Abstract
Delivery of microbial products into the mammalian cell cytosol by bacterial secretion systems is a strong stimulus for triggering pro-inflammatory host responses. Here we show that Salmonella enterica serovar Typhi (S. Typhi), the causative agent of typhoid fever, tightly regulates expression of the invasion-associated type III secretion system (T3SS-1) and thus fails to activate these innate immune signaling pathways. The S. Typhi regulatory protein TviA rapidly repressed T3SS-1 expression, thereby preventing RAC1-dependent, RIP2-dependent activation of NF-κB in epithelial cells. Heterologous expression of TviA in S. enterica serovar Typhimurium (S. Typhimurium) suppressed T3SS-1-dependent inflammatory responses generated early after infection in animal models of gastroenteritis. These results suggest that S. Typhi reduces intestinal inflammation by limiting the induction of pathogen-induced processes through regulation of virulence gene expression.
Author Summary
Bacterial pathogens translocate effector proteins into the cytoplasm of host cells to manipulate the mammalian host. These processes, e.g. the stimulation of small regulatory GTPases, activate the innate immune system and induce pro-inflammatory responses aimed at clearing invading microbes from the infected tissue. Here we show that strict regulation of virulence gene expression can be used as a strategy to limit the induction of inflammatory responses while retaining the ability to manipulate the host. Upon entry into host tissue, Salmonella enterica serovar Typhi, the causative agent of typhoid fever, rapidly represses expression of a virulence factor required for entering tissue to avoid detection by the host innate immune surveillance. This tight control of virulence gene expression enables the pathogen to deploy a virulence factor for epithelial invasion, while preventing the subsequent generation of pro-inflammatory responses in host cells. We conclude that regulation of virulence gene expression contributes to innate immune evasion during typhoid fever by concealing a pattern of pathogenesis.
Introduction
One function of the innate immune system in the intestinal tract is to generate temporary inflammatory responses against invasive enteric pathogens while avoiding detrimental overreaction against harmless commensal bacteria under homeostatic conditions. In contrast to commensal microbes, pathogenic microbes express an array of virulence factors to manipulate host cell functions. Pathogen-induced processes, also known as patterns of pathogenesis [1], activate specific pathways of the innate immune system, enabling the host to distinguish virulent microbes from ones with lower disease-causing potential. By detecting pathogen-induced processes the host can escalate innate immune responses to levels that are appropriate to the threat [2].
Salmonella enterica serovar Typhimurium (S. Typhimurium), an invasive enteric pathogen associated with human gastroenteritis, triggers acute intestinal inflammation in the terminal ileum and colon, thereby producing symptoms of diarrhea and abdominal pain within less than one day after ingestion [3]. The inflammatory infiltrate in the affected intestinal tissue is dominated by neutrophils [4], [5]. Similarly, neutrophils are the primary cell type in the stool during acute illness [6]–[8]. In contrast, individuals infected with serovar Typhi (S. Typhi) develop a febrile illness (typhoid fever) with systemic dissemination of the organism. In contrast to Salmonella-induced gastroenteritis, only a third of patients develop diarrhea that is characterized by a dominance of mononuclear cells in the stool [6]. The dominant cell type in intestinal infiltrates is mononuclear, while neutrophils are infrequent [9]–[11]. Unlike S. Typhi, interaction of S. Typhimurium with intestinal model epithelia induces hepoxilin A3-dependent transmigration of neutrophils [12]. Moreover, infection of human colonic tissue explants with S. Typhimurium results in the increased production of the neutrophil-attracting chemokine IL-8, while S. Typhi does not elicit this response [13]. These observations suggest that invasion of the intestinal mucosa by S. Typhimurium is accompanied by a rapid escalation of host responses leading to acute, purulent inflammation, while S. Typhi elicits little intestinal inflammation during early stages of infection, however the molecular mechanisms underlying these apparent differences are poorly defined.
One pathogen-induced processes that triggers pro-inflammatory immune responses is the transfer of bacterial molecules into the host cell cytosol by secretion systems. The invasion-associated type III secretion system (T3SS-1) expressed by all Salmonella serovars and delivers effector proteins into the cytosol of epithelial cells [14]. A subset of these translocated effector proteins activate Rho-family GTPases [15]–[18], thereby triggering alterations in the host cell cytoskeleton that result in bacterial invasion of epithelial cells [19]. Excessive stimulation of Rho-family GTPases activates the transcription factor nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and promotes the subsequent release of proinflammatory cytokines and chemokines [15], [20], [21]. In a bovine model of S. Typhimurium-induced gastroenteritis, the rapid induction of intestinal inflammation and diarrhea requires the T3SS-1 apparatus as well as the effector proteins SipA, SopA, SopB, SopD, and SopE2 [22]–[24]. Similarly, in a murine model of Salmonella induced colitis, SipA, SopE and SopE2 can independently induce intestinal inflammation [25] and mutants lacking a functional T3SS-1 are unable to initiate neutrophil recruitment to the intestinal mucosa during early infection [25], [26]. These findings indicate that T3SS-1-mediated effector translocation induces innate immune responses during S. Typhimurium-induced colitis.
Similar to S. Typhimurium, invasion of cultured intestinal epithelial cells by S. Typhi is mediated by the T3SS-1 [27]. Replacement of S. Typhimurium T3SS-1 effector proteins with their S. Typhi orthologues does not attenuate inflammatory responses elicited by S. Typhimurium in the intestinal mucosa of calves [28], demonstrating that S. Typhi T3SS-1 effector proteins can exhibit intrinsic pro-inflammatory properties in vivo. Thus, the molecular basis for the absence of T3SS-1-dependent innate immune responses early during S. Typhi infection remains unclear.
Results
In contrast to S. Typhimurium, S. Typhi fails to activate the NF-κB signaling pathway in human epithelial cells
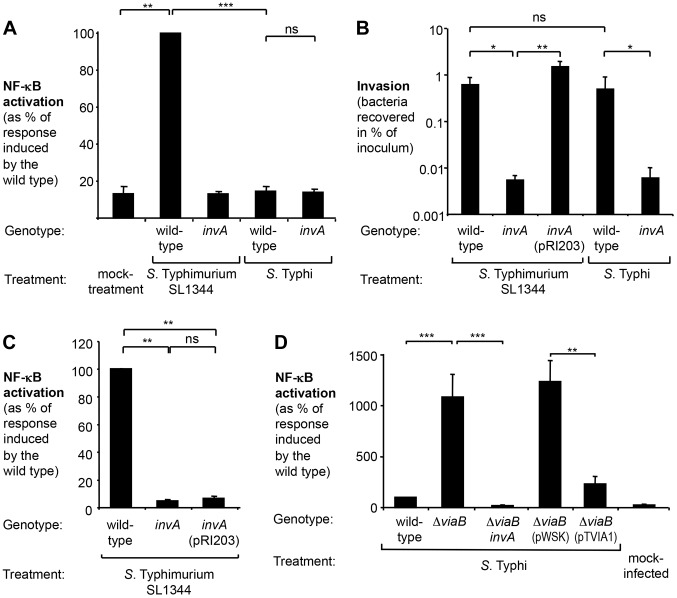
To study the induction of pro-inflammatory signaling pathways upon infection with S. Typhimurium and S. Typhi, we employed a human epithelial cell line permanently transfected with a NF-κB-dependent luciferase reporter (HeLa 57A) [29]. Infection with the S. Typhimurium wild-type strain SL1344 resulted in a significant increase (7-fold; P<0.01) in luciferase activity compared to mock-infected cells (Fig. 1A), while a derivative of S. Typhimurium SL1344 carrying a mutation in the T3SS-1 apparatus gene invA (SW767) did not elicit NF-κB signaling [20], [30]. In contrast to the S. Typhimurium wild type, the S. Typhi wild-type strain Ty2 failed to trigger NF-κB activation (Fig. 1A), suggesting that S. Typhi is a poor activator of T3SS-1-dependent inflammatory processes in human epithelial cells.
Figure 1. S. Typhi does not elicit inflammatory responses in epithelial cells.
Human epithelial cells permanently transfected with a NF-κB-luciferase reporter system (HeLa 57A) were infected with the indicated Salmonella strains at a multiplicity of infection of 5 or mock treated (bacterial growth media alone). (A) Cells were infected with the S. Typhimurium wild type SL1344, an isogenic invA mutant (SW767), the S. Typhi wild type Ty2, an isogenic invA mutant (SW222). Luciferase activity as a measure of NF-κB activation was determined after 5 h (N = 3). (B) Monolayers of cells were infected for 1 h with the indicated Salmonella strains. Bacterial numbers recovered after 90 min of Gentamicin treatment were standardized to the number of the bacteria in the inoculum (N = 4). Plasmid pRI203 encodes the Y. pseudotuberculosis invasin. (C) Luciferase activity exhibited by Salmonella-infected HeLa57A cells was determined as described above (N = 4). (D) Cells were infected the S. Typhi wild-type strain Ty2, a ΔviaB mutant (SW347), an invA ΔviaB mutant (STY4), a ΔviaB mutant (STY2) harboring the cloning plasmid pWSK29 (pWSK), a ΔviaB mutant (STY2) expressing the tviA gene (pTVIA1) and luciferase activity determined 5 h after infection (N = 5). Bars represent geometric means ± standard error. *, P<0.05; **, P<0.01; ***, P<0.001; ns, not statistically significant.
Effect of the viaB operon on T3SS-1 mediated inflammatory responses in epithelial cells
The T3SS-1 mediates invasion of non-phagocytic cells. S. Typhi has been reported to differ from S. Typhimurium with regards to invasion of human epithelial cells [31]–[33], thus raising the possibility that the observed differential activation of the NF-κB signaling pathway could be due to varying degrees of invasiveness. To test this hypothesis, HeLa cells were infected with S. Typhimurium and S. Typhi strains and a gentamicin protection assay was performed (Fig. 1B). The S. Typhimurium wild type SL1344 and the S. Typhi wild type Ty2 were recovered in similar numbers, while the respective isogenic invA-deficient mutants displayed significantly reduced invasiveness. T3SS-1 activity has to two functional consequences: manipulation of host signaling pathways and subsequent bacterial uptake. To discern between effects mediated directly by the T3SS-1 or indirectly by increasing the intracellular bacterial load, we next sought to reinstate invasiveness of the S. Typhimurium invA mutant without restoring T3SS-1 function. Expression of the Yersinia pseudotuberculosis invasin, encoded by the plasmid pRI203, raised invasiveness of the S. Typhimurium invA mutant comparable to the wild type strain (Fig. 1B), but failed to restore the ability to induce NF-κB activation in epithelial cells (Fig. 1C) [34]. Taken together, these observations indicate that immune evasion by S. Typhi did not directly correlate with the intracellular bacterial load or invasiveness.
