Abstract
Long-term exposure to benzene causes several adverse health effects, including an increased risk of acute myeloid leukemia. This study was to identify genetic alternations involved in pathogenesis of leukemia in benzene-exposed workers without clinical symptoms of leukemia. This study included 33 shoe-factory workers exposed to benzene at levels from 1 ppm to 10 ppm. These workers were divided into 3 groups based on the benzene exposure time, 1- < 7, 7- < 12, and 12- < 24 years. 17 individuals without benzene exposure history were recruited as controls. Cytogenetic analysis using Affymetrix Cytogenetics Array found copy-number variations (CNVs) in several chromosomes of benzene-exposed workers. Expression of targeted genes in these altered chromosomes, NOTCH1 and BSG, which play roles in leukemia pathogenesis, was further examined using real-time PCR. The NOTCH1 mRNA level was significantly increased in all 3 groups of workers, and the NOTCH1 mRNA level in the 12- < 24 years group was significantly higher than that in 1- < 7 and 7- < 12 years groups. Compared to the controls, the BSG mRNA level was significantly increased in 7- < 12 and 12- < 24 years groups, but not in the 1- < 7 years group. These results suggest that CNVs and leukemia-related gene expression might play roles in leukemia development in benzene-exposed workers.
Benzene, an aromatic hydrocarbon compound, is a primary industrial chemical produced at high levels in the manufacture of plastics, resins, and dyes. Occupational exposure to benzene occurs in the oil, shipping, automobile repair, shoe manufacture, and other industries1. Benzene is also a well-known environmental pollutant present in cigarette smoke and motor vehicle exhaust2. It has been widely studied that exposure to benzene leads to several adverse health effects. Benzene's toxic effects on the blood and bone marrow include increasing the risk of acute myeloid leukemia (AML), myelodysplastic syndrome, and other hematological malignancies, such as non-Hodgkin lymphoma3,4,5. Benzene also exerts hematotoxic effect at relatively low levels of exposure6.
Studies regarding the mechanisms by which benzene increases the risk of AML reveal that benzene's metabolites induce chromosomal aberrations7,8,9. Benzene exposure has been associated with high levels of AML-related chromosomal changes, and these AML-related chromosomal changes have also been observed in human cell cultures treated with benzene metabolites9,10,11,12,13. Further studies have shown that target genes regulated by benzene's metabolites are critical for hematopoiesis in hematopoietic stem cells during leukemia development. It has been reported that 29 known genes in peripheral blood mononuclear cells are related to high benzene exposure14. Other studies including a larger number of samples have demonstrated that global gene expression changes involved in signaling pathways mediating AML, inflammatory and stress responses are associated with occupational benzene exposure15,16.
The copy-number variations (CNVs), which may serve as a major cause for changes in RNA expression17, is defined as a deletion, duplication, or size inversion of DNA fragments from one kilobase pair to several megabase pairs18. CNVs are common cytogenetic aberrations in healthy individuals, but they more frequently occur in cancers, including AML19,20,21,22. CNVs can influence gene expression by disrupting coding sequences, perturbing long-range gene regulation, or altering gene dosage. These effects contribute to phenotypic variations or increase disease risks23,24,25,26. Microarray technology is widely used in toxicological studies of CNVs17,24,25,26,27,28. However, information of CNVs in benzene-exposed workers is limited.
We show here that genetic alterations, such as long-segment copy-number variations in several chromosomes are found in blood cells of benzene-exposed workers without clinical symptoms of leukemia. Targeted genes of these altered chromosomes, which are related to development of leukemia, have been further evaluated by real-time PCR analysis. The NOTCH1 and BSG mRNA levels are increased in workers with benzene exposure. These results suggest that CNVs and leukemia-related gene expression might play roles in leukemia development in benzene-exposed workers.
