Background: Promyelocytic leukemia zinc finger (PLZF) is transcription repressor that recruits nuclear co-repressors at the target promoters. PLZF is overexpressed in various human solid tumors, such as clear cell renal carcinoma, glioblastoma, and seminoma.
Results: PLZF represses transcription of CDKN1A by inhibition of p53 acetylation, Sp1 binding.
Conclusion: PLZF causes cellular transformation and increases cell proliferation by repressing transcription of CDKN1A.
Significance: PLZF can act as a proto-oncogene depending on the cell types.
Keywords: Cell Cycle, p53, Specificity Protein 1 (Sp1), Transcription Regulation, Transcription Repressor, PLZF, p21WAF/CDKN1A
Abstract
Promyelocytic leukemia zinc finger (PLZF) is a transcription repressor that was initially isolated as a fusion protein with retinoic acid receptor α. PLZF is aberrantly overexpressed in various human solid tumors, such as clear cell renal carcinoma, glioblastoma, and seminoma. PLZF causes cellular transformation of NIH3T3 cells and increases cell proliferation in several cell types. PLZF also increases tumor growth in the mouse xenograft tumor model. PLZF may stimulate cell proliferation by controlling expression of the genes of the p53 pathway (ARF, TP53, and CDKN1A). We found that PLZF can directly repress transcription of CDKN1A encoding p21, a negative regulator of cell cycle progression. PLZF binds to the proximal Sp1-binding GC-box 5/6 and the distal p53-responsive elements of the CDKN1A promoter to repress transcription. Interestingly, PLZF interacts with Sp1 or p53 and competes with Sp1 or p53. PLZF interacts with corepressors, such as mSin3A, NCoR, and SMRT, thereby deacetylates Ac-H3 and Ac-H4 histones at the CDKN1A promoter, which indicated the involvement of the corepressor·HDACs complex in transcription repression by PLZF. Also, PLZF represses transcription of TP53 and also decreases p53 protein stability by ubiquitination. PLZF may act as a potential proto-oncoprotein in various cell types.
Introduction
The POZ-domain Krüppel-like zinc finger (POK)2 family proteins have an N-terminal POZ domain and Krüppel-like (C2H2) zinc finger domains at the C-terminal. The POK proteins are major determinants in apoptosis, differentiation, development, transcription, tumor suppression, and oncogenesis (1). The oncogenic members of the POK family include promyelocytic leukemia zinc finger (PLZF)-retinoic acid receptor α (2), B cell lymphoma 6 (BCL-6) (3), and FBI-1 (factor that binds to the inducer of short transcripts of human immunodeficiency virus-1) (4–6). PLZF has a variety of regulatory functions and is one of the few well characterized POK family transcription factors. PLZF was originally identified as a fusion protein with retinoic acid receptor α in acute promyelocytic leukemia, a disease characterized by an accumulation of undifferentiated myeloid blasts (2).
PLZF is a transcription repressor that recruits nuclear co-repressors to establish a silent chromatin structure at the target promoters (7–10). Plzf is required to regulate self-renewal and stem cell pool maintenance in testis (11, 12). Plzf also plays roles in development by repressing Hox and bone morphogenetic protein gene expression. Plzf controls segment patterning and establishes proper Hox gene expression boundaries (13, 14). PLZF is expressed in CD34+ hematopoietic progenitors, suggesting it may play a role in lineage determination (15). PLZF has been implicated in the development of the megakaryocytic (16) and NKT cell lineages (17, 18). Ectopic PLZF inhibited proliferation and differentiation in myeloid cell lines (19–21).
Overexpression of PLZF has been shown to induce cell cycle arrest at the G1 to S transition and represses the expression of pro-proliferative genes, such as Cyclin A, CCNA2, and MYC (19, 22, 23). The cyclin-dependent kinase involved during the G1 to S transition (CDK2) phosphorylates PLZF at two consensus sites found within the PEST domain in the hinge region. The phosphorylation triggers ubiquitination and subsequent degradation of PLZF, which antagonizes its growth inhibitory effects and may be relevant for cell cycle progression during human cancer development (23). Tumor xenograft experiment showed that Plzf reduces melanoma tumor growth, suggesting PLZF has a suppressor function in melanoma solid tumors (24). PLZF knock-out mice study showed that PLZF can act as a growth inhibitor and proapoptotic factor in limb bud (13). PLZF has been shown to promote apoptosis in cervical cancer and Jurkat T-cell leukemic cells (25).
However, the function of PLZF on either anti-proliferation or apoptosis was obscured by the following observations. Plzf knock-out mice show increased expression of p21 and p53 in spermatogonia (Gene expression omnibus analysis: www.ncbi.hlm.nih.gov/geo). More recent publications also indicate that PLZF might stimulate cell proliferation. Costaya et al. (12) reported that, in Plzf knock-out mice, testis size and mass were significantly reduced. Expression of Cyclin D1, a marker of mitotic spermatogonia, and BrdU incorporation were decreased. The number of spermatogonia was decreased (12). PLZF was shown to down-regulate apoptosis by inhibiting expression of the proapoptotic BID protein in lymphocytes (26). These data suggest that PLZF might stimulate cell proliferation. In some cancer tissues, such as clear cell renal cell carcinoma, glioblastoma, and seminoma, PLZF expression is increased and might contribute to cellular transformation and proliferation (Oncomine database; www.ncbi.nlm.nih.gov/geo).
p21, encoded by the CDKN1A gene, is a major regulator of cell cycle arrest (27, 28). CDKN1A is primarily regulated at the transcription level (29). Whereas induction of p21 predominantly leads to cell cycle arrest, repression of CDKN1A gene expression may have a variety of outcomes, including cell proliferation, depending on the cell context (29). The CDKN1A gene is regulated by p53 induced by DNA-damaging agents and plays a crucial role in mediating G1, G2, and S phase growth arrest (28, 29). In addition to p53, Sp1-family transcription factors (30, 31) are major regulators that affect CDKN1A gene expression, and they bind to the proximal promoter. Sp1 can interact with basal transcription machinery, other transcription factors, co-activators and corepressors, including Myc, p53, Rb, TATA-binding protein, p300, HDAC, and SMRT/NCoR. These interactions and direct binding competition between Sp1 family and POK family transcription factors are important for transcription regulation of the CDKN1A gene (4, 5, 29–34).
Although there are a number of publications on PLZF, little is known on how PLZF regulates cell cycle or proliferation. We investigated how expression of the tumor suppressor p21 can be controlled by PLZF. Our data showed that PLZF represses transcription of CDKN1A, stimulates cellular transformation and proliferation in several cell lines, and increases tumor growth in mice xenograft tumor model.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents
Various promoter-reporter gene fusion plasmids and pcDNA3-p53, pcDNA3-Sp1, and co-repressor expression vectors were either reported elsewhere or prepared by us (4, 5). The pcDNA3.1-PLZF plasmid was prepared by cloning a full-length cDNA into pcDNA3.1 (Invitrogen). To prepare the recombinant GST-PLZF-ZF proteins, cDNA fragments encoding the zinc fingers (amino acids 404 to 673) were cloned into pGEX4T3 (Amersham Biosciences). All plasmid constructs were verified by DNA sequencing. Antibodies against p21, p53, HDAC1, HDAC3, MDM2, PLZF, Sp1, GAPDH, Myc tag, Ac-H3, Ac-H4, mSin3A, NCoR, and SMRT were purchased from Upstate (Charlottesville, VA), Chemicon (Temecula, CA), Cell Signaling Technology (Beverly, MA), Calbiochem, and Santa Cruz Biotechnology (Santa Cruz, CA). Most of the chemical reagents were purchased from Sigma.
Cell Cultures
HEK293, HeLa, HCT116 p53+/+, and HCT116 p53−/− cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen). Saos-2 cells were cultured in McCoy's 5A medium (Invitrogen) supplemented with 15% FBS. Caki-1, H460, and Jurkat T-cells were cultured in RPMI medium (Invitrogen) supplemented with 10% FBS.
Immunohistochemistry
Histologic analyses of mice were conducted by following standard protocols. The 4-μm formalin-fixed, paraffin-embedded tissue sections were stained with hematoxylin and eosin reagent for counterstaining. 3,3′-Diaminobenzidine immunohistochemical staining was done according to the manufacturer's instructions after incubation with primary antibody (R.T.U. VECTASTAIN ABC Kit; Vector Laboratories).
Preparation of Recombinant Adenovirus shRNA against PLZF mRNA
To prepare recombinant adenovirus expressing shRNA against PLZF, annealed shRNA DNA sequences (sense, 5′-GATCCCGATGTTTGAGATCCTCTTCTTTTTCAAGAGA(loop)-CTACAAACTCTAGGAGAAGTTTTTTTGGAA(loop)-A-3′); antisense, 5′-AGCTTTTCCAAAAA(loop)-AAGATGTTTGAGATCCTCTTCTTCTCTTGAA(loop)-AACTACAAACTCTAGGAGAAGGG-3′) were cloned into pSilencer 2.0-U6 (Ambion, Austin, TX) and subcloned into the pΔE1sp1A vector. The pΔE1sp1A-U6-shPLZF vector and the adenovirus vector vmdl324Bst were linearized by restriction enzyme digestion. The linearized pΔE1sp1A-U6-shPLZF was cotransformed into Escherichia coli BJ518 with the vmdl324Bst vector for homologous recombination. Homologous recombinant adenoviral plasmid was digested with PacI and transfected into HEK293A cells to generate the adenovirus shRNA against PLZF (dE1-k35/shPLZF).
