Summary
Background
Electronic health records (EHRs) play an important role in the treatment of chronic diseases such as diabetes mellitus. Although the interoperability and selected functionality of EHRs are already addressed by a number of standards and best practices, such as IHE or HL7, the majority of these systems are still monolithic from a user-functionality perspective. The purpose of the OntoHealth project is to foster a functionally flexible, standards-based use of EHRs to support clinical routine task execution by means of workflow patterns and to shift the present EHR usage to a more comprehensive integration concerning complete clinical workflows.
Objectives
The goal of this paper is, first, to introduce the basic architecture of the proposed OntoHealth project and, second, to present selected functional needs and a functional categorization regarding workflow-based interactions with EHRs in the domain of diabetes.
Methods
A systematic literature review regarding attributes of workflows in the domain of diabetes was conducted. Eligible references were gathered and analyzed using a qualitative content analysis. Subsequently, a functional workflow categorization was derived from diabetes-specific raw data together with existing general workflow patterns.
Results
This paper presents the design of the architecture as well as a categorization model which makes it possible to describe the components or building blocks within clinical workflows. The results of our study lead us to identify basic building blocks, named as actions, decisions, and data elements, which allow the composition of clinical workflows within five identified contexts.
Conclusions
The categorization model allows for a description of the components or building blocks of clinical workflows from a functional view.
Keywords: Electronic health records, medical records, diabetes mellitus, workflow, semantics
1. Introduction
1.1 Scientific Background
Health care is facing an ever-increasing number of multimorbid patients worldwide. This is due to various reasons, such as the aging population or the increasing number of patients with chronic non-communicable diseases in Western countries [1]. Higher patient volumes, complex cases, and the proliferation of technology in health care are leading to a serious increase in health care expenses. Diabetes mellitus, for example, results in economic costs that rose from $174 billion (2007) to $245 billion (2012) in the United States [2]. Hence, there is an urgent need to keep costs low while at the same time guaranteeing or even improving the quality of patient care.
Information systems and, specifically, the electronic health record (EHR) play an important role in achieving such an ambitious goal by e.g. fostering information exchange between various health care providers or by increasing the transparency of information and patient flows within the health care system [3]. The concept of an EHR is to share institution-specific patient data between various health care providers in order to establish a comprehensive and longitudinal collection of the patient’s health and health care data. This intention requires the active participation of the patient in his/her treatment by adding and providing health-related self-documented data.
An obvious requirement of and major challenge to EHRs is interoperability at all layers of communication between the different EHR systems as well as their involved local information systems. With regard to technical and semantic interoperability and information semantics, important standards and best practices have already been developed over the last few years. One of these is the Integrating the Healthcare Enterprise (IHE) initiative [4]. IHE describes best practices based on available standards or de-facto standards, such as CEN13606 [5], openEHR [6], and Health Level 7 (HL7) Clinical Document Architecture (CDA) [7], to gain maximum interoperability of health care IT. IHE is used by many projects worldwide, including the Austrian electronic health record (ELGA) [8], epSOS [9], and EHR4CR [10].
However, one problem that has not been tackled sufficiently is the domain of functionality perceived by users of trans-institutional information systems such as EHRs. Similar to local information systems, EHR systems are still regarded as monolithic systems from the user-functionality perspective. This means that interaction with the EHR system is based on single and individual queries performed manually and for specific purposes by the medical personnel. These queries are usually executed within a clinical workflow conducted by the medical personnel in order to obtain a result that is relevant for a given medical activity. Hence, supporting complete clinical workflows and executing all needed queries in a semi-automatic way within such workflows would lead to an improvement in efficiency and would support health care providers in their daily tasks.
Based on a service-orientated approach, the OntoHealth project [11] establishes a functionally flexible layer that connects on top of standards-based EHR systems and makes it possible to answer and solve clinical problems by means of customizable workflow patterns which are generally not limited to a certain organization or EHR system. Although the basic framework and components of the system will be generic, clinical functionalities of the system will be specifically designed for the diabetes domain. Diabetes mellitus (DM) is a chronic illness whose prevalence has increased in recent years. Some estimations indicate that 7.7% of the world’s adult population will suffer from diabetes by the year 2030 [12]. Although diagnostics and treatment of diabetes is well-established, diabetes care requires complex clinical information from various medical domains.
1.2 Objectives
The objectives of this paper are to: 1) introduce the underlying concept of the OntoHealth project by presenting the architecture of the intended system; 2) present a functional categorization of workflows and building blocks regarding workflow-based interactions with EHRs in the domain of diabetes.
The remainder of the document is organized as follows: Section 2 presents the state of the art regarding the implementation of clinical workflows and main benefits of service composition for this aim. Methods used for the systematic literature review as well as considerations regarding the OntoHealth architecture are presented in Section 3. A schematic overview of the modules that compose the OntoHealth architecture together with the results from the systematic literature review is presented in Section 4. Finally, methods and results as well as their clinical relevance are discussed.