Methods
Study population
Methods used in this study were carried out in accordance with the approved guideline by the Committees for Ethical Review of Research involving Human Subjects at Tianjin Medical University. Informed consents were obtained from all subjects. This study included 33 workers from a shoe factory who were exposed to benzene levels from 1 ppm (the American occupational exposure level) to 10 ppm for various times. Assessment for the level of benzene exposure was performed as described previously29. Air monitoring was conducted every month for 3–4 months prior to biological sample collection. These workers were divided into 3 groups based on the benzene exposure time, 1- < 7, 7- < 12, and 12- < 24 years. All benzene-exposed workers had normal levels of hemoglobin, RBCs, WBCs, platelets, and alanine aminotransferase (Table 1). These workers had no leukemia symptoms or other health problem at the time when blood samples were collected.
Table 1. Characteristics of the control subjects and workers exposed to benzene.
| Benzene exposure time (year) | ||||
|---|---|---|---|---|
| Control | 1 - < 7 | 7 - < 12 | 12 - < 24 | |
| Male | 9 | 3 | 9 | 2 |
| Female | 8 | 13 | 3 | 3 |
| Age | 34.7 ± 6.01 | 39.4 ± 5.6 | 35.5 ± 5.6 | 40.2 ± 4.3 |
| Hemoglobin (g/dl) | 14.6 ± 1.9 | 13.3 ± 1.6 | 15.1 ± 1.5 | 14.9 ± 2.5 |
| Red blood cells (RBCs, ×106/ml) | 5.3 ± 1.3 | 4.4 ± 0.4 | 4.9 ± 0.4 | 5.61 ± 1.1 |
| White blood cells (WBCs, ×1000/ml) | 4.9 ± 0.5 | 6.3 ± 1.8 | 6.4 ± 1.5 | 4.74 ± 0.6 |
| Platelets (×1000/ml) | 253.8 ± 48.3 | 250.3 ± 38.2 | 226.7 ± 31.8 | 244.2 ± 46.3 |
| Alanine aminotransferase (ALT, U/L) | 24.94 ± 9.4 | 19.0 ± 12.6 | 23.4 ± 10.7 | 21.6 ± 15.2 |
Values are mean ± SD.
Normal range of blood tests: Hemoglobin (g/dl): 13.5–16.5 (male), 12.0–15.0 (female). RBCs (×106/ml): 4.5–5.5 (male), 4.0–4.9 (female). WBCs (ml): 4,500–10,000. Platelets (ml): 100,000–450,000. ALT (U/L): 10–40 (male), 7–35 (female).
17 unexposed subjects were recruited from factories in the same region without benzene exposure as controls. In the control region, benzene and toluene were not detected in the air. The unexposed subjects were matched with the exposed workers for age and smoking history (Table 1).
Information was obtained from answering a questionnaire by all subjects include working history, past and current tobacco and alcohol use and medical history, including infections and ionizing radiation exposure, medication, and family disease history.
Blood sample collection
Fresh blood specimens were collected from the median cubital vein. EDTA-K2 was added to blood samples to prepare fresh anticoagulation blood for DNA extraction. Blood tests, including hemoglobin, RBCs, WBCs, platelets, and Alanine aminotransferase, were performed.
Cytogenetic microarray
Blood samples were collected from 6 benzene exposed-workers (3 female and 3 male, age: female: 39.7 ± 6.7, male: 44.7 ± 5.0, benzene exposure time: female: 6 ± 1 years, male: 6 ± 2) for microarray Affymetric cytogenetic microarray analysis.
Genomic DNA was isolated from peripheral blood cells using a Gentra Puregene blood kit (Santa Clara, California, USA) according to the manufacturer's instructions. Genomic DNA (0.1 mg) was labeled using an Affymetrix Cytogenetics Reagent Kit, and the labeled DNA was applied to an Affymetrix Cytogenetics Whole-Genome Array, including 2.7 million probes for the detection of copy number variation (Affymetrix Inc., Santa Clara, California, USA) according to the manufacturer's instructions. The array was scanned and the data were analyzed using the Affymetrix Chromosome Analysis Suite. The control group included 35 healthy Asian people (17 male and 18 female). The chromosomal structure of this control group was used as the standard, and array data from benzene-exposed workers were compared to this standard. Significant chromosomal changes were chosen by selecting the area with size >200 kbp, confidence level > 88%, and mean marker distance <2.5 kbp.
