Abstract
MicroRNAs (miRNAs) play important roles in multiple biological processes and have attracted much scientific attention recently. Their expression can be altered by environmental factors (EFs), which are associated with many diseases. Identification of the phenotype-genotype relationships among miRNAs, EFs, and diseases at the network level will help us to better understand toxicology mechanisms and disease etiologies. In this study, we developed a computational systems toxicology framework to predict new associations among EFs, miRNAs and diseases by integrating EF structure similarity and disease phenotypic similarity. Specifically, three comprehensive bipartite networks: EF-miRNA, EF-disease and miRNA-disease associations, were constructed to build predictive models. The areas under the receiver operating characteristic curves using 10-fold cross validation ranged from 0.686 to 0.910. Furthermore, we successfully inferred novel EF-miRNA-disease networks in two case studies for breast cancer and cigarette smoke. Collectively, our methods provide a reliable and useful tool for the study of chemical risk assessment and disease etiology involving miRNAs.
MicroRNA (miRNA) is a newly identified type of small non-coding RNA that downregulates gene expression at the post-transcriptional level by inhibiting translation of mRNA or degrading mRNA1,2,3,4. As important regulators of at least 60% of all protein-coding gene expression, miRNA networks have become an important research field of the systems biology5. miRNA expression profiles can be altered by toxic environmental factors (EFs), such as radiation6, pollution7, cigarette smoke8, and others. The gene networks targeted by miRNAs may change with altered miRNA expression. These changes ultimately cause diverse diseases, such as cancer9, neurological diseases10 and cardiovascular diseases11. Thus, miRNA networks bridge the toxicology mechanism gap between EFs and diseases, providing useful information for interpreting EF toxicity and disease etiology12,13,14,15. For example, in one study, miR-31 expression in normal respiratory epithelia and lung cancer cells was induced by cigarette smoke, resulting in lung cancer16. In another study, two well-known endocrine disrupting compounds, bisphenol A (BPA) and dichlorodiphenyltrichloroethane (DDT), could alter the miRNA expression profiles of MCF-7 breast cancer cells including estrogen-regulated onco-miR-21. This displays the toxicology mechanisms of xenoestrogens and the pathology of breast cancer in a new perspective17. Although investigations of the associations among EFs, miRNAs and diseases are gaining increasing attention and becoming a hot research field, experimental studies are time-consuming and costly due to the huge number of EFs available for analysis.
As the number of experimental data has increased rapidly, computational models provide useful tools for identifying new human health hazards associated with EFs. Computational methods can be divided into classic quantitative structure-activity relationships (QSARs) and computational systems toxicology approaches. The latter has advantages against classic QSAR models, such as the OECD QSAR Toolbox (http://www.oecd.org/chemicalsafety/risk-assessment/theoecdqsartoolbox.htm) and admetSAR18. In our previous study, we developed predictive toxicogenomics-derived models (PTDMs) to predict chemical-gene-disease associations using the network-based inference (NBI) algorithm19. Other computational systems toxicology approaches have also been published to study the disease etiologies caused by proteins20 and chemical metabolism21. However, the toxicology mechanisms of EF exposure and disease etiology remain a major topic of research today22. The recent appearance of miRNAs has provided huge opportunities for the development of computational models from a systems biology perspective, and computational methods have been developed to predict potential associations in miRNA related networks. Qiu et al. uncovered a number of biological patterns of EF-miRNA interactions and proposed a computational model to predict new EF-disease associations23. Jiang et al. constructed cancer specific networks to identify the biological links between small molecules and miRNAs24. Chen et al. reported a method named miREFScan to predict disease-related EF-miRNA associations using a semi-supervised classifier25. Currently, there is still a great need for feasible, effective and/or efficient models.
In this study, we developed a computational systems toxicology framework to predict miRNA networks by systematic integration of EF structure similarity and disease phenotypic similarity. Specially, we constructed three high-quality bipartite networks: EF-miRNA, EF-disease and miRNA-disease associations, to build predictive computational systems toxicology models. High predictive performance was achieved in 10-fold cross validation. Furthermore, two case studies were performed to illustrate the predictive capability of the constructed framework. Collectively, the developed computational model provides new useful tools to elucidate the mechanisms of environmental toxicity and disease etiologies at the miRNA level.
