [HbA1ci-min, HbA1ci-min] together form the entire range (∞, ∞+) of all possible HbA1c values. Then, we sorted all the upper and lower bounds in the ascending order and used them to divide the entire value range into connected but non-overlapping smaller bins of varying width. For each bin, we calculated the number of trials that include the corresponding HbA1c value range in their HbA1c eligibility criterion. One trial may cover multiple bins. Then, we drew a curve whose X-axis is HbA1c value and whose Y-axis is the number of trials divided by the total, 1,761, representing the percentage of trials falling into this bin.
Later, we used Local Polynomial Regression Fitting to smoothen the distribution curve. This function further divided the bins of varying width to smaller bins of equal width. For example, the bin (8%, 17%] was expanded to 10 smaller bins, including (8%, 9%], (9%, 10%], (10%, 11%], (11%, 12%], (12%, 13%], (13%, 14%], (14%, 15%], (15%, 16%], (16%, 17%], (17%, 18%]. All these bins have the equal likelihood to enroll patients. According to ►Table 1, if one trial falls into the bin of (8%, 17%], then each of the small bin has one trial. The interpretation is “if there exists one trial enrolling patients whose A1c is >8 and ≤17, then there is one trial enrolling patients whose A1c is >8 and ≤9, one trial enrolling patients whose A1c ≤9 and <10, and so on.”
Step 2.2 Creating histograms of patients over age and HbA1c respectively
For each Type 2 diabetes patient in the EHR, we obtained all the HbA1c values and date of birth; the latter was used to calculate the patient’s age at the earliest, middle, and latest HbA1c measurement times. For example, if a patient had 5 HbA1c measurements at ages of 10, 11, 15, 17, 20 respectively, the earliest, middle, and latest measurement times were 10, 15, and 20 respectively. Then, we drew histograms for age and HbA1c respectively for the patient population, where the X-axis represents the earliest, latest, mean, median, and middle-measurement-time value of HbA1c or age at the earliest, latest, and median measurements, and the Y-axis is the percentage of patients with that value.
Step 2.3 Juxtaposition of the distributions of trials and patients
In the trials histogram, the Y-axis represents the percentage of trials that include the value on X-axis in the inclusion criteria, whereas in the patient population histogram, the Y-axis represents the percentage of patients with the value on X-axis. We juxtaposed the trial and patient distributions for both variables and for both patient groups.
We developed the Generalizability Index for Study Traits (GIST) as a metric to evaluate the relative generalizability of a study characteristic from a set of clinical trials to a real-world population. GIST is the sum across all intervals of the proportion of trials including patients in that interval, multiplied by the proportion of patients in the real-world population observed in that interval.
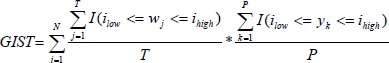 |
Where N is the number of distinct intervals of the study trait, T is the number of trials, P is the number of patients in the population, wj is the inclusion interval for the jth study, such that an indicator I can be defined when the jth study interval subsumes the ith interval low and high boundary threshold, and yk is the observed value of the characteristic for the kth patient such that an indicator I can be defined when the kth patient’s value falls within the ith interval.
The GIST metric is on a 0 to 1 scale that characterizes the proportion of real-world population that would be eligible across the clinical trial studies, with 1 being perfectly generalizable (all patients would be eligible for all studies), and 0 being completely not generalizable (no real-world patients would be eligible for any studies).
As a motivating example, assume we wanted to estimate the generalizability of two studies for a treatment of a disease, that both were restricted to patients aged 20–50. If in the real-world population of 100 patients, we found all patients with that disease fell between the ages of 20–50, we would say the study was perfectly generalizable on the study trial of age; GIST = 1 because there is only one interval (20–50), 100% of trials cover this interval and 100% of patients fall within this interval. In contrast, if all the real-world patients were found to be >50, we would say the study results could not be generalized to the real-world population on the basis of age; GIST = 0 because there are two intervals (20–50, 51-inf), 100% of trials cover the 20–50 interval, but 0% of trials cover the 51-inf, whereas 0% of patients fall within the 20–50 interval, and 100% of patients fall in the 51-inf interval. Now assume one study imposed a restriction on age of 20–50, but the other study only imposed a restriction of age ≤20, and assume 10 patients were <20, 60 patients were 20–50, and 30 patients were >50; in this case, there were three observed intervals: 0–20, 20–50; 50-inf. The trial coverage would be 0%, 100%, 50% across these three intervals. The real-world population proportions would be 10%, 60%, 30% across the same intervals. Accordingly, GIST = 0%*10% + 100%*60% + 30*50% = 0.75.