Real-time PCR assay
Real-time PCR analyses were performed using 2× EVaGreen qPCR Master Mix in a 20 μl reaction mixture containing forward and reverse primers (10 μM each) and 25 ng of DNA. Reactions were run on ABI Stepone plus real-time PCR as following: 95°C for 10 min, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Double-stranded DNA fluorescence was repeatedly detected at the end of the elongation phase of each PCR cycle. The forward and reverse primers used were as following: 5′-GGATCAGCAAGCAGGAGTATG-3′ and 5′-CAATCTCATCTTGTTTTCTGCG-3′ (135 bp) for β-actin; 5′- CCATGCTGGTCTGCAAGTCAG -3′ and 5′- CCGTTCATGAGGGCCTTGTC-3′ (194 bp) for BSG; 5′-GACAACGCCTACCTCTGCTTC-3′ and 5′-ACAGTCATCCAGGTTGATCTCG-3′ (154 bp) for NOTCH1.
Statistical analysis
Statistical significance was determined by one-way ANOVA followed by Newman-Keuls analysis using Prism 5.0 (GraphPad Software, Inc. San Diego, CA) for multiple comparisons. Data obtained from each group were expressed as the mean ± standard deviation. P < 0.05 was considered statistically significant.
Results
Study subjects and exposure assessment
Since the incident rate of leukemia is significantly increased in individuals with long-term benzene exposure as compared to that in the un-exposed people30, the goal of this study was to identify whether there are early effects of benzene exposure on genetic alterations in workers. This study included 33 benzene-exposed workers from a shoe manufacturer with benzene exposure time from 1 to 12 years. They did not have any clinical symptoms of leukemia. Their blood test results, including hemoglobin, RBCs, WBCs, platelets, and alanine aminotransferase, were among the normal ranges (Table 1). Thus, the health status of these workers was normal when this study was performed.
Effects of benzene exposure on chromosome alterations
The Affymetrix Cytogenetics microarray was applied to examine whether these benzene-exposed workers have chromosome alterations. Genomic DNA samples were prepared from blood cells of 6 benzene exposed-workers. Affymetrix Gene Chips (Human March 2006 (hg18) assembly) was used to screen the chromosomal structure. After filtering the area by using the criteria, size <200 kbp, confidence level < 88%, and mean marker distance >2.5 kbp, we found that chromosome gain with long-segment CNVs regions amplified from 200 kbp to 400 kbp in chromosomes 1, 2, 8, 9, 10, 13, 16, 19, 22 and chromosome X (Table 2). In addition, a 311–354 kbp deletion in chromosome 19 and a 458 kbp deletion in chromosome 7 were found (Table 2). These results suggest that chromosomal structural aberrations are related to occupational exposure to benzene in workers before symptoms of leukemia occurs.
Table 2. Genetic alterations in workers exposed to benzene.