Results
Overview of the computational systems toxicology framework
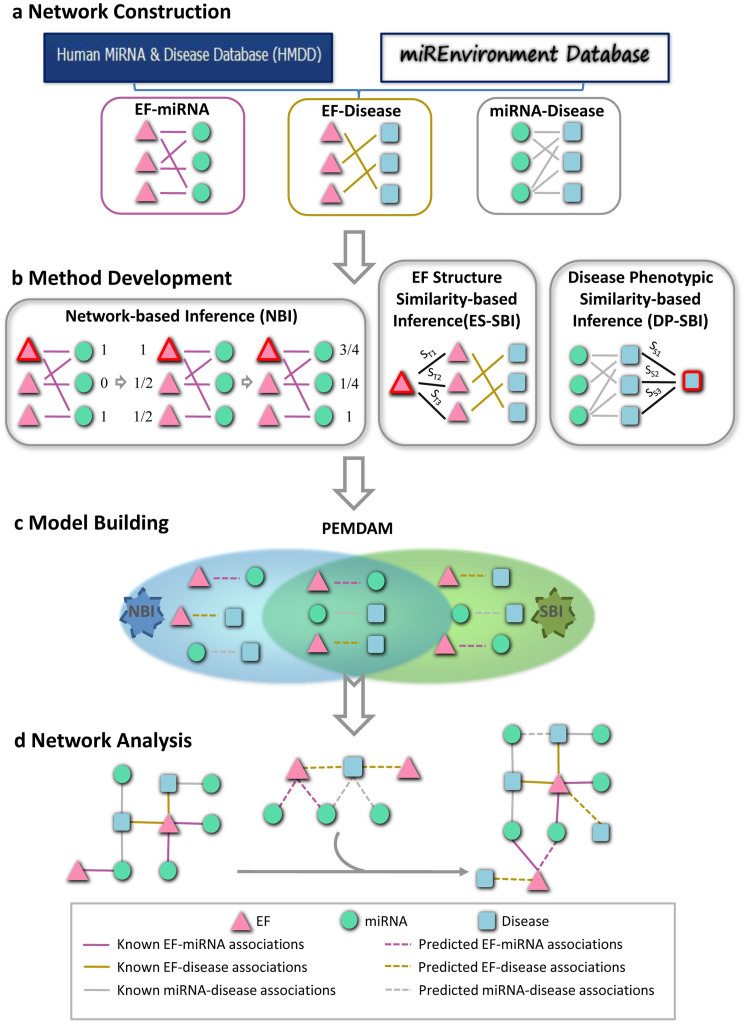
We proposed a new computational systems toxicology framework to predict putative EF-miRNA-disease associations. As shown in Figure 1, three bipartite networks: EF-miRNA association (EMA), EF-disease association (EDA) and miRNA-disease association (MDA), were constructed. The EMA network included 1,770 associations between 184 EFs and 395 miRNAs, while the MDA network consisted of 6,466 associations connecting 569 miRNAs and 396 diseases. The EDA network contained 320 associations linking 171 EFs and 115 diseases (Table 1). More detailed information is provided in Supplementary Table S1. Next, we used three network-based methods, including network-based inference (NBI)26, EF structure similarity-based inference (ES-SBI) and disease phenotypic similarity-based inference (DP-SBI), to build a predictive EF-miRNA-disease association model (PEMDAM). Finally, the PEMDAM was validated using 10-fold cross validation and applied to two case studies on breast cancer and cigarette smoke.
Figure 1. Diagram of the computational systems toxicology framework.
(a) The original data were collected from the Human MiRNA Disease Database and miREnvironment Database, and used to construct three bipartite networks: the EF-miRNA association (EMA), EF-disease association (EDA), and miRNA-disease association (MDA) networks. (b) Three methods, network-based inference (NBI), EF structure similarity-based inference (ES-SBI) and disease phenotypic similarity-based inference (DP-SBI), were developed to build the predictive model designated the predictive EF-miRNA-disease association model (PEMDAM). (c) The PEMDAM was built using the intersection of both of the prioritized lists from NBI and SBI. (d) Network visualization and analysis. EF: the environmental factor; ST: the Tanimoto similarity between two EFs; SS: the phenotypic similarity between two diseases.
Table 1. Datasets of the known EMAs, MDAs and EDAs used in this study.
| Numbers | EMAs | MDAs | EDAs |
|---|---|---|---|
| NE | 184 | / | 171 |
| Nm | 395 | 569 | / |
| ND | / | 396 | 115 |
| NA | 1770 | 6466 | 320 |
EMAs: EF-miRNA associations; MDAs: miRNA-disease associations; EDAs: EF-disease associations; NE: the number of EFs; Nm: the number of miRNAs; ND: the number of diseases; NA: the number of associations.
Network characteristics of the known EF-miRNA-disease association network
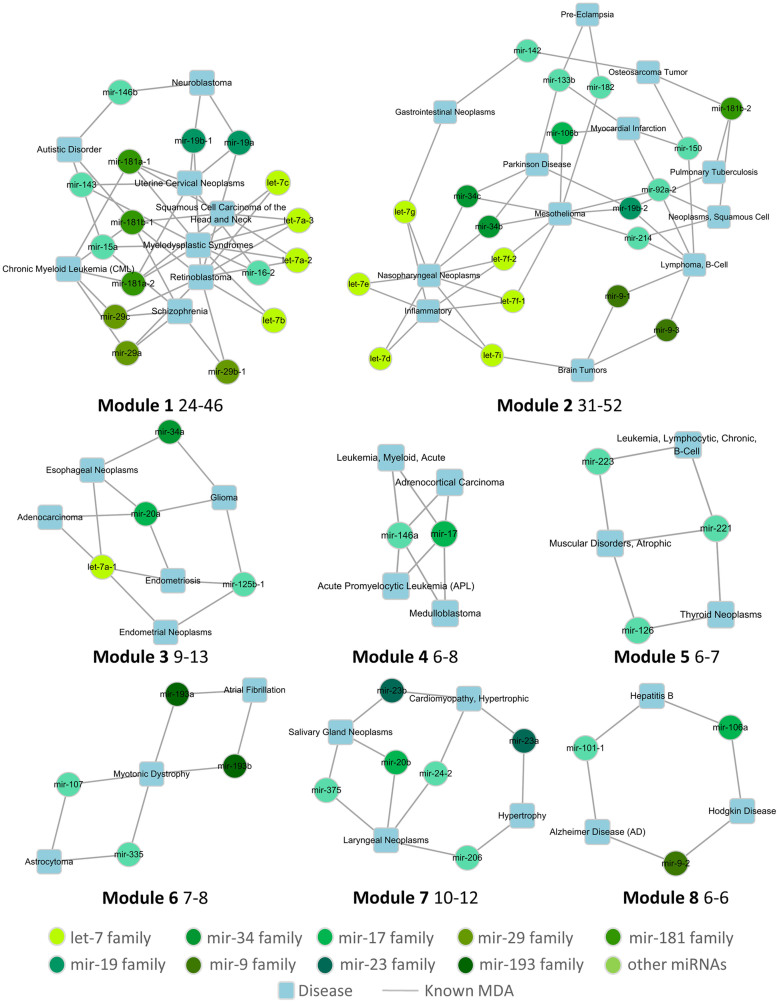
The MDA network displays the miRNA signatures of specific diseases, which is helpful for studying the pathological mechanisms of these diseases. We identified eight modules with sizes ranging from 31 to 6 based on the MDA network using the Cytoscape plugin MCODE27 (Figure 2). In these modules, the common miRNA signatures between diseases were displayed. For example, as shown in module 1, two psychiatric diseases, schizophrenia and autistic disorder, shared mir-15a, which was confirmed to target genes, such as regulator of G-protein signaling 4 (RGS4), glutamate receptor metabotropic 7 (GRM7), glutamate receptor subunit 3A (GRIN3A) and visinin-like 1 (VSNL1)28. Furthermore, the miRNAs from different families were depicted in various colors, which illustrates that miRNAs in the same family share the same important seed-pairing region and consequently tend to have similar functions. The most obvious miRNA family found is the let-7 family that has four members in module 1 and six members in module 2. In module 1, the four let-7 members cooperate with each other in three diseases: myelodysplastic syndromes, head & neck squamous cell carcinomas and retinoblastomas. In module 2, all six of the let-7 members play important roles in inflammation and nasopharyngeal neoplasms. In addition, the members of the mir-193 family function together in both chronic atrial fibrillation and myotonic dystrophy, as shown in module 6. Other miRNA family members, mir-9, mir-19, mir-29, mir-34, and mir-181, were also found to cooperate in specific diseases.