PLZF Action on Tumor Growth in a Xenograft Tumor Model in Mice
Caki-1 tumor cells were implanted under the abdominal skin of male BALB/c-nu mice. Once tumors reached 100 to 120 mm3 in volume, mice were injected intratumorally 3 times at 2-day intervals with either control dE1-k35 or dE1-k35/shPLZF adenovirus (1 × 108 pfu). Tumor growth was monitored by measuring the length and width of the tumor 3 times a week using a caliper. Tumor volume was calculated as 0.523 Lw2, in which L is the length and w is the width in mm.
FACS Analysis
HEK293 cells were transfected with either a PLZF expression vector or PLZF siRNA. The cells were washed, fixed with methanol, and stained with a solution containing propidium iodide (50 μg/ml) and ribonuclease A (100 μg/ml) for 30 min at 37 °C in the dark. The DNA content, cell cycle profiles, and forward scatter were analyzed using a FACSCalibur (BD Biosciences) flow cytometer with excitation at 488 nm and detection at 575 nm (peak emission). The data were analyzed using ModFit LT 2.0 (Verity Software House, Inc.) and WindMDI 2.8 (Joseph Trotter, Scripps Research Institute, CA).
Foci Formation Assays
NIH3T3 cells (1 × 105 cells/well) were transfected with either pcDNA3 or pcDNA3.1-PLZF (0.5 μg/μl). Transfected cells were cultured for 2 weeks, and the colonies resistant to G418 (800 μg/ml) selection were stained with crystal violet (0.5% in 20% ethanol).
MTT Assays
Confluent HEK293, Caki-1, HCT116, and H460 cells grown on 10-cm culture dishes were transfected with either a PLZF expression vector or PLZF siRNA, transferred to 6-well culture dishes and grown for 0–4 days. At days 0, 1, 2, 3, and 4, the cells were incubated for 1 h at 37 °C with 20 μl/well of MTT (2 mg/ml). Precipitates were dissolved with 1 ml of dimethyl sulfoxide. The level of cellular proliferation was determined by MTT conversion to formazan using a SpectraMAX 250 (Molecular Device Co., Sunnyvale, CA) at 570 nm.
Cell Cultures and Transcription Analysis of ARF, MDM2, TP53, p21WAF/CDKN1A, and p53 Responsive Promoters
The cells (HEK293, HCT116, H460, and Saos-2) were cultured in the medium recommended by ATCC (Rockville, MD). The reporter fusion plasmids pGL2-ARF-Luc, pGL2-MDM2-Luc, pGL2-TP53-Luc, pG13-Luc, pG5–5x(GC-box)-Luc, and pGL2-CDKN1A-Luc, as well as pcDNA3.1-PLZF, pcDNA3-p53, and pCMV-LacZ in various combinations were transiently transfected into cell lines using Lipofectamine Plus reagent (Invitrogen) and analyzed as described elsewhere (4, 5)
Knock-down of PLZF mRNA by siRNA
The following two siRNA against PLZF mRNA were designed and purchased from Bioneer (Taejeon, Korea): PLZF siRNA-1, sense, 5′-GGC CAACCAGAUGCGGCUGUU-3′, and antisense, 5′-UUCCGGUUGGUCUACGCCGAC-3′; and PLZF siRNA-2, sense, 5′-GAUGUUUGAGAUCCUCUUCUU-3′, and antisense, 5′-UUCUACAAACUCUAGGAGAAG-3′. The siRNA (200 pmol) was transfected into HEK293 cells using Lipofectamine RNAiMAX (Invitrogen). After transfection, the cells were harvested, total RNA was prepared, and mRNA RT-quantitative PCR (qPCR) analysis was performed as described above.
RT-qPCR of PLZF, MDM2, p53, GAPDH, and p21 mRNA Expression
Total RNA was isolated from the cells using TRIzol reagent (Invitrogen). cDNA was synthesized using 5 μg of total RNA, random hexamer (10 pmol), and Superscript reverse transcriptase II (200 units) (Invitrogen). RT-qPCR was performed using SYBR Green Master Mix (Applied Biosystems) and the following RT-qPCR oligonucleotide primers sets: PLZF-1 forward, 5′-AACCACAAGGCTGACGCTGTA-3′ and reverse, 5′-CATAGGTGCTGAAGTCCATGGA-3′; PLZF-2 forward, 5′-CGGGACTTTGTGCGATGTG-3′ and reverse, 5′-GCGGTGGAAGAGGATCTCAA-3′; MDM2 forward, 5′-CCCCTTAATGCCATTGAACCT-3′ and reverse, 5′-ACTGGGCAGGGCTTATTCCT-3′; p53 forward, 5′-CCTGAGGTTGGCTCTGACTGTA-3′, and reverse, 5′-AAAGCTGTTCCGTCCCAGTAGA-3′; p21 forward, 5′-AGGGGACAGCAGAGGAAG-3′ and reverse, 5′-GCGTTTGGAGTGGTAGAAATCTG-3′; GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′.
Western Blot Analysis
Cells were harvested and lysed in RIPA buffer (50 mm Tris-HCl, pH 8.0, 1% Nonidet P-40, 0.25% sodium deoxycholic acid, 150 mm NaCl, 1 mm EGTA, complete Mini-Protease mixture). Cell extracts (40 μg) were separated using 10% SDS-PAGE, transferred onto ImmunoBlotTM PVDF membranes (Bio-Rad), and blocked with 5% skim milk (BD Biosciences). Blotted membranes were incubated with antibodies against Myc tag (Cell Signal, MA), GAPDH (Chemicon, CA), p21, p53, HDAC1, NcoR, HDAC3, PLZF, and SMRT and then incubated with either an anti-mouse or rabbit secondary antibody conjugated with HRP (Vector Laboratory). Protein bands were visualized using an ECL solution (PerkinElmer Life Sciences).
Chromatin Immunoprecipitation-qPCR Assays
HCT116 and HEK293 cells were transfected with an increasing amount of PLZF expression vector, pcDNA3.1-PLZF. In these cells, the molecular interaction between PLZF and either p53 or Sp1 at the endogenous CDKN1A promoter and histone modification at the CDKN1A proximal promoter was analyzed using the standard ChIP assay protocol, as described elsewhere (4, 5).
The levels of PLZF binding at p53RE-1 and -2, as well as at the Sp1-binding GC-boxes 5/6, were analyzed using an anti-PLZF antibody (Santa Cruz Biotechnology). The levels of endogenous Sp1 and p53 protein binding were analyzed using polyclonal antibodies against p53 and Sp1 (Santa Cruz). As a negative control for the qChIP assays, IgG was used.
The following oligonucleotide primer sets, designed to amplify the upstream regulatory regions surrounding the p53-binding sites, the CDKN1A proximal promoter region, and the 3′-UTR region, were used: p53RE-1 binding primers (bp, −2307 to −1947), forward, 5′-CTGTGGCTCTGATTGGCTTT-3′, and reverse, 5′-GGGTCTTTAGAGGTCTCCTGTCT-3′; p53RE-2 binding primers (bp, −1462 to −1128), forward, 5′-CCACAGCAGAGGAGAAAGAAG-3′, and reverse, 5′-GCTGCTCAGAGTCTGGAAATC-3′; CDKN1A proximal promoter primers (bp, −133 to +30), forward, 5′-GCGCTGGGCAGCCAGGAGCCT-3′, and reverse, 5′-CTGACTTCGGCAGCTGCTCAC-3′; CDKN1A 3′-UTR (bp, +869 to +1046), forward, 5′-TCCTTCCCATCGCTGTCACA-3′, and reverse, 5′-GTCACCCTGCCCAACCTTAG-3′. To analyze histone H3 and H4 modifications at the CDKN1A proximal promoter (bp, −131 to +100), forward, 5′-GATCGGTACCGCGCTGGGCAGCCAGGAGCCT-3′, and reverse, 5′-TCGTCACCCGCGCACTTAGA-3′, primers were used.
Immunoprecipitation Assays
HCT116 and HEK293 cells (transfected with an expression vector, if necessary) were washed, pelleted, and resuspended in a lysis buffer supplemented with protease inhibitors (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1% Triton X-100). The cell lysate was precleared, the supernatant was incubated overnight with an anti-PLZF (or anti-p53, anti-Sp1 anti-Myc, anti-p300, and anti-corepressor) antibody on a rotating platform at 4 °C and then incubated with protein A-Sepharose Fast Flow beads. The beads were collected, washed, and resuspended in equal volumes of 5× SDS loading buffer. The immunoprecipitated proteins were separated using 12% SDS-PAGE. The Western blot assay was performed as described above using the appropriate antibodies.
GST Fusion Protein Purification
Recombinant GST, GST-POZ-PLZF, and GST-ZF-PLZF fusion proteins were prepared from E. coli BL21(DE3) grown for 4 h at 37 °C in medium containing 1 mm isopropyl 1-thio-β-d-galactopyranoside. The E. coli were lysed and purified using glutathione-agarose 4 bead affinity chromatography (Peptron, Taejeon, Korea). The purified proteins were then resolved using 12% SDS-PAGE to quantify and assess purity.
Ubiquitination Assay
p53 null H1299 cells were transfected with pcDNA3-p53 and pcDNA3-His-Ubiquitin in the presence or absence of pcDNA3.1-PLZF. 24 h after transfection, the cells were treated with 20 μm MG132 for 3 h and harvested. Cell pellets were resuspended in RIPA buffer and cell lysates were incubated with MagneHisTM nickel particles for 1 h at 4 °C. The precipitated pellets were washed, resuspended in 2× SDS sample buffer, resolved by 10% SDS-PAGE, and analyzed by Western blotting using anti-p53 antibody.