Despite causing disparate disease entities, the genomes of S. Typhimurium and S. Typhi display remarkable similarity. Chromosomal DNA sequences of both serovars are highly syntenic, with mostly minor inversions, deletions and insertions [35], [36]. One DNA region that is present in S. Typhi but absent from S. Typhimurium is the Salmonella pathogenicity island 7 (SPI-7). Situated within SPI-7 is the viaB locus, an operon encoding regulatory (tviA), biosynthesis (tviBCDE), and export (vexABCDE) genes involved in the production of the virulence (Vi) capsular polysaccharide of S. Typhi [37] (Fig. S1A). The viaB locus has been shown to suppress Toll-like receptor (TLR) signaling pathways [13], [38], [39]. We therefore explored the contribution of the viaB locus on diminishing NF-κB activation in epithelial cells (Fig. 1D and S1B). Deletion of the entire viaB locus in S. Typhi (ΔviaB mutant; SW347) markedly increased the ability to activate NF-κB in epithelial HeLa cells (P<0.001). Akin to the findings with S. Typhimurium, NF-κB signaling induced by the S. Typhi viaB mutant was independent of invasiveness (Fig. S1C) but required a functional T3SS-1 since inactivation of invA in the viaB mutant background (ΔviaB invA mutant, STY4) completely abolished luciferase activity (P<0.001) (Fig. S1D). These results supported the idea that the viaB locus attenuates T3SS-1-induced, pro-inflammatory signaling pathways in human epithelial cells.
The viaB locus has been shown to alter interaction of S. Typhi with host cells through multiple distinct mechanisms (reviewed in [40]). The Vi capsular polysaccharide prevents complement deposition, phagocytosis, and TLR4 activation, while the regulatory protein TviA is known to dampen TLR5 signaling. We therefore wanted to discern whether the absence of NF-κB signaling in human epithelial cells is due to the production of the Vi capsule or due to altered gene expression mediated by TviA. To this end, the tviA gene cloned into a low copy number plasmid (pTVIA1) was introduced into a S. Typhi viaB mutant (STY2). Expression of tviA under control of the native promoter significantly lowered NF-κB activation (P<0.01) in comparison to cells infected with the S. Typhi viaB mutant carrying the empty vector control (pWSK29). Remarkably, expression of tviA reduced inflammatory responses to levels comparable to the S. Typhi wild-type strain (Fig. 1D and S1B), suggesting that the regulatory protein TviA is involved in dampening inflammatory responses in cultured human epithelial cells.
TviA reduces T3SS-1-mediated inflammation in the bovine ligated ileal loop model
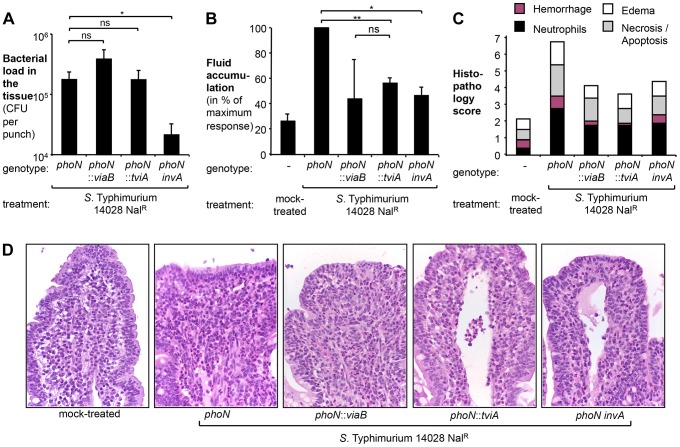
We had recently demonstrated that a S. Typhimurium strain carrying the S. Typhi viaB locus on a plasmid elicits less mucosal inflammation in a bovine ligated ileal loop model than the isogenic S. Typhimurium wild type ATCC14028 [38], raising the possibility that TviA might be involved in suppressing inflammatory responses in vivo. To delineate the relative contribution of the Vi capsule and the regulator TviA to reducing inflammatory responses in the bovine ligated ileal loop model [23], we repeated these studies with derivatives of S. Typhimurium strain ATCC 14028 in which the phoN gene in the chromosome had been replaced with the entire S. Typhi viaB locus (phoN::viaB mutant, TH170) or the tviA gene only (phoN::tviA mutant, SW474). In these strains, transcription of tviA and the downstream genes is solely controlled by the native S. Typhi promoter [41], [42]. This strategy was chosen to ensure that attenuation of intestinal inflammation in this model was not caused by introduction of the viaB locus on a multi-copy plasmid [38].
We compared the phoN::viaB mutant and the phoN::tviA mutant to a strain carrying an antibiotic resistance gene inserted chromosomally in the phoN gene (phoN mutant, AJB715). The phoN::viaB mutant, the phoN::tviA mutant, and the isogenic phoN mutant were recovered in equal numbers from gentamycin-treated tissue samples five hours after inoculation (Fig. 2A), suggesting that neither the tviA gene nor the entire viaB locus interfered with tissue invasion. Consistent with our previous observations [38], the phoN::viaB mutant elicited less fluid accumulation (Fig. 2B) and less pathological changes in the mucosa (Fig. 2C and D) than the isogenic phoN mutant. Remarkably, expression of tviA alone (phoN::tviA mutant) significantly reduced fluid accumulation and inflammation compared to the phoN mutant (P<0.01). The responses elicited by the phoN::tviA mutant and the phoN::viaB mutant were indistinguishable, suggesting that the viaB-mediated attenuation of inflammatory responses five hours after inoculation of bovine ligated ileal loops with S. Typhimurium was mostly attributable to the action of the TviA regulatory protein. Taken together, these data suggested that gene regulation mediated by TviA could dampen inflammatory processes in vivo.
Figure 2. Expression of TviA in S. Typhimurium reduces early host responses.
Bovine ligated ileal loops (N = 4 animals) were mock treated (LB broth) or infected with a S. Typhimurium ATCC14028 phoN mutant (AJB715), a phoN::viaB mutant (TH170), a phoN::tviA mutant (SW474), or a phoN invA mutant (SW737) pre-cultured in LB broth. Samples were collected 5 h after infection. (A) Bacterial load in the tissue was determined by treating tissue biopsies with Gentamicin and plating on selective media. Bars represent geometric means of bacterial loads ± standard error. (B) Fluid accumulation recorded 5 h after infection. Data is expressed as a percent of the response observed in loops infected with the phoN mutant (maximum response). Bars represent geometric means ± standard error. (C) Pathological changes in the ileal mucosa. Formalin-fixed ileal tissue was scored by the following criteria: hemorrhage (purple bars), infiltration with neutrophils (black bars), presence of edema (white bars), and necrosis/apoptosis (grey bars). Bars represent the average obtained from 4 animals. (D) Representative images of hematoxilin and eosin-stained sections of the ileal mucosa (magnification 60×). *, P<0.05; **, P<0.01; ns, not statistically significant.
TviA represses transcription of regulatory, structural, and effector proteins of the S. Typhi T3SS-1
A functional T3SS-1 is required for the induction of intestinal host responses in cattle [22], [24], [43]. A S. Typhimurium strain carrying a mutation in the T3SS-1 apparatus gene invA (invA phoN mutant, SW737) was significantly less invasive than a phoN mutant (Fig. 2A) (P<0.05). Interestingly, inactivation of invA (invA phoN mutant) reduced fluid accumulation (Fig. 2B) and intestinal inflammation (Fig. 2C and D) by a magnitude that was similar to that observed for the phoN::tviA mutant. This finding was consistent with the idea that TviA reduces T3SS-1-dependent host responses in vivo, prompting us to further investigate the mechanism by which TviA inhibits T3SS-1 gene expression.
TviA is a key activator of the tviBCDEvexABCDE operon but can also control transcription of genes outside its own operon (Fig. S2A). Expression of TviA results in diminished motility and flagellin secretion due to downregulation of the flagellar regulon by repressing transcription of the flhDC genes [42], [44]. FlhDC, the master regulator of flagellar gene expression, activates transcription of class II flagellar genes, such as fliA and fliZ [45], [46]. FliA is a positive regulator of class III flagellar genes, including flagellin [45], [47]. To determine whether reduced motility or diminished flagellin production could account for the TviA-dependent reduction in NF-κB activation, we inactivated the fliC gene encoding the sole flagellin of the monophasic serovar Typhi, thereby rendering strains carrying these mutations aflagellate and non-motile. Deletion of the entire viaB operon (ΔviaB ΔfliC mutant, SW483) in the fliC background (ΔfliC mutant, SW359) significantly increased NF-κB signaling in infected HeLa and HEK293 epithelial cells (Fig. S2B and S2C). Expression of TviA from a plasmid (pTVIA1) in a viaB fliC mutant reduced luciferase activity to levels comparable to the fliC mutant (Fig. S2B and S2C), demonstrating that TviA-dependent repression of NF-κB activation was flagellin-independent.
Gene expression profiling experiments suggest that TviA affects transcription of T3SS-1 genes through the following signaling cascade [42]: By repressing transcription of flhDC, TviA downregulates expression of FliZ. The regulatory protein FliZ is an activator of hilA [48]–[50], the master regulator of T3SS-1 genes [51], [52], thus placing T3SS-1 gene expression under negative control of TviA (Fig. S2A). We therefore analyzed the effect of TviA on the transcription of a subset of regulatory, structural, and effector proteins in S. Typhi (Fig. S3). Consistent with previous findings, deletion of the Vi capsule biosynthesis genes alone (ΔtviB-vexE mutant, SW74) did not alter transcription of T3SS-1 genes [42], [44]. In contrast, concomitant deletion of tviA and capsule biosynthesis genes (ΔviaB mutant, SW347) significantly enhanced transcription of the regulatory genes flhD, hilA, and invF, the structural component gene prgH, as well as the effector genes sipA and sopE (Fig. S3).
In the absence of TviA, the S. Typhi T3SS-1 effector protein SopE is the major inducer of NF-κB activation in epithelial cells
We next determined which T3SS-1 effector proteins contributed to pro-inflammatory responses elicited by S. Typhimurium and S. Typhi. Previous work has demonstrated that SopE, SopE2, SopB, and SipA contribute to NF-κB activation in epithelial cells [15]–[17], [53]. The bacteriophage-encoded sopE gene is present in S. Typhi Ty2 but absent from S. Typhimurium strain ATCC 14028. To better model the contribution of TviA on attenuating T3SS-1-induced host responses, we chose to continue our studies using the S. Typhimurium strain SL1344, an isolate that carries the sopE gene. Consistent with previous reports [15]–[17], [53], we found that simultaneous inactivation of sopE, sopE2, sopB, and sipA (sopE sopE2 sopB sipA mutant, SW868) reduced the ability of the S. Typhimurium strain SL1344 to induce NF-κB activation to levels observed in an isogenic S. Typhimurium strain unable to translocate effector proteins (invA mutant; SW767) (Fig. S4). A S. Typhimurium strain only expressing SopE (sopE2 sopB sipA mutant, SW867) elicited considerable NF-κB activation. A moderate NF-κB activation was also observed with S. Typhimurium strains only expressing SopB (sopE sopE2 sipA mutant, SW972) or only expressing SipA (sopE sopE2 sopB mutant, SW940) (Fig. S4). Essentially no response was observed in cells infected with a SL1344 derivative that only expressed SopE2 (sopE sopA sopB mutant, SW973). Collectively, these data suggested that SopE was the most potent inducer of pro-inflammatory responses in this tissue culture model, while the contributions of SopB and SipA were more modest.
