Background: PlaB is the most prominent phospholipase A of Legionella pneumophila.
Results: PlaB possesses high activity at nanomolar but low activity at micromolar concentrations where it forms tetramers.
Conclusion: PlaB tetramerization inhibits and oligomer dissociation activates phospholipase activity.
Significance: Our data highlight the first example of concentration-dependent phospholipase inactivation by tetramerization, which may be a mechanism for self-protection.
Keywords: Bacteria, Enzyme Purification, Pathogenesis, Phospholipase A, Protein Structure
Abstract
The intracellularly replicating lung pathogen Legionella pneumophila consists of an extraordinary variety of phospholipases, including at least 15 different phospholipases A (PLA). Among them, PlaB, the first characterized member of a novel lipase family, is a hemolytic virulence factor that exhibits the most prominent PLA activity in L. pneumophila. We analyzed here protein oligomerization, the importance of oligomerization for activity, addressed further essential regions for activity within the PlaB C terminus, and the significance of PlaB-derived lipolytic activity for L. pneumophila intracellular replication. We determined by means of analytical ultracentrifugation and small angle x-ray scattering analysis that PlaB forms homodimers and homotetramers. The C-terminal 5, 10, or 15 amino acids, although the individual regions contributed to PLA activity, were not essential for protein tetramerization. Infection of mouse macrophages with L. pneumophila wild type, plaB knock-out mutant, and plaB complementing or various mutated plaB-harboring strains showed that catalytic activity of PlaB promotes intracellular replication. We observed that PlaB was most active in the lower nanomolar concentration range but not at or only at a low level at concentration above 0.1 μm where it exists in a dimer/tetramer equilibrium. We therefore conclude that PlaB is a virulence factor that, on the one hand, assembles in inactive tetramers at micromolar concentrations. On the other hand, oligomer dissociation at nanomolar concentrations activates PLA activity. Our data highlight the first example of concentration-dependent phospholipase inactivation by tetramerization, which may protect the bacterium from internal PLA activity, but enzyme dissociation may allow its activation after export.
Introduction
Phospholipases are important bacterial virulence factors that may either cause massive host cell destruction or modulate the host more subtly by means of hijacking signaling cascades (1, 2). Legionella pneumophila, an important intracellular lung pathogen that primarily thrives in environmental amoebae, possesses a variety of phospholipases (1–3). So far, phospholipases A (PLA)2/lysophospholipases A (LPLA) comprising 15 different proteins are the most prominent ones, although three additional phospholipases C and one phospholipase D have been described (1, 2, 4, 5). The PLA enzymes fall into three major groups as follows: the GDSL-like (three members), the patatin-like (10 or 11 members), and the PlaB-like (one member) proteins. The GDSL and patatin-like lipase families were also analyzed in other bacteria (1, 2, 6–12). Importantly, PlaB is the only experimentally characterized member of a recently discovered protein family (13, 14). Yet uncharacterized PlaB homologs are found in Pseudomonas aeruginosa and other water-associated bacteria, and all genome-sequenced strains of the genus Legionella possess a plaB gene (13, 15–21). Both the GDSL and patatin-like hydrolases show homology to eukaryotic proteins in higher plants and the latter to cytosolic PLA2 (1, 2, 6). In contrast, PlaB does not possess similarities to eukaryotic proteins suggesting that rather the other mentioned families might mimic their eukaryotic counterparts during Legionella infection.
Nevertheless, PlaB is a virulence factor of L. pneumophila as shown in in vivo guinea pig infections in which the knock-out mutants confer attenuated replication in the lung and reduced dissemination to the spleen (22). PlaB consists of 474 amino acids corresponding to a calculated molecular mass of about 54 kDa. It is a cell-associated and hemolytic protein hydrolyzing predominantly phosphatidylglycerol (PG) and phosphatidylcholine (PC), where the hemolytic activity is determined by the ability to hydrolyze PC (13, 14). Previous data suggested that PlaB is displayed on the bacterial cell surface and that it is transported independently of several L. pneumophila protein secretion systems based on the detection of enzyme activity (22).
Cell-associated PLA activity was not only found for L. pneumophila but also in some non-pneumophila species, such as Legionella gormanii and Legionella spiritensis, which are seldom or are not associated with Legionnaires disease (23, 24). Interestingly, the PLA activity of the non-pneumophila species tested includes a different phospholipid substrate spectrum as compared with L. pneumophila, which may restrict hemolytic activity of the former (13, 14).
PlaB possesses the most prominent PLA/LPLA activity to date found in in vitro grown L. pneumophila as well as during host cell infection. It is about 100-fold more active than the PLAs released into the culture supernatant. Important amino acids for catalytic activity, such as the potential catalytic triad Ser-85, Asp-203, and His-251, have been identified by mutagenesis of individual amino acids of conserved residues within the novel family of lipases. Protein truncation studies have pointed to the importance of the C-terminal 15 amino acids for lipolytic activity (13).