Site-directed Mutagenesis of the p21WAF/CDKN1A Gene Promoter
To investigate the role of each Sp1 binding site, mutations were introduced into the CDKN1A gene proximal promoter sequence using the QuikChange site-directed mutagenesis kit (Stratagene). To introduce mutations into the core binding sequences of GC-box sites, the following oligonucleotides were used (note that only top strands are shown): mSp1–2, 5′-CCCGGGCGGCGCGGTTTTCCGAGCGCGGGTCCCG-3′; mSp1–3, 3′-CCCGCCTCAAGGAGGCGGGAAAAGCGCTCGGCCC-5′; mSp1–4, 5′-CCCGCCTCCTTGAGGCTTTCCCGGGCGGGGCGGT-3′; and mSp1–5/6′-TGAGGCGGGCCCGGGCTTTGCGGTTGTATATCAG-3′. For site-directed mutagenesis, 18 PCR cycles with denaturation at 94 °C for 30 s, hybridization at 55 °C for 1 min, and extension at 68 °C for 10 min per cycle were used. Amplified mixtures were treated with DpnI (Stratagene) at 37 °C for 1 h, and aliquots were used to transform competent E. coli. All of the plasmid constructs were confirmed by DNA sequencing using an ABI automatic DNA sequencer (Ramlsey, MN).
Oligonucleotide Pulldown Assays
HEK293 cells were lysed in HKMG buffer (10 mm HEPES, pH 7.9, 100 mm KCl, 5 mm MgCl2, 10% glycerol, 1 mm DTT, and 0.5% Nonidet P-40). Cellular extracts were incubated with 1 μg of biotinylated double-stranded oligonucleotides (p53RE-1, p53RE-2, and Sp1–5/6) for 16 h. The oligonucleotide sequences were as follows (only the top strands are shown): Sp1–5/6, 5′-CCTTGAGGCGGGCCCGGGCGGGGCGGTTGTATATCAGGGC-3′; p53RE-1, 5′-GTCAGGAACATGTCCCAACATGTTGAGCTC-3′; and p53RE-2, 5′-TAGAGGAAGAAGACTGGGCATGTCTGGGCA-3′; 3′-UTR, 5′-TCCTGGAGCAGACCACCCCGC-3′. To collect the DNA-bound proteins, the mixtures were incubated with streptavidin-agarose beads for 2 h, washed with HKMG buffer, and precipitated by centrifugation. The precipitate was analyzed using Western blots and antibodies against PLZF, Sp1, p53, and GAPDH, as described above.
EMSA (Electrophoretic Mobility Shift Assay)
The EMSAs were performed following standard protocols. The probe sequences for the Sp1 response elements at the CDKN1A proximal promoter used in the EMSAs were as follows (only the top strands are shown): GC-box 1, 5′-GATCGGGAGGGCGGTCCCG-3′; GC-box 2, 5′-GATCTCCCGGGCGGCGCG-3′; GC-box 3, 5′-GATCCGAGCGCGGGTCCCGCCTC-3′; GC-box 4, 5′-GATCCTTGAGGCGGGCCCG-3′; GC-box 5/6, 5′-GATCGGGCGGGGCGGTTGTATATCA-3′.
Jurkat T-cell FACS Analysis, Apoptosis Analysis, and Western Blotting
Jurkat T-cells (5 × 106) were transfected with either a pcDNA3 or pcDNA3.1-PLZF expression plasmid in a 0.4-cm cuvette by applying five pulses of 270 V for 5 ms using a Gene Pulser X cell electroporation system (Bio-Rad). Cells were recovered in fresh media for 24 h and apoptosis was measured using an apoptosis detection kit (BD Pharmingen) by flow cytometry. Both early apoptotic (Annexin V-positive, PI-negative) and late apoptotic (Annexin V-positive and PI-positive) cells were included in cell death determination. Cell cycle analysis was independently performed by staining cells with propidium iodide, and immediately analyzing them using flow cytometry. The expression levels of PLZF, p53, and p21 were detected by Western blotting.
Statistical Analysis
Student's t test was used for the statistical analyses.
RESULTS
PLZF Expression Is Higher in Human Solid Tumors Compared with Normal Tissues
We investigated a relationship between PLZF expression and cancer types using the data available from the Oncomine database of tumor expression profiles. PLZF expression is significantly increased in tumor tissues such as clear cell renal cell carcinoma, glioblastoma, and seminoma, compared with control tissues. PLZF expression is increased by an average of 3.2–4.2-fold in three types of cancers shown (Fig. 1A). An immunohistochemical analysis of human tissue microarrays (AccuMax array, A301V) of paired normal and cancer tissues using an antibody against PLZF showed that PLZF expression was significantly higher in kidney, brain, and testis cancer tissues compared with paired normal tissues (Fig. 1B). The microscopy images were analyzed by ImmunoRatio program. The mean percentage of PLZF-positive cells in normal kidney, brain, and testis tissues was 20.2–25.1%, and that of clear cell renal cell carcinoma, glioblastoma, and seminoma tissues with high PLZF expression was 42.3–86.6%, suggesting a high correlation between PLZF expression and cancer of kidney, brain, and testis (Fig. 1C).
FIGURE 1.
PLZF expression is increased in human clear cell renal cell carcinoma, testis seminoma, and brain glioblastoma. A, Oncomine data for PLZF mRNA expression in three cancer tissues. PLZF expression is increased in clear cell renal cell carcinoma (4.2-fold), glioblastoma (3.7-fold), and seminoma (3.2-fold), compared with control tissues. y-axis, log2 PLZF mRNA expression. Kidney cancer, control, three normal fetal kidneys. Sample, 14 clear cell renal cell carcinomas; brain cancer. Control, average of PLZF mRNA expression of three neural stem cell lines. Sample, average of 22 glioblastoma; testis cancer. Control, three testes. Sample, three testicular seminomas. B, immunohistochemical analysis of PLZF expression in paired normal (N) and cancer tissues (C#1, #2) using an antibody against PLZF. Magnification at ×40. Insets, ×80. Scale bar, 100 μm. C, ImmunoRatio Program analyses of the microscopy images of PLZF expression in normal and cancer tissues. *, p < 0.05; **, p < 0.001, t test.
Knock-down of PLZF mRNA Inhibits Tumor Growth in a Mouse Xenograft Model
As PLZF is overexpressed in human cancer tissues, PLZF may stimulate tumor growth by stimulating cell proliferation. We tested whether knock-down of PLZF mRNA can inhibit tumor growth in mouse (BALB/c-nu) xenografted with human clear cell renal cell carcinoma Caki-1 cells. The recombinant adenovirus dE1-k35/shPLZF expressing shRNA against PLZF inhibited tumor growth by 49% compared with the control infected with dE1-k35 adenovirus. At day 49 post-adenoviral infection, the tumors treated with PBS showed an average size of 1824.9 mm3. Tumors infected with adenovirus dE1-k35 or dE1-k35/shPLZF showed an average size of 923.3 and 241.5 mm3, respectively (Fig. 2, A and B).
FIGURE 2.
Knockdown of PLZF mRNA inhibits tumor growth in mice xenograft models. A, tumor growth curve. Implanted subcutaneous tumors derived from Caki-1 cells were injected with PBS (♦), adenovirus dE1-k35 (■), or dE1-k35/shPLZF (▴). PBS, a negative control; dE1-k35, control adenovirus; dE1-k35/shPLZF, adenovirus overexpressing shRNA against PLZF. Tumor volume was measured every other day after injection with adenovirus. The arrows indicated the days that adenovirus was injected intratumorally (1 × 108 pfu per mouse). The number of animals per group is indicated in parentheses. Values represent mean ± S.E. for eight animals per group. p < 0.001 versus dE1-k35. B, representative images of nude mice with tumor xenografts at 49 days of post-treatment with PBS, dE1-k35, or dE1-k35/shPLZF.
PLZF Stimulates Cellular Transformation and Proliferation
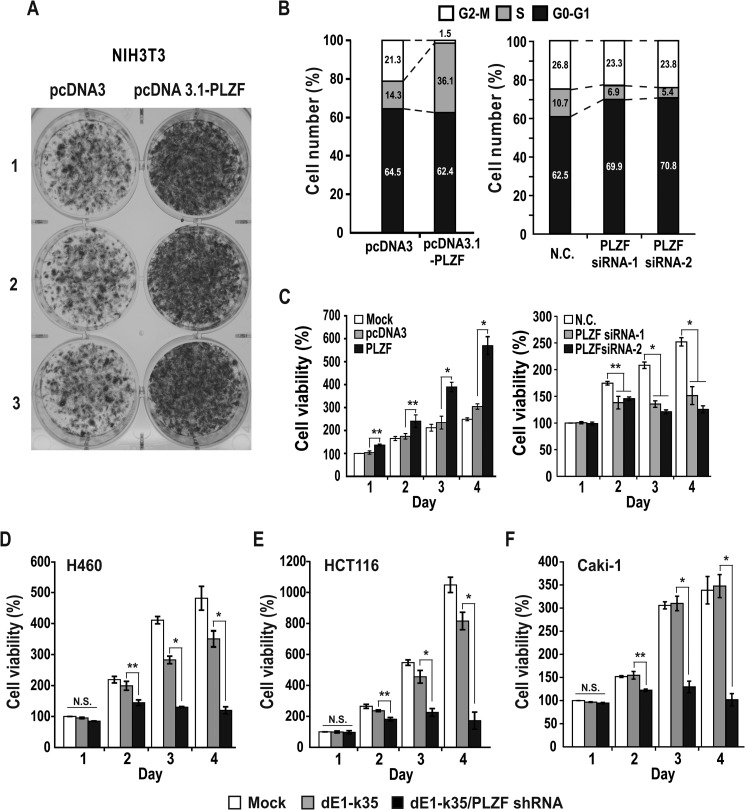
As PLZF is overexpressed in human cancer tissues and knock-down of PLZF inhibits tumor growth in a mice xenograft model, PLZF may stimulate cell proliferation. We tested whether PLZF can cause cellular transformation and promote cell proliferation. The NIH3T3 cells transfected with a PLZF expression vector formed a substantial number of large, transformed foci, suggesting that PLZF caused NIH3T3 cells to transform into faster growing cells (Fig. 3A). FACS analysis of the HEK293 cells transfected with a PLZF expression plasmid showed that PLZF stimulated cell cycle progression and increased the number of S phase HEK293 cells (14.3% in control versus 36.1% in HEK293-PLZF) (Fig. 3B). The cells transfected with PLZF siRNA showed a decrease in the number of cells in S phase (10.7% in negative control versus 5.4–6.9% in PLZF siRNA). MTT assays showed that ectopic PLZF significantly increased cell proliferation and knock-down of PLZF expression by siRNA decreased cell proliferation in HEK293 cells (Fig. 3C).