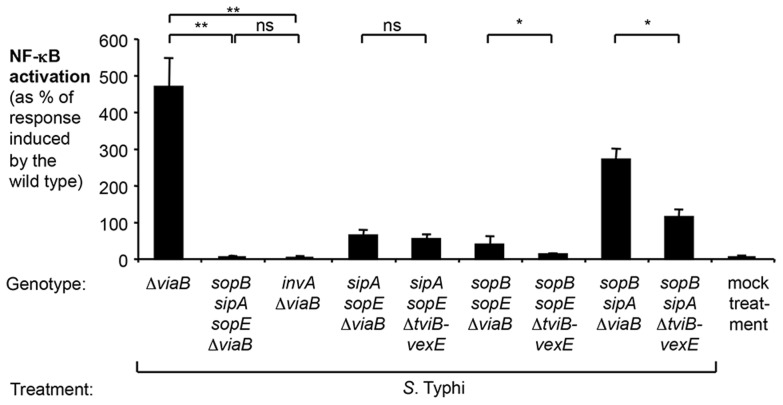
We next determined the potential contribution of the S. Typhi orthologues of these effectors to the induction of NF-κB signaling in the absence of the tviA gene (S. Typhi ΔviaB mutant, SW347) (Fig. 3). The sopE2 gene is a pseudogene in S. Typhi Ty2 and was not further analyzed. Concomitant inactivation of sopE, sipA and sopB in the S. Typhi viaB mutant (sopB sipA sopE ΔviaB mutant, SW1217) completely abolished NF-κB-driven luciferase activity (Fig. 3). This indicated that, akin to the findings with the S. Typhimurium strain SL1344, SopE, SipA, and SopB are critical for the induction of inflammatory responses in epithelial cells upon infection with S. Typhi. A S. Typhi viaB sopB sipA mutant (SW1211) elicited pronounced NF-κB activation, but a more modest NF-κB activation was also observed with the S. Typhi viaB sopE sipA mutant (SW1214) and the viaB sopE sopB mutant (SW1216) (Fig. 3). These data suggested that SopE was the most potent inducer of pro-inflammatory responses in S. Typhi strains lacking the tviA gene while SopB and SipA contributed moderately. In contrast, diminished NF-κB activation was observed with S. Typhi tviB-vexE mutant (carrying the tviA gene) and its derivatives (Fig. 3). This intricate comparison between derivatives of the viaB mutant and the tviB-vexE mutant allowed us to preclude any confounding effects expression of the Vi antigen might have on gene regulation: both the viaB mutant and the tviB-vexE mutant are non-encapsulated and only differ in their capability of expressing tviA. In contrast, a simple tviA mutant would exhibit a pleiotropic effect, i.e. it would lack the regulatory TviA protein but at the same time exhibit virtually no production of the Vi antigen [37]. Collectively, these data suggested that TviA-mediated gene regulation reduced T3SS-1 effector-triggered NF-κB activation.
Figure 3. Effect of the regulator TviA on NF-κB activation triggered by S. Typhi T3SS-1 effectors.
HeLa cells permanently transfected with a NF-κB-luciferase reporter system (HeLa 57A) were either mock treated (bacterial growth media alone) or infected with the S. Typhi wild type Ty2, a ΔviaB mutant (SW347), a sopB sipA sopE ΔviaB mutant (SW1217), an invA ΔviaB mutant (STY4), a sipA sopE ΔviaB mutant (SW1214), a sipA sopE ΔtviB-vexE mutant (SW1215), a sopB sopE ΔviaB mutant (SW1216), a sopB sopE ΔtviB-vexE mutant (SW1213), a sopB sipA ΔviaB mutant (SW1211), or a sopB sipA ΔtviB-vexE mutant (SW1212). After 5 h, luciferase activity was measured (N = 4). Bars represent geometric means ± standard error. *, P<0.05; **, P<0.01; ns, not statistically significant.
TviA reduces activation of the Rac1 and NOD1/2 signaling pathway
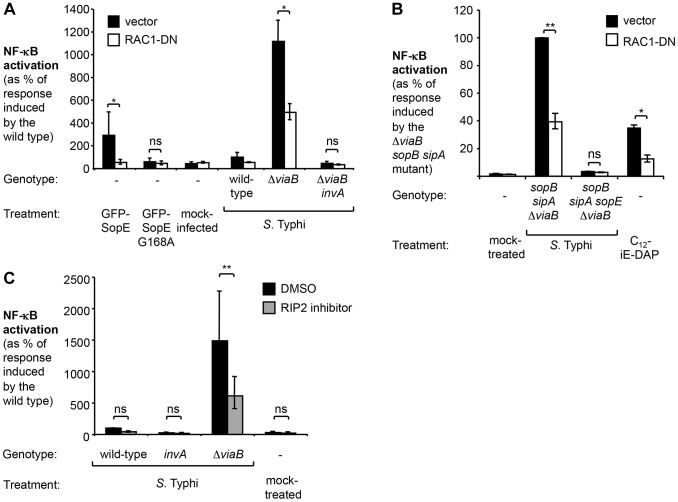
Since SopE triggered the most pronounced host responses in the absence of tviA, we focused our further analysis on this signaling pathway. Mechanistic studies in cultured epithelial cells have revealed that the bacterial guanine nucleotide exchange factor (GEF) SopE activates the Rho-family GTPase Ras-related C3 botulinum toxin substrate 1 (RAC1) [15]. Excessive stimulation of RAC1 by bacterial effectors is sensed by the nucleotide-binding oligomerization domain-containing protein 1 (NOD1) [30]. Activation of NOD1 leads to phosphorylation of the receptor-interacting serine/threonine-protein kinase 2 (RIP2) and activation of NF-κB signaling in epithelial cells [15], [20], [30], [54]. The NOD1/2 signaling pathway in HeLa cells can also be triggered by SipA [34], although this pathway plays a lesser role in the SopE-encoding strain SL1344 (Fig. S4). Taken together, these findings raised the possibility that TviA-mediated downregulation of SopE allows S. Typhi to abate immune recognition by the RAC1-NOD1/2-RIP2 signaling pathway. To test this hypothesis, we abrogated RAC1 and RIP2 signaling by either ectopically expressing a dominant negative form of RAC1 (RAC1-DN) [30], [55] or by treating cells with the RIP2 inhibitor (SB203580) (Fig. 4). Consistent with previous reports, ectopic expression of a GFP-SopE fusion protein alone was sufficient to induce NF-κB activation while no upregulation of this signaling pathway was observed with a GFP-SopE construct lacking GEF activity (GFP-SopE G168A) [30], [56]. Simultaneous expression of the GFP-SopE fusion protein and a RAC1-DN construct abrogated NF-κB signaling (Fig. 4A).
Figure 4. In the absence of tviA, S. Typhi induces NF-κB activation in a RAC1- and RIP2-dependent manner.
(A and B) HeLa cells permanently transfected with a NF-κB luciferase reporter system (HeLa 57A) were transfected with pCMV-myc (vector, black bars) or pRAC1-DN (RAC1-DN, white bars) (A) Cells were transfected with the indicated GFP-SopE constructs or infected with the indicated S. Typhi strains for 5 h (N = 3). (B) Cells were mock-treated, infected with the indicated S. Typhi trains, or treated with the NOD1 agonist C12-iE-DAP (an acylated derivative of γ-D-Glu-mDAP) for 5 h. (C) HeLa 57A cells were either treated with dimethyl sulfoxide (DMSO) or RIP2 inhibitor SB203580 dissolved in DMSO and subsequently infected with the indicated S. Typhi strains or were mock treated (bacterial growth media alone) (N = 3). Bars represent geometric means ± standard error. *, P<0.05; **, P<0.01; ns, not statistically significant.
Infection of HeLa cells with the S. Typhi wild type or the T3SS-1-deficient viaB invA mutant did not result in a statistically significant increase in NF-κB activation and abrogation of RAC1 or RIP2 signaling did not further impact signaling (Fig. 4A, B, and C). In marked contrast, infection with the S. Typhi viaB mutant led to a substantial upregulation of NF-κB-driven responses. Abrogation of RAC1 or RIP2 activity significantly blunted the induction of NF-κB responses in cells infected with the viaB mutant. Moreover, NF-κB activation in cells infected with a viaB sopB sipA mutant was inhibited when cells were transfected with a plasmid construct encoding RAC1-DN (Fig. 4C), suggesting that SopE, translocated into host cells in the absence of TviA, could activate NF-κB signaling in a RAC1-dependent manner. Treatment with the RIP2 inhibitor did not impact T3SS-1-mediated invasion of S. Typhi strains towards epithelial cells (Fig. S5), excluding the possibility that the RIP2 inhibitor inadvertently interfered with the function of the T3SS-1 machinery. Collectively, these data supported the idea that TviA restricts activation of the RAC1-NOD1/2-RIP2 signaling pathway in S. Typhi-infected epithelial cells.
Heterologous expression of TviA in S. Typhimurium blunts T3SS-1-dependent responses in vivo
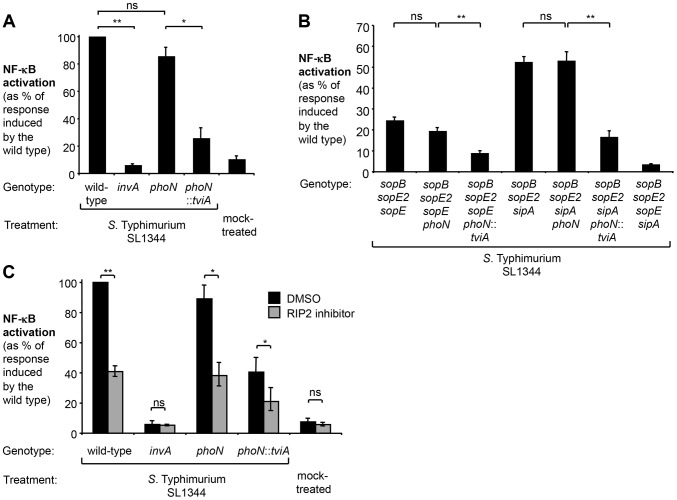
In addition to repressing T3SS-1 genes, TviA also suppresses flagella expression (Fig. S2A) [57]. Flagellin is known to induce pro-inflammatory responses by activating TLR5 [58] and the NLRC4- (nucleotide-binding oligomerization domain [NOD]-like receptor [NLR] family caspase-associated recruitment domain [CARD]-containing protein 4-) inflammasome [59], [60]. While our initial experiments in the bovine ligated ileal loop model suggest that TviA could mitigate mucosal inflammation (Fig. 2), it is conceivable that TviA-mediated gene regulation of flagellar biosynthesis could have affected flagellin-dependent innate immune pathways. To better study consequences of the expression of TviA on the RAC1-NOD1/2-RIP2 signaling pathway in an animal model, we therefore generated a phoN::tviA mutant in the S. Typhimurium SL1344 background (SW760). Akin to the findings with S. Typhi, expression of TviA in S. Typhimurium reduced transcription of T3SS-1 genes (Fig. S3) and the phoN::tviA mutant elicited significantly less (P<0.05) NF-κB activation than the phoN control strain (Fig. 5A and S6). We next introduced the tviA gene into SL1344 derivatives that only expressed the most potent inducers of the NF-κB pathway, SipA (sopE sopE2 sopB phoN::tviA mutant; SW809) and SopE (sopE2 sopB sipA phoN::tviA mutant; SW807). Upon infection of HeLa cells (Fig. 5B), strains carrying the phoN::tviA insertion elicited significantly less luciferase activity than the respective phoN mutants (P<0.01), indicating that TviA is able to reduce the NF-κB activation elicited by the S. Typhimurium orthologues of SopE and SipA. Inhibition of RIP2 significantly reduced NF-κB activation levels induced by the wild-type strain or the phoN mutant (Fig. 5C). The modest response induced by the phoN::tviA mutant was further blunted by inhibition of RIP2 signaling (P<0.05) (Fig. 5C), suggesting that TviA-mediated regulation of T3SS-1 is partially able to avoid induction of the NOD1/2-RIP2 pathway in vitro.