Figure 2. Modules obtained from the miRNA-disease association (MDA) network.
The first number behind a module code denotes the node number in that module, while the latter number denotes the edge number, for example, there are 24 nodes and 46 edges in Module 1.
In addition, the three classical network parameters connectivity (K), clustering coefficient (C) and betweenness (B) were calculated to measure the topological features of the EMA, EDA and MDA networks, respectively (Supplementary Fig. S1). Most bionetworks are scale-free networks whose connectivity follows a power-law distribution29. In our bipartite networks, the minority nodes have high degrees while the majority nodes have low degrees. The disease with the highest connectivity is breast cancer, which is associated with 287 miRNAs in the MDA network and 26 EFs in the EMA network. The most studied EFs are radiation, hypoxia and 17beta-estradiol. The clustering coefficient measures the local density of links and their tendency to form clusters or communities of nodes. The average clustering coefficients in our study ranged from 0.087 to 0.206. Although the EDA network is comparatively smaller than the MDA network, the component nodes connect closely with each other, thus their clustering coefficients are relatively high. A node's betweenness is defined by the fraction of all of the shortest paths between all nodes in the network that pass through the node. In all three networks, only a few nodes have high betweenness values while many nodes have very low betweenness values. Collectively, the EMA, EDA and MDA networks are similar to other bionetworks; however, they are relatively sparse and not well defined, which leaves plenty of room for research and reveals a need to find new methods to predict miss-links in the networks.
Performance of the computational systems toxicology model
miRNA-disease association prediction
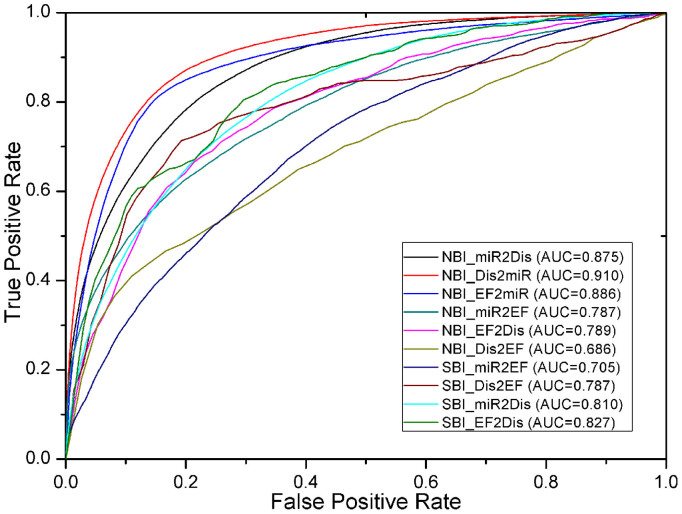
The prediction of new candidate MDAs is the basis for studying individual miRNA roles in disease pathogenesis. A comprehensive MDA network supported by experimental evidence was collected from the HMDD and miREnvironment databases. In the PEMDAM, the predicted list of new candidate diseases linked to miRNAs was obtained using NBI algorithm, while the prediction of new candidate miRNAs linked to diseases was found by combining NBI with DP-SBI. The prediction of putative diseases linked to miRNAs (NBI_Dis2miR) achieved an AUC of 0.910. A high AUC of 0.875 was also achieved when prioritizing new candidate miRNAs linked to diseases using NBI (NBI_miR2Dis) versus 0.810 by DP-SBI (SBI_miR2Dis). These results showed the high predictive accuracy of our PEMDAM toward the prediction of new candidate MDAs.
EF-disease association prediction
New EDA predictions could enhance our knowledge about how EFs affect our health. To this end, known EDA data were extracted from the miREnvironment database. Prediction of EDAs involved prioritizing new candidate EFs linked to diseases and also prioritizing new candidate diseases linked to EFs. When prioritizing new candidate EFs linked to diseases, NBI and DP-SBI were applied (NBI_EF2Dis, SBI_EF2Dis). In addition, NBI and ES-SBI were used to predict new candidate diseases linked to EFs (NBI_Dis2EF, SBI_Dis2EF). Heat maps of EF structure similarity and disease phenotypic similarity are given in Supplementary Figure S2. AUC values of 0.789, 0.686, 0.827, and 0.787 were obtained for NBI_EF2Dis, NBI_Dis2EF, SBI_EF2Dis, and SBI_Dis2EF, respectively. As shown in Figure 3, integrating EF structure similarity and disease phenotypic similarity with the NBI algorithm would greatly improve the performance of the PEMDAM.
Figure 3. The receiver operating characteristic (ROC) curves of NBI and SBI.