FIGURE 3.
PLZF causes cellular transformation and promotes cell proliferation in HEK293, H460, HCT116, and Caki-1 cells. A, foci formation assays. NIH3T3 cells transfected with PLZF expression vector were selected using G418. Cells were fixed and stained with crystal violet. B, FACS analysis of the HEK293 cells transfected with either a pcDNA3.1-PLZF plasmid or PLZF siRNA-1 or -2. C, MTT assay of cell proliferation. HEK293 cells were transfected with either pcDNA3.1-PLZF plasmid or PLZF siRNA-1 and -2, grown 1–4 days, and analyzed for viability by MTT to formazan conversion using colorimetry at 540–600 nm. D–F, MTT assay of cell proliferation. H460, HCT116, and Caki-1 cells infected with either a recombinant control adenovirus (dE1-k35) or adenovirus expressing PLZF shRNA-1 (dE1-k35/PLZF shRNA) were grown 1–4 days and analyzed as described above for C. *, p < 0.001. Mean values of three independent experiments are shown. Error bars represent S.D.
We also tested whether PLZF stimulates cell proliferation also in other cell types such as human colon cancer HCT116, lung cancer H460, and clear cell renal cell carcinoma Caki-1 (Fig. 3, D–F). Knock-down of PLZF expression by recombinant adenovirus expressing shRNA against PLZF resulted in a decrease in cell proliferation, suggesting PLZF can stimulate cell proliferation in HEK293, H460, HCT116, and Caki-1 cells.
PLZF Is a Transcription Repressor of TP53 and CDKN1A
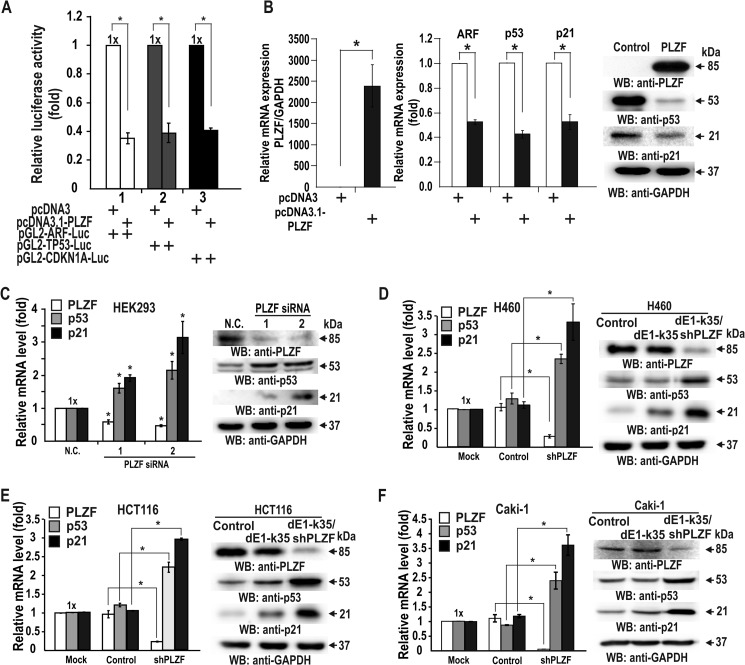
As PLZF is expressed high in human cancer tissues compared with normal tissues, PLZF may stimulate cell proliferation by controlling expression of the genes (ARF, TP53, and CDKN1A) of the p53 pathway, in particular the CDKN1A gene. Transient transcription assays showed that PLZF represses not only the gene promoters of the p53 pathway fused with the luciferase gene in HEK293 cells but also the expression of endogenous p53, and p21 at mRNA and protein levels (Fig. 4, A and B). Additionally, knockdown of PLZF mRNA in HEK293, H460, HCT116, and Caki-1 cells by either transfection with PLZF siRNA or infection with recombinant adenovirus expressing PLZF shRNA increased expression of endogenous genes (TP53 and CDKN1A) of the p53 pathway (Fig. 4, C–F).
FIGURE 4.
PLZF represses transcription of the CDKN1A and TP53 genes in HEK293, H460, HCT116, and Caki-1 cells. A, transient transcription assays of ARF, TP53, and CDKN1A. PLZF expression vector and promoter-luciferase fusion reporter plasmids were transiently co-transfected into HEK293 cells, and luciferase activity was measured. B, RT-qPCR and Western blot (WB) analysis of endogenous p53 and p21 expression in HEK293 cell lysates with ectopic PLZF expression. GAPDH, control. C–F, RT-qPCR and Western blot analysis of endogenous PLZF, p53, and p21 expression in HEK293, H460, HCT116, and Caki-1 cells transfected with PLZF siRNA-1 or -2 or treated with recombinant adenovirus overexpressing PLZF shRNA. 18S RNA, mRNA normalization control. GAPDH, protein loading control. N.C., scrambled RNA. dE1-k35, control adenovirus. dEl-k35/PLZF shRNA, recombinant adenovirus overexpressing PLZF shRNA. *, p < 0.001. Mean values of three independent experiments are shown. Error bars represent S.D.
PLZF Interacts with p53 and Decreases p53 Acetylation, which Result in Ubiquitination of p53
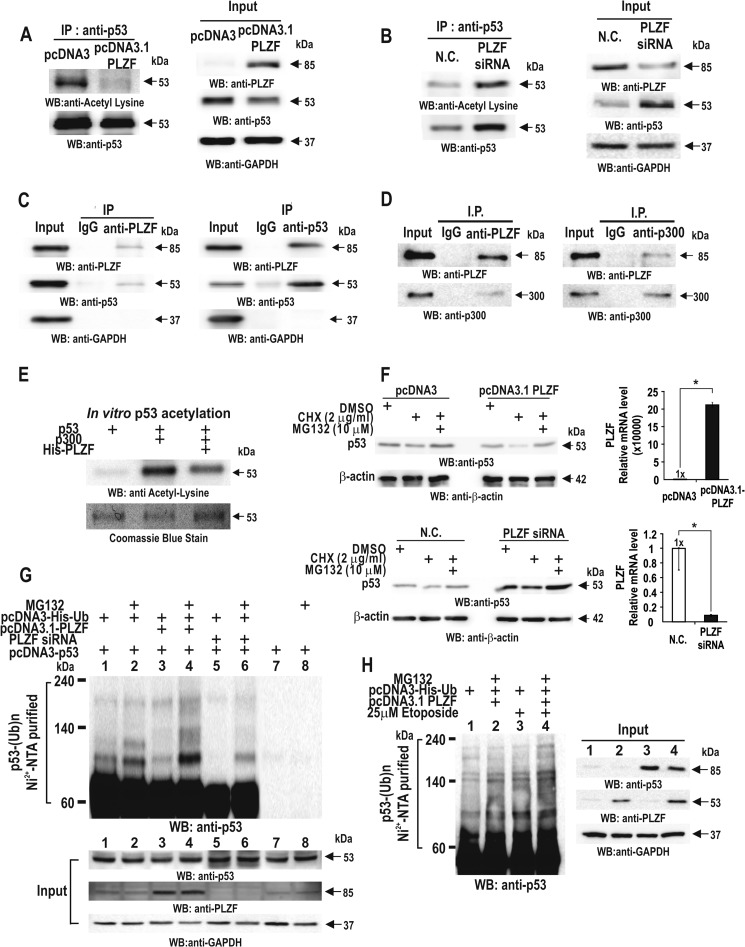
PLZF repressed transcription of TP53. We further investigated whether PLZF can regulate p53 expression at the post-translational level. Ectopic PLZF decreased not only the p53 protein level but also acetylation of p53 in the HCT116 cells. Also, knockdown of PLZF resulted in an increase in acetylated p53 and p53 expression (Fig. 5, A and B).
FIGURE 5.
PLZF interacts with p53 and decreases p53 protein stability by decreasing its acetylation and increasing its ubiquitination. A and B, immunoprecipitation of p53, followed by Western blot (WB) assays of acetylated p53 of the immunoprecipitates using anti-lysine antibody. HCT116 cells were transfected with PLZF expression vector (A) or PLZF siRNA (B), and lysates were immunoprecipitated using anti-p53 antibody. C and D, co-immunoprecipitation of PLZF and p300. HCT116 cell lysates were immunoprecipitated using anti-PLZF antibody and analyzed by Western blot using an anti-p300 or anti-p53 antibody. Reverse co-immunoprecipitation was also performed. E, in vitro p53 acetylation assay. p53 was incubated with mock, p300, or p300+PLZF. Acetylated p53 was detected by Western blot using anti-acetylated-lysine antibody. F, PLZF decreases p53 expression or stability by ubiquitin-mediated proteasomal protein degradation. HCT116 cells were transfected with pcDNA3.1-PLZF or PLZF siRNA. The cells were treated with cyclohexamide for 12 h with or without MG132 treatment for 3 h, and the cell lysates were then analyzed by Western blotting using anti-p53 antibody. β-actin, control. MG132, an inhibitor of proteasome activity, inhibited enhanced degradation of existing p53 by PLZF. G, p53 ubiquitination assay. H1299 p53-null cells were transfected with pcDNA3-p53, pcDNA3-His-Ubiquitin, pcDNA3.1-PLZF, or PLZF siRNA in the various combinations indicated. The cells were treated with MG132 for 3 h, the cell lysates then incubated with the MagneHisTM nickel particles, and the precipitated pellets were analyzed by Western blotting using anti-p53 antibody. GAPDH, control. H, p53 ubiquitination assay. HCT116 cells were transfected with pcDNA3-His-Ubiquitin, pcDNA3.1-PLZF in the combinations indicated. The cells were treated with etoposide to induce p53 expression and MG132 for 3 h. GAPDH, control.