2. Electronic Health Records, Clinical Workflows and Service Composition
Structuring of the functional level of electronic health records is still a problem that has not been sufficiently tackled by the majority of current solutions in the health care sector. Electronic health records are still very often regarded solely as records allowing basic access to data, such as viewing and retrieving existing documents, and not as integrated, complex systems of data and functionality. Besides digital formats and interoperability, “meaningful use” of EHRs requires improving their functionality, supporting the implementation of customizable workflows, and fostering their integration within daily routine care in the clinical setting [13, 14]. Also, the integration of EHRs within common clinical workflows has been pointed out as a major prerequisite for their adoption and implementation [14]. A clinical workflow can be defined as a set of activities conducted in the hospital setting that lead to the achievement of a clinical result related with the assessment, diagnosis, or general supervision of the patient. Hence, EHR usage can be integrated as part of these activities and can also be used to execute implementation in an efficient and flexible manner.
Experiences in other domains, such as the business domain, have demonstrated that service orientation is a suitable means for structuring complex distributed software systems, reducing cost, and easing the composition of new or adapted applications by using existing services [15]. The latter, in particular, commonly known as service composition, is one key feature of service orientation. Service composition typically requires combining multiple existing services, which in turn can be combined even further to result in higher-level functionalities. In this way, a particular goal which cannot be fulfilled by each of the services alone can be jointly fulfilled as part of the composition. A service composition contains execution constraints on how services are to be combined in order to fulfill the particular goal. Such constraints can be created manually, semi-automatically, or automatically [16]. In the manual approach, a human expert creates the composition; in a semi-automatic approach, she/he is supported by a tool; while in automatic composition, the composition is created without any human interaction. The semi-automatic and automatic compositions bring more flexibility and enable easy adaptation to changes. Semantics have been recognized as an enabler for (semi-)automation of various service-related tasks, including service composition, leading to an entire field of research known as Semantic Web Services [17]. By semantically specifying service interfaces and how they can be consumed, intelligent service composition algorithms can create new services with minimal user support. Furthermore, if some of the services in the composition are failing, they can be dynamically replaced by other similar services.
Semantically modeled and operated workflows, as well as (semi-)automatic composition based on semantics, which brings flexibility and scalable adaptation, could help to solve a common problem for standard eHealth infrastructures, i.e. the lack of proper solutions for on-the-fly, painless integration of services in order to provide new functionalities.
3. Methods
3.1 OntoHealth Architecture
The OntoHealth architecture aims at supporting the dynamic composition and execution of clinical workflows related to EHR interaction focused on the domain of diabetes mellitus. Hence, in order to implement this functionality, we based our design on tools and methods for composition and integration of services in a clinical environment.
OntoHealth builds on standards and solutions from composition and workflows that are available in the eHealth domain extended with semantics. In OntoHealth, we take a semi-automatic approach in which generic predefined composition templates created by a user, assisted by a machine, can be selected and customized to realize service compositions useful for specific domains and situations. The project delivers flexible support for the modification of the workflow processes by utilizing rules and decision logics. Flexibility of the composition process can be reached through incremental composition of services based on the reaction of the user at each step of the composition.
3.2 Systematic Literature Review
As a precondition to the development of the final architecture, as well as specific services for the domain of diabetes, it is necessary to identify and analyze health care professionals’ clinical workflows as well as their informational or functional needs regarding the treatment of patients. Although fostering the quality of care by using EHRs involves various stakeholders, including the patient him/herself, the OntoHealth project focuses on the meaningful interaction between EHRs and physicians in their daily practice of diabetes care.
To achieve this goal, a systematic literature review was conducted based on the PRISMA Statement [18] combining scientific literature and other pertinent sources, such as e.g. clinical guidelines, best practices, and legislation. We decided to analyze both sources independently, beginning with the scientific literature described in the present paper.
According to PRISMA, the initial search in the Identification phase was conducted to retrieve papers from the selected databases given in ►Table 1. The search terms were built using the defined keywords or derived MeSH terms supplied by the National Center for Biotechnology Information (NCBI). Each search considered title and abstract for selection. The databases ACM Digital, EMBASE, IEEExplore, Citeseer, and DARE did not allow using an advanced search for MeSH terms or abstract lookup. These were searched using the predefined keywords. Only papers which were published between the years 1995 and 2013 were considered for further analysis. All duplicates were removed. In order to manage the complete review process, a shared Excel sheet was used containing at least title and author(s) of all papers.
Table 1.
Setup for the systematic review according to PRISMA, comprising inclusion and exclusion criteria for every phase.
| Identification | |
|---|---|
| Databases | MEDLINE/PubMed, Cochrane, ACM Digital, IEEExplore, EMBASE, Taylor & Francis, Citeseer, DARE, BioMed Central, ScienceDirect |
| Keywords | Diabetes; electronic health record; EHR; clinical workflow; health information technology; information system |
| MeSH Terms | „Diabetes Mellitus“[Mesh], „Workflow“[Mesh], „Guidelines as Topic“[Mesh], „Guideline“ [Publication Type], „Practice Guideline“ [Publication Type], „Health Planning Guidelines“[Mesh], „Medical Informatics“[Mesh], „Information Systems“[Mesh], „Medical Records Systems, Computerized“[Mesh], „Medical Records, Problem-Oriented“[Mesh], „Health Records, Personal“[Mesh], „Electronic Health Records“[Mesh], „Medical Record Administrators“[Mesh], „Medical Record Linkage“[Mesh], „Medical Records“[Mesh] |
| Screening | |
| Exclusion Criteria | • Papers not written in English • Symposium reports • Opinion papers • Letters, interviews • Commentaries and discussion reports • Documents that used abbreviations that stood for other terms than our purpose (e.g. author’s name) |
| Eligibility | |
| Inclusion Criteria | • General statements or tasks regarding functional or non-functional requirements for workflows regarding the interaction with EHRs or other electronic information systems in the context of diabetes or tasks that are included in a clinical workflow. • Organizational requirements |
| Exclusion Criteria | • Pure medical studies about diabetes e.g. concerning medical errors or new findings • Studies in emergency departments • Outpatient cases • Workflows out of the physician’s office or medical consultation |
| Included | |
| Inclusion Criteria | Same inclusion criteria as for eligibility |
The Screening phase excluded all documents that did not fulfill a certain document type (►Table 1) or were not written in English.