| Chromosome locus | Gene targets | Confidence | |
|---|---|---|---|
| Gain | |||
| 1 | p36.33 | PUSL1, MRPL20, CCNL2, TTLL10, AURKAIP1, SDF4, TNFRSF18, TAS1R3 | 0.88 |
| 2 | q21.1 | FAM128A, TUBA3D, CCDC74A | 0.90 |
| 8 | q24.3 | C8ORFK29, LRRC14, NFKBIL2, GPT, FOXH1, MGC70857, DGAT1, FBXL6, CYHR1 | 0.88 |
| 9 | q34.3 | NOTCH1, EGFL7, RECGL4 | 0.89 |
| 10 | p15.3 | F108, DIP2C | 0.88 |
| 13 | q34 | RASA3, FAM70b | 0.89 |
| 16 | q22.3 | CLEC18B, PSMD7, GLG1 | 0.90 |
| 16 | q24.3 | C16, FAM38A, GALNS, CDT1, APRT, RNF166, CBFA2T3, TRAPPC2L | 0.88 |
| 19 | p13.3 | BSG, SHC2 | 0.89 |
| 22 | q11.23 | LRP5L, IGLL3 | 0.88 |
| X | q26.3 | NCRNA00086, DDX26B, ZNF449 | 0.92 |
| Loss | |||
| 7 | q11.21 | ZNF92, INTS4L | 0.89 |
| 19 | q13.31 | PSG6, PSG8, PSG10, PSG11 | 0.90 |
Effects of benzene exposure on expression of genes associated with leukemia
We next used real-time PCR assay to detect expression of genes in these altered segments of chromosomes which are associated with leukemia. We detected expression of 2 genes, BSG (also known as CD147) and NOTCH1, which have been shown to be associated with occurrence and development of leukemia31,32.
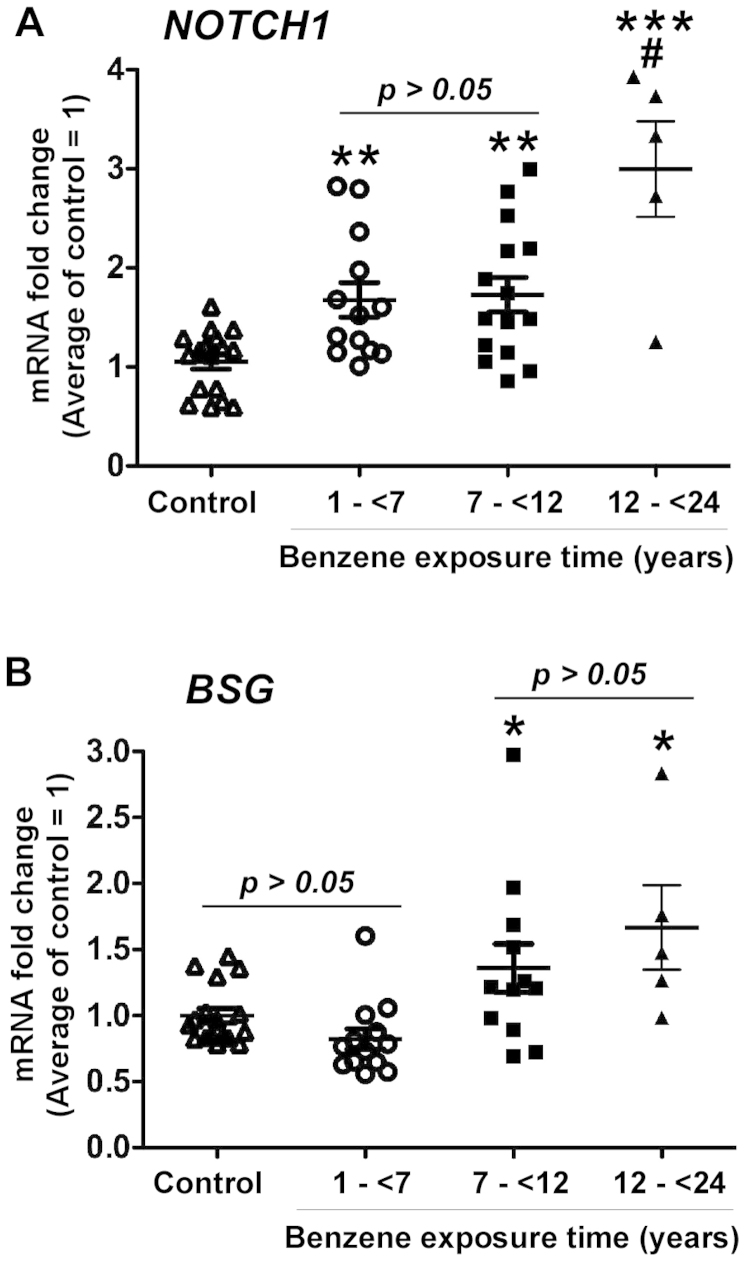
For this study, we categorized exposure groups based on the exposure time, 1- < 7 years, 7- < 12 years, and 12- < 24 years groups. Compared to the control group, the NOTCH1 mRNA level was significantly increased in workers in 1- < 7 years (p < 0.01), 7- < 12 years (p < 0.01) and 12- < 24 years groups (p < 0.001, Figure 1A). The NOTCH1 mRNA level in 12- < 24 years group was significantly higher than that in 1- < 7 years (p < 0.001), and 7- < 12 years groups (p < 0.001, Figure 1A). The BSG mRNA level was significantly increased in the 7- < 12 years and 12- < 24 years groups (p < 0.05), but not in the 1- < 7 years group (p > 0.05, Figure 1B). These results are in consistent with previous finding that cytogenetic alterations which can lead to altered gene expressions33. Benzene exposure up-regulates leukemia-associated gene expression in workers and the longer benzene exposure time correlates with higher mRNA expression levels of leukemia-associated genes.
Figure 1. Up-regulation of NOTCH1 and BSG gene expression in benzene-exposed workers.