ROC curves were generated by 100 simulations of 10-fold cross validation. miR2Dis is the abbreviation for the predicting putative miRNAs to diseases, and the other abbreviations can be deduced similarly. NBI: network-based inference; SBI: similarity-based inference, including ES-SBI (miR2EF and Dis2EF) and DP-SBI (miR2Dis and EF2Dis).
EF-miRNA association prediction
Carcinogens and drugs are two major types of EFs. Prediction of new EMAs will help to understand the underlying mechanisms of xenobiotic toxicity. The PEMDAM was built based on a known EF-miRNA bipartite network collected from the miREnvironment database. The prioritization of new candidate EFs linked to miRNAs was obtained by NBI (NBI_EF2miR), while the prediction of new candidate miRNAs linked to EFs was found by combining NBI (NBI_miR2EF) and ES-SBI (SBI_miR2EF). NBI_EF2miR achieved an AUC of 0.886, and the prioritization of new candidate miRNAs linked to EFs obtained an AUC of 0.787 by NBI, and an AUC of 0.705 by SBI. Collectively, our PEMDAM was verified to be reliable for predicting new candidate EMAs.
Case study 1: discovery of new risks for breast cancer
Breast cancer is the most common neoplasm in women and caused 458,503 deaths worldwide in 200830. Moreover, the breast cancer phenotype is the most studied disease on the miRNA level31, having the highest degrees in both the EMA and MDA networks. The dataset used to build this predictive model contained >300 associations related to breast cancer supported by ~300 experimental documents. Prioritizing new potent EFs and miRNAs linked to breast cancer would improve our knowledge of breast cancer etiology. Thus, the predicted lists for breast cancer were extracted from the final prioritized lists from our PEMDAM as a case study, and a sub-network was constructed with Cytoscape for network analysis.
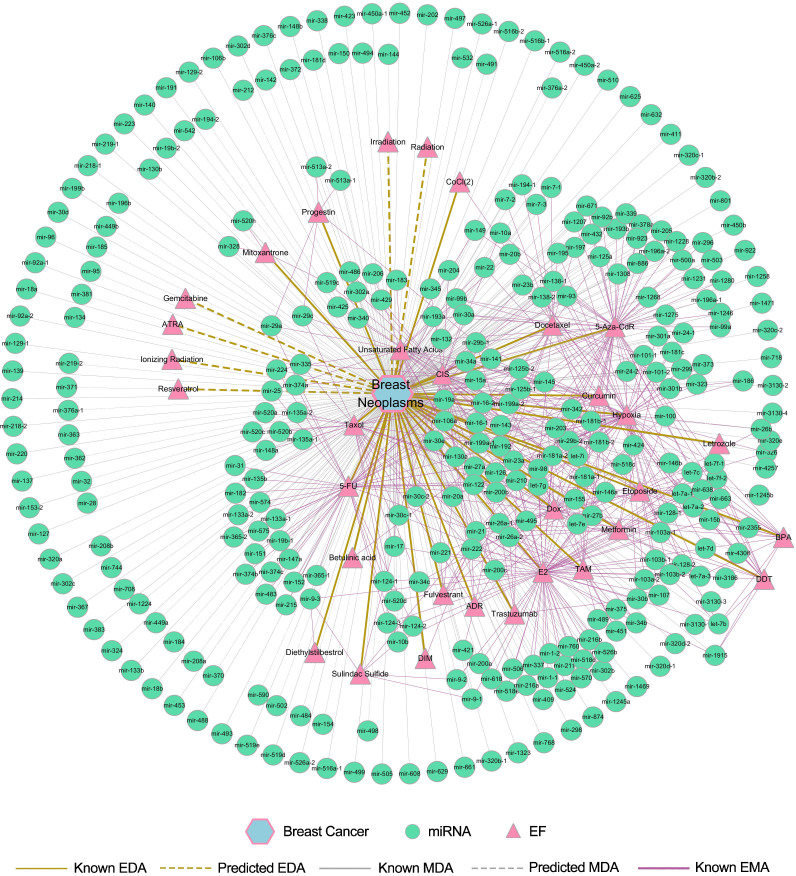
Six new candidate EFs associated with breast cancer were predicted based on the common top 10 candidates using both NBI and DP-SBI methods. Interestingly, all of the predicted EFs (6/6, 100%) related to breast cancer were found to be supported by experimental evidence in the literature (Supplementary Table S2). Due to research bias, these EFs haven't been studied with respect to miRNA expression changes related to breast cancer. However, this information can be discovered using the PEMDAM. Information about the associated miRNAs of the six new candidate EFs prioritized for breast cancer were extracted from known networks. In total, 40 potential miRNAs for breast cancer were obtained through utilizing the common candidates of the top 50 lists by both NBI and DP-SBI. Among the 40 new candidate miRNAs prioritized for breast cancer, 39 (97.5%) miRNAs were validated by databases or newly published literature (Supplementary Table S3). For these validated miRNAs, the EFs that can alter their expression were also extracted from the entire network. The putative lists shown in Supplementary Tables S2 and S3 are very promising for further study. For example, radiation may alter the expression of 32 breast cancer related miRNAs, and mir-181b may be another miRNA that plays an important role in the tobacco related pathology of breast cancer. Figure 4 shows a global breast cancer network constructed with known and predicted EMAs, MDAs and EDAs. The network includes 32 EFs and 327 miRNAs related to breast cancer. In the center of the network, 219 miRNAs are specific for EFs, thus, these miRNAs may be developed as biomarkers of breast cancer for people who are exposed to these toxic EFs. Although the miRNAs in the periphery are not defined to be associated with specific EFs, they are quite important for understanding the pathology of breast cancer.
Figure 4. The discovered EF-miRNA-disease association network for breast cancer.
Breast cancer is shown as a hexagon. The network includes the associations between breast cancer and 287 known miRNAs, 26 known EFs, 40 predicted miRNAs and 6 predicted EFs as well as the known associations between these miRNAs and EFs.