Figure 5. Expression of TviA in S. Typhimurium SL1344 reduces T3SS-1-driven NF-κB activation in epithelial cells.
HeLa 57A cells were infected with S. Typhimurium or mock treated with bacterial growth media (mock treatment) and luciferase activity determined 5 h after infection (N = 4). (A) The SL1344 wild type, an invA mutant (SW767), a phoN mutant (SW759), and a phoN::tviA mutant (SW760) were used to infect monolayers of HeLa 57A cells. (B) Cells were infected with the SL1344 wild type, an isogenic sopB sopE2 sopE sipA mutant (SW868), a sopB sopE2 sopE mutant (SW940), a sopB sopE2 sipA mutant (SW867), and derivatives thereof carrying a phoN (SW808; SW806) or a phoN::tviA (SW809; SW807) mutation, respectively. (C) Prior to infection with the indicated S. Typhimurium strains, cells were either treated with DMSO or the RIP2 inhibitor (SB203580) dissolved in DMSO. Bars represent geometric means ± standard error. *, P<0.05; **, P<0.01; ns, not statistically significant.
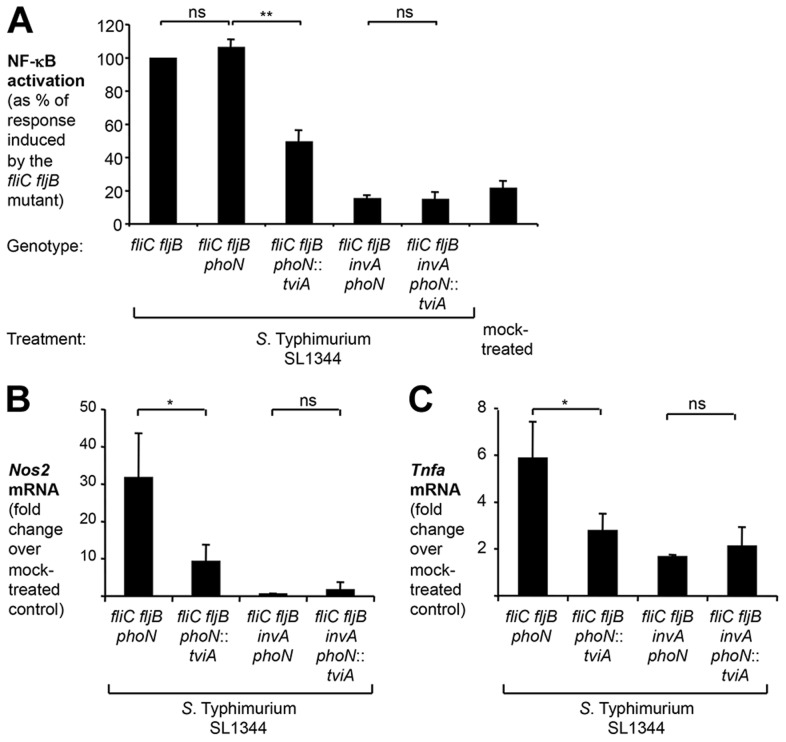
To exclude any effects of TviA on flagellin-dependent pathways, we introduced the phoN::tviA mutation into a non-motile S. Typhimurium strain lacking phase 1 and 2 flagellins, FliC and FljB (fliC fljB mutant, SW762) (Fig. 6A). Both the fliC fljB mutant and the fliC fljB phoN mutant (SW793) elicited significant levels of NF-κB activation in cultured epithelial cells (Fig. 6A), while this response was greatly reduced in cells infected with the fliC fljB phoN::tviA mutant (SW764). Inactivation of the essential T3SS-1 gene invA completely abolished the ability to induce NF-κB signaling (Fig. 6A).
Figure 6. TviA reduces T3SS-1-dependent, flagellin-independent inflammatory responses in the cecal mucosa.
(A) HeLa 57A cells were infected with an aflagellate SL1344 fliC fljB mutant (SW762), a fliC fljB phoN mutant (SW793), a fliC fljB phoN::tviA mutant (SW764) and derivatives carrying an additional mutation in invA (SW794; SW766). NF-κB activation was determined as described above. (B and C) Groups of streptomycin-pretreated C57BL/6 mice were intragastrically inoculated with either a S. Typhimurium SL1344 fliC fljB phoN mutant (N = 8), a fliC fljB phoN::tviA mutant (N = 8), a fliC fljB phoN invA mutant (N = 4), a fliC fljB phoN::tviA invA mutant (N = 4), or LB broth (mock-treated; N = 3). 12 h after infection, the relative abundance of Nos2 (B) and Tnfa (C) mRNA was determined by qRT-PCR. Bars represent geometric means ± standard error. *, P<0.05; **, P<0.01; ns, not statistically significant.
To directly assess the ability of TviA to impede inflammatory processes in the intestinal mucosa, we used the Streptomycin pre-treated mouse model [61]. In this model, detection of cytosolic access by the S. Typhimurium T3SS-1 through the NOD1/2 signaling pathway contributes to intestinal inflammation early during infection [30], [34], [62]. Compared to mock infected mice, transcript levels of the pro-inflammatory genes Nos2, encoding inducible nitric oxide synthase (iNOS), and Tnfa, encoding tumor necrosis factor (TNF)-α, were significantly (P<0.05) elevated in the cecal mucosa at 12 hours after infection with a non-flagellated S. Typhimurium phoN fliC fljB mutant (SW793) (Fig. 6B and C). Introduction of the S. Typhi tviA gene into a S. Typhimurium fliC fljB mutant (phoN::tviA fliC fljB mutant, SW764) significantly (P<0.05) reduced pro-inflammatory gene expression (Fig. 6B and 6C), but not bacterial numbers recovered from intestinal contents or Peyer's patches (Fig. S7). Inflammatory responses observed in the cecal mucosa at this early time point were T3SS-1-dependent, because introduction of a mutation in invA abrogated the ability of S. Typhimurium to elicit pro-inflammatory gene expression. Collectively, these data suggested that TviA represses T3SS-1-dependent, early inflammatory responses in vivo through a flagellin-independent mechanism.
Discussion
S. Typhi invades the intestinal mucosa without triggering the massive neutrophil influx observed during gastroenteritis caused by non-typhoidal serovars. Here we show that one mechanism for attenuating host responses is a TviA-mediated repression of T3SS-1, a virulence factor known to induce potent inflammatory host responses. Effector molecules translocated by the T3SS-1 into the host cell cytosol activate Rho-family GTPases [15]–[17]. The activation of Rho-family GTPases is a pathogen-induced process that is sensed by NOD1 [21], [30], which ultimately results in the activation of pro-inflammatory responses in vitro [15], [20], [54] and in vivo [30], [62]. However, S. Typhi requires a functional T3SS-1 to invade the intestinal epithelium during infection [27]. Our data suggest that S. Typhi might have evolved to invade the intestinal epithelium without inducing a potent antibacterial inflammatory response by regulating T3SS-1 expression in a TviA-dependent manner. Osmoregulation prevents expression of TviA in the intestinal lumen, which renders S. Typhi invasive [42] (Fig. 2A). However, TviA expression is rapidly upregulated upon entry into tissue [63], resulting in repression of T3SS-1 and flagella expression while biosynthesis of the Vi capsule is induced [42], [44]. Here we show that TviA prevented NF-κB activation in epithelial cells by reducing T3SS-1-dependent activation of RAC1. Furthermore, the TviA-mediated reduction of T3SS-1-dependent inflammatory responses elicited at early time points in animal models was independent of flagella and the Vi capsule. These data support the hypothesis that TviA attenuates inflammation because it rapidly turns off T3SS-1 expression upon entry into tissue, thereby concealing a pathogen-induced process from the host.
Bovine ligated ileal loops are suited to model the initial 12 hours of host pathogen interaction, a time period during which inflammatory responses are largely T3SS-1-dependent [22], [23], [64]. Similarly, in the mouse colitis model, inflammatory responses elicited in the cecum at early time points (i.e. during the first 2 days) after infection are largely T3SS-1-dependent [61], [65], [66]. However, mechanisms independent of T3SS-1 are responsible for cecal inflammation observed at later time points (i.e. at days 4 and 5 after infection) in the mouse colitis model [65]. Expression of the S. Typhi Vi capsular polysaccharide in S. Typhimurium leads to an attenuation of these T3SS-1-independent inflammatory responses in the mouse colitis model [41], by reducing complement activation and TLR4 signaling [67], [68]. Thus the viaB locus reduces intestinal inflammation by multiple different mechanisms (Fig. S2A). A TviA-mediated repression of T3SS-1 reduces early inflammatory responses while the Vi capsular polysaccharide attenuates responses generated through T3SS-1-independent mechanisms at later time points. It is tempting to speculate that the result of these immune evasion mechanisms is a reduction in the intestinal inflammatory response that could contribute to differences in disease symptoms caused by typhoidal and non-typhoidal serotypes.
Materials and Methods
Ethics statement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol on mouse experiments was approved by the Institutional Animal Care and Use Committee of the University of California, Davis (Permit Number: 16179). The protocol on calf experiments was approved by the Institutional Committee at the Universidade Federal de Minas Gerais, Brazil (Permit Number: CETEA 197/2008).
Bacterial strains and culture conditions
The bacterial strains, including relevant properties, are listed in table 1. Unless noted otherwise, bacteria were aerobically grown at 37°C in Luria-Bertani (LB) broth (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl) or LB agar (15 g/l agar). To induce expression of tviA and Vi capsule biosynthesis genes, an overnight culture in LB broth was diluted 1∶50 in tryptone yeast extract (TYE) broth (10 g/l tryptone, 5 g/l yeast extract) or Dulbecco's modified Eagle's medium (DMEM) as indicated and incubated aerobically at 37°C for 3 h. When appropriate, antibiotics were added to LB broth cultures or LB agar plates at the following concentrations: carbenicillin (0.1 mg/ml), chloramphenicol (0.03 mg/ml), kanamycin (0.05 mg/ml), nalidixic acid (0.05 mg/ml), and tetracycline (0.01 mg/ml).