Because acetylation of p53 is important for p53 stability by inhibiting ubiquitination-mediated proteosomal degradation, we investigated whether PLZF interacts with p53 and affects p53 stability by decreasing its acetylation. Co-immunoprecipitation and Western blot assays of HCT116 cell lysates revealed that endogenous PLZF interacts with both p53 and the HAT protein p300 (Fig. 5, C and D). In vitro acetylation assays using p53, PLZF, and p300 also showed that PLZF inhibits p53 acetylation by p300 (Fig. 5E).
Because PLZF both decreases acetylation of p53 and p53 expression, we investigated whether ectopic overexpression or knockdown of PLZF affected p53 expression, and also whether the alteration in p53 expression was blocked by MG132, an inhibitor of ubiquitination-mediated proteasomal degradation. HCT116 cells were treated with cyclohexamide for 12 h and further treated with or without MG132 for 3 h. Like above, ectopic PLZF decreased p53 expression, whereas knockdown of PLZF increased p53 expression. These changes were more evident in the presence of cyclohexamide, an inhibitor of de novo protein synthesis (Fig. 5F).
Furthermore, we investigated whether PLZF increased ubiquitination of ectopic p53 in H1299 p53-null cells, showing PLZF increased p53 ubiquitination, whereas PLZF knockdown decreased p53 ubiquitination (Fig. 5G). Also, Western blot assays of cellular extracts prepared from etoposide-treated HCT116 cells showed that PLZF increased ubiquitination of endogenous p53 induced by DNA damage (Fig. 5H). These results suggest that PLZF may decrease p53 protein stability by ubiquitination-mediated proteosomal degradation.
PLZF Represses Transcription of CDKN1A by Decreasing p53 Binding to the Distal p53 Binding Elements of the CDKN1A
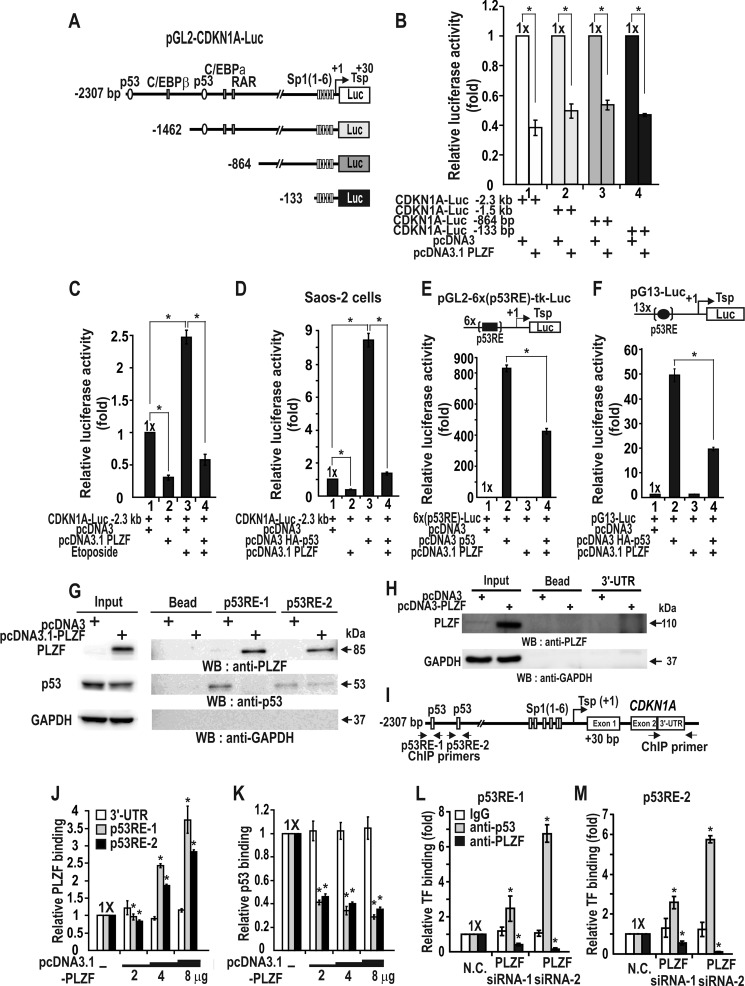
Because PLZF repressed transcription of TP53 and decreased p53 stability, we investigated whether transcription of the p53 target genes (e.g. endogenous CDKN1A and reporter constructs) can be regulated by PLZF. As PLZF repressed transcription of the CDKN1A, we examined which region of the CDKN1A promoter is important for transcription repression by PLZF. PLZF repressed transcription of all four different CDKN1A promoter-reporter gene fusion constructs by 55–62% (Fig. 6, A and B). The data indicated that PLZF repressed transcription by acting at the proximal promoter, which has six Sp1-binding GC-boxes. The transcription repression by PLZF was slightly more potent with a −2.4-kb promoter containing the distal p53 binding element. The results suggested that PLZF may decrease transcription of CDKN1A by binding competition with p53 or by decreasing p53 available at the p53RE at both transcription and protein level.
FIGURE 6.
PLZF represses CDKN1A transcription by decreasing p53 binding to the distal p53 binding elements in vitro and in vivo. A, structures of various CDKN1A promoter constructs tested. B, transcription assays. HEK293 cells were transiently co-transfected with a PLZF expression vector and pGL2-CDKN1A-Luc reporter plasmids and were analyzed for luciferase activity as described under ”Experimental Procedures.“ C, transcription analysis of HCT116 cells. Etoposide treatment of the cells increased CDKN1A gene expression, which was repressed by PLZF. D, transcription analysis. Saos-2 cells lacking p53 were transiently co-transfected with combinations of expression vectors for p53, PLZF, and pGL2-CDKN1A-Luc WT, −2.3 kb, and luciferase activity was measured. E and F, transcription assays. PLZF repressed transcriptional activation of pGL2–6x(p53RE)-Luc and pG13-Luc by ectopic p53 in Saos-2 cells. p53RE, distal p53-binding element of the CDKN1A promoter. pG13 contains 13 copies of the putative p53-binding element. G, oligonucleotide pulldown assay for PLZF binding to p53-binding elements. The precipitate was analyzed using a Western blot (WB) assay with antibodies against PLZF and p53. H, oligonucleotide pulldown assay for PLZF binding to a CDKN1A 3′-UTR probe as a negative control (G). I–L, ChIP-qPCR assays of p53 and PLZF binding to the distal p53-binding regions of the endogenous CDKN1A gene in HCT116 cells transfected with an increasing amount of PLZF expression vector or PLZF siRNA-1 or -2. Antibodies against PLZF and p53 were used in the ChIP assays. IgG and 3′-UTR, ChIP assay controls. *, p < 0.001. Mean values of three independent experiments are shown. Error bars represent S.D.
We investigated whether transcription activation of the CDKN1A reporter construct by p53 induced by etoposide or ectopic p53 can be inhibited by PLZF. In HCT116 cells, transcription activation of CDKN1A and TP53 by etoposide was inhibited by PLZF (Figs. 6C and 7, A–D). Also, transcription activation of the CDKN1A by ectopic p53 in the p53 null Saos-2 cells or HCT116 p53−/− cells can be repressed by PLZF (Figs. 6D and 7, E–H). The data also suggested that PLZF can also repress transcription of CDKN1A independent of p53 (Figs. 6D and 7, E and F). Additional assays using pG5–6x(p53RE)-Luc with six copies of the distal p53RE of the CDKN1A and pG13-Luc with 13 copies of the putative p53RE showed similar results (Fig. 6, E and F).
FIGURE 7.
Transcriptional activation of CDKN1A by either etoposide-induced p53 or ectopic p53 can be repressed by PLZF. A–D, RT-qPCR and Western blot assays. HCT116 cells transfected with PLZF expression vector or PLZF siRNA were treated with etoposide and analyzed for p53 and p21 expression at the mRNA and protein levels. PLZF repressed p53 and p21 expression in the presence or absence of the DNA damaging agent etoposide. E–H, RT-qPCR and Western blot assays. HCT116 p53−/− cells transfected with PLZF expression vector and/or p53 expression vector and analyzed for p53 and p21 expression at the mRNA and protein levels. PLZF repressed p53 and p21 expression in the presence or absence of p53 expression vector, whereas ectopic p53 did not affect ectopic PLZF expression or vice versa. Also, the same cells were transfected with PLZF siRNA and analyzed for p53 and p21 expression (G and H). *, p < 0.001. Mean values of three independent experiments are shown. Error bars represent S.D. WB, Western blot; N.C., scrambled RNA.