During the third step (Eligibility), a group discussion of three of the authors was held to define a set of inclusion and exclusion criteria common to our objectives. These criteria were used to select meaningful documents by reviewing title and abstract. The inclusion and exclusion criteria which were applied are depicted in Table 1. This step was collaboratively performed by three of the authors. The quantity of included and excluded papers were collected in the aforementioned Excel sheet as well. All papers were divided into two groups, which were then reviewed by the authors separately. The authors revised title and abstract and assigned the appropriate exclusion or inclusion criteria. A control sample was shared between the authors to validate the assignment of criteria to each paper within the control sample.
In the last phase (Included), the remaining set of papers was full-text analyzed using a quantitative content analysis as described in Mayring [19] and Bortz [20]. All eligible papers were separated and reviewed by two authors finding pertinent statements concerning the inclusion criteria from Table 1. The utilization of MAXQDA software allowed for the coding of all found statements on-the-fly and building a first categorization system. The software describes a created categorization as “code” and an assigned statement to a certain category as “coding”. After reviewing 25% of all included papers, the categorization system was redefined and generalized by further group discussion.
4. Results
4.1 OntoHealth Architecture
Built upon the principles of service composition and semantic integration, ►Figure 1 depicts a schematic overview of the OntoHealth architecture and its consisting modules. The modules of the basic architecture can be described according to its three main functionalities: 1) Workflow definition: GUI + workflow generator, 2) Workflow composition and execution: orchestrator + semantic bus service 3) Workflow execution: EHR module.
Fig. 1.
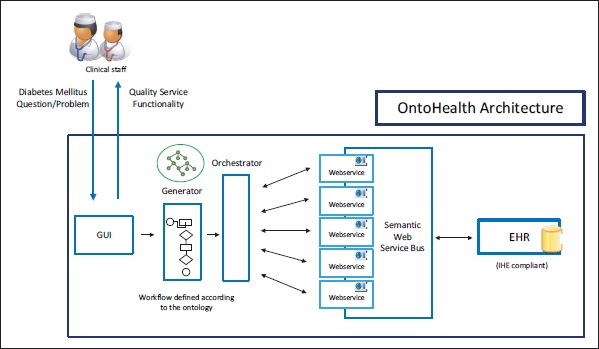
Overview of the general OntoHealth architecture
The interaction between the health care professional and the architecture is established by means of a graphical user interface (GUI). This GUI allows the physician to execute different clinical workflows that may require several interactions with the EHR in an easy and fast manner. For example, a physician could obtain information regarding side effects of a medication given the particular conditions of the evaluated patient and then come up with a better alternative in a one-step interaction. In addition, the GUI allows the definition and composition of new clinical workflows using predefined building blocks associated to common tasks in clinical daily routines. Hence, the GUI enables the communication between health care professionals and the workflow generator module. According to a predefined ontology, this module provides a workflow instance that is compliant with the ontology and filled according to the health care professional’s requirements. The specification of this compound of sequence actions for a specific patient’s case is denoted as clinical workflow specification, the generalization of these specifications has been denoted as clinical workflow pattern. As shown in Figure 1, this information serves as an input for the orchestrator module.
The orchestrator module is responsible for selecting the most suitable service available in the semantic web service bus for the execution of each of the tasks that comprises the clinical workflow given as an input. For this task, the module examines each part of the workflow, communicates to the available services, retrieves semantic information, and matches both descriptions for the best selection.
The orchestrator module relies on an underlying linked data infrastructure which enables the integration of different data sources [21]. It exploits the integration of various data types, instances, and links between the data sets of the linked data in order to figure out relations between data produced and consumed by services. In this way, output data of a service can be related to input data of another service and thus a composition of the two can be identified as possible. Composition and orchestration is thus based on the integration of linked data and services via linked data services [22] using lightweight semantic descriptions based on standards (e.g. RDF [23], SPARQL [24]).
The semantic web service bus to be developed within the project facilitates the task of composing and orchestrating services and will enable a flexible interaction with EHRs. It relies on a linked data infrastructure which enables the integration of different data sources. The semantic web service bus provides a semantic abstraction over the EHR’s functionalities. It includes formal descriptions of the EHR’s functionalities based on semantics enabling agents to orchestrate and compose these functionalities based on their semantic description (see the Orchestrator module).