Table 1. Bacterial strains and plasmids used in this study.
| Strain designation | Relevant characteristicsa/Genotype | Source/Reference |
| S. Typhi | ||
| Ty2 | Wild-type strain, Vi+ | ATCC 700931b |
| STY2 | Ty2 ΔviaB::KanR | [13] |
| STY4 | Ty2 ΔviaB::KanR invA::pINV5 (CmR) | [13] |
| SW74 | Ty2 ΔtviB-vexE::CmR, Vi− | [57] |
| SW222 | Ty2 invA::pINV5 (CmR) | [57] |
| SW347 | Ty2 ΔviaB, Vi− | [42] |
| SW359 | Ty2 ΔfliC | [42] |
| SW398 | Ty2 ΔviaB ΔfliC invA::pSW127 | This study |
| SW483 | Ty2 ΔviaB ΔfliC | [42] |
| SW611 | Ty2 ΔtviB-vexE::CmR invA::pSW127 | This study |
| SW904 | Ty2 ΔtviB-vexE, Vi− | This study |
| SW1207 | Ty2 ΔviaB sopB::MudJ | This study |
| SW1208 | Ty2 tviB-vexE sopB::MudJ | This study |
| SW1209 | Ty2 ΔviaB ΔsopE | This study |
| SW1210 | Ty2 tviB-vexE ΔsopE | This study |
| SW1211 | Ty2 ΔviaB sopB::MudJ ΔsipA | This study |
| SW1212 | Ty2 ΔtviB-vexE sopB::MudJ ΔsipA | This study |
| SW1213 | Ty2 ΔtviB-vexE sopB::MudJ ΔsopE | This study |
| SW1214 | Ty2 ΔviaB ΔsopEΔsipA | This study |
| SW1215 | Ty2 ΔtviB-vexE ΔsopEΔsipA | This study |
| SW1216 | Ty2 ΔviaB sopB::MudJ ΔsopE | This study |
| SW1217 | Ty2 ΔviaB sopB::MudJ ΔsipA ΔsopE | This study |
| S. Typhimurium | ||
| AJB715 | IR715 phoN::KanR | [73] |
| CS019 | ATCC14028 phoN::Tn10dCm | [74] |
| IR715 | ATCC14028 NalR | [75] |
| SL1344 | StrepR | [76] |
| SPN305 | IR715 ΔfliC::pSPN29 | [42] |
| SW284 | IR715 phoN: CmR | [42] |
| SW399 | IR715 invA::pSW127 | [71] |
| SW474 | IR715 phoN::tviA-CmR | [42] |
| SW562 | IR715 ΔinvA::TetR | [77] |
| SW737 | IR715 phoN::KanR ΔinvA::TetR | This study |
| SW751 | IR715 phoN::pSW208 | This study |
| SW759 | SL1344 phoN::CmR | This study |
| SW760 | SL1344 phoN::tviA-CmR | This study |
| SW761 | SL1344 ΔfliC | This study |
| SW762 | SL1344 ΔfliC fljB5001::MudJ | This study |
| SW764 | SL1344 ΔfliC fljB5001::MudJ phoN::tviA-CmR | This study |
| SW766 | SL1344 ΔfliC fljB5001::MudJ phoN::tviA-CmR ΔinvA::TetR | This study |
| SW767 | SL1344 ΔinvA::TetR | This study |
| SW793 | SL1344 ΔfliC fljB5001::MudJ phoN::pSW208 | This study |
| SW794 | SL1344 ΔfliC fljB5001::MudJ phoN::pSW208 ΔinvA::TetR | This study |
| SW798 | SL1344 sopB::MudJ | [30] |
| SW800 | SL1344 sopE2::pSB1039 | [30] |
| SW806 | SL1344 ΔsipA sopB::MudJ sopE2::pSB1039 phoN::Tn10dCm | This study |
| SW807 | SL1344 ΔsipA sopB::MudJ sopE2::pSB1039 phoN::tviA-CmR | This study |
| SW808 | SL1344 ΔsopE sopB::MudJ sopE2::pSB1039 phoN::Tn10dCm | This study |
| SW809 | SL1344 ΔsopE sopB::MudJ sopE2::pSB1039 phoN::tviA-CmR | This study |
| SW839 | SL1344 ΔsipA::pSW244 | This study |
| SW867 | SL1344 ΔsipA sopB::MudJ sopE2::pSB1039 | [30] |
| SW868 | SL1344 ΔsipAΔsopE sopB::MudJ sopE2::pSB1039 | [30] |
| SW940 | SL1344 ΔsopE sopB::MudJ sopE2::pSB1039 | This study |
| SW972 | SL1344ΔsipA ΔsopE sopE2::pSB1039 | This study |
| SW973 | SL1344 ΔsipA ΔsopE sopB::MudJ | [30] |
| SW974 | IR715 ΔsipA::pSW244 | [30] |
| SW976 | SL1344 ΔsopE | [78] |
| SW977 | SL1344 ΔsopE::pSW245 | This study |
| SW1009 | SL1344ΔsipA ΔsopE | [30] |
| TH170 | IR715 phoN::viaB | [41] |
| E. coli | ||
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrepR) endA1 nupG | Life Technologies |
| DH5α λpir | F− endA1 hsdR17 (r−m+) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 Φ80lacZ Δ M15 λpir | Laboratory strain collection |
| S17-1 λpir | recA1 thi pro hsdR (r−m+) zxx::RP4 2-(TetR::Mu) (KanR::Tn7) λpir | [79] |
CmR: Chloramphenicol resistance; KanR: Kanamycin resistance; NalR: Nalidixic acid resistance; StrepR: Streptomycin resistance; TetR: Tetracycline resistance (tetRA).
American Type Culture Collection, Manassas, VA.
Construction of plasmids
Standard cloning techniques were performed to generate the plasmids listed in table 2. Cloning vectors and ori(R6K)-based suicide plasmids were routinely maintained in E. coli TOP10 and DH5α λpir, respectively.
Table 2. Plasmids used in this study.
| Plasmid designation | Relevant characteristicsa/Genotype | Source/Reference |
| pCMV-myc | ori(pMB1) bla P CMVIE myc-tag | Clontech |
| pCR2.1 | Cloning vector | Life Technologies |
| pEP185.2 | ori(R6K) mobRP4 cat | [80] |
| pEGFP-C1 | ori(pMB1) KanR P CMVIE EGFP | Clontech |
| pGFP-SopE | sopE cloned into pEGFP-C1; N-terminal GFP tag | [30] |
| pGFP-SopE-G168A | G168A amino acid substitution in SopE in pEGFP-C1; N-terminal GFP tag | [30] |
| pNFkB-luc | NF-κB -responsive luciferase reporter plasmid | [81] |
| pRAC1-DN | Dominant-negative form of hRAC1 cloned into pCMV-myc; T17N amino acid substitution; N-terminal myc tag. | [30] |
| pRDH10 | ori(R6K) mobRP4 cat tetC sacRB | [82] |
| pRI203 | Y. pseudotuberculosis invasin gene in pREG153 | [83] |
| pSW28 | Upstream and downstream regions of the tviBCDEvexABCDE region of S. Typhi Ty2 in pGP704 | [57] |
| pSW208 | Internal fragment of the S. Typhimurium phoN gene cloned into pEP185.2 | This study |
| pSW233 | Upstream and downstream regions of the tviBCDEvexABCDE region of S. Typhi Ty2 in pRDH10 | This study |
| pSW245 | Upstream and downstream region of the S. Typhimurium sopE gene in pRDH10 | [78] |
| pTK-LacZ | Normalization of transfection efficiency | [84] |
| pTVIA1 | tviA under control of its native promoter in pWSK29 | [38] |
| pWSK29 | ori(pSC101) bla | [85] |
An internal fragment of the phoN coding sequence was PCR amplified from the S. Typhimurium IR715 chromosome using the primers listed in table 3, subcloned into pCR2.1 (TOPO TA cloning kit, Life Technologies), and cloned into pEP185.2 utilizing the unique XbaI and SacI restriction sites to give rise to pSW208. To generate pSW233, pSW28 was digested with EcoRI and the DNA fragment comprising the joint upstream- and downstream regions of the tviB and vexE genes, respectively, was cloned into the EcoRI site of pRDH10.
Table 3. Primers used in this study.
| Target | Sequencec | Reference |
| Mutagenesis | ||
| S. Typhimurium phoN | 5′-TCTAGACGATGGAAACAAGCTGC-3′ | This study |
| 5′-GAGCTCTACTAATGCCAGAAGTGT-3′ | ||
| Real time PCR | ||
| Salmonella gmk | 5′-TTGGCAGGGAGGCGTTT-3′ | [86] |
| 5′-GCGCGAAGTGCCGTAGTAAT-3′ | ||
| Salmonella flhD | 5′-ACAGCGTTTGATCGTCCAG-3′ | [63] |
| 5′-GTTTGCCATCTCTTCGTTGA-3′ | ||
| Salmonella hilA | 5′-ATTAAGGCGACAGAGCTGGA-3′ | [42] |
| 5′-GAATAGCAAACTCCCGACGA-3′ | ||
| Salmonella invF | 5′-GTTGTCGCACCAGTATCAGG-3′ | This study |
| 5′-TCGGATTCAGCATATGTCGT-3′ | ||
| Salmonella prgH | 5′-CACTGAACGGCTGTGAGTTT-3′ | This study |
| 5′-CGGCAGGTATATCAGGGAGT-3′ | ||
| Salmonella sipA | 5′-TTCAAATAATGTCGCCGGTA-3′ | This study |
| 5′-TTCATCAGTAGCGTCTTCGC-3′ | ||
| Salmonella sopE | 5′-CAACACACTTTCACCGAGGA-3′ | This study |
| 5′-ATCATTGAGCGTTTGAAGCA-3′ | ||
| Murine Gapdh | 5′-TGTAGACCATGTAGTTGAGGTCA-3′ | [87] |
| 5′-AGGTCGGTGTGAACGGATTTG-3′ | ||
| Murine Tnfa | 5′-AGCCAGGAGGGAGAACAGAAAC-3′ | [68] |
| 5′-CCAGTGAGTGAAAGGGACAGAACC-3′ | ||
| Murine Nos2 | 5′-TTGGGTCTTGTTCACTCCACGG-3′ | [88] |
| 5′-CCTCTTTCAGGTCACTTTGGTAGG-3′ | ||
restriction endonuclease cleavage sites are underlined.
Generation of mutants by allelic exchange
Plasmids were introduced into S17-1 λpir and conjugation performed as described previously [57]. The unmarked S. Typhi ΔtviB-vexE mutant SW904 was constructed by inserting the plasmid pSW233 into the STY2 mutant chromosome, selecting for single crossover events (creating merodiploids) on LB agar plates containing Cm and Kan. Sucrose selection was performed as described previously [69] to select for a second crossover event, thus effectively deleting the tviBCDEvexABCDE genes, yielding SW904. The deletion was confirmed by PCR. To facilitate transduction of the unmarked ΔsopE mutation, pSW245 was introduced in this locus in the SW976 chromosome by conjugation with S17-1 λpir as the donor strain, creating SW977 as an intermediate.
Construction of mutants by P22-mediated generalized phage transduction
Phage P22 HT int-105 was utilized for generalized phage transduction in S. Typhimurium as described previously [70]. For S. Typhi recipients, a similar protocol was followed except the multiplicity of infection (MOI) was increased to 100.
A phage lysate of SW399 was used to transduce the invA::pSW127 mutation into SW483 and SW74, thus generating the S. Typhi ΔviaB ΔfliC invA mutant (SW398) and the ΔtviB-vexE invA mutant (SW611). SW1207 and SW1208 were created by transducing the sopB::MudJ mutation from SW798 into the ΔviaB mutant (SW347) and the ΔtviB-vexE mutant (SW904), respectively. The S. Typhi ΔviaB ΔsopE (SW1209), ΔtviB-vexE ΔsopE (SW1210), ΔviaB sopB::MudJ ΔsopE (SW1216), and ΔtviB-vexE sopB::MudJ ΔsopE (SW1213) mutants were constructed by transducing the ΔsopE::pSW245 mutation from SW977 into SW347, SW904, SW1207, and SW1208, respectively. Subsequent sucrose selection allowed selecting for mutants that had lost the plasmid by allelic exchange and generated a clean ΔsopE mutation, thus creating SW1209, SW1211, SW1216, and SW1213, respectively. Similarly, a P22 lysate of SW839 was used to transduce the ΔsipA::pSW244 mutation (SW839) into SW1207, SW1208, SW1209, and SW1210. The intermediates were subjected to sucrose selection, thus creating the clean ΔsipA mutation of strains SW1211, SW1212, SW1214, and SW1215, respectively. The ΔviaB sopB::MudJ ΔsipA ΔsopE mutant (SW1217) was generated through transduction of the ΔsopE::pSW245 mutation from SW977 into the SW1211 chromosome and sucrose selection.