We further investigated the molecular event among PLZF, p53, and p53REs in vivo and in vitro by oligonucleotide pulldown and ChIP assays. PLZF binds the p53REs and decreases p53 binding to the elements, which is particularly effective for p53RE-1 (Fig. 6, G–K). Knockdown of PLZF expression increased p53 binding, which also suggested the binding competition between p53 and PLZF at p53REs or increase in p53 expression and stability (Fig. 6, L and M). PLZF may repress the transcription of CDKN1A by binding competition with p53, transcription repression of TP53, and decreasing of p53 stability.
Identification of a PLZF Binding Proximal Promoter Element of p21WAF/CDKN1A Critical in Transcription Repression by PLZF and PLZF Competes with Sp1 to Repress Transcription
The above data suggested that PLZF can repress transcription of CDKN1A independent of p53 (Figs. 6D and 7, E and F). In addition, transient transcription assays showed that PLZF can repress transcription by acting on the CDKN1A short promoter region (bp −131 to +30) (Fig. 6B). The proximal promoter region (bp −131 to +1) is GC-rich, contains six Sp1 binding sites, and is important in transcription activation by Sp1. PLZF is also a GC-box binding transcription factor, and it represses transcription activation of the short CDKN1A promoter by Sp1, which may suggest binding competition between Sp1 and PLZF at Sp1 binding GC-boxes (Fig. 8A). Sp1 activates transcription of the reporter pG5–5x(GC)-Luc with 5 copies of the Sp1-binding GC-box, and PLZF represses transcription activation by Sp1 (Fig. 8B).
FIGURE 8.
PLZF represses CDKN1A transcription by competition with Sp1 for binding to its proximal promoter GC-boxes 5/6 in vitro and in vivo. A, transient transcription analysis in HEK293 cells. Cells were transiently co-transfected with a mixture of expression vectors for Sp1 and/or PLZF, as well as pGL2-CDKN1A-Luc WT, −131 bp, and luciferase activity was measured. B, transient transcription assays. PLZF repressed transcriptional activation of pG5–5x(GC box)-tk*-Luc by Sp1 in HEK293 cells. The GC-box was derived from the well defined Sp1-binding consensus sequence. C, EMSA. PLZF binds to the proximal GC-box 5/6 in the CDKN1A promoter. Five [α-32P]dATP-labeled Sp1 binding GC-box probes were incubated with GST-PLZF-ZF (0.5 μg) and separated by 4% nondenaturing PAGE. PLZF-ZF, zinc finger DNA binding domain of PLZF. D, oligonucleotide pulldown assay of PLZF binding to the GC-box 5/6 probe. HEK293 cell extracts were incubated with biotinylated double-stranded oligonucleotides and analyzed as described in the legend to Fig. 6G. 3′-UTR, negative control. E, mutagenesis and transcription assays of the CDKN1A proximal promoter in HEK293 cells. The proximal GC-box 5/6 in the CDKN1A promoter element functions in CDKN1A transcriptional repression by PLZF. F, ChIP-qPCR assays of Sp1 and PLZF competitive binding at the endogenous CDKN1A proximal promoter in HEK293 cells. The cells were transfected with an increasing amount (0–8 μg) of PLZF expression vector or PLZF siRNA and analyzed for transcription factor (TF) binding using antibodies against PLZF, Sp1, and IgG. IgG and 3′-UTR, controls of ChIP antibody and TF binding region. The endogenous CDKN1A gene structure is shown above. Arrows indicate the binding positions of the ChIP-qPCR oligonucleotide primers. Tsp(+1), transcription start site. *, p < 0.001. Mean values of three independent experiments are shown. Error bars represent S.D. WB, Western blot; N.C., scrambled RNA.
We tested which GC-box at the CDKN1A promoter is important for transcription repression by PLZF. EMSA showed that PLZF binds to the GC-box 5/6 element (Fig. 8C). Oligonucleotide pulldown assays also indicated that PLZF binds the element and decreases Sp1 binding without affecting Sp1 expression (Fig. 8D). Mutagenesis and transient transcription assays showed that GC-box 5/6 is functionally significant in mediating transcription repression by PLZF (Fig. 8E). These results suggested that PLZF and Sp1 may compete with each other to bind to the GC-box 5/6 element. ChIP assays of PLZF and Sp1 binding at the endogenous CDKN1A promoter in the presence of ectopic PLZF or PLZF siRNA, showed not only that PLZF bound to the proximal promoter but also that PLZF and Sp1 compete with each other. PLZF significantly decreases Sp1 binding at the GC-box 5/6 to repress endogenous CDKN1A transcription (Fig. 8F).
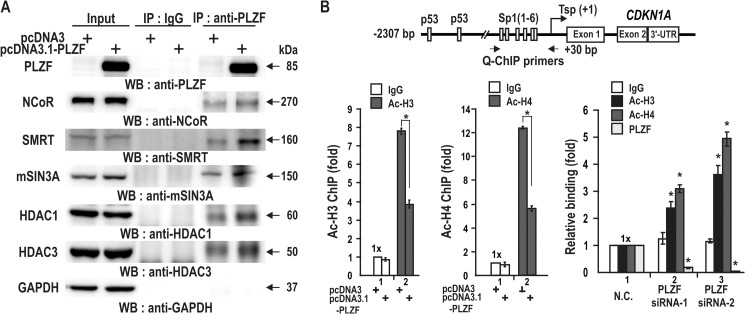
PLZF Interacts with Corepressor·HDAC Complex and Decreased Acetylated Histones H3 and H4 at the Endogenous CDKN1A Proximal Promoter
PLZF represses transcription by decreasing p53 binding at the distal p53 binding elements and also by binding competition with Sp1 (Figs. 6 and 8). Transcription repressors often repress transcription by interacting with corepressor·HDAC complexes containing, for example, SMRT/NCoR, mSin3A, and BCoR (9, 10). Co-immunoprecipitation and Western blot analysis of either HEK293 cell extracts or HEK293 cell extracts transfected with a PLZF expression vector revealed that PLZF interacts with NCoR, SMRT, mSin3A, HDAC1, and HDAC3 in vivo (Fig. 9A). This indicates that PLZF may inhibit transcription at the CDKN1A proximal promoter by interacting with the coepressor·HDAC complex as demonstrated for SMRT-HDACs by others (10).
FIGURE 9.
PLZF interacts with corepressors SMRT, NCoR, mSin3A, and HDACs. PLZF·corepressor·HDAC complex deacetylates histones Ac-H3 and Ac-H4 at the proximal promoter. A, co-immunoprecipitation (IP) of PLZF, NCoR, SMRT, mSin3A, HDAC1, and HDAC3. Cell lysates prepared from HEK293 cells transfected with either PLZF expression vector or pcDNA3 control vector were immunoprecipitated using anti-PLZF antibody and analyzed for corepressors or HDACs by Western blot (WB) using anti-corepressors, HDAC1, and HDAC3 antibodies. B, qChIP assays of histone modifications at the endogenous CDKN1A gene proximal promoter using antibodies against PLZF, Ac-H3, and Ac-H4. Cells were transfected with PLZF expression vector or PLZF siRNA and immunoprecipitated with the indicated antibodies, IgG, anti-Ac-H3, or anti-Ac-H4. IgG, control of ChIP antibody. *, p < 0.001. Mean values of three independent experiments are shown. Error bars represent S.D. N.C., scrambled RNA.
The corepressor complexes recruited by transcription repressor PLZF contain HDAC1 and HDAC3, which could deacetylate the histones of the nucleosomes around the proximal promoter. Indeed, ChIP assays showed that PLZF·corepressor·HDAC complexes significantly decreased acetylation of histones H3 and H4 at the CDKN1A proximal promoter by 40–65%. In addition, knockdown of PLZF expression resulted in an increase in acetylation of the histone H3 and H4 (Fig. 9B). These data indicated the involvement of the corepressor·HDACs complex in transcription repression by PLZF bound at the proximal promoter of CDKN1A in vivo.
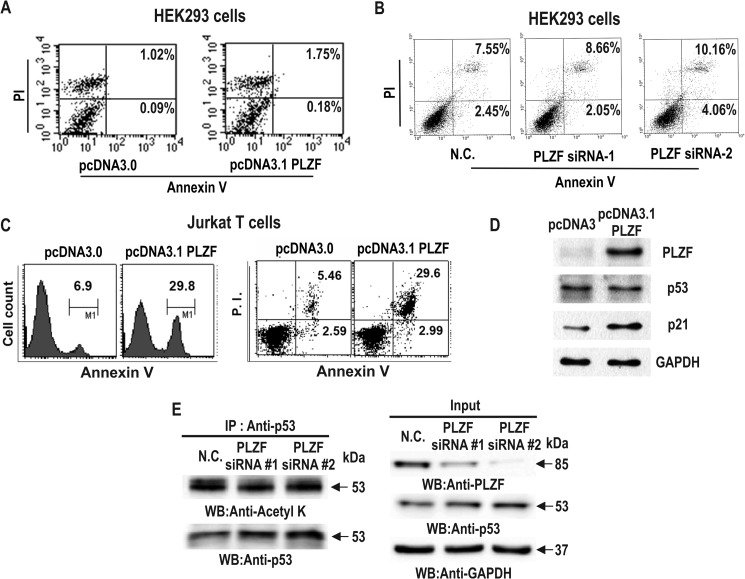
PLZF Has Little Effect on Apoptosis of HEK293 Cells, but Can Induce Apoptosis and Increase p21 Expression in Jurkat Cells
PLZF has been considered to be an inhibitor of cell proliferation in hematopoietic cells such as NB4, U937, and Jurkat T-cells. In contrast, our results convincingly indicate that PLZF increases cell proliferation in the cell types we tested. Accordingly, we tested whether PLZF could induce cell cycle arrest and apoptosis in human T-lymphocytic Jurkat T-cells. Although PLZF had little effect on apoptosis in HEK293 cells, PLZF significantly induced apoptosis and increased p21 expression in Jurkat cells, as reported by others (Fig. 10, A–D). Interestingly, in Jurkat cells, PLZF decreased p53 expression very weakly, whereas knockdown of PLZF expression slightly increased p53 expression but had very little effect on p53 acetylation (Fig. 10, D and E). These results suggest that PLZF regulates expression of p53 and p21 quite differently, depending on the cell type.