Here, we addressed protein oligomerization, the influence of oligomerization on activity, analyzed the contribution of the PlaB C terminus to lipolytic activity and oligomerization, and the significance of PlaB-derived lipolytic activity for L. pneumophila intracellular replication. We conclude that PlaB is a potent PLA and virulence factor that assembles in inactive tetramers at micromolar concentrations, and oligomer dissociation at lower concentrations activates PLA activity.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
L. pneumophila sg1 strain Corby (25) was used as the wild type control and for gene cloning and expression. All cloning experiments and plasmid propagations were performed in Escherichia coli strains DH5α or Top10 (Invitrogen); recombinant expression of PlaB protein was done in E. coli BL21. The L. pneumophila strains as well as the different E. coli strains used in this study are listed in Table 1 and were grown on buffered charcoal yeast extract agar (BCYE) and in buffered yeast extract (BYE) broth (for Legionella) or on Luria-Bertani (LB) agar and in LB broth (for E. coli), as described previously (26–28). When needed, media were supplemented with antibiotics at final concentrations as follows: 25 μg/ml kanamycin for L. pneumophila or 50 μg/ml for E. coli, 9 μg/ml chloramphenicol for L. pneumophila or 30 μg/ml for E. coli, and 100 μg/ml ampicillin for E. coli.
TABLE 1.
Overview of strains used in this study
The abbreviations used are as follows: term., terminal; aa, amino acid; L.pn, L. pneumophila Corby.
| Organism | Mutation(s)/plasmid | Tags | Selection marker | Source/Ref. |
|---|---|---|---|---|
| L.pn. | Wild type L. pneumophila Sg1 Corby | 25 | ||
| L.pn. | dotA knock-out E01 | KmR | This studya | |
| L.pn. plaB− | plaB::km insertion mutant (= plaB1) | KmR | 14 | |
| L.pn. plaB− | pJB04 = pBCKS plaB (complementing strain) | CmR | 13 | |
| L.pn. plaB− | pKK50 = pBCKS plaB (aa 1–459) | CmR | This study | |
| L.pn. plaB− | pJB12 = pBCKS plaB D203N | CmR | 13 | |
| L.pn. plaB− | pJB06 = pBCKS plaB S85A | CmR | 13 | |
| E. coli BL21 | pKK19 = pGP172 plaB | N-term. Strep tag | AmpR | This study |
| E. coli BL21 | pKK25 = pGP172 plaB (aa 1–469) | N-term. Strep tag | AmpR | This study |
| E. coli BL21 | pKK26 = pGP172 plaB (aa 1–464) | N-term. Strep tag | AmpR | This study |
| E. coli BL21 | pKK27 = pGP172 plaB (aa 1–459) | N-term. Strep tag | AmpR | This study |
a P. Aurass, S. Tröller, and A. Flieger, unpublished data.
DNA Techniques and Sequence Analysis
E. coli DH5α or Top10 was used for the propagation of recombinant plasmid DNA with backbones of the following vectors: pBCKS+ (Stratagene) and pGP172 (kindly provided by S. Halbedel, Robert Koch-Institut) (29). Plasmid DNA was prepared, amplified, and sequenced according to standard protocols. Primers were obtained from Eurofins MWG Operon (see Table 2). Restriction enzymes were purchased from New England Biolabs. Foreign DNA was introduced into L. pneumophila strains by electroporation with an Invitrogen cell porator according to the manufacturer's specifications as described previously (26). Foreign DNA was transformed into chemo-competent E. coli cells according to standard protocols. Nucleotide and translated protein sequences were analyzed using the DNASTAR package, the NCBI website, and ExPASy.
TABLE 2.
Overview of primers used in this study
The abbreviations used are as follows: term., terminal; aa, amino acid.