The S. Typhimurium SL1344 derivatives SW759 and SW760 were established by transducing the phoN::CmR and phoN::tviA-CmR mutations from SW284 and SW474 into the SL1344 wild type. Transduction of the ΔfliC::pSPN29 from SPN305 into the SL1344 wild type and subsequent sucrose selection gave rise to the SL1344 fliC deletion mutant SW761. Subsequent introduction of the fljB5001::MudJ into this strain led to the SL1344 ΔfliC fljB5001::MudJ mutant (SW762). To construct SW764 and SW793, the phoN::tviA-CmR (SW474) and phoN::pSW208 (SW751) mutations were transduced separately into SW762. Invasion-deficient derivatives of these strains were generated by transducing the invA::TetR mutation from SW562 into SW764, SW793, and SL1344, thus creating strains SW766, SW794, and SW767, respectively. SW806, SW807, SW808, and SW809 were generated by transducing the phoN::Tn10dCm (CS019) or phoN::tviA-CmR (SW474) into SW867 or SW940. The ΔsipA::pSW244 mutation (SW974) was moved into the SL1344 wild type to create SW839. SW940 was established by transduction of the sopB::MudJ mutation (SW798) into SW976 and subsequent introduction of the sopE2::pSB1039 mutation (SW800). A P22 phage lysate of SW800 was used to create SW972 using SW1009 as the recipient strain. The phoN::KanR mutation from AJB715 was transduced into SW562 to give rise to the phoN::KanR invA::TetR mutant (SW737).
Tissue culture experiments
HeLa 57A cells [29], [34] were generously provided by R. T. Hay (the Wellcome Trust Centre for Gene Regulation and Expression, College of Life Sciences, University of Dundee, United Kingdom). HEK-293 cells were obtained from ATCC (ATCC CRL-1573). Both cells lines were routinely cultured at 37°C in a 5% CO2 atmosphere in DMEM containing 10% fetal bovine serum (FBS) (Life Technologies). For NF-κB activation and invasion experiments, cells were seeded in 24-well plates and 48-well plates (Corning) at densities of 1×105 cells/well and 2×105 cells/well, respectively, and incubated for 24 h prior to subsequent experiments.
Measurement of NF-κB activation in epithelial cells
S. Typhi and S. Typhimurium strains were pre-cultured in TYE broth as described above. HeLa 57A cells or HEK-293 cells transfected with a NF-κB -luciferase reporter construct were infected with the indicated strains at a final concentration of approximately 106 colony forming units (CFU)/ml. To synchronize the infection, plates were centrifuged for 5 min at 500 g at room temperature. After 3 h, cells were washed with DPBS and incubated at 37°C for an additional 2 h in the presence of DMEM containing 10% FBS. Cells were washed in DPBS, lysed in 0.1 ml of reporter lysis buffer (Promega), and firefly luciferase activity was measured using the luciferase assay system (Promega) in a FilterMax3 microplate reader (Molecular Devices). Results are expressed as percentage of maximum signal elicited in each individual assay. In some experiments, cells were treated 30 min prior to infection until the end of the experiment with either DMSO (vehicle control) or the RIP2-inhibitor SB203580 at a final concentration of 10 µM dissolved DMSO. The NOD1 agonist C12-iE-DAP (Invivogen) was added a final concentration of 100 ng/ml.
For transfection assays [34], HeLa 57A cells were grown to a confluency of about 60% and transiently transfected with a total of 250 ng of plasmid DNA, consisting of 50 ng of the β-galactosidase-encoding vector pTK-LacZ, and either 200 ng of pCMV-myc (control vector) or 100 ng pRAC1-DN and 100 ng of control vector. For co-transfection with pGFP-SopE constructs, 50 ng of pTK-LacZ, 10 ng of the pGFP-SopE plasmid, 90 ng of pEGFP (empty vector), and 100 ng of either pCMV-myc or pRAC1-DN was added. HEK-293 cells were transfected with 25 ng of pTK-LacZ and 25 ng of pNFkB-luc. 48 h after transfection, cells were infected with the indicated Salmonella strains or mock-treated (LB broth) as described above. Efficiency of transfection was normalized by adjusting luciferase values to β-galactosidase values.
Invasion assays
Invasiveness of the indicated Salmonella strains was determined using a Gentamicin protection assay as described previously [71]. Briefly, HeLa 57A cells were infected at a MOI of 5 with Salmonella strains pre-cultured in TYE broth. After 1 h, cells were washed and media containing 0.1 mg/ml Gentamicin was added for 90 min. Diluted cell lysates (0.5% Triton-X-100) were spread on LB agar plates to determine the number of CFU per well. Invasiveness was calculated as percentage of recovered bacteria compared to the inoculum.
Bacterial gene expression analysis
Overnight cultures of the indicated S. Typhi and S. Typhimurium strains were diluted 1∶50 in TYE broth and incubated at 37°C for 3 h. Total RNA was extracted from approximately 2×109 CFU using the Aurum Total RNA Mini Kit (Biorad). 1 µg of total RNA was subjected to an additional DNase treatment (DNA-free kit, Life Technologies) and converted to cDNA using MuLV reverse transcriptase (Life Technologies) in a 25 µl volume as described previously [71]. 4 µl of this cDNA was used as the template for real time PCR analysis with the primers listed in table 3. Data was acquired on a ViiA 7 real-time PCR instrument (Life Technologies). Relative target gene expression was normalized to mRNA levels of the house keeping gene gmk, encoding guanylate kinase (ΔΔCt method). DNA contamination was less than 1% for all amplicons as determined by a separate RT-PCR mock reaction lacking reverse transcriptase.
Bovine ligated ileal loop model
Salmonella Typhimurium was cultured in LB broth at 37°C under agitation, followed by subculture in fresh LB (without antibiotics) for 3 hours, at 37°C under agitation. Four 3–4 week-old male healthy Salmonella-free Holstein calves were used in this study. Ligated ileal loops were surgically prepared as previously described [23]. Ligated loops were mock treated with intraluminal injection of sterile LB broth or inoculated with 3 ml of suspensions containing 1×108 CFU of the S. Typhimurium ATCC14028 phoN mutant (AJB715), a phoN::viaB mutant (TH170), a phoN::tviA mutant (SW474), or a phoN invA mutant (SW737). Ligated loops were surgically removed at 5 h after infection for tissue sampling and measurement of intraluminal fluid accumulation. Samples containing the intestinal mucosa and the associated lymphoid tissue were collected with a 6 mm biopsy punch. Each intestinal biopsy was kept in sterile PBS with 50 µg/ml of gentamicin for 1 h, homogenized in 2 ml of PBS, serially diluted, and plated on LB agar plates containing nalidixic acid. Additional biopsies were fixed by immersion in 10% buffered formalin, processed for paraffin embedding, cut and stained with hematoxylin and eosin. Histopathologic changes including hemorrhage, neutrophilic infiltration, edema, and necrosis and/or apoptosis were scored from 0 to 3 (0 for absence of lesions, and 1, 2, or 3 for mild, moderate, or severe lesion, respectively) for a combined total score ranging from 0 to 12.
Mouse colitis model
Animals were obtained from The Jackson Laboratory (Bar Harbor), housed under specific-pathogen-free conditions and provided with water and food ad libitum. Groups of female, 9–12 week old C57BL/6 mice were orally treated with 20 mg Streptomycin. After 24 h, these mice were inoculated as described previously [61] with either 0.1 ml LB broth (mock treatment) or 1×109 CFU of the S. Typhimurium SL1344 fliC fljB phoN mutant (SW793), the fliC fljB phoN::tviA mutant (SW764), the fliC fljB phoN invA mutant (SW794), or the fliC fljB phoN::tviA invA mutant (SW766) suspended in 0.1 ml LB broth. 12 h after infection, animals were euthanized and tissues were collected. The bacterial load was determined by spreading serial 10-fold dilutions of homogenates on LB agar plates containing the appropriate antibiotics. Flash-frozen cecal tissue was homogenized in a Mini-beadbeater (Biospec Products) and RNA was extracted by the TRI reagent method (Molecular Research Center). cDNA was generated using MuLV reverse transcriptase and reverse transcription reagents (Life Technologies). SYBR Green (Life Technologies)-based real-time PCR was performed as described previously [72] using the primers listed in table 3. Data was acquired by a ViiA 7 real-time PCR system (Life Technologies) and analyzed using the comparative Ct method (ΔΔCt method). Murine target gene transcription within each sample was normalized to the respective levels of Gapdh mRNA.
Statistical analysis
Data obtained from tissue culture experiments, bacterial gene transcription experiments, and the bovine ligated ileal loop model was log-transformed prior to analysis with a paired Student's t-test. To determine statistical significance for relative mucosal mRNA transcription and tissue bacterial load between treatment groups, an unpaired Student's t-test was employed.
Supporting Information
TviA reduces T3SS-1-induced inflammatory responses independent of bacterial entry into host cells. (A) Genetic organization of the viaB operon in S. Typhi Ty2. (B) The S. Typhi Ty2 wild-type strain, a ΔviaB mutant (SW347), a ΔtviB-vexE mutant (SW74), a ΔviaB invA mutant (STY4), and a ΔtviB-vexE invA mutant (SW611) cultured in DMEM were used to infect HeLa57 cells. NF-κB activation was determined after 5 h (N = 4). (C and D) HeLa 57A cells were infected with the S. Typhi Ty2 wild-type strain, a ΔviaB mutant (SW347), a ΔviaB invA mutant (STY4), or a ΔviaB invA mutant harboring pRI203 (N = 3) precultured in TYE broth. (C) Cells were infected at a multiplicity of infection of 5 for 1 h and extracellular bacteria killed by treatment with Gentamicin for 90 min. Recovered bacterial numbers were standardized to the number of the bacteria in the inoculum. (D) To determine NF-κB activation, luciferase activity measured 5 h after infection (N = 4). Bars represent geometric means ± standard error. **, P<0.01; ns, not statistically significant.
(TIF)
TviA reduces T3SS-1-induced NF-κB activation independent of flagellin expression. (A) Schematic representation of the TviA regulatory network in S. Typhi and effect on host signaling pathways. (B and C) HeLa 57A cells (B) or HEK-293 cells transiently transfected with a NF-κB-dependent reporter plasmid (pNFkB-luc) (C) were infected with a S. Typhi ΔfliC mutant (SW359), a ΔfliC ΔviaB mutant (SW483), derivatives carrying the cloning plasmids pWSK29 (pWSK) or the plasmid pTVIA1, and a ΔfliC ΔviaB invA mutant (SW398). Luciferase activity was quantified 5 h after infection to determine NF-κB activation levels (N = 3). Bars represent geometric means ± standard error. *, P<0.05; **, P<0.01; ***, P<0.001.