Interestingly, some of the EFs are drugs. Studies about associations among drugs, miRNAs and diseases will help to increase our knowledge about polypharmacology and personalized medicine. Breast cancers were classified into two major subtypes: luminal and basal subtypes. Here, we tried to make predictions for drug-disease associations based on the above two breast cancer subtypes. 5 known associations among subtypes and specific drugs were collected from published literatures32,33 and added into our computational framework. Predicted lists were obtained by the top 10 lists using the NBI algorithm (Supplementary Table S4). As there are not enough known data about subtypes, predicted lists here need more experiments for validation. With sufficient compound-disease associations based on specific disease subtypes collected, our computational approaches will perform better.
Collectively, the predictive computational systems toxicology model developed here is valuable and can reliably predict potential new EF exposure risks and miRNA biomarkers to help increase our understanding of breast cancer etiology. Moreover, our computational program showed predictive capability for subtype specific drug-disease associations.
Case study 2: discovery of new hazards from cigarette smoke
Approximately 1.3 billion people smoke cigarettes, which results in 5 million preventable deaths per year34. Cigarette smoke contains many toxic components and has been found to alter a number of genetic factors, including miRNAs. These miRNAs may be used as biomarkers for the diagnosis and progression of the diseases of tobacco smokers35 and help to elucidate the biological mechanisms of tobacco toxicity. In this study, two of the major carcinogens in cigarettes: nicotine and benzo(a)pyrene (BaP), were included in addition to tobacco. In total, 58 miRNAs were found to be experimentally altered by cigarette smoke and contributed to the pathology of seven smoking-related diseases. Among them, mir-128 was strongly affected by cigarette smoke and played an important role in the host response by regulating the target gene MAFG36. miR-31 was verified as an oncomiR during lung cancer progression and its expression can be induced by cigarette smoke16. miRNA expression changes were also related to maternal cigarette use during pregnancy and poor fetal outcome37. An increasing amount of research has been focused on the changes in miRNA expression caused by tobacco smoke.
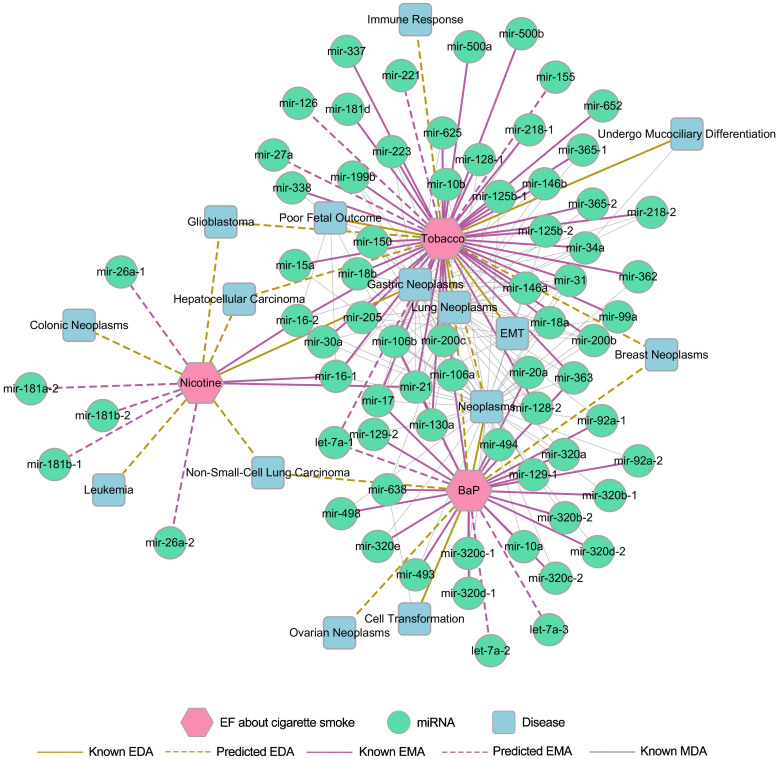
In order to further examine how tobacco influences human health at the miRNA level, predicting new candidate miRNAs and new disease risks for tobacco use were performed using the PEMDAM. Because tobacco is a mixture without a specific structure, the predicted lists were obtained only by NBI. Predicted lists for nicotine and benzo(a)pyrene were generated by both the NBI and ES-SBI methods. Supplementary Tables S5 and S6 list the top 5 miRNAs and top 5 diseases for tobacco prioritized by NBI. In addition, 5 potential miRNAs and 5 potential diseases were prioritized for nicotine, while 4 new candidate miRNAs and 4 new candidate diseases were predicted for benzo(a)pyrene by the common top 10 lists in the NBI and ES-SBI methods. Related diseases were extracted from the whole network for the potential miRNAs that were predicted to be altered by cigarette smoke. Meanwhile, the known MDAs were also extracted from our model for the candidate diseases prioritized for cigarette use. Collectively, inferring new miRNA biomarkers could improve our understanding of the relationships between cigarette smoke and smoking-related diseases. The predicted associations among tobacco smoke, miRNAs and diseases (Supplementary Tables S5 and S6) provide potential candidates for further experimental validation. For example, tobacco was predicted to alter the expression of mir-155, mir-221, let-7a-1 and mir-126, which play important roles in lung neoplasm pathology. Although there are some newly published8,38 studies for tobacco smoke, there are still not enough data to validate the performance of the PEMDAM. The entire network of tobacco smoke (Figure 5) was constructed with the known and predicted EMAs, MDAs and EDAs. This network contains 58 miRNAs and 7 diseases, which were confirmed to be associated with cigarette smoke by experimental studies. 14 predicted EMAs and 14 prioritized EDAs related to cigarette smoke were also included.
Figure 5. The discovered EF-miRNA-disease association network for Tobacco, nicotine and benzo(a)pyrene (BaP).