The GIST metric can be used to evaluate the generalizability of one study or a collection of studies and can be applied to any study characteristic that is observed in the real-world population, including binary, categorical, and continuous-valued attributes. We applied GIST to two study traits: HbA1c and age, for the collection of all Type 2 diabetes studies, as well as the subset of studies classified as Phase I, Phase II, Phase III, and Phase IV.
Results
Of the 1,761 trials, 21.2% did not have phase information, 0.3% were phase 0, 10.7% were phase I, 21.6% were phase II, 27.9% were phase III, and 18.2% were phase IV; 95.6% (N = 1,683) were interventional and the rest (N = 81) observational; 96.6% recruited patients of both genders, 2% recruited only male patients, and 1.3% recruited only female patients. Only 10 trials had restrictions for race or ethnicity; all others recruited patients regardless of their race and ethnicity. Therefore, study type, gender, and race (or ethnicity) are not likely to affect the results; accordingly, we stratified the 1,761 trials only by significant phases (i.e., phase I, II, III, and IV).
We generated two figures to illustrate how the patient population compares with trial populations: one for HbA1c (►Figure 1) and the other for the age (►Figure 2). They show the peak values and percentages of trials at the peak values for the distributions of Type 2 diabetes trials of four phases over the value ranges for HbA1c and age respectively.
Fig. 2.
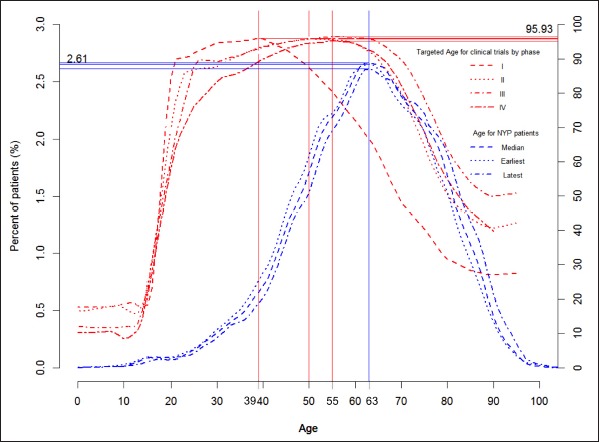
For each age value on X, the red solid line indicates the percentage of trials of different phases whose eligibility criteria include that value and the blue lines indicate the percentage of patients who were that age at the earliest, median, or latest HbA1c measurements.
Independent from diabetes medication status, a person 44 years old has the best chance to be included in most (90.8%) Type 2 diabetes trials. A person with HbA1c value of 8.2 has the best chance to be eligible for most (88.3%) Type 2 diabetes trials. The greatest percentage of Phase I trials (81.9%) recruit patients with an HbA1c of 8.0, the greatest percentage of Phase II trials (82.5%) recruit most frequently with an HbA1c value of 7.8, and the greatest percentage of Phase III trials (87.6%) and Phase IV trials (78.6%) recruit most frequently with an HbA1c value of 8.1. ►Figure 1 shows that between 40% and 50% of Phase I or II trials recruit patients whose HbA1c is less than or equal to 6.4, while between 25% or 30% Phases III and IV trials recruit patients of the same HbA1c value range, indicating that a larger portion of Phases I and II trials define their target population younger and healthier than Phases III and IV trials do.
In ►Figure 2 the age distribution of Phase I trials is narrower and to the left of that of Phase II, III, and IV trials, which almost align, demonstrating that most (95.9%) Phase I trials recruit significantly younger patients (with peak value around age 39) than most (96%) Phases II, III, and IV trials (with peak value of 50 or 55). ►Figure 2 shows that about 20% of Phases I or II trials recruit patients younger than 20 years old, while only about 10% Phases III or IV trials recruit patients of the same age range. From age 50 to 63, the percentage of Phase I trials recruiting patients of this age range drops markedly from 90% to 65%, while the percentages of Phases II, III, and IV trials recruiting the same patient group remain stable at around 96%. These results indicate that Phases I and II trials recruit younger and healthier patients than Phases III and IV trials. The age distributions for trials of different phases are all wider than the age distributions of the patients and are consistently to the left of the age distributions of patients, showing that trials attempt to recruit participants younger than Type 2 diabetes patients, especially in Phases I and II trials.