Finally, the EHR module allows for the retrieval of required information and storage of information when appropriate for all involved services within a workflow from EHRs. The EHR module will be based on relevant IHE profiles comprising Cross-Enterprise Document Sharing (XDS), Audit Trail and Node Authentication (ATNA), and Cross-Enterprise Document Workflow (XDW), which are all part of the IHE IT Infrastructure Technical Framework [25]. Hence, the OntoHealth architecture sets up an IHE-based information system that allows native, dynamic workflow support regardless of the type of clinical workflow.
4.2 Categorization Model
►Figure 2 summarizes the literature review with respect to the papers selected for each step of the review. As shown in the figure, the 543 initially found papers from the database search led to 87 papers that met the predefined inclusion criteria based on the analysis of abstract and title. The following content analysis was then conducted to find statements related to tasks and complete clinical workflows in order to create a categorization model for defining and structuring building blocks and characteristics of physicians’ workflows in the domain of diabetes care. According to Niazkhani et al. [26], a clinical workflow is defined as “the allocation of multiple tasks of a provider or of co-working providers in the processes of care and the way they collaborate”. Using this definition in combination with the inclusion criteria, diverse findings concerning tasks and complete clinical workflows could be obtained from the raw data.
Fig. 2.
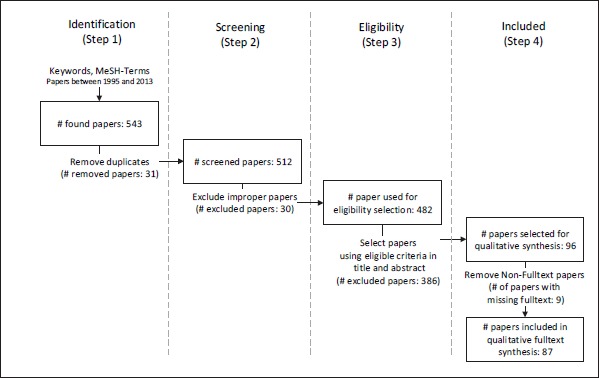
Results of each step in the literature review.
In total, 802 single statements were identified from the data, which were generalized and categorized in a three-part identification model. Using this model, a workflow and its containing components can be classified by 1) Actions, 2) Data Elements and 3) Contexts. An overview of the categorization is depicted in►Figure 3, which also includes the main levels of subcategories. The full categorization can be obtained from the authors.
Fig. 3.
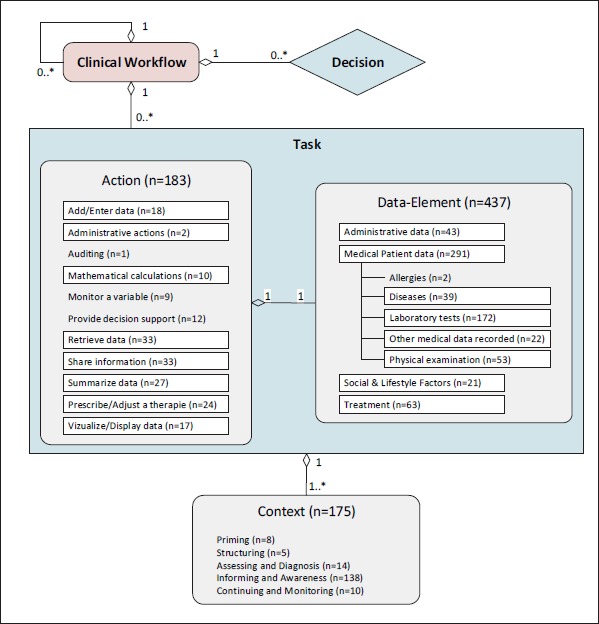
Description and categorization model of clinical workflows including cardinalities between the components. Actions, Data Elements and Context show contained items at which framed items form subcategories holding further elements. Numbers show assigned codings to the categories.
Every clinical workflow consists of several tasks and decisions which define a closed action or activity executed by a health care professional or a system. The tasks do not need to be atomic, but they are also able to represent complex actions by building another sub-workflow. This hierarchical structure establishes a dynamic use of the proposed categorization model. Each task consists of a generic Action that belongs to a certain Data Element which describes the specific application content of the action. For example, the action “Retrieve data” can be applied over the data element “Laboratory tests”. The data element “HbA1c”, as a sub-item of “Laboratory tests”, can then form a task “Retrieve HbA1c”. Finally, the Context defines the environment a certain task is executed in. For example, “Retrieve HbA1c” in the context of “Continuing & Monitoring” shows the purpose of the task. The decisions are part of the process flow within a workflow. They will be determined in the next step of the literature review using other literature sources as clinical guidelines. We found different diabetes-related guidelines in the analyzed literature. All 91 guideline codings build examples for tasks, consisting of action and data element, decisions, or complete workflows, but they were not considered during this study.
A complete sample workflow could be “Retrieve the summary of laboratory measurements of a certain diabetes patient”. Using the proposed categorization model, this workflow comprises one task for each laboratory value (e.g. “HbA1c”, “foot examination results”, “eye examination results”, “physical exercise adherence”) using a “retrieve” action and is followed by the task “Display summary” to visualize the requested results.
Statements found within the groups Action, Data Element and Context were further classified according to different subcategories.