Tobacco, nicotine and BaP are shown as magenta hexagons. The network contains 58 known miRNAs, 7 known diseases, 12 predicted miRNAs, 8 predicted diseases, and the known associations between these miRNAs and diseases.
Discussion
miRNA network analysis will open up new avenues for the understanding of environmental toxicity and disease etiology. In addition, miRNA networks have several advantages over other types of bionetworks. miRNAs are located upstream of gene signal transduction, thus changes in miRNA expression are more sensitive and occur before changes in proteins. Furthermore, because miRNAs can be easily detected in circulation, they are suitable as sensitive indicators of toxic exposure or novel biomarkers for the prevention, diagnosis and progression of EF-related diseases39.
Our predictive computational systems toxicity model obtained a high accuracy in prioritizing the potential associations among EFs, miRNAs and diseases. This high performance is likely due to three factors: the data quality, the design of the algorithm and the workflow strategy. Firstly, the data used to build the predictive model were obtained from highly reliable databases and supported by experimental data40,41. In network analysis, including topological features and modules, it is necessary not only to have an overall understanding of the dataset used but also to ensure that these known networks conform to the inherent nature of bionetworks, which are small world42, scale-free29. These network topological characteristics are of great importance for the algorithms we used. Secondly, the NBI and SBI algorithms used in this paper were well defined and have already been proven to be successful for predicting drug-target interactions26,43 and chemical-gene-disease associations19. Only two models were needed to predict the associations in one bipartite bionetwork, thus the computational workload was greatly reduced. Last but not least, the PEMDAM has the advantages of both NBI and SBI because the final prediction results were obtained by utilizing the common lists of both NBI and SBI. For NBI, only the network topology structure similarity was needed, which was easily obtained, while SBI was only applied when specific similarities like structural similarity and phenotypic similarity are available. However, SBI performed better than NBI in small networks, such as the EDA network. Thus, using the common prioritized lists made the predicted results more reliable than using a single algorithm.
There are some limitations and room for improvement in our current methods. First, the present model can only predict new associations among known EFs, miRNAs and diseases. Our current model is unable to predict brand new EFs, miRNAs and diseases without having known association information in the training set. This could be improved by adding similarities to homogeneous nodes in a bipartite network. Based on its similarity to other nodes, the initial resource could be defined to include nodes without known links. Furthermore, our methods focused on nodes and their relationships in bipartite networks. Thus, it was a simplified model that ignored detailed mechanisms of interaction, which differs from real and complicated biological systems. EFs alter miRNA expression in directional ways, positively or negatively. There have also been inconsistencies in miRNA expression changes under the same experimental conditions. For example, in MCF-7 (ER+) breast cancer cells, oncomiR-21 was found to be down-regulated by estradiol44,45 in one study, but was found to be up-regulated by estradiol in another46. Expression profiles of the same miRNA can also vary across different samples of the same disease. As the underlying mechanisms are revealed, a directional network of interactions among EFs, miRNAs and diseases will be up for consideration. Finally, there is also room to improve our algorithm in handling small networks or sub-networks. The similarity of miRNAs, for example, their functional similarity47, could be integrated into SBI.
All of the methods applied in this paper are data-driven approaches that depend on the quantity and quality of the evaluation datasets48 for good performance. Currently, the known information about miRNA networks, especially involving environmental toxicity, is notably sparser than other networks. As more experiments are carried out, there will be enough data for the external validation and literature verification of further case studies. It will then be possible to compare different predicted miRNA results using various computational programs49. As the experimental dataset becomes enriched, computational systems toxicology programs will perform better, resulting in the development of experimental studies. We generated a comprehensive prediction list, the ‘PEMDAM lists', that includes all of the potential MDAs, EDAs and MDAs found by our computational program. Researchers interested in EF-miRNA-disease associations can download the profile for further experimental validation (www.lmmd.org/database/pemdam).
Methods
Construction of the miRNA networks
Data preparation
Three association datasets, EMA, EDA and MDA, were collected from the miREnvironment database40 (September, 2012) and the Human MicroRNA Disease Database (HMDD)41 (September, 2012). Only data tested on humans was kept. Because the same EFs, diseases or miRNAs might have different names in the databases, all of the EF and disease terms were annotated with the most commonly used vocabularies of the Unified Medical Subject Headings (MeSH)50, and the miRNAs were named according to miRBase51. After removing duplicated data, the remaining data were integrated to construct the network.
Network construction
The complete network of EFs, miRNAs and diseases was transformed into three bipartite networks: EMA, EDA and MDA. The three networks were further transformed into quantitatively descriptive matrices. The EF set was denoted as E = {e1,e2,…,en}, while M = {m1,m2,…,mn} and D = {d1,d2,…,dn} represented the miRNA and disease sets, respectively. The EMA bipartite pairs were then represented as N(E,M,A), where A = {aij: ei∈E, mj∈M}, the EDA network pairs were represented as N(E,D,A), where A = {aij: ei∈E, dj∈D}, and the MDA network pairs were represented as N(M,D,A), where A = {aij: mi∈M, dj∈D}. In this way, the EMA, EDA and MDA bipartite networks were represented as n × m adjacent matrices, where aij = 1 if direct experimental data exists in the above two databases, and 0 otherwise.
Measurement of the network topology
In order to gain a full understanding of the constructed networks, the Cytoscape plugin MCODE27 was applied to define the modules in the MDA network, and NetworkX (http://networkx.lanl.gov/, version 1.8.1) was used to calculate three classical topological features, connectivity (k), clustering coefficient (C) and betweenness (B), for the EMA, EDA and MDA networks.