FIGURE 10.
Ectopic PLZF or knockdown of PLZF expression does not affect apoptosis in HEK293 cells, but PLZF does induce apoptosis in human Jurkat T-cells. A and B, flow cytometry analysis of HEK293 transfected with either a PLZF expression plasmid or PLZF siRNA. The cells were stained with Annexin-V and propidium iodide (PI) 48 h after transfection. x-axis, Annexin-V-FITC staining; y-axis, PI staining. The number represents the percentage of apoptotic cells in each condition (right quadrants). C, flow cytometry analysis of Jurkat T-cells transfected with either a pcDNA3 or pcDNA3.1-PLZF expression plasmid. The cells were stained with Annexin-V with or without PI 48 h after transfection. x-axis, Annexin-V-FITC staining; y-axis, cell count or PI staining (right). The number represents the percentage of apoptotic cells in each right-hand quadrant. D, Western blot (WB) of Jurkat T-cells transfected with either a pcDNA3 or pcDNA3.1-PLZF expression plasmid. PLZF increased p21 expression and weakly decreased p53 expression. E, Western blot of Jurkat T-cells transfected with control scrambled RNA or PLZF siRNA. Knockdown of PLZF increases p53 expression weakly and shows little effect on acetylation of p53.
DISCUSSION
PLZF has been shown to induce cell cycle arrest and represses the expression of pro-proliferative genes (35). However, there are some reports indicating that PLZF might stimulate cell proliferation. Plzf knock-out mice showed increased expression of p21 and p53 in spermatogonia, suggesting that Plzf is important in maintaining proliferative spermatogonia. Also, PLZF was shown to down-regulate apoptosis by inhibiting expression of the proapoptotic BID protein in lymphocytes (26). In this article, we were able to show that PLZF expression is increased in some cancers, like clear cell renal cell carcinoma, glioblastoma, and seminoma. Our results indicated that not only PLZF can transform NIH3T3 cells into faster growing cells (Fig. 3A) but also PLZF stimulates cancer cell proliferation and tumor growth in a mouse xenograft model (Fig. 2).
Our investigation of CDKN1A transcription regulation by PLZF revealed several interesting features of PLZF as a key regulator of p21 and p53 expression. PLZF is a GC-box-binding transcription factor and can compete with Sp1 for binding at the proximal GC-box 5/6 of CDKN1A. PLZF inhibited p53 activity by repressing TP53 gene transcription and decreasing p53 protein stability through deacetylation and ubiquitination-mediated proteosomal degradation. PLZF can also compete with p53 for binding at the distal p53-responsive elements. PLZF binds to the key regulatory elements at both the proximal and distal regions, and abrogates the synergistic communication between Sp1 and p53. The PLZF activities on p53 expression might play roles in blocking the DNA damage response mediated by p53, such as cell cycle arrest, DNA repair, and apoptosis. Indeed, in the several cell lines tested, PLZF stimulates cell proliferation and PLZF showed no effect on apoptosis (Fig. 10).
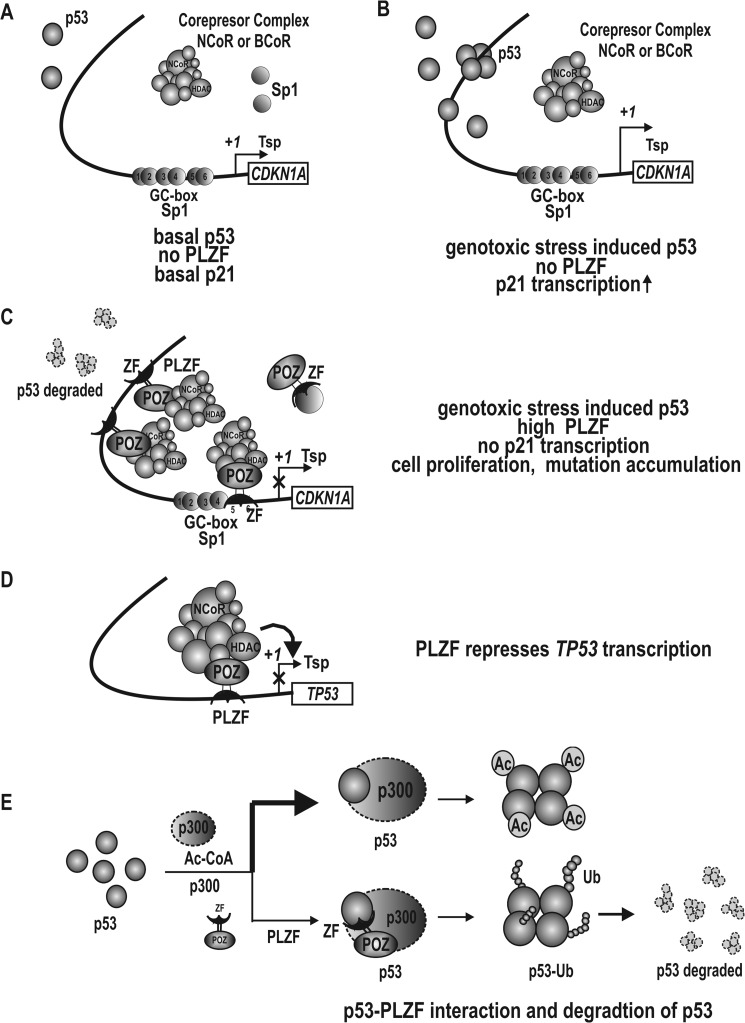
We propose a hypothetical model for CDKN1A transcription regulation by PLZF (Fig. 9). Under normal cellular conditions, where p53 levels are low and no PLZF is present, p21 is expressed at a low level and cells proliferate normally (Fig. 11A). When cells are exposed to genotoxic stress, p53 is induced and activates CDKN1A transcription by interacting with Sp1 bound at the proximal promoter (Fig. 11B). The induced p21 arrests cell cycle progression and allows cells to either repair DNA damage or undergo apoptosis. When cells are under genotoxic stress and PLZF expression is high in cancer tissues, PLZF represses transcription by directly binding both the proximal GC-box 5/6 and the distal p53 binding elements of CDKN1A to block the transcription activation contributed by Sp1 and p53. Furthermore, PLZF bound at the proximal promoter recruits corepressor-HDAC complex to silence CDKN1A expression epigenetically (Fig. 11C).
FIGURE 11.
Hypothetical model of transcription regulation for the cell cycle arrest gene, CDKN1A, by PLZF under three different cellular conditions. A, in the absence of or under low levels of PLZF, the CDKN1A gene is transcribed at a basal level, primarily activated by Sp1 family transcription factors. B, when cells are challenged with genotoxic stress, p53 expression is induced and activates transcription synergistically by interacting with Sp1, which is bound at the CDKN1A proximal promoter. C, in cancer cells with relatively high PLZF expression, PLZF represses CDKN1A transcription by competing with p53 for binding to the two distal p53 binding elements and also competes with Sp1 for binding to the proximal GC-box 5/6 element. PLZF recruits co-repressor-HDAC complexes, which deacetylate histones Ac-H3 and Ac-H4 at the CDKN1A proximal promoter to repress transcription. D and E, PLZF represses transcription of TP53. PLZF interacts with p53 and inhibits acetylation of p53 by p300, leading to ubiquitination and degradation of p53. PLZF indirectly represses transcription of CDKN1A by repressing TP53 transcription and inducing p53 protein degradation. Ac, acetylation; Tsp(+1), transcription start site; Ub, ubiquitination; x, transcription repression; ZF, zinc finger DNA-binding domain.
Although p53 expression can be induced by genotoxic stress, PLZF potently represses transcription of TP53 and decreases p53 stability (Fig. 11, D and E). Also, existing p53 has to compete with PLZF to bind the distal p53 binding elements (Fig. 11C). Decrease in p53 expression and molecular competition between p53 and PLZF significantly lower expression of CDKN1A and other p53 target genes important in apoptosis such as APAF-1, BAX, and NOXA.3
PLZF represses transcription of CDKN1A not only by binding competition between PLZF and p53 on the p53REs, but also by decreasing p53 expression by repressing transcription of TP53 and decreasing p53 stability (Figs. 4–6). Lack of apoptotic activity in the several cell lines we tested may be the result of potent transcription repression of TP53 and p53 degradation by PLZF. In the presence of high PLZF expression, mutations are accumulated in cells exposed to genotoxic stress, and the cells likely undergo oncogenic cellular transformations and proliferate rapidly.
PLZF has been considered an inhibitor of cell proliferation, based on the studies in hematopoietic cells such as NB4, U937, and Jurkat T-cells. For example, PLZF suppresses cell growth by repression of c-Myc expression in U937 acute promyelocytic leukemic cells (19). PLZF was thought to increase p21 expression by repressing c-Myc expression (19). In contrast, our results convincingly indicate that PLZF increases cell proliferation. Because PLZF can act opposite the role in control of cell proliferation depending on the cell types, we tested whether PLZF can induce cell cycle arrest and apoptosis in human T-lymphocytic Jurkat T-cells (Fig. 10). Although PLZF has little effect on apoptosis in HEK293 cells, PLZF can induce apoptosis and increase p21 expression in Jurkat cells, as reported by others in hematopoietic cells (Fig. 10) (36). Interestingly in Jurkat cells, PLZF repressed TP53 very weakly and also showed little effect on acetylation of p53. Knockdown of PLZF expression slightly increased p53 expression and very little effect on acetylation of p53. These results suggested that PLZF regulates expression of p53 and p21 quite differently depending on the cell types.