Actions
In total, 183 statements were classified as Actions. These statements were again classified according to different activities of medical routine. Most of them are related to “Retrieve data” (n = 33), which describes all actions of obtaining any kind of data. Especially for the diabetes domain, these actions could be subdivided into “Retrieve patient’s self-documentation” and “Collect relevant clinical data”. The latter comprises “Retrieve single data element” as it could be e.g. for a laboratory result and “multiple data elements” which describes e.g. a summary or report of multiple laboratory results. Depending on the point of view of a certain action, “multiple data elements” will rather relate to complete further workflows.
Data Elements
For the group Data Element, 437 codes were identified in the content analysis. The most prominent sub-items are “Medical patient data” (n = 291), “Treatment” (n = 63), and “Administrative data” (n = 43). Some of the Data Elements’ subcategories relate to one object (e.g. a patient) or more objects (e.g. group of patients) depending on the situation. For an overview of all categories and subcategories, see ►Figure 3. Further subdivisions were available in all framed categories but only “Medical patient data” displays them in the figure. Holding subdivisions is not restricted to assigning elements to the category itself, which means that “Administrative data” could also be a valid data item within a certain task. ►Table 2 shows examples of selected categories with found statements and relevant references.
Table 1.
Selected sample categorizations for Data Element and Context including statements found and references.
| Component | Category | Example Source Statement | Assigned References |
|---|---|---|---|
| Data Element | HbA1c | “The records of Type 2 diabetics were searched for … glycosylated haemoglobin (HbA1c).” [28] | [28–54] |
| Patient-reported data | “… interpret the results of blood glucose self-monitoring …” [43] | [27, 43, 49] | |
| Context | Informing and Awareness | “… some providers combined patient data sets in order to help patients understand the health consequences” [27] | [27, 29, 30, 32, 33, 35, 39, 41, 43, 50, 51, 55–62] |
| Continuing and Monitoring | “… doctors monitor patients’ levels of adherence to care processes (e.g. annual assessment, review visits, education sessions, laboratory tests) and self management as well as their status of attainment of treatment targets.” [55] | [27, 28, 55, 56, 62–65] |
Contexts
Extending the classification proposed by Veinot et al. [27] for classifying patient-centered EHR use in clinical consultations, context of use can be categorized by: “Priming” (n = 8), “Structuring” (n = 5), “Assessing & Diagnosis” (n = 14), “Informing and Awareness” (n = 138), and “Continuing and Monitoring” (n = 10). Examples of selected statements concerning a certain Context are shown in ►Table 2.
5. Discussion
5.1 Methods
The categorization builds on acquired statements from a qualitative content analysis. Literature sources are related to well-known online literature databases. The search terms were carefully determined in group discussion, but it cannot be assured that all pertinent papers concerning the requested content were retrieved. Also, a search for gray literature was not included during this study. Coding differences from authors during the content analysis were solved by discussion. However, a qualitative approach always involves some subjectivity.
5.2 Results
The integration of the flexible use of electronic health records is a crucial factor in successfully supporting a physician’s daily practice [14]. The developed categorization makes it possible to describe all components or building blocks within clinical workflows. The results presented in this paper are essential for both the design of the services included in the semantic service bus and the clinical workflow models. The latter govern the adequate execution of workflows over the services included in the bus. In order to establish helpful constraints in the vast medical domain, the study is limited to the domain of diabetes. It might be possible to generalize the proposed categorization model, but further investigations need to be done to prove this. The dynamic model using tasks as single actions connected with a certain data element or as further workflows enables a categorization in a hierarchal and flexible manner. Furthermore, tasks could either be specific or generic depending on the assignment of a certain data element or not.
Note that the given categorization does not describe any workflow process rules. The model only provides a tool describing the components or building blocks of a decomposed workflow from a functional view. Using another way, Hsieh [66] e.g. focused more on describing the detailed process related to clinical workflows using Petri nets. This offers workflow-related details that are important for process execution (detailed identification, time of execution, etc.) but does not classify the particular components according to their functional meaning.
Other studies, such as Brindisi et al. [67], try to adjust a certain clinical application system to the workflows a physician is used to. In contrast, OntoHealth tries to offer a context-aware functionality independent of a specific system by building on EHR standards and best practices. Therefore, the categorization model provides a possibility to describe building blocks of clinical workflows from a user perspective without relating to any specific application system.
Acknowledgments
This work was supported by the Austrian Science Foundation (FWF), project number P 25895-B24.
Footnotes
Outlook and Clinical Relevance
The conducted scientific review is followed by involving other pertinent literature sources to gather further requirements in order to extend or modify the present categorization model. As a next step, direct observations in the hospital setting will be held in order to identify executed workflows by shadowing diabetes specialists. The combined result of literature review and observations will be used for the following expert interviews.
The project OntoHealth is utilizing the gathered categorization model in further steps for implementing the semantic service grid and service prototypes for diabetes care. The detailed work and progress of the OntoHealth project can be followed online [11].
The establishment of a standards-based flexible EHR usage to a more comprehensive integration within complete clinical workflows by (semi-)automatic service orchestration allows the physician to be the main actor in defining and executing information requests within an EHR system. This could be compared to “apomediation” as referred to in [68], which describes that there is no need for an intermediary when an actor is accessing or retrieving information or executing any other health care workflow. The OntoHealth project creates a functionality access to EHR systems which establishes this direct access for health care professionals to patient-related data.
Conflict of Interest
The authors declare that they have no conflicts of interest in the research.
Human Subjects Protections
Human and/or animal subjects were not included in the project.