Method development
Network-based inference (NBI)
Network-based inference is an algorithm that allocates known initial resources to obtain predictive lists. Figure 1 shows a simple EMA example to illustrate how to use this network-based inference algorithm to prioritize unknown miRNAs linked to EFs. The initial resources for a given EF ei in the bipartite network N (EMA) are located in the miRNAs, which are associated with ei. Each miRNA averages its resources to all of its neighbors, and they immediately redistribute these resources to every neighboring miRNA. Finally, the miRNAs that are not connected with ei are assigned the end resources, which is their score. In theory, the higher score a candidate miRNA gets, the more likely it is to be associated with ei. The initial resources of aij between ei (the yellow triangle) and mj (the green circle) was found as follows: by denoting F0n × m as the initial resource and setting F0ij = aij, Rn × n as the total resources (degrees) of each miRNA and 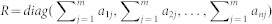 , Hm × m as the total resources of each EF and
, Hm × m as the total resources of each EF and 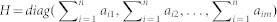 , the resource matrix was obtained as F1n × m, and F1 = F0Wm × mor
, the resource matrix was obtained as F1n × m, and F1 = F0Wm × mor 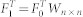 , where the transfer matrix Wm × m = (F0H−1)T(R−1F0) or Wn × n = (R−1F0)(F0H−1)T.
, where the transfer matrix Wm × m = (F0H−1)T(R−1F0) or Wn × n = (R−1F0)(F0H−1)T.
Mathematically, an algorithm to predict other associations among the EFs, miRNAs and diseases in the EF-miRNA, EF-disease and miRNA-disease partite networks can be similarly deduced.
EF structure similarity-based inference (ES-SBI)
The hypothesis underlying this method is that if an EF ei associates with miRNAs or diseases by experimental evidence, then other EFs similar to ei tend to be linked with these ei-associating miRNAs or diseases. For an unknown EMA, the linkage between ei and mj is determined by the predictive scoring function in formula (1). The association-predicting score for unknown EDAs is shown in formula (2).
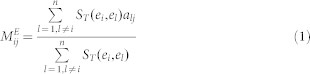 |
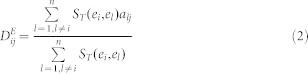 |
ST(ei,el) indicates the Tanimoto similarity of the 2D chemical structures between EFs ei and el. Detailed information about Tanimoto similarity can be found in Willett's work52. aij is adjacency matrix of N(E,M,A) in 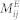 , and N(E,D,A) in
, and N(E,D,A) in 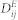 . The structures of the EFs were transformed to MACCS keys using the OpenBabel software53. However, a small portion of the EFs could not be identified with structures, for example, pathogens, radiation and pollutants. The prediction lists for these cases were generated only by NBI.
. The structures of the EFs were transformed to MACCS keys using the OpenBabel software53. However, a small portion of the EFs could not be identified with structures, for example, pathogens, radiation and pollutants. The prediction lists for these cases were generated only by NBI.
Disease phenotypic similarity-based inference (DP-SBI)
This method was designed based on the hypothesis that diseases in the same phenotypic classification tend to be associated with similar EFs and miRNAs. The phenotypic similarity of two diseases was measured by finding their relative positions in the MeSH disease directed acyclic graph (more details are given in Wang et al.47) Formulas (3) and (4) describe the predicted scores of the unknown EDAs and MDAs, respectively, where Ss(di,dl) denotes the phenotypic similarity between two diseases di & dl and aij represents the adjacency matrix of N(E,D,A) in 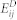 , and N(M,D,A) in
, and N(M,D,A) in 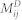 .
.
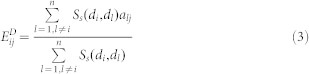 |
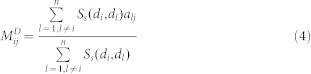 |
Performance assessment
Performance of all the models was evaluated by 10-fold cross validation. For each dataset, all links in the EMA, EDA and MDA networks were randomly divided into 10 parts of equal size. Each part was used as the validation set in turn, while the remaining nine parts served as the training set. To eliminate the error caused by separating datasets, all of the results were produced by a simulation of 100 independent tests, and the receiver operating characteristic (ROC) curves were used. Due to random partitioning of the data, some EFs, miRNAs or diseases only existed in the test set without seed information in the training set. Links among these nodes were not considered in the performance assessment.
Network visualization and analysis
The final predicted associations among EFs, miRNAs and diseases were obtained by the common prioritized lists of NBI and SBI. To visualize the relationships among the EFs, miRNAs and diseases, networks were constructed using Cytoscape 3.054 with the known associations generated by data integration and the predicted links found by the PEMDAM. The associations regarding breast cancer and cigarette smoke were then extracted to build the subnetworks during the case study analysis.
Author Contributions
Y.T., F.C. and J.L. designed the study and wrote the manuscript; J.L. and Z.W. performed the study; W.L. and G.L. participated in data analysis and model building.
Supplementary Material
Supplimentary Information
Supplimentary Table S1
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant 81373329), the 863 Project (Grant 2012AA020308), the Shanghai Tobacco Group Co. Ltd. Research Fund (Grant K2013-1-044P), the Fundamental Research Funds for the Central Universities (Grant WY1113007), and the 111 Project (Grant B07023).