Regardless of trial stratification by phase, patient stratification by HbA1c measurement time or type, and patient stratification by medication order status, the distributions of trials are always to the right of the distributions of patients, implying that trials attempt to recruit patients with higher HbA1c values than are found among Type 2 diabetes patients. In contrast, the distributions of trials are always to the left of the distributions of patients, implying that trials attempt to recruit patients younger than are found among Type 2 diabetes patients. ►Figure 1 and ►Figure 2 together show that Type 2 diabetes trials tend to recruit young and sick (i.e., with high HbA1c values) diabetes patients.
►Table 2 shows the GIST for HbA1c and age across all trials and within the individual phases. The Type 2 diabetes clinical trials had a GIST of 0.51 for HbA1c overall, with the index decreasing from 0.59 to 0.50 from Phase I to Phase III studies. The study were observed to have greater generalizability for age (i.e., an overall GIST of 0.75), with GIST = 0.67 for Phase 1 increasing to GIST = 0.86 for Phase III studies.
Table 2.
GIST for Type 2 Diabetes Trials for HbA1c and Age in Different Phases
| GIST | Variable | |
|---|---|---|
| Clinical Trials | HbA1c | Age |
| All (N = 1761) | 0.51 | 0.75 |
| I (N = 188) | 0.59 | 0.67 |
| II (N = 380) | 0.55 | 0.82 |
| III (N = 491) | 0.50 | 0.86 |
| IV (N = 321) | 0.50 | 0.82 |
Discussion
Implications
This study contributes an early data-driven approach for illustrating the differences between aggregated target populations for Type 2 diabetes trials, represented by selected eligibility criteria variables for Type 2 diabetes trials, and diabetes patient populations that are available for study, represented by their corresponding EHR data. It also shows the feasibility of using electronic data, such as ClinicalTrials.gov and EHRs, for comparing clinical trial target populations and clinical patients.
Empirically, HbA1c is directly proportional to age [44, 45]; therefore, it should be easier to recruit older patients with high HbA1c values or younger patients with low HbA1c values. However, ►Figure 1, ►Figure 2 and ►Table 2 together demonstrate that most diabetes trials target younger and sicker diabetes patients (those with high HbA1c values). Although our study does not address the clinical optimality of inclusion criteria of diabetes trials, it reveals the misalignment between aggregated clinical trial target populations and patient populations, suggesting that in certain patient subgroups, the amount of research investment is not commensurate with the size of patient populations by recruiting overrepresented or underrepresented patients from clinical settings.
We developed GIST as a single metric to provide a relative assessment of generalizability of a given study attribute. We found diabetes studies are more generalizable with respect to age than with regards to HbA1c. This is consistent with clinical expectations, since there is general consensus that Type 2 diabetes can inflict patients 18 years and over and all ages may be eligible for treatment, whereas there is greater debate about the proper thresholds for HbA1c that should be used for diagnosis and a targeted maintenance range. Also, clinical trials may opt to be more restrictive with HbA1c than age, because study sponsors may think that patients with lower HbA1c values do not have sufficient blood glucose imbalance or that patients with high values may be too far out-of-control to be considered the primary targeted candidates for a given intervention. The differences in GIST across phases were also informative. The GIST values for age increased from Phase I to Phase III, potentially because study sponsors may be more conservative with inclusion of older patients during early studies aimed at establishing safety and initial efficacy, whereas broaden their scope during Phase III when the targeted indication is under investigation. In contrast, we observed the GIST values for HbA1c decreased from Phase I to Phase III, perhaps because study designers sought tighter control of the HbA1c range to test efficacy in the larger studies.
There can be many reasons behind the differences, including a possible good clinical rationale. As Altman pointed out, in general, but not always, we do not expect treatment to do much for patients who already have an excellent prognosis, nor for those with a dire prognosis [46]. Still, it is important to show such differences and provide a means for trial designers to gain a transparent overview of research distributions and for predicting trial representativeness of the general patient population. Understanding what clinical rationale justifies the differences could be a separate study, yet we see the opportunity of using EHR data to inform rational clinical trial eligibility criteria designs in the future.
Limitations
The major limitations of this study are its use of a small number of structured data elements from EHR, ClinicalTrials.gov data quality problems, and EHR data complexity and quality problems.