| Plasmid | Gene | Tag | Primer name | Primer sequence (5′ to 3′) |
|---|---|---|---|---|
| pKK19 | plaB | N-term. Strep tag | plaBkpnIf | -CAGGGTACCATGATTGTTATC- |
| plaB_SmaIr | -CTACCCGGGTCAATCTATCTTT- | |||
| pGP172XmaIf | -CTACCCGGGGATCCGGCT- | |||
| pGP172KpnIr | -GATGGTACCGGCGCCTTTTTCG- | |||
| pKK25 | plaB (aa 1–469) | N-term. Strep tag | pGP172plaB-5f | -GAAAGCCAACTTGACCCGGGGATCCGGC- |
| pGP172plaB-5r | -CCGGGTCAAGTTGGCTTTCCACTGATTTTTGCCGGAG- | |||
| pKK26 | plaB (aa 1–464) | N-term. Strep tag | pGP172plaB-10f | -GCAAAAATCTGACCCGGGGATCCGGC- |
| pGP172plaB-10r | -CCGGGTCAGATTTTTGCCGGAGTAAGGTTGTTGC- | |||
| pKK27 | plaB (aa 1–459) | N-term. Strep tag | pGP172plaB-15f | -CAACAACCTTTGACCCGGGGATCCGGC- |
| pGP172plaB-15r | -CCGGGTCAAAGGTTGTTGCTGATACGGAAAACTGTCC- | |||
| pKK50 | plaB (aa 1–459) | pJB04–15Ctf | -CAACAACCTTTGATACCTCTTTCGTTTGATGC- | |
| pJB04–15Ctr | -GAGGTATCAAAGGTTGTTGCTGATACGGAAAACTG- |
Cloning of L. pneumophila plaB into Vectors for Recombinant Expression in E. coli and in L. pneumophila plaB Mutants
For recombinant overexpression of the N-terminally Strep-tagged versions of PlaB, the corresponding gene was amplified using a proofreading polymerase (Pfu Polymerase, Fermentas; see Table 2 for primers). For expression of N-terminally Strep-tagged PlaB, the purified PCR product was ligated into the SmaI/KpnI site of pGP172 resulting in pKK19. Truncated versions of PlaB and Strep-tagged PlaB were generated by amplifying pJB04 or pKK19 with Phusion polymerase (New England Biolabs, see Table 2 for primers), respectively. All mentioned plasmids were confirmed by sequencing prior to use.
Expression of Recombinant Strep-PlaB and Variants and Purification of Strep-PlaB
Miniprep DNA of plaB as well as its truncated variants expressing plasmids were transformed into chemo-competent E. coli BL21 cells and grown in LB broth supplemented with 100 μg/ml ampicillin. After overnight incubation at 37 °C, cultures were diluted 100-fold in fresh LB broth and were grown to an OD600 of 0.6 at 37 °C. Strep-PlaB expression was induced by addition of 2 mm isopropyl 1-thio-β-d-galactopyranoside and transferred to 20 °C for overnight expression. For purification of Strep-tagged variants, cell pellets were solubilized at 4 °C in 100 mm Tris, pH 8.0, 100 mm NaCl, 1 mm EDTA supplemented with Complete Protease Inhibitor Mixture (Roche Diagnostics). Cells were lysed via three passages through an Emulsi-Flex-C3 homogenizer (Avestin). Lysates were cleared by centrifugation at 37,000 × g for 20 min at 4 °C. Strep-tagged proteins were purified at 4 °C using Strep-Tactin Superflow high capacity matrix (IBA) according to the manufacturer's recommendations.
Determination of Lipolytic Activities
Incubation of rStrep-PlaB and protein versions with different lipids from Avanti Polar Lipids, Inc. (dipalmitoylphosphatidylcholine (PC), dipalmitoylphosphatidylglycerol (PG), 1-monopalmitoyl-lysophosphatidylcholine, and 1-monopalmitoyl-lysophosphatidylglycerol), was performed as described previously (13).
Size Exclusion Chromatography (SEC)
Purified recombinant protein was analyzed by size exclusion chromatography using an analytical Superdex 200 10/300 GL column (GE Healthcare) connected to an Äkta explorer system (GE Healthcare). The column equilibrated with Tris buffer (100 mm Tris, pH 8.0, 100 mm NaCl) or HEPES buffer (50 mm HEPES, pH 7.5, 100 mm NaCl) was loaded with Strep-tagged protein in a volume of 0.5 ml with a concentration of 1 mg/ml (18 μm). The elution was carried out with the same buffer by applying 1 column volume (∼24 ml) at a flow rate of 0.5 ml/min. Elution fractions were analyzed by 12.5% SDS-PAGE. The Superdex 200 column was calibrated with protein standards purchased from Sigma to determine the molecular weight of Strep-PlaB and its variants.
Analytical Ultracentrifugation
Sedimentation velocity (SV) experiments were carried out in a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge at 10 °C using an An-50 Ti rotor at 40,000 rpm and ProteomeLab XL-I GUI version 6.0 (firmware 5.7). Data analysis was performed with the program SEDFIT providing a model for diffusion-deconvoluted differential sedimentation coefficient distributions (c(s) distributions) (30). Partial specific volume, buffer density, and viscosity were calculated by the program SEDNTERP (31) and were used to correct the experimental s values to standard conditions (20 °C, water) yielding s20, w. Sedimentation equilibrium experiments were performed at 4 °C in an An-50 Ti rotor using a Beckman Optima XL-A analytical ultracentrifuge equipped with XL-A Data Acquisition and Analysis Software version 3.0. The samples were spun at 4,000, 6,000, and 8,000 rpm until no change in concentration gradients was observed for at least 12 h. Data analysis was performed using the program BPCFIT as described previously (32). Experiments were performed in standard 3- or 12-mm double sector centerpieces, and concentration profiles were measured at 280 or 230 nm using the UV absorption scanning optics of the centrifuges. For SV at high protein concentrations (5.4 to 24.4 μm), rStrep-PlaB was dialyzed against 100 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 1 mm EDTA. Prior to SV at low protein concentrations (0.13–1.1 μm) and sedimentation equilibrium experiments, rStrep-PlaB was subjected to SEC in the same buffer but without EDTA using a Superdex 200 column (GE Healthcare). Protein concentrations were determined spectrophotometrically using the absorption coefficients at 280 nm as calculated from amino acid composition with the program SEDNTERP (31) and are given in monomers throughout the text.