Blood cell samples were collected from benzene-exposed workers and controls for DNA isolation and Real-time PCR analysis to detect the mRNA levels of NOTCH1 (A) and BSG (B). The average of mRNA expression level in the control group was set as 1, and the mRNA expression level in each control subject and worker was compared to the average. * p < 0.05, ** p < 0.01, and *** p < 0.001 compared to the control group. # p < 0.01 compared to the 1 - < 7 years and 7 - < 12 years groups.
Discussion
Long-term exposure to benzene has been shown to lead to an increased risk of acute myeloid leukemia and myelodysplastic syndrome. Benzene-induced decreases in blood cells could be observed within a few months after benzene exposure. However, there is a lag time of years between initial benzene exposure and the development of leukemia30. In this study, we focused on investigating the potential effects of benzene exposure on leukemia-associated gene expression in workers who showed no clinical evidence of leukemia. We identified CNVs in chromosomes of benzene-exposed workers. By analyzing functions of genes in altered chromosomes, we further confirmed that the levels of BSG and NOTCH1, which are associated with the occurrence and development of leukemia, were increased in these workers. These genetic and gene expression changes might be used as biomarkers for evaluation of the risk of leukemia in benzene-exposed workers.
A study showed that the benzene metabolites induce the binding of chlorine to DNA. Halogenated DNA can induce both genetic and epigenetic changes that contribute to carcinogenesis. Halogenative stress may account for benzene-induced bone marrow disorders and myeloid leukemia34. Thus, CNVs found in this study may represent a form of the halogenated DNA resulting from genetic changes.
NOTCH1 is crucial in T-cell differentiation and proliferation. NOTCH1 mutations appear in approximately 50% of acute T-lymphoblastic leukemia cases35. It has been reported that the significantly high expression level of NOTCH1 is positively correlated with acute T-lymphocyte leukemia (ATLL)36. NOTCH1 mutations occur in 10% of patients with chronic lymphocytic leukemia and are associated with poor prognosis37,38,39. Our results show that the copy number of NOTCH1 in the benzene exposed workers exhibits amplified changes. Thus, the NOTCH1 gene may be a target of benzene-affecting gene.