In summary, our investigation showed that PLZF causes cellular transformation of NIH3T3 cells and increases cell proliferation in several cell types. Along with the data on high PLZF expression in cancer tissues and tumor growth inhibition by shRNA against PLZF in a mouse xenograft model, PLZF may act as a potential proto-oncoprotein in various cell types.
This work was supported by Grant 00001561 (to M.-W. H.) from the National R & D Program for Cancer Control, Korean Ministry for Health, Welfare and Family affairs and DOYAK Research Grant 2011-0028817 from the Korea Research Foundation of the Korea Ministry of Education, Science and Technology.
W.-I. Choi and M.-W. Hur, unpublished data.
- POK
- poxviruses and zinc finger (POZ) and Krüppel
- POZ
- poxvirus and zinc finger
- ARF
- alternative reading frame gene
- MDM2
- human analogue of mouse double minute oncogene
- NCoR
- nuclear receptor corepressor
- PLZF
- promyelocytic leukemia zinc finger protein
- SMRT
- silencing mediator for retinoid and thyroid receptors
- Sp1
- specificity protein 1
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- qPCR
- quantitative PCR
- HDAC
- histone deacetylase.
REFERENCES
- 1. Kelly K. F., Daniel J. M. (2006) POZ for effect–POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 11, 578–587 [DOI] [PubMed] [Google Scholar]
- 2. Chen Z., Brand N. J., Chen A., Chen S. J., Tong J. H., Wang Z. Y., Waxman S., Zelent A. (1993) Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-α locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 12, 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phan R. T., Dalla-Favera R. (2004) The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432, 635–639 [DOI] [PubMed] [Google Scholar]
- 4. Choi W. I., Jeon B. N., Yun C. O., Kim P. H., Kim S. E., Choi K. Y., Kim S. H., Hur M. W. (2009) Proto-oncogene FBI-1 represses transcription of p21CIP1 by inhibition of transcription activation by p53 and Sp1. J. Biol. Chem. 284, 12633–12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi W. I., Jeon B. N., Yoon J. H., Koh D. I., Kim M. H., Yu M. Y., Lee K. M., Kim Y., Kim K., Hur S. S., Lee C. E., Kim K. S., Hur M. W. (2013) The proto-oncoprotein FBI-1 interacts with MBD3 to recruit the Mi-2/NuRD-HDAC complex and BCoR and to silence p21WAF/CDKN1A by DNA methylation. Nucleic Acids Res. 41, 6403–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maeda T., Merghoub T., Hobbs R. M., Dong L., Maeda M., Zakrzewski J., van den Brink M. R., Zelent A., Shigematsu H., Akashi K., Teruya-Feldstein J., Cattoretti G., Pandolfi P. P. (2007) Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science 316, 860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li J. Y., English M. A., Ball H. J., Yeyati P. L., Waxman S., Licht J. D. (1997) Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J. Biol. Chem. 272, 22447–22455 [DOI] [PubMed] [Google Scholar]
- 8. Hong S. H., David G., Wong C. W., Dejean A., Privalsky M. L. (1997) SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 94, 9028–9033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin R. J., Nagy L., Inoue S., Shao W., Miller W. H., Jr., Evans R. M. (1998) Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391, 811–814 [DOI] [PubMed] [Google Scholar]
- 10. Grignani F., De Matteis S., Nervi C., Tomassoni L., Gelmetti V., Cioce M., Fanelli M., Ruthardt M., Ferrara F. F., Zamir I., Seiser C., Grignani F., Lazar M. A., Minucci S., Pelicci P. G. (1998) Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature 391, 815–818 [DOI] [PubMed] [Google Scholar]
- 11. Buaas F. W., Kirsh A. L., Sharma M., McLean D. J., Morris J. L., Griswold M. D., de Rooij D. G., Braun R. E. (2004) Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 36, 647–652 [DOI] [PubMed] [Google Scholar]
- 12. Costoya J. A., Hobbs R. M., Barna M., Cattoretti G., Manova K., Sukhwani M., Orwig K. E., Wolgemuth D. J., Pandolfi P. P. (2004) Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 36, 653–659 [DOI] [PubMed] [Google Scholar]
- 13. Barna M., Hawe N., Niswander L., Pandolfi P. P. (2000) Plzf regulates limb and axial skeletal patterning. Nat. Genet. 25, 166–172 [DOI] [PubMed] [Google Scholar]
- 14. Barna M., Merghoub T., Costoya J. A., Ruggero D., Branford M., Bergia A., Samori B., Pandolfi P. P. (2002) Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev. Cell 3, 499–510 [DOI] [PubMed] [Google Scholar]
- 15. Reid A., Gould A., Brand N., Cook M., Strutt P., Li J., Licht J., Waxman S., Krumlauf R., Zelent A. (1995) Leukemia translocation gene, PLZF, is expressed with a speckled nuclear pattern in early hematopoietic progenitors. Blood 86, 4544–4552 [PubMed] [Google Scholar]
- 16. Labbaye C., Spinello I., Quaranta M. T., Pelosi E., Pasquini L., Petrucci E., Biffoni M., Nuzzolo E. R., Billi M., Foà R., Brunetti E., Grignani F., Testa U., Peschle C. (2008) A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat. Cell Biol. 10, 788–801 [DOI] [PubMed] [Google Scholar]
- 17. Kovalovsky D., Uche O. U., Eladad S., Hobbs R. M., Yi W., Alonzo E., Chua K., Eidson M., Kim H. J., Im J. S., Pandolfi P. P., Sant'Angelo D. B. (2008) The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9, 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Savage A. K., Constantinides M. G., Han J., Picard D., Martin E., Li B., Lantz O., Bendelac A. (2008) The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McConnell M. J., Chevallier N., Berkofsky-Fessler W., Giltnane J. M., Malani R. B., Staudt L. M., Licht J. D. (2003) Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-Myc expression. Mol. Cell. Biol. 23, 9375–9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaknovich R., Yeyati P. L., Ivins S., Melnick A., Lempert C., Waxman S., Zelent A., Licht J. D. (1998) The promyelocytic leukemia zinc finger protein affects myeloid cell growth, differentiation, and apoptosis. Mol. Cell Biol. 18, 5533–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ward J. O., McConnell M. J., Carlile G. W., Pandolfi P. P., Licht J. D., Freedman L. P. (2001) The acute promyelocytic leukemia-associated protein, promyelocytic leukemia zinc finger, regulates 1,25-dihydroxyvitamin D3-induced monocytic differentiation of U937 cells through a physical interaction with vitamin D3 receptor. Blood 98, 3290–3300 [DOI] [PubMed] [Google Scholar]
- 22. Yeyati P. L., Shaknovich R., Boterashvili S., Li J., Ball H. J., Waxman S., Nason-Burchenal K., Dmitrovsky E., Zelent A., Licht J. D. (1999) Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene 18, 925–934 [DOI] [PubMed] [Google Scholar]
- 23. Costoya J. A., Hobbs R. M., Pandolfi P. P. (2008) Cyclin-dependent kinase antagonizes promyelocytic leukemia zinc-finger through phosphorylation. Oncogene 27, 3789–3796 [DOI] [PubMed] [Google Scholar]
- 24. Felicetti F., Bottero L., Felli N., Mattia G., Labbaye C., Alvino E., Peschle C., Colombo M. P., Carè A. (2004) Role of PLZF in melanoma progression. Oncogene 23, 4567–4576 [DOI] [PubMed] [Google Scholar]
- 25. Cheung M., Pei J., Pei Y., Jhanwar S. C., Pass H. I., Testa J. R. (2010) The promyelocytic leukemia zinc-finger gene, PLZF, is frequently down-regulated in malignant mesothelioma cells and contributes to cell survival. Oncogene 29, 1633–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parrado A., Robledo M., Moya-Quiles M. R., Marín L. A., Chomienne C., Padua R. A., Alvarez-López M. R. (2004) The promyelocytic leukemia zinc finger protein down-regulates apoptosis and expression of the proapoptotic BID protein in lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 101, 1898–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]
- 28. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 29. Gartel A. L., Radhakrishnan S. K. (2005) Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 65, 3980–3985 [DOI] [PubMed] [Google Scholar]
- 30. Koutsodontis G., Tentes I., Papakosta P., Moustakas A., Kardassis D. (2001) Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21WAF1/Cip1 gene by the p53 tumor suppressor protein. J. Biol. Chem. 276, 29116–29125 [DOI] [PubMed] [Google Scholar]
- 31. Kaczynski J., Cook T., Urrutia R. (2003) Sp1- and Krüppel-like transcription factors. Genome Biol. 4, 206.1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gartel A. L., Ye X., Goufman E., Shianov P., Hay N., Najmabadi F., Tyner A. L. (2001) Myc represses the p21WAF1/CIP1 promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. U.S.A. 98, 4510–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kardassis D., Papakosta P., Pardali K., Moustakas A. (1999) c-Jun transactivates the promoter of the human p21WAF1/Cip1 gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J. Biol. Chem. 274, 29572–29581 [DOI] [PubMed] [Google Scholar]
- 34. Lomberk G., Urrutia R. (2005) The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem. J. 392, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hobbs R. M., Pandolfi P. P. (2010) Shape-shifting and tumor suppression by PLZF. Oncotarget 1, 3–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernardo M. V., Yelo E., Gimeno L., Campillo J. A., Parrado A. (2007) Identification of apoptosis-related PLZF target genes. Biochem. Biophys. Res. Commun. 359, 317–322 [DOI] [PubMed] [Google Scholar]