References
- 1.Danaei G, Finucane MM, Lu Y, Singh GM, Cowan MJ, Paciorek CJ, et al. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980 : systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2·7 million participants. Lancet 2011; 378(9785): 31–40 [DOI] [PubMed] [Google Scholar]
- 2.American Diabetes Association. Economic costs of diabetes in the U.S. in 2012. Diabetes Care 2013; 36(4): 1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waegemann C.EHR vs. CPR vs. EMR. Healthc Informatics Online 2003; 1: 1–4 [Google Scholar]
- 4.IHE International. About IHE. Available from: http://www.ihe.net/About_IHE/ [Google Scholar]
- 5.EN 13606 Association. CEN/ISO EN1 3606. Available from: http://www.en13606.org/the-cenisoen13606-standard [Google Scholar]
- 6.openEHR Foundation. What is openEHR? [cited 2014 Jan 22]. Available from: http://www.openehr.org/what_is_openehr [Google Scholar]
- 7.Dolin R, Alschuler L, Boyer S, Beebe C, Behlen FM, Biron P V, et al. HL7 clinical document architecture, release 2. J Am Med Informatics Assoc 2006; 13(1): 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ELGA GmbH. ELGA – die elektronische Gesundheitsakte. [cited 2014 Mar 10]. Available from: http://www.elga.gv.at/ [Google Scholar]
- 9.epSOS Project. epSOS. [cited 2014 Mar 10]. Available from: http://www.epsos.eu/ [Google Scholar]
- 10.EHR4CR – Electronic Health Records for Clinical Research. [cited 2014 Mar 25]. Available from: http://www.ehr4cr.eu [Google Scholar]
- 11.UMIT – University for Health Sciences Medical Informatics Technology STI – Semantic Technology Insitute at University of Innsbruck. OntoHealth project. [cited 2014 Jan 23]. Available from: http://www.ontohealth.org [Google Scholar]
- 12.Shaw JE, Sicree RA, Zimmet PZ.Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract 2010; 87(1): 4–14 [DOI] [PubMed] [Google Scholar]
- 13.D’Avolio L, Ferguson R, Goryachev S, Woods P, Sabin T, O’Neil J, et al. Implementation of the Department of Veterans Affairs’ first point-of-care clinical trial. J Am Med Informatics Assoc 2012; 19(1): 170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowens FM, Frye PA, Jones WA.Health information technology: integration of clinical workflow into meaningful use of electronic health records. Perspect Heal Inf Manag 2010; 7: 1d. [PMC free article] [PubMed] [Google Scholar]
- 15.Binder W, Bonetta D, Pautasso C, Peternier A, Milano D, Schuldt H, et al. Towards Self-Organizing Service-Oriented Architectures. IEEE World Congress on Services. IEEE; 2011: 115–121 [Google Scholar]
- 16.Dustdar S, Schreiner W.A survey on web services composition. Int J Web Grid Serv 2005; 1(1): 1–30 [Google Scholar]
- 17.Fensel D, Facca FM, Simperl E, Toma I.Semantic Web Services. Springer; 2011 [Google Scholar]
- 18.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. PLoS Med 2009; 6(7): 1–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayring P.Qualitative Inhaltsanalyse: Grundlagen und Techniken. 8th ed Weinheim und Basel: Beltz Verlag; 2000 [Google Scholar]
- 20.Bortz J, Döring N.Forschungsmethoden und Evaluation: Für Human- und Sozialwissenschaftler. 3rd ed Berlin, Heidelberg, New York: Springer-Verlag; 2003 [Google Scholar]
- 21.Bizer C, Heath T, Berners-Lee T.Linked Data – The Story So Far. Int J Semant Web Inf Syst 2009; 5: 1–22 [Google Scholar]
- 22.Pedrinaci C, Domingue J.Toward the Next Wave of Services: Linked Services for the Web of Data. J Univers Comput Sci 2010; 16(13): 1694–1719 [Google Scholar]
- 23.W3C. RDF/XML Syntax Specification (Revised). 2004[cited 2014 Mar 28]. Available from: http://www.w3.org/TR/REC-rdf-syntax/ [Google Scholar]
- 24.W3C. SPARQL 1.1 Overview. 2013[cited 2014 Mar 28]. Available from: http://www.w3.org/TR/ sparql11-overview/ [Google Scholar]
- 25.Integrating the Healthcare Enterprise. IHE IT (ITI) Technical Framework. Volume 1 (ITI TF-1) Integration Profiles 2013[cited 2014 Mar 15]. Available from: http://www.ihe.net/uploadedFiles/Documents/ITI/IHE_ITI_TF_Vol1.pdf [Google Scholar]
- 26.Niazkhani Z, Pirnejad H, Berg M, Aarts J.The impact of computerized provider order entry systems on inpatient clinical workflow: a literature review. J Am Med Informatics Assoc 2009; 16(4): 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veinot T, Zheng K, Lowery J.Using electronic health record systems in diabetes care: emerging practices. IHI ’10 Proceedings of the 1st ACM International Health Informatics Symposium 2010: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lusignan S, Sismanidis C, Carey IM, De Wilde S, Richards N, Cook DG.Trends in the prevalence and management of diagnosed type 2 diabetes 1994–2001 in England and Wales. BMC Fam Pract 2005; 6(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciemins EL, Coon PJ, Fowles JB, Min S.Beyond health information technology: critical factors necessary for effective diabetes disease management. J Diabetes Sci Technol 2009; 3(3): 452-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Browne ED, Schrefl M, Warren JR.Goal-focused Self-Modifying Workflow in the Healthcare Domain. Proceedings of the 37th Hawaii International Conference on System Sciences 2004: 1-10 [Google Scholar]