(TIF)
Effect of TviA on bacterial gene expression in vitro . The S. Typhi wild type Ty2 (WT), a ΔviaB mutant (SW347), a ΔtviB-vexE mutant (SW74), the S. Typhimurium wild-type SL1344, a SL1344 phoN mutant (SW759), a SL1344 phoN::tviA mutant (SW760), the S. Typhimurium 14028 NalR wild type (IR715), a 14028 NalR phoN mutant (AJB715), and a 14028 NalR phoN::tviA (SW474) were cultured in TYE broth for 3 h. RNA was extracted and qRT-PCR performed to determine the relative abundance of flhD (A), hilA (B), invF (C), prgH (D), sipA (E), and sopE (F) mRNA. Data presented is fold change over the abundance of mRNA recovered from the respective wild-type strain after standardization to the housekeeping gene gmk. The dotted line indicates no change in gene expression. Bars represent geometric means from 3 (S. Typhimurium) or 4 (S. Typhi) independent experiments ± standard error. *, P<0.05; **, P<0.01; ***, P<0.001; ns, not statistically significant.
(TIF)
Contribution of SopE, SipA, SopB, and SopE2 to NF-κB activation in human epithelial cells. HeLa 57A cells were treated with media only (mock treatment) or infected with the indicated S. Typhimurium SL1344 derivatives. Certain Salmonella strains lacked defined T3SS-1 effector proteins to analyze the responses induced by SopB (light grey bar), SipA (white bar), and SopE (dark grey bar). NF-κB activation was assessed 5 h after infection based on a NF-κB-driven luciferase reporter system (N = 4). Bars represent geometric means ± standard error. **, P<0.01; ns, not statistically significant.
(TIF)
Inhibition of RIP2 signaling does not affect invasiveness of S. Typhi strains. HeLa 57A cells pretreated with DMSO or RIP2 inhibitor (SB203580; dissolved in DMSO) were infected with the indicated S. Typhi strains at a MOI of 10 and invasion determined by a Gentamicin protection assay. Bars represent geometric means ± standard error. ns, not statistically significant.
(TIF)
Expression of TviA in S. Typhimurium 14028 NalR reduces T3SS-1-driven NF-κB activation in epithelial cells. HeLa 57A cells were infected with the S. Typhimurium 14028 NalR derivatives or treated with bacterial growth media (mock treatment). Luciferase activity determined 5 h after infection (N = 4). Bars represent geometric means ± standard error. **, P<0.01;
(TIF)
Bacterial colonization in the mouse colitis model. (A and B) Streptomycin-pretreated mice were infected with the indicated S. Typhimurium SL1344 derivatives as described in Figure 6. The bacterial load in the colon content (A) and the Peyer's patches (B) was determined 12 h after infection. Bars represent geometric means ± standard error. ns, not statistically significant.
(TIF)
Funding Statement
This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR12088-01 from the National Center for Research Resources, National Institutes of Health. This work was supported by American Heart Association Grant 12SDG12220022 to AMK, Public Health Service Grants AI044170 to AJB and AI103248 to SEW as well as a fellowship from the John Simon Guggenheim Memorial Foundation to RLS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vance RE, Isberg RR, Portnoy DA (2009) Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6: 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tukhvatulin AI, Gitlin II, Shcheblyakov DV, Artemicheva NM, Burdelya LG, et al. (2013) Combined stimulation of Toll-like receptor 5 and NOD1 strongly potentiates activity of NF-kappaB, resulting in enhanced innate immune reactions and resistance to Salmonella enterica serovar Typhimurium infection. Infect Immun 81: 3855–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Glynn JR, Palmer SR (1992) Incubation period, severity of disease, and infecting dose: evidence from a Salmonella outbreak. Am J Epidemiol 136: 1369–1377. [DOI] [PubMed] [Google Scholar]
- 4. Day DW, Mandal BK, Morson BC (1978) The rectal biopsy appearances in Salmonella colitis. Histopathology 2: 117–131. [DOI] [PubMed] [Google Scholar]
- 5. McGovern VJ, Slavutin LJ (1979) Pathology of salmonella colitis. Am J Surg Pathol 3: 483–490. [DOI] [PubMed] [Google Scholar]
- 6. Harris JC, Dupont HL, Hornick RB (1972) Fecal leukocytes in diarrheal illness. Ann Intern Med 76: 697–703. [DOI] [PubMed] [Google Scholar]
- 7. Alvarado T (1983) Faecal leucocytes in patients with infectious diarrhoea. Trans R Soc Trop Med Hyg 77: 316–320. [DOI] [PubMed] [Google Scholar]
- 8. Guyot J, Gonvers JJ, Pyndiah N, Heitz M (1984) [Value of fecal leukocyte studies in cases of acute diarrhea]. Schweiz Med Wochenschr 114: 634–636. [PubMed] [Google Scholar]
- 9. Sprinz H, Gangarosa EJ, Williams M, Hornick RB, Woodward TE (1966) Histopathology of the upper small intestines in typhoid fever. Biopsy study of experimental disease in man. Am J Dig Dis 11: 615–624. [DOI] [PubMed] [Google Scholar]
- 10. Mukawi TJ (1978) Histopathological study of typhoid perforation of the small intestines. Southeast Asian J Trop Med Public Health 9: 252–255. [PubMed] [Google Scholar]
- 11. Nguyen QC, Everest P, Tran TK, House D, Murch S, et al. (2004) A clinical, microbiological, and pathological study of intestinal perforation associated with typhoid fever. Clin Infect Dis 39: 61–67. [DOI] [PubMed] [Google Scholar]
- 12. McCormick BA, Miller SI, Carnes D, Madara JL (1995) Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect Immun 63: 2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, et al. (2005) The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun 73: 3367–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fu Y, Galan JE (1998) The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol 27: 359–368. [DOI] [PubMed] [Google Scholar]
- 15. Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE (1998) S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93: 815–826. [DOI] [PubMed] [Google Scholar]
- 16. Friebel A, Ilchmann H, Aepfelbacher M, Ehrbar K, Machleidt W, et al. (2001) SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell. J Biol Chem 276: 34035–34040. [DOI] [PubMed] [Google Scholar]
- 17. Zhou D, Chen LM, Hernandez L, Shears SB, Galan JE (2001) A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol Microbiol 39: 248–259. [DOI] [PubMed] [Google Scholar]
- 18. Patel JC, Galan JE (2006) Differential activation and function of Rho GTPases during Salmonella-host cell interactions. The Journal of cell biology 175: 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frances CL, Ryan TA, Jones BD, Smith SJ, Falkow S (1993) Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature 364: 639–642. [DOI] [PubMed] [Google Scholar]
- 20. Hobbie S, Chen LM, Davis RJ, Galan JE (1997) Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J Immunol 159: 5550–5559. [PubMed] [Google Scholar]
- 21. Keestra AM, Baumler AJ (2013) Detection of enteric pathogens by the nodosome. Trends Immunol 35: 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang S, Santos RL, Tsolis RM, Stender S, Hardt W-D, et al. (2002) SipA, SopA, SopB, SopD and SopE2 act in concert to induce diarrhea in calves infected with Salmonella enterica serotype Typhimurium. Infect Immun 70: 3843–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Santos RL, Tsolis RM, Zhang S, Ficht TA, Baumler AJ, et al. (2001) Salmonella-induced cell death is not required for enteritis in calves. Infect Immun 69: 4610–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsolis RM, Adams LG, Ficht TA, Baumler AJ (1999) Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67: 4879–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, et al. (2004) Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72: 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sekirov I, Gill N, Jogova M, Tam N, Robertson M, et al. (2010) Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut microbes 1: 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elsinghorst EA, Baron LS, Kopecko DJ (1989) Penetration of human intestinal epithelial cells by Salmonella: molecular cloning and expression of Salmonella typhi invasion determinants in Escherichia coli . Proc Natl Acad Sci USA 86: 5173–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raffatellu M, Sun YH, Wilson RP, Tran QT, Chessa D, et al. (2005) Host restriction of Salmonella enterica serotype Typhi is not caused by functional alteration of SipA, SopB, or SopD. Infect Immun 73: 7817–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodriguez MS, Thompson J, Hay RT, Dargemont C (1999) Nuclear retention of IkappaBalpha protects it from signal-induced degradation and inhibits nuclear factor kappaB transcriptional activation. J Biol Chem 274: 9108–9115. [DOI] [PubMed] [Google Scholar]
- 30. Keestra AM, Winter MG, Auburger JJ, Frassle SP, Xavier MN, et al. (2013) Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature 496: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bishop A, House D, Perkins T, Baker S, Kingsley RA, et al. (2008) Interaction of Salmonella enterica serovar Typhi with cultured epithelial cells: roles of surface structures in adhesion and invasion. Microbiology 154: 1914–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weinstein DL, O'Neill BL, Hone DM, Metcalf ES (1998) Differential early interactions between Salmonella enterica serovar Typhi and two other pathogenic Salmonella serovars with intestinal epithelial cells. Infect Immun 66: 2310–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mills SD, Finlay BB (1994) Comparison of Salmonella typhi and Salmonella typhimurium invasion, intracellular growth and localization in cultured human epithelial cells. Microb Pathog 17: 409–423. [DOI] [PubMed] [Google Scholar]
- 34. Keestra AM, Winter MG, Klein-Douwel D, Xavier MN, Winter SE, et al. (2011) A Salmonella virulence factor activates the NOD1/NOD2 signaling pathway. MBio 2: e00266–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, et al. (2001) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413: 852–856. [DOI] [PubMed] [Google Scholar]
- 36. Baker S, Dougan G (2007) The genome of Salmonella enterica serovar Typhi. Clin Infect Dis 45 Suppl 1: S29–33. [DOI] [PubMed] [Google Scholar]
- 37. Virlogeux I, Waxin H, Ecobichon C, Popoff MY (1995) Role of the viaB locus in synthesis, transport and expression of Salmonella typhi Vi antigen. Microbiology 141 Pt 12: 3039–3047. [DOI] [PubMed] [Google Scholar]
- 38. Raffatellu M, Santos RL, Chessa D, Wilson RP, Winter SE, et al. (2007) The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype Typhi. Infect Immun 75: 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jansen AM, Hall LJ, Clare S, Goulding D, Holt KE, et al. (2011) A Salmonella Typhimurium-Typhi genomic chimera: a model to study Vi polysaccharide capsule function in vivo. PLoS Pathog 7: e1002131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wangdi T, Winter SE, Baumler AJ (2012) Typhoid fever: “you can't hit what you can't see”. Gut Microbes 3: 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haneda T, Winter SE, Butler BP, Wilson RP, Tukel C, et al. (2009) The capsule-encoding viaB locus reduces intestinal inflammation by a Salmonella pathogenicity island 1-independent mechanism. Infect Immun 77: 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Winter SE, Winter MG, Thiennimitr P, Gerriets VA, Nuccio SP, et al. (2009) The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol Microbiol 74: 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wallis TS, Wood M, Watson P, Paulin S, Jones M, et al. (1999) Sips, Sops, and SPIs but not stn influence Salmonella enteropathogenesis. Adv Exp Med Biol 473: 275–280. [DOI] [PubMed] [Google Scholar]
- 44. Arricau N, Hermant D, Waxin H, Ecobichon C, Duffey PS, et al. (1998) The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol Microbiol 29: 835–850. [DOI] [PubMed] [Google Scholar]
- 45. Kutsukake K, Ohya Y, Iino T (1990) Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol 172: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, et al. (2006) Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol 188: 2233–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohnishi K, Kutsukake K, Suzuki H, Iino T (1990) Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet 221: 139–147. [DOI] [PubMed] [Google Scholar]