BSG encodes a protein called Basigin. This protein is a single transmembrane glycoprotein with a high degree of glycosylation and is a member of the immunoglobulin superfamily of adhesion molecules. Basigin has diverse biological functions and is involved in tissue repair, reproductive development, as well as energy metabolism. Basigin is up-regulated in tumors40. This protein promotes tumor invasion and metastasis by inducing the expression of matrix metalloproteinases41,42,43,44 and the degradation of the extracellular matrix45. Basigin is an ideal target for cancer therapy as a new tumor marker. BSG was recently identified as a part of a gene-expression signal associated with the recurrence of childhood acute lymphoblastic leukemia46,47. In ATLL patients, most T-cells express significantly higher CD147 levels48. In this study, increased copy number of BSG was found in benzene-exposed workers. Thus, benzene may affect the lymphocytes of exposed individuals, which eventually leads to the occurrence of leukemia by the amplification of BSG.
Other genes found in altered chromosomes in benzene-exposed workers are involved in normal human functions. APRT encodes adenine phosphoribosyltransferase. This enzyme catalyzes the formation of AMP and inorganic pyrophosphate from adenine and 5-phosphoribosyl-1-pyrophosphate. It also produces adenine as a byproduct of the polyamine biosynthetic pathway. A deficiency in this enzyme is associated with urolithiasis. GALNS, encodes N-acetylgalactosamine-6-sulfatase. Mutations of this gene, including point, missense, and nonsense mutations, may lead to a lysosomal storage disorder. AGPAT2 encodes lysophosphatidic acid acyltransferase β. This protein is found within the endoplasmic reticulum membrane and converts lysophosphatidic acid to phosphatidic acid, which is involved in phospholipid biosynthesis. RECQL4 encodes a DNA helicase that belongs to the RecQ helicase family. Mutations in this gene may lead to chromosomal instability.
In summary, occupational exposure to benzene can cause CNVs in several chromosomes and increased NOTCH1 and BSG expression in workers without symptoms of leukemia. Further investigations, including recruitment of additional workers and prolonged follow-up time are required to determine whether these changes can serve as the mechanisms underlying berzene exposure-induced leukiame and can be used as biomarkers for evaluation of the risk of leukemia development in benzene-exposed workers.
Author Contributions
L.K., J.Y., Y.C., L.S., Z.Y., H.X., L.F. and H.J. designed and carried out the experiments. L.K., J.Y., Y.C. and L.G. analyzed the data and wrote the manuscript. All authors reviewed the manuscript.
Acknowledgments
This study was supported by National Natural Science Foundation of China (21177091), Tianjin Science and Technology Support Program (12ZCZDSY03400), and National High Technology Development Project (2012AA021003). We gratefully acknowledge Wanxin Zhao at the Department of Occupational Disease Prevention at Liaoning Province Hospital for helping sample collection and monitoring benzene levels.
References
- McHale C. M., Zhang L. & Smith M. T. Current understanding of the mechanism of benzene-induced leukemia in humans: implications for risk assessment. Carcinogenesis 33, 240–252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur S. et al. ATSDR evaluation of potential for human exposure to benzene. Toxicol. Ind. Health. 24, 399–442 (2008). [DOI] [PubMed] [Google Scholar]
- Khalade A., Jaakkola M. S., Pukkala E. & Jaakkola J. J. Exposure to benzene at work and the risk of leukemia: a systematic review and meta-analysis. Environ. Health. 9, 31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. T. Advances in understanding benzene health effects and susceptibility. Ann. Rev. Public Health. 31, 133–148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmaus C., Smith A. H., Jones R. M. & Smith M. T. Meta-analysis of benzene exposure and non-Hodgkin lymphoma: biases could mask an important association. Occup. Environ. Med. 65, 371–378 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q. et al. Hematotoxicity in Workers Exposed to Low Levels of Benzene. Science 306, 1774–1776 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuba V., Rozgaj R. & Sentija K. Cytogenetic changes in subjects occupationally exposed to benzene. Chemosphere 40, 307–310 (2000). [DOI] [PubMed] [Google Scholar]