Methodology Limitations
Various algorithms have been proposed for identifying diabetic patients from EHRs, some of which use clinical notes and medication information. Our reliance on ICD-9 diagnosis codes may have missed the undiagnosed diabetes population or included misdiagnosed non-diabetic patients. Moreover, we included only EHR data for two variables, age and HbA1c, to profile diabetes patients from one hospital. Other variables, such as glucose, body mass index, comorbidities, and medications, are either more complex with contextual measurements or exist as free text and, as such, are difficult to parse and quantify. It would be helpful to use more disease biomarkers to generate temporal profiles for patients, but we believe that modeling the relationships of variables disease biomarkers is an independent research topic that warrants further studies that are beyond the scope of this feasibility study.
To ensure a highly accurate dataset, for this study, we manually extracted age and HbA1c from ClinicalTrials.gov. Use of natural language processing would improve the efficiency of this task [20, 22]. The manually created dataset serves as an important gold standard for us to develop a separate automated algorithm for extracting numerical expressions from ClinicalTrials.gov. The design and evaluation of that algorithm for assisting with automated aggregation of clinical trial target populations are beyond the scope of this paper but we are preparing a manuscript about a generalizable rule-based algorithm for extracting numerical expressions from eligibility criteria text so that others who want to replicate this study’s results or apply the method on other diseases or other numerical variables do not have to perform manual processes for data extraction and review. This algorithm is available online: http://columbiaelixr.appspot.com/valx. Its preliminary precision and recall for HbA1c using the gold standard created from this study are 98.9% and 97.1%, respectively.
Formal comparison over categorical or binary variables needs to overcome challenges for aggregating heterogeneous semantic concept representations in EHRs. Our simplified aggregate analysis with one binary variable showed that about 38% Type 2 diabetes trials exclude patients with renal failures or kidney diseases, while 3% include patients with related kidney conditions. In contrast, these kidney conditions are prevalent in about 22% of the Type 2 diabetic population in our EHR.
Data Quality issues in ClinicalTrials.gov
While ClinicalTrials.gov is a tremendous resource and utility for the community, the nature of reporting and free text form that much of the information it provides presents several data quality challenges to research. It is likely that the “Conditions” field’s information was not 100% accurate so that we may have missed some Type 2 diabetes trials. In addition, the eligibility criteria summaries may be incomplete [47]. Consequently, we may have mistakenly excluded some diabetes studies that did not have HbA1c values on ClinicalTrials.gov but had that information in the corresponding protocols, which may affect our trial distribution analysis. We also noticed some inconsistency in the eligibility criteria on ClinicalTrials.gov. For example, one source in a study may indicate “inclusion criteria: gender: both” and another “exclusion criterion: male”. We also did not consider the heterogeneity of clinical trial interventions and outcomes when we aggregated these trials’ target populations. We did not stratify trials by their purposes and intervention types, partially because such information may be incomplete and inaccurate. For example, we only retrieved 187 trials using the search “comparative effectiveness” as of July 2013. Again, such granular analysis requires sophisticated and specialized NLP support that we do not have now.
Data Quality issues in EHR
Data quality issues in EHR provide an additional limitation of this study. A large fraction of patients did not have HbA1c values even though they had ICD-9 diagnosis codes for Type 2 diabetes. This can happen with patients who visited Columbia University Medical Center occasionally to treat diseases more severe than Type 2 diabetes, such as cancer or heart attack, and whose primary care providers were not at Columbia University Medical Center so that their HbA1c were not regularly collected. Also, as shown in ►Supplement Figure 1 (“histogram of HbA1c measures for the Type 2 diabetes patients”), among patients who had HbA1c values, most did not have regular measurements documented in their EHRs even though HbA1c is a standard measure of Type 2 diabetes. This result is consistent with our recently published paper on EHR data completeness for secondary use [35], where we reported that of the patients with data in the clinical data warehouse, 29.3% had at least one visit with a recorded laboratory result (20.0% glucose, 23.0% hemoglobin), 12.6% had at least one with a medication order, and 44.5% had at least one with a diagnosis. Either improving clinical documentation with these targeted areas or using predictive methods to impute values for these variables may overcome these problems.