- 31.Cleveringa FGW, Minkman MH, Gorter KJ, van den Donk M, Rutten GEHM.Diabetes Care Protocol: effects on patient-important outcomes. A cluster randomized, non-inferiority trial in primary care. Diabet Med 2010; 27(4): 442-450 [DOI] [PubMed] [Google Scholar]
- 32.Grant RW, Wald JS, Poon EG, Schnipper JL, Gandhi TK, Volk LA, et al. Design and Implementation of a Web-Based Patient Portal Linked to an Ambulatory Care Electronic Health Record: Patient Gateway for Diabetes Collaorative Care. Diabetes Technol Ther 2006; 8(5): 576-586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa BM, Fitzgerald KJ, Jones KM, Dunning Am T.Effectiveness of IT-based diabetes management interventions: a review of the literature. BMC Fam Pract 2009; 10: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Hagan C, De Vito G, Boreham CAG.Exercise prescription in the treatment of type 2 diabetes mellitus/?/: current practices, existing guidelines and future directions. Sports Med 2013; 43(1): 39-49 [DOI] [PubMed] [Google Scholar]
- 35.Friedman NM, Gleeson JM, Kent MJ, Foris M, Rodriguez DJ, Cypress M.Management of diabetes mellitus in the Lovelace Health Systems’ EPISODES OF CARE program. Am Coll Physicians-American Soc Intern Med 1998; 1(1): 5-11 [PubMed] [Google Scholar]
- 36.Grundel B, White GL, Eichold BH.Diabetes in the managed care setting: a prospective plan. South Med J 1999; 92(5): 459-464 [DOI] [PubMed] [Google Scholar]
- 37.Holbrook A, Thabane L, Keshavjee K, Dolovich L, Bernstein B, Chan D, et al. Individualized electronic decision support and reminders to improve diabetes care in the community: COMPETE II randomized trial. Can Med Assoc J 2009; 181(1-2): 37-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hetlevik I, Holmen J, Krüger O, Kristensen P, Iversen H, Furuseth K.Implementing clinical guidelines in the treatment of diabetes mellitus in general practice. Evaluation of effort, process, and patient outcome related to implementation of a computer-based decision support system. Int J Technol Assess Health Care 2000; 16(1): 210-227 [DOI] [PubMed] [Google Scholar]
- 39.Kaufman DR, Pevzner J, Rodriguez M, Cimino JJ, Ebner S, Fields L, et al. Understanding workflow in telehealth video visits: observations from the IDEATel project. J Biomed Inform 2009; 42(4): 581-592 [DOI] [PubMed] [Google Scholar]
- 40.Khan S, Mac Lean C, Littenberg B.The effect of the Vermont Diabetes Information System on inpatient and emergency department use: results from a randomized trial. Health Outcomes Res Med 2010; 1(1): 1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khattab MS, Swidan AM, Farghaly MN, Swidan HM, Ashtar MS, Darwish EA, et al. Quality improvement programme for diabetes care in family practice settings in Dubai. East Mediterr Heal J 2007; 13(3): 492-503 [PubMed] [Google Scholar]
- 42.Mac Lean CD, Littenberg B, Gagnon M, Reardon M, Turner PD, Jordan C.The Vermont Diabetes Information System (VDIS): study design and subject recruitment for a cluster randomized trial of a decision support system in a regional sample of primary care practices. Clin Trials 2004; 1(6): 532-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mc Culloch DK, Price MJ, Hindmarsh M, Wagner EH.A population-based approach to diabetes management in a primary care setting: early results and lessons learned. Eff Clin Pract 1995; 1(1): 12-22 [PubMed] [Google Scholar]
- 44.Maclean CD, Gagnon M, Callas P, Littenberg B.The Vermont diabetes information system: a cluster randomized trial of a population based decision support system. J Gen Intern Med 2009; 24(12): 1303-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller CD, Phillips LS, Tate MK, Porwoll JM, Rossman SD, Cronmiller N, et al. Meeting American Diabetes Association guidelines in endocrinologist practice. Diabetes Care 2000; 23(4): 444-448 [DOI] [PubMed] [Google Scholar]
- 46.O’Connor P, Pronk N.Integrating population health concepts, clinical guidelines, and ambulatory medical systems to improve diabetes care. J Ambul Care Manage 1998; 21(1): 67-73 [DOI] [PubMed] [Google Scholar]
- 47.O’Connor P, Sperl-Hillen JM, Rush WA, E. Johnson P, Amundson GH, Asche SE, et al. Impact of electronic health record clinical decision support on diabetes care: a randomized trial. Ann Fam Med 2011; 9(1): 12-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward MM, Yankey JW, Vaughn TE, BootsMiller BJ, Flach SD, Welke KF, et al. Physician process and patient outcome measures for diabetes care: relationships to organizational characteristics. Med Care 2004; 42(9): 840-850 [DOI] [PubMed] [Google Scholar]