- Zhang L. et al. Aberrations in chromosomes associated with lymphoma and therapy-related leukemia in benzene-exposed workers. Environ. Mol. Mutagen. 48, 467–474 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang L., Eastmond D. A. & Smith M. T. The nature of chromosomal aberrations detected in humans exposed to benzene. Crit Rev Toxicol. 32, 1–42 (2002). [DOI] [PubMed] [Google Scholar]
- Smith M. T. et al. Increased translocations and aneusomy in chromosomes 8 and 21 among workers exposed to benzene. Cancer Res. 58, 2176–2181 (1998). [PubMed] [Google Scholar]
- Zhang L. et al. Benzene increases aneuploidy in the lymphocytes of exposed workers: a comparison of data obtained by fluorescence in situ hybridization in interphase and metaphase cells. Environ. Mol. Mutagen. 34, 260–268 (1999). [PubMed] [Google Scholar]
- Smith M. T. et al. Hydroquinone, a benzene metabolite, increases the level of aneusomy of chromosomes 7 and 8 in human CD34-positive blood progenitor cells. Carcinogenesis 21, 1485–1490 (2000). [PubMed] [Google Scholar]
- Stillman W. S., Varella-Garcia M. & Irons R. D. The benzene metabolite, hydroquinone, selectively induces 5q31- and -7 in human CD34+CD19- bone marrow cells. Exp. Hematol. 28, 169–176 (2000). [DOI] [PubMed] [Google Scholar]
- Forrest M. S. et al. Discovery of novel biomarkers by microarray analysis of peripheral blood mononuclear cell gene expression in benzene exposed workers. Environ. Health Perspect. 113, 801–807 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale C. M. et al. Changes in the peripheral blood transcriptome associated with occupational benzene exposure identified by cross comparison on two microarray platforms. Genomics 93, 343–349 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale C. M. et al. Global gene expression profiling of a population exposed to a range of benzene levels. Environ. Health Perspect. 119, 628–634 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L., Carson A. R. & Scherer S. W. Structural variation in the humangenome. Nat. Rev. Genet. 7, 85–97 (2006). [DOI] [PubMed] [Google Scholar]
- Kim K. I. et al. Copy number variations in normal karyotype acute myeloid leukemia and their association with treatment response. Basic Clin. Pharmacol. Toxicol. 111, 317–324 (2012). [DOI] [PubMed] [Google Scholar]
- Burkhardt B. et al. Loss of heterozygosity on chromosome 6q14-q24 isassociated with poor outcome in children and adolescents with T-cell lymphoblasticlymphoma. Leukemia 20, 1422–1429 (2006). [DOI] [PubMed] [Google Scholar]
- Huse K. et al. Geneticvariants of the copy number polymorphic beta-defensin locus are associated with sporadic prostate cancer. Tumour Biol. 29, 83–92 (2008). [DOI] [PubMed] [Google Scholar]
- Lin L. J. et al. Integrated analysis of copy number alterations and loss of heterozygosity in human pancreatic cancer using a high-resolution, single nucleotidepolymorphismarray. Oncology 75, 102–112 (2008). [DOI] [PubMed] [Google Scholar]
- Wang Z. C. et al. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res. 64, 64–71 (2004). [DOI] [PubMed] [Google Scholar]
- Yang T. et al. Genome-wide Copy Number Variation Study: Identified a Susceptibility Gene, UGT2B17, for Osteoporosis. Am J Hum Genet. 83, 663–674 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland P. R. Polymorphically duplicated genes: their relevance to phenotypic variation in humans. Ann. Med. 35, 308–315 (2003). [DOI] [PubMed] [Google Scholar]
- Nguyen D. Q., Webber C. & Ponting C. P. Bias of selection on human copy-number variants. PLoS Genet. 2, e20 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S. A. et al. Common deletion polymorphisms in the human genome. Nat. Genet. 38, 86–92 (2006). [DOI] [PubMed] [Google Scholar]