Conclusion
We contribute an original data-driven, distribution-based method for comparing aggregated clinical trial target populations with their intended patient populations. A key merit of this scalable approach is its use of electronic data resources to analyze a large number of clinical trials and patients simultaneously, a scale not possible using existing non-electronic methods, to show the transparency of the generalizability of clinical trials. Making clinical trial populations generalizable to real-world patients is important. This method can potentially bring us one step closer to this goal. Future studies are needed to evaluate the GIST measure.
Supplementary Material
Footnotes
Funding
This study was sponsored by National Library of Medicine grants R01LM009886, R01LM010815, and R01LM006910, and by National Center for Advancing Translational Sciences grant UL1TR000040.
Contributorship
CW initiated the concept, supervised data collection and analysis, and wrote the manuscript. YL and PR analyzed data and helped write the data analysis methodology. YZ and FL collected data for clinical trials. JG contributed ideas for data analysis. PR invented GIST. PR, GH and JTB advised CW and made substantial methodology contributions.
Clinical Relevance Statement
This study contributes an informatics approach to assess the generalizability of clinical trials and to increase the transparency of clinical trial generalizability for the public and healthcare providers. It has significance for evidence-based medicine and learning health systems.
Conflicts of Interest
None
Protection of Human and Animal Subjects
The study was performed in compliance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research Involving Human Subjects, and was approved by the Columbia University Medical Center Institutional Review Board.
References
- 1.Rothwell PM.External validity of randomised controlled trials: „to whom do the results of this trial apply?”. Lancet 2005; 365(9453): 82–93 [DOI] [PubMed] [Google Scholar]
- 2.Fuks A, Weijer C, Freedman B, Shapiro S, Skrutkowska M, Riaz A.A study in contrasts: Eligibility criteria in a twenty-year sample of NSABP and POG clinical trials. Journal of Clinical Epidemiology 1998; 51(2): 69–79 [DOI] [PubMed] [Google Scholar]
- 3.Van Spall HGC, Toren A, Kiss A, Fowler RA.Eligibility criteria of randomized controlled trials published in high-impact general medical journals: A systematic sampling review. Journal of the American Medical Association 2007; 297(11): 1233–1240 [DOI] [PubMed] [Google Scholar]
- 4.Williams L, Huang J, Bargh J.The Scaffolded Mind: Higher mental processes are grounded in early experience of the physical world. European Journal of Social Psychology 2009; 39(7): 1257–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hertwig R, Barron G, Weber E, Erev I.Decisions from experience and the effect of rare events. Psychological Science 2004; 15: 534–539 [DOI] [PubMed] [Google Scholar]
- 6.Cheng P, Holyoak K.Pragmatic reasoning schemas. Cognitive Psychology 1985; 17: 391–416 [DOI] [PubMed] [Google Scholar]
- 7.Weisberg R.Memory, Thought, and Behavior. New York: Oxford University Press.; 1980 [Google Scholar]
- 8.Ommaya Ak KJ.Challenges facing the us patient-centered outcomes research institute. JAMA: The Journal of the American Medical Association 2011; 306(7): 756–757 [DOI] [PubMed] [Google Scholar]
- 9.Schoenmaker N, Van Gool WA.The age gap between patients in clinical studies and in the general population: a pitfall for dementia research. The Lancet Neurology 2004; 3(10): 627–630 [DOI] [PubMed] [Google Scholar]
- 10.Masoudi FA, Havranek EP, Wolfe P, Gross CP, Rathore SS, Steiner JF, et al. Most hospitalized older persons do not meet the enrollment criteria for clinical trials in heart failure. American Heart Journal 2003; 146(2): 250–257 [DOI] [PubMed] [Google Scholar]
- 11.Etulain J, Negrotto S, Carestia A, Pozner RG, Romaniuk MA, D’Atri LP, et al. Acidosis downregulates platelet haemostatic functions and promotes neutrophil proinflammatory responses mediated by platelets. Thrombosis and haemostasis 2012; 107(1): 99–110 PubMed PMID: 22159527. Epub 2011/12/14 [DOI] [PubMed] [Google Scholar]
- 12.Sokka T, Pincus T.Eligibility of patients in routine care for major clinical trials of anti–tumor necrosis factor á agents in rheumatoid arthritis. Arthritis & Rheumatism 2003; 48(2): 313–318 [DOI] [PubMed] [Google Scholar]