- 49.Rodbard HW, Schnell O, Unger J, Rees C, Amstutz L, Parkin CG, et al. Use of an automated decision support tool optimizes clinicians’ ability to interpret and appropriately respond to structured self-monitoring of blood glucose data. Diabetes Care 2012; 35(4): 693-698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roshanov PS, Misra S, Gerstein HC, Garg AX, Sebaldt RJ, Mackay J a, et al. Computerized clinical decision support systems for chronic disease management: a decision-maker-researcher partnership systematic review. Implement Sci. BioMed Central Ltd 2011; 6(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas B.Improving blood pressure control among adults with CKD and diabetes: provider-focused quality improvement using electronic health records. Adv Chronic Kidney Dis 2011; 18(6): 406-411 [DOI] [PubMed] [Google Scholar]
- 52.Dinneen SF, Bjornsen SS, Bryant SC, Zimmerman BR, Gorman CA, Knudsen JB, et al. Towards an optimal model for community-based diabetes care: design and baseline data from the Mayo Health System Diabetes Translation Project. J Eval Clin Pract 2000; 6(4): 421-429 [DOI] [PubMed] [Google Scholar]
- 53.Sidorenkov G, Haaijer-Ruskamp FM, de Zeeuw D, Denig P.A longitudinal study examining adherence to guidelines in diabetes care according to different definitions of adequacy and timeliness. PLoS One 2011; 6(9): e24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oh S-W, Lee HJ, Chin HJ, Hwang J-I.Adherence to clinical practice guidelines and outcomes in diabetic patients. Int J Qual Heal Care 2011; 23(4): 413-419 [DOI] [PubMed] [Google Scholar]
- 55.Ko GT, So W-Y, Tong PC, Le Coguiec F, Kerr D, Lyubomirsky G, et al. From design to implementation-- the Joint Asia Diabetes Evaluation (JADE) program: a descriptive report of an electronic web-based diabetes management program. BMC Med Inform Decis Mak 2010; 10: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shea S, Starren J, Weinstock RS, Knudson PE, Teresi J, Holmes D, et al. Columbia University’s Informatics for Diabetes Education and Telemedicine (IDEATel) Project: J Am Med Informatics Assoc 2002; 9(1): 49-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beliakov G, Warren J, Noone J.Mixed-initiative Decision Support for Care Planning Based on Clinical Practice Guidelines. J Decis Syst 2001; 10(3-4): 429-448 [Google Scholar]
- 58.Goldberg HI, Lessler DS, Mertens K, Eytan TA, Cheadle AD.Case Study in Brief Self-Management Support in a Web-Based Medical Record⊠: A Pilot Randomized Controlled Trial. Jt Comm J Qual Saefty 2004; 30(11): 629-635 [DOI] [PubMed] [Google Scholar]
- 59.Peleg M, Shachak A, Wang D, Karnieli E.Using multi-perspective methodologies to study users’ interactions with the prototype front end of a guideline-based decision support system for diabetic foot care. Int J Med Inform 2009; 78(7): 482-493 [DOI] [PubMed] [Google Scholar]
- 60.Sequist T, Gandhi T, Karson A, Fiskio JM, Bugbee D, Sperling M, et al. A randomized trial of electronic clinical reminders to improve quality of care for diabetes and coronary artery disease. J Am Med Informatics Assoc 2005; 12(4): 431-438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russell K, Rosenzweig J.Improving outcomes for patients with diabetes using Joslin Diabetes Center’s Registry and Risk Stratification system. J Healthc Inf Manag 2007; 21(2): 26-33 [PubMed] [Google Scholar]
- 62.Eccles M, Hawthorne G, Whitty P, Steen N, Vanoli A, Grimshaw J, et al. A randomised controlled trial of a patient based Diabetes Recall and Management System: the DREAM trial: a study protocol [ISRCTN32042030]. BMC Health Serv Res 2002; 2(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maser RE, Lenhard MJ, Henderson BC, Cobb RS, Hands KE.Detection of subsequent episodes of gestational diabetes mellitus: a need for specific guidelines. J Diabetes Complications 2004; 18(2): 86-90 [DOI] [PubMed] [Google Scholar]
- 64.Dorr D, Bonner L, Cohen A.Informatics systems to promote improved care for chronic illness: a literature review. J Am Med Informatics Assoc 2007; 14(2): 156-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crosson JC, Obman-Strickland PA, Cohen DJ, Clark EC, Crabtree BF.Typical Electronic Health Record Use in Primary Care Practices and the Quality of Diabetes Care. Ann Fam Med 2012; 10(3): 221-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsieh F.Context-aware workflow driven resource allocation for e-healthcare. e-Health Networking, Application and Services, 2007 9th International Conference on. 2007: 34-39 [Google Scholar]
- 67.Brindisi M, Bui R, Mazo J, Marcus OZY, Vleet S Van, Bailey R.Redesigning clinic workflows for electronic medical record integration. Proceedings of the 2013 IEEE Systems and Information Engineering Design Symposium. USA2013: 133-138 [Google Scholar]
- 68.O’Connor D.The apomediated world: regulating research when social media has changed research. J Law Med Ethics 2013; 41(2): 470-483 [DOI] [PubMed] [Google Scholar]