References
- Lee R. C., Feinbaum R. L. & Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993). [DOI] [PubMed] [Google Scholar]
- Lee R. C. & Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864 (2001). [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature 431, 350–355 (2004). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Friedman R. C., Farh K. K.-H., Burge C. B. & Bartel D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19, 92–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemoeller O. M. et al. MicroRNA expression profiles in human cancer cells after ionizing radiation. Radiat Oncol 6, 29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim M. J. MicroRNAs: implications for air pollution research. Mutat Res-Fund Mol M 717, 38–45 (2011). [DOI] [PubMed] [Google Scholar]
- Graff J. W. et al. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PLoS One 7, e44066 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A. & Croce C. M. MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–866 (2006). [DOI] [PubMed] [Google Scholar]
- Lukiw W. J. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport 18, 297–300 (2007). [DOI] [PubMed] [Google Scholar]
- Van Rooij E. & Olson E. N. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest 117, 2369–2376 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A. & Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr 21, 243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudder A. & Novak R. F. miRNAs: effectors of environmental influences on gene expression and disease. Toxicol Sci 103, 228–240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. & Cui Q. H. Specific roles of microRNAs in their interactions with environmental factors. J Nucleic Acids 2012, 978384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. The role of microRNA in chemical carcinogenesis. J Environ Sci Heal C 28, 89–124 (2010). [DOI] [PubMed] [Google Scholar]
- Xi S. C. et al. Cigarette smoke induces C/EBP-β-mediated activation of miR-31 in normal human respiratory epithelia and lung cancer cells. PLoS One 5, e13764 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman S. L. et al. Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PLoS One 7, e32754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. X. et al. admetSAR: a comprehensive source and free tool for assessment of chemical admet properties. J Chem Inf Model 52, 3099–3105 (2012). [DOI] [PubMed] [Google Scholar]
- Cheng F. X. et al. Prediction of human genes and diseases targeted by xenobiotics using predictive toxicogenomic-derived models (PTDMs). Mol BioSyst 9, 1316–1325 (2013). [DOI] [PubMed] [Google Scholar]
- Audouze K. et al. Deciphering diseases and biological targets for environmental chemicals using toxicogenomics networks. PLoS Comput Biol 6, e1000788 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audouze K. & Grandjean P. Application of computational systems biology to explore environmental toxicity hazards. Environ Health Persp 119, 1754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. X., Li W. H., Liu G. X. & Tang Y. In silico ADMET prediction: recent advances, current challenges and future trends. Curr Top Med Chem 13, 1273–1289 (2013). [DOI] [PubMed] [Google Scholar]
- Qiu C. X., Chen G. & Cui Q. H. Towards the understanding of microRNA and environmental factor interactions and their relationships to human diseases. Sci Rep 2, 10.1038/srep00318 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. et al. Identification of links between small molecules and miRNAs in human cancers based on transcriptional responses. Sci Rep 2, 10.1038/srep00282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu M. X., Cui Q. H. & Yan G. Y. Prediction of disease-related interactions between microRNAs and environmental factors based on a semi-supervised classifier. PLoS One 7, e43425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. X. et al. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol 8, e1002503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G. D. & Hogue C. W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge N. J., Gardiner E., Carroll A. P., Tooney P. A. & Cairns M. J. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatr 15, 1176–1189 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A. L. & Albert R. Emergence of scaling in random networks. Science 286, 509–512 (1999). [DOI] [PubMed] [Google Scholar]
- Boyle P. & Levin B. World Cancer Report 2008. (IARC Press, InteRNAtional Agency for Research on Cancer, 2008). [Google Scholar]
- Dvinge H. et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 497, 378–382 (2013). [DOI] [PubMed] [Google Scholar]
- Zaman N. et al. Signaling network assessment of mutations and copy number variations predict breast cancer subtype-specific drug targets. Cell Rep 5, 216–223 (2013). [DOI] [PubMed] [Google Scholar]
- Heiser L. M. et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A 109, 2724–2729 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. The facts about smoking and health. (2006).
- Banerjee A. & Luettich K. MicroRNAs as potential biomarkers of smoking-related diseases. Biomark Med 6, 671–684 (2012). [DOI] [PubMed] [Google Scholar]
- Schembri F. et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A 106, 2319–2324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani M. A. et al. Maternal cigarette smoking during pregnancy is associated with downregulation ofmiR-16, miR-21, and miR-146a in the placenta. Epigenetics 5, 583–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. K. et al. Nicotine alters microRNA expression and hinders human adult stem cell regenerative potential. Stem Cells Dev 22, 781–790 (2012). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18, 997–1006 (2008). [DOI] [PubMed] [Google Scholar]
- Yang Q. Q., Qiu C. X., Yang J., Wu Q. & Cui Q. H. miREnvironment database: providing a bridge for microRNAs, environmental factors and phenotypes. Bioinformatics 27, 3329–3330 (2011). [DOI] [PubMed] [Google Scholar]
- Lu M. et al. An analysis of human microRNA and disease associations. PLoS One 3, e3420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D. J. & Strogatz S. H. Collective dynamics of ‘small-world' networks. Nature 393, 440–442 (1998). [DOI] [PubMed] [Google Scholar]
- Cheng F. X., Zhou Y. D., Li W. H., Liu G. X. & Tang Y. Prediction of chemical-protein interactions network with weighted network-based inference method. PLoS One 7, e41064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe N. S. et al. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res 37, 2584–2595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcuklu S. D., Donoghue M. T., Kerin M. J. & Spillane C. Regulatory interplay between miR-21, JAG1 and 17beta-estradiol (E2) in breast cancer cells. Biochem Bioph Res Co 423, 234–239 (2012). [DOI] [PubMed] [Google Scholar]
- Bhat-Nakshatri P. et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res 37, 4850–4861 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Wang J., Lu M., Song F. & Cui Q. H. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics 26, 1644–1650 (2010). [DOI] [PubMed] [Google Scholar]
- Lusher S. J. et al. Data-driven medicinal chemistry in the era of big data. Drug Discov Today 19, 10.1016/j.drudis.2013.12.004 (2014). [DOI] [PubMed] [Google Scholar]
- Witkos T. M., Koscianska E. & Krzyzosiak W. J. Practical aspects of microRNA target prediction. Curr Mol Med 11, 93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb C. E. Medical subject headings (MeSH). Bull Med Libr Assoc 88, 265–266 (2000). [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res 32, D109–D111 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discov Today 11, 1046–1053 (2006). [DOI] [PubMed] [Google Scholar]
- O'Boyle N. M. et al. Open Babel: An open chemical toolbox. J Cheminformatics 3, 1–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplimentary Information
Supplimentary Table S1