- Redon R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski J. R. Retrotransposition and structural variation in the human genome. Cell 356: 141, 1110–1112 (2010). [DOI] [PubMed] [Google Scholar]
- Vermeulen R. et al. Detailed exposure assessment for a molecular epidemiology study of benzene in two shoe factories in China. Ann. Occup. Hyg. 48, 105–116 (2004). [DOI] [PubMed] [Google Scholar]
- Snyder R. Leukemia and Benzene. Int J Environ Res Public Health 9, 2875–2893 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlierberghe P. V. & Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J. Clin. Invest. 122, 3398–3406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beesley A. H. et al. The gene expression signature of relapse in paediatric acute lymphoblastic leukaemia: implications for mechanisms of therapy failure. Br. J. Haematol. 131, 447–456 (2005). [DOI] [PubMed] [Google Scholar]
- Schoch C. et al. Genomic gains and losses influence expression levels of genes located within the affected regions: a study on acute myeloid leukemias with trisomy 8, 11, or 13, monosomy 7, or deletion 5q. Leukemia 19, 1224–1228 (2005). [DOI] [PubMed] [Google Scholar]
- Etienne W. et al. Comparison of mRNA gene expression by RT-PCR and DNA microarray. Biotechniques 36, 618–626 (2004). [DOI] [PubMed] [Google Scholar]
- Grabher C., Boehmer H. V. & Look A. T. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 6, 347–359 (2006). [DOI] [PubMed] [Google Scholar]
- López C. et al. Different distribution of NOTCH1 mutations in chronic lymphocytic leukemia with isolated trisomy 12 or associated with other chromosomal alterations. Genes Chromosomes Cancer 51, 881–889 (2012). [DOI] [PubMed] [Google Scholar]
- Lin C. et al. Mutations increased overexpression of Notch1 in T-cell acute lymphoblastic leukemia. Cancer Cell Int. 12, 13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri G. et al. Analysis of the chronic lymphocytic leukemia coding genome: Role of NOTCH1 mutational activation. J. Exp. Med. 208,1389–13401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Put N. et al. Translocation t(14;18) is not associated with inferior outcome in chronic lymphocytic leukemia. Leukemia 23, 1201–1204 (2009). [DOI] [PubMed] [Google Scholar]
- Rossi D. et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 119, 521–529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W. C. et al. Increasing EMMPRIN and matriptase expression in hepatocellularcarcinoma: tissue microarrayanalysis of immunohistochemical scores with clinic opathological parameters. Histopathology 49, 388–395 (2006). [DOI] [PubMed] [Google Scholar]
- Biswas C. Tumor cell stimulation of collagenase production by fibroblasts. Biochem. Biophys. Res. Commun. 109, 1026–1034 (1982). [DOI] [PubMed] [Google Scholar]
- Biswas C. et al. The human tumor cell-derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res. 1995; 55, 434–439 (1995). [PubMed] [Google Scholar]
- Guo H. et al. Stimulation of matrix metalloproteinase production by recombinant extracellular matrix metalloproteinase inducer from transfected Chinese hamster ovary cells. J. Biol. Chem. 272, 24–27 (1997). [PubMed] [Google Scholar]
- Kataoka H., DeCastro R., Zucker S. & Biswas C. Tumor cell-derived collagenase stimulatory factor increases expression of interstitial collagenase, stromelysin, and 72-kDa gelatinase. Cancer. Res. 53, 3154–3158 (1993). [PubMed] [Google Scholar]
- DeCastro R. et al. Human keratinocytes express EMMPRIN, an extracellular matrix metalloproteinase inducer. J. Invest. Dermatol. 106, 1260–1265 (1996). [DOI] [PubMed] [Google Scholar]
- Beesley A. H. et al. The gene expression signature of relapse in paediatric acute lymphoblasticleukaemia: implications for mechanisms of therapy failure. Br. J. Haematol. 131, 447–456 (2005). [DOI] [PubMed] [Google Scholar]
- Beesley A. H., Weller R. E. & Kees U. R. The role of BSG (CD147) in acute lymphoblastic leukaemia and relapse. Br. J. Haematol. 142, 1000–1002 (2008). [DOI] [PubMed] [Google Scholar]