- 13.Davis KL, Thal LJ, Gamzu ER, Davis CS, Woolson RF, Gracon SI, et al. A Double-Blind, Placebo-Controlled Multicenter Study of Tacrine for Alzheimer’s Disease. New England Journal of Medicine 1992; 327(18): 1253–1259 [DOI] [PubMed] [Google Scholar]
- 14.Weng C, Bigger J, Busacca L, Wilcox A, Getaneh A, editors Comparing the Effectiveness of a Clinical Data Warehouse and a Clinical Registry for Supporting Clinical Trial Recruitment: A Case Study. Proceeding of American Medical Informatics Association Fall Symposium 2010 [PMC free article] [PubMed] [Google Scholar]
- 15.Thadani SR, Weng C, Bigger JT, Ennever JF, Wajngurt D.Electronic Screening Improves Efficiency in Clinical Trial Recruitment. Journal of the American Medical Informatics Association 2009; 16(6): 869–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng C, Batres C, Borda T, Weiskopf N, Wilcox A, Bigger J, et al. A real-time screening alert improves patient recruitment efficiency. Proceedings of American Medical Informatics Association Fall Symposium 2011. p.1489–1498 [PMC free article] [PubMed] [Google Scholar]
- 17.Albers DJ, Hripcsak G.A statistical dynamics approach to the study of human health data: resolving population scale diurnal variation in laboratory data. Phys Lett A 2010; 374(9): 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hripcsak G, Albers DJ, Perotte A.Exploiting time in electronic health record correlations. Journal of the American Medical Informatics Association 2011; 18(Suppl. 1): i109-i115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Souri M, Sugiura-Ogasawara M, Saito S, Kemkes-Matthes B, Meijers JC, Ichinose A.Increase in the plasma levels of protein Z-dependent protease inhibitor in normal pregnancies but not in non-pregnant patients with unexplained recurrent miscarriage. Thrombosis and haemostasis 2012; 107(3). PubMed PMID: 22274138. Epub 2012/01/26 [DOI] [PubMed] [Google Scholar]
- 20.Luo Z, Johnson SB, Lai AM, Weng C.Extracting temporal constraints from clinical research eligibility criteria using conditional random fields. Proceedings of American Medical Informatics Association Fall Symposium 2011. p.843–52 [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Z, Miotto R, Weng C.A human–computer collaborative approach to identifying common data elements in clinical trial eligibility criteria. J Biomed Inform 2013; 46(1): 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Z, Yetisgen-Yildiz M, Weng C.Dynamic categorization of clinical research eligibility criteria by hierarchical clustering. Journal of Biomedical Informatics 2011; 44(6): 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng C, Tu SW, Sim I, Richesson R.Formal representation of eligibility criteria: a literature review. Journal of Biomedical Informatics 2010; 43(3): 451–467 PubMed PMID: 20034594. Pubmed Central PMCID: 2878905. Epub 2009/12/26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boland MR, Miotto R, Gao J, Weng C.Feasibility of feature-based indexing, clustering, and search of clinical trials. A case study of breast cancer trials from ClinicalTrials.gov. Methods of information in medicine 2013; 52(5): 382–394 PubMed PMID: 23666475. Pubmed Central PMCID: 3796134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boland MR, Miotto R, Weng C.A method for probing disease relatedness using common clinical eligibility criteria. Studies in health technology and informatics 2013; 192: 481–485 PubMed PMID: 23920601. Pubmed Central PMCID: 3803102 [PMC free article] [PubMed] [Google Scholar]
- 26.Miotto R, Jiang S, Weng C.eTACTS: a method for dynamically filtering clinical trial search results. J Biomed Inform 2013; 46(6): 1060–1067 PubMed PMID: 23916863. Pubmed Central PMCID: 3843999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miotto R, Weng C.Towards dynamic and interactive retrieval of clinical trials using common eligibility features. AMIA Summits on Translational Science proceedings AMIA Summit on Translational Science. 2013; 2013: 182 PubMed PMID: 24303261. Pubmed Central PMCID: 3845761 [PubMed] [Google Scholar]
- 28.Miotto R, Weng C.Unsupervised mining of frequent tags for clinical eligibility text indexing. J Biomed Inform 2013; 46(6): 1145–1151 PubMed PMID: 24036004. Pubmed Central PMCID: 3843986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchena PJ, Nieto JA, Guil M, Garcia-Bragado F, Rabunal R, Boccalon H, et al. Long-term therapy with low-molecular-weight heparin in cancer patients with venous thromboembolism. Thrombosis and haemostasis 2012; 107(1): 37–43 PubMed PMID: 22116496. Epub 2011/11/26 [DOI] [PubMed] [Google Scholar]