- 48. Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, et al. (2000) Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar typhimurium. J Bacteriol 182: 1872–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kage H, Takaya A, Ohya M, Yamamoto T (2008) Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease. J Bacteriol 190: 2470–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin D, Rao CV, Slauch JM (2008) The Salmonella SPI1 type three secretion system responds to periplasmic disulfide bond status via the flagellar apparatus and the RcsCDB system. J Bacteriol 190: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bajaj V, Hwang C, Lee CA (1995) hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Molecular microbiology 18: 715–727. [DOI] [PubMed] [Google Scholar]
- 52. Lee CA, Jones BD, Falkow S (1992) Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proceedings of the National Academy of Sciences of the United States of America 89: 1847–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Figueiredo JF, Lawhon SD, Gokulan K, Khare S, Raffatellu M, et al. (2009) Salmonella enterica Typhimurium SipA induces CXC-chemokine expression through p38MAPK and JUN pathways. Microbes and infection/Institut Pasteur 11: 302–310. [DOI] [PubMed] [Google Scholar]
- 54. Chen LM, Hobbie S, Galan JE (1996) Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science 274: 2115–2118. [DOI] [PubMed] [Google Scholar]
- 55. Coso OA, Chiariello M, Yu JC, Teramoto H, Crespo P, et al. (1995) The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell 81: 1137–1146. [DOI] [PubMed] [Google Scholar]
- 56. Schlumberger MC, Friebel A, Buchwald G, Scheffzek K, Wittinghofer A, et al. (2003) Amino acids of the bacterial toxin SopE involved in G nucleotide exchange on Cdc42. J Biol Chem 278: 27149–27159. [DOI] [PubMed] [Google Scholar]
- 57. Winter SE, Raffatellu M, Wilson RP, Russmann H, Baumler AJ (2008) The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell Microbiol 10: 247–261. [DOI] [PubMed] [Google Scholar]
- 58. Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL (2001) Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol 167: 1882–1885. [DOI] [PubMed] [Google Scholar]
- 59. Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, et al. (2006) Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol 7: 576–582. [DOI] [PubMed] [Google Scholar]
- 60. Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, et al. (2006) Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol 7: 569–575. [DOI] [PubMed] [Google Scholar]
- 61. Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, et al. (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71: 2839–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, et al. (2009) Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS pathogens 5: e1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Winter SE, Winter MG, Godinez I, Yang H-J, Russmann H, et al. (2010) A Rapid Change in Virulence Gene Expression during the Transition from the Intestinal Lumen into Tissue Promotes Systemic Dissemination of Salmonella. PLoS Pathogens 6: e1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang S, Adams LG, Nunes J, Khare S, Tsolis RM, et al. (2003) Secreted effector proteins of Salmonella enterica serotype typhimurium elicit host-specific chemokine profiles in animal models of typhoid fever and enterocolitis. Infect Immun 71: 4795–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ, et al. (2005) The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174: 1675–1685. [DOI] [PubMed] [Google Scholar]
- 66. Keestra AM, Godinez I, Xavier MN, Winter MG, Winter SE, et al. (2011) Early MyD88-Dependent Induction of Interleukin-17A Expression during Salmonella Colitis. Infection and immunity 79: 3131–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wilson RP, Winter SE, Spees AM, Winter MG, Nishimori JH, et al. (2011) The Vi capsular polysaccharide prevents complement receptor 3-mediated clearance of Salmonella enterica serotype Typhi. Infection and immunity 79: 830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wilson RP, Raffatellu M, Chessa D, Winter SE, Tukel C, et al. (2008) The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol 10: 876–890. [DOI] [PubMed] [Google Scholar]
- 69. Lawes M, Maloy S (1995) MudSacI, a transposon with strong selectable and counterselectable markers: use for rapid mapping of chromosomal mutations in Salmonella typhimurium. J Bacteriol 177: 1383–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schmieger H (1972) Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet 119: 75–88. [DOI] [PubMed] [Google Scholar]
- 71. Winter SE, Thiennimitr P, Nuccio SP, Haneda T, Winter MG, et al. (2009) Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype typhimurium infection. Infect Immun 77: 1904–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, et al. (2013) Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339: 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, et al. (2003) Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun 71: 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Miller SI, Kukral AM, Mekalanos JJ (1989) A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A 86: 5054–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stojiljkovic I, Baumler AJ, Heffron F (1995) Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177: 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hoiseth SK, Stocker BA (1981) Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291: 238–239. [DOI] [PubMed] [Google Scholar]
- 77. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, et al. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467: 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, et al. (2012) Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio 3: pii: e00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Simon R, Priefer U, Puhler A (1983) A Broad Host Range Mobilization System for Invivo Genetic-Engineering - Transposon Mutagenesis in Gram-Negative Bacteria. Bio-Technology 1: 784–791. [Google Scholar]
- 80. Kinder SA, Badger JL, Bryant GO, Pepe JC, Miller VL (1993) Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene 136: 271–275. [DOI] [PubMed] [Google Scholar]
- 81. Keestra AM, de Zoete MR, Bouwman LI, van Putten JP (2010) Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J Immunol 185: 460–467. [DOI] [PubMed] [Google Scholar]
- 82. Kingsley RA, Reissbrodt R, Rabsch W, Ketley JM, Tsolis RM, et al. (1999) Ferrioxamine-mediated Iron(III) utilization by Salmonella enterica. Appl Environ Microbiol 65: 1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Isberg RR, Falkow S (1985) A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317: 262–264. [DOI] [PubMed] [Google Scholar]
- 84. Keestra AM, de Zoete MR, van Aubel RA, van Putten JP (2007) The central leucine-rich repeat region of chicken TLR16 dictates unique ligand specificity and species-specific interaction with TLR2. J Immunol 178: 7110–7119. [DOI] [PubMed] [Google Scholar]
- 85. Wang RF, Kushner SR (1991) Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100: 195–199. [PubMed] [Google Scholar]
- 86. Bohez L, Ducatelle R, Pasmans F, Botteldoorn N, Haesebrouck F, et al. (2006) Salmonella enterica serovar Enteritidis colonization of the chicken caecum requires the HilA regulatory protein. Vet Microbiol 116: 202–210. [DOI] [PubMed] [Google Scholar]
- 87. Overbergh L, Giulietti A, Valckx D, Decallonne R, Bouillon R, et al. (2003) The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Tech 14: 33–43. [PMC free article] [PubMed] [Google Scholar]
- 88. Godinez I, Haneda T, Raffatellu M, George MD, Paixao TA, et al. (2008) T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect Immun 76: 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TviA reduces T3SS-1-induced inflammatory responses independent of bacterial entry into host cells. (A) Genetic organization of the viaB operon in S. Typhi Ty2. (B) The S. Typhi Ty2 wild-type strain, a ΔviaB mutant (SW347), a ΔtviB-vexE mutant (SW74), a ΔviaB invA mutant (STY4), and a ΔtviB-vexE invA mutant (SW611) cultured in DMEM were used to infect HeLa57 cells. NF-κB activation was determined after 5 h (N = 4). (C and D) HeLa 57A cells were infected with the S. Typhi Ty2 wild-type strain, a ΔviaB mutant (SW347), a ΔviaB invA mutant (STY4), or a ΔviaB invA mutant harboring pRI203 (N = 3) precultured in TYE broth. (C) Cells were infected at a multiplicity of infection of 5 for 1 h and extracellular bacteria killed by treatment with Gentamicin for 90 min. Recovered bacterial numbers were standardized to the number of the bacteria in the inoculum. (D) To determine NF-κB activation, luciferase activity measured 5 h after infection (N = 4). Bars represent geometric means ± standard error. **, P<0.01; ns, not statistically significant.
(TIF)
TviA reduces T3SS-1-induced NF-κB activation independent of flagellin expression. (A) Schematic representation of the TviA regulatory network in S. Typhi and effect on host signaling pathways. (B and C) HeLa 57A cells (B) or HEK-293 cells transiently transfected with a NF-κB-dependent reporter plasmid (pNFkB-luc) (C) were infected with a S. Typhi ΔfliC mutant (SW359), a ΔfliC ΔviaB mutant (SW483), derivatives carrying the cloning plasmids pWSK29 (pWSK) or the plasmid pTVIA1, and a ΔfliC ΔviaB invA mutant (SW398). Luciferase activity was quantified 5 h after infection to determine NF-κB activation levels (N = 3). Bars represent geometric means ± standard error. *, P<0.05; **, P<0.01; ***, P<0.001.
(TIF)
Effect of TviA on bacterial gene expression in vitro . The S. Typhi wild type Ty2 (WT), a ΔviaB mutant (SW347), a ΔtviB-vexE mutant (SW74), the S. Typhimurium wild-type SL1344, a SL1344 phoN mutant (SW759), a SL1344 phoN::tviA mutant (SW760), the S. Typhimurium 14028 NalR wild type (IR715), a 14028 NalR phoN mutant (AJB715), and a 14028 NalR phoN::tviA (SW474) were cultured in TYE broth for 3 h. RNA was extracted and qRT-PCR performed to determine the relative abundance of flhD (A), hilA (B), invF (C), prgH (D), sipA (E), and sopE (F) mRNA. Data presented is fold change over the abundance of mRNA recovered from the respective wild-type strain after standardization to the housekeeping gene gmk. The dotted line indicates no change in gene expression. Bars represent geometric means from 3 (S. Typhimurium) or 4 (S. Typhi) independent experiments ± standard error. *, P<0.05; **, P<0.01; ***, P<0.001; ns, not statistically significant.
(TIF)
Contribution of SopE, SipA, SopB, and SopE2 to NF-κB activation in human epithelial cells. HeLa 57A cells were treated with media only (mock treatment) or infected with the indicated S. Typhimurium SL1344 derivatives. Certain Salmonella strains lacked defined T3SS-1 effector proteins to analyze the responses induced by SopB (light grey bar), SipA (white bar), and SopE (dark grey bar). NF-κB activation was assessed 5 h after infection based on a NF-κB-driven luciferase reporter system (N = 4). Bars represent geometric means ± standard error. **, P<0.01; ns, not statistically significant.
(TIF)
Inhibition of RIP2 signaling does not affect invasiveness of S. Typhi strains. HeLa 57A cells pretreated with DMSO or RIP2 inhibitor (SB203580; dissolved in DMSO) were infected with the indicated S. Typhi strains at a MOI of 10 and invasion determined by a Gentamicin protection assay. Bars represent geometric means ± standard error. ns, not statistically significant.
(TIF)
Expression of TviA in S. Typhimurium 14028 NalR reduces T3SS-1-driven NF-κB activation in epithelial cells. HeLa 57A cells were infected with the S. Typhimurium 14028 NalR derivatives or treated with bacterial growth media (mock treatment). Luciferase activity determined 5 h after infection (N = 4). Bars represent geometric means ± standard error. **, P<0.01;
(TIF)
Bacterial colonization in the mouse colitis model. (A and B) Streptomycin-pretreated mice were infected with the indicated S. Typhimurium SL1344 derivatives as described in Figure 6. The bacterial load in the colon content (A) and the Peyer's patches (B) was determined 12 h after infection. Bars represent geometric means ± standard error. ns, not statistically significant.
(TIF)