- 30.Lin C-C, Li C-I, Hsiao C-Y, Liu C-S, Yang S-Y, Lee C-C, et al. Time trend analysis of the prevalence and incidence of diagnosed type 2 diabetes among adults in Taiwan from 2000 to 2007: a population-based study. BMC Public Health 2013; 13(1): 318 PubMed PMID: doi:10.1186/1471–2458–13–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klompas M, Eggleston E, McVetta J, Lazarus R, Li L, Platt R.Automated Detection and Classification of Type 1 Versus Type 2 Diabetes Using Electronic Health Record Data. Diabetes Care 2013; 36(4): 914–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei W-Q, Leibson CL, Ransom JE, Kho AN, Caraballo PJ, Chai HS, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. Journal of the American Medical Informatics Association 2012; 19(2): 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudyakov R, Bowen J, Ewen E, West S, Daoud Y, Fleming N, et al. Electronic health record use to classify patients with newly diagnosed versus preexisting type 2 diabetes: infrastructure for comparative effectiveness research and population health management. Popul Health Manag 2012; 15(1): 3–11 [DOI] [PubMed] [Google Scholar]
- 34.Weiskopf NG, Weng C.Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc 2013; 20(1): 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiskopf NG, Hripcsak G, Swaminathan S, Weng C.Defining and measuring completeness of electronic health records for secondary use. Journal of Biomedical Informatics, in press, http://dx.doi.org/10.1016/j.jbi.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. Journal of the American Medical Informatics Association 2012; 19(e1): e162-e169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlo L, Chase HS, Weng C.Aligning Structured and Unstructured Medical Problems Using UMLS. AMIA Annu Symp Proc 2010; 2010: 91–5 PubMed PMID: 21346947. Pubmed Central PMCID: 3041294. Epub 2011/02/25 [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Chase HS, Patel CO, Friedman C, Weng C.Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study. AMIA Annu Symp Proc 2008: 404–408 PubMed PMID: 18999285. Pubmed Central PMCID: 2656007. Epub 2008/11/13 [PMC free article] [PubMed] [Google Scholar]
- 39.Rhodes ET, Laffel LMB, Gonzalez TV, Ludwig DS.Accuracy of Administrative Coding for Type 2 Diabetes in Children, Adolescents, and Young Adults. Diabetes Care 2007; 30(1): 141–143 [DOI] [PubMed] [Google Scholar]
- 40.Ding EL, Song Y, Manson JE, Pradhan AD, Buring JE, Liu S.Accuracy of Administrative Coding for Type 2 Diabetes in Children, Adolescents, and Young Adults: Response to Rhodes et al. Diabetes Care 2007; 30(9): e98. [DOI] [PubMed] [Google Scholar]
- 41.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. Journal of the American Medical Informatics Association 2012; 19(2): 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, et al. Robust Replication of Genotype-Phenotype Associations across Multiple Diseases in an Electronic Medical Record. The American Journal of Human Genetics 2010; 86(4): 560–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson S.Generic data modeling for clinical repositories. Journal of the American Medical Informatics Association 1996; 3(5): 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manzano-Fernandez S, Cambronero F, Caro-Martinez C, Hurtado-Martinez JA, Marin F, Pastor-Perez FJ, et al. Mild kidney disease as a risk factor for major bleeding in patients with atrial fibrillation undergoing percutaneous coronary stenting. Thrombosis and haemostasis 2012; 107(1): 51–58 PubMed PMID: 22072287. Epub 2011/11/11 [DOI] [PubMed] [Google Scholar]
- 45.Catherine C., Cowie, Rust KF, Byrd-Holt DD, Eberhardt MS, Flegal KM, Engelgau MM, et al. Prevalence of Diabetes and Impaired Fasting Glucose in Adults in the U.S. Population. Diabetes Care 2006; 29: 1263–1268 [DOI] [PubMed] [Google Scholar]
- 46.Altman DG.Practical Statistics for Medical Research: Chapman and Hall/CRC; 1st ed edition(November 22, 1990); 1990. Hardcover: 624.pages p [Google Scholar]
- 47.Bhattacharya S, Cantor MN.Analysis of eligibility criteria representation in industry-standard clinical trial protocols. Journal of Biomedical Informatics 2013; 46(5): 805–813 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