Small Angle X-ray Scattering (SAXS)
SAXS data were recorded at the BioSAXS P12 station of the EMBL Hamburg at the synchrotron storage ring PETRA III at DESY. The protein solution sample was loaded by an automated sample changer to a quartz capillary mounted in vacuum. The x-ray energy was set to 12 keV, and the s axis for the scattering vector was calibrated with a silver-behenate standard. To check for radiation damage, 10 frames of 100 ms were recorded. Using the first frame of every series as reference, the following frames were compared, and only those frames without any sign of radiation damage were averaged. The resulting scattering functions were normalized for exposure time and sample transmission. For further analysis, corresponding buffers were subtracted by an automated procedure, and the scattering function was normalized against the sample concentration. Initial SAXS parameters such as radius of gyration, the molecular mass of the protein, and the pair distance distribution function p(r) were derived by automated standard procedures (33). The SAXS curves of wild type rStrep-PlaB and three variants truncated by 5, 10, and 15 amino acids were recorded at concentrations of 1 mg/ml. The good quality of these data was verified by manual checks and directed toward further SAXS modeling procedures. Twenty low resolution models of PlaB were calculated with the program DAMMIF (34) and used for the construction of a representative ab initio model by using the programs DAMAVER and SUPCOMB (35). The models had a mean normalized spatial discrepancy (NSD) of 1.119 and a variation of NSD of 0.216. One model showed an NSD of greater than 1.551 (= mean NSD + 2· variation of NDS) and was hence excluded from the calculation of the ab initio model.
L. pneumophila Infection of RAW 264.7 Mouse Macrophages
RAW 264.7 cells, a murine macrophage cell line (ATCC CRL-24), are susceptible to Legionella infection (2, 8, 36, 37) and were maintained in high glucose DMEM (GE Healthcare/PAA) supplemented with 10% heat-inactivated fetal calf serum (GE Healthcare/PAA) at 37 °C and 5% CO2. To assess intracellular growth, cells were harvested in DMEM with 10% fetal calf serum transferred to 24-well tissue culture plates (Greiner Bio-One) at a concentration of 5 × 105/ml. After 18 h, the cells were infected with wild type bacteria, isogenic mutants, or complementing strains at multiplicities of infection of 1 in high glucose DMEM. After 2 h of incubation, nonadhering bacteria were removed by three washing steps with DMEM. The cfu determination by means of plating serial dilutions on BCYE agar was performed at the indicated time points after host cell lysis using a final concentration of 0.1% saponin (Roth).
RESULTS
Establishment of an Expression and Purification Protocol for L. pneumophila PlaB
To analyze activity and structural features of purified L. pneumophila PlaB, we recombinantly expressed PlaB as a fusion protein together with the following N-terminal tags: His6, GST, MBP, and Strep. Only the two latter constructs allowed expression of sufficient amounts of soluble protein. Cleavage of the MBP tag was not efficiently obtained, and therefore this approach was not continued. Expression of the eight amino acid short StrepTag construct yielded good protein levels, and further purification by means of affinity chromatography resulted in suitable amounts (about 5 mg of protein/liter of culture volume) of soluble and active rStrep-PlaB (Fig. 1A). The minor amount of further protein bands at about 60 and 70 kDa belonged to the co-purifying chaperones DnaK and GroEL. We tested the protein for PLA activity at a concentration of 0.01 μg/ml (0.186 nm), and the most prominent activity was determined toward PG and 1-monopalmitoyl-lysophosphatidylglycerol (Fig. 1B). In summary, we successfully recombinantly expressed and purified active soluble PlaB.
FIGURE 1.

Affinity chromatographic purification of rStrep-PlaB and PLA activity. A, Coomassie-stained 12.5% SDS-PAGE of affinity chromatography fractions (1-ml Strep-Tactin column) from rStrep-PlaB expressed from a 1-liter culture of E. coli BL21 (pKK19). Lane 1, soluble supernatant after cell disruption; lane 2, flow-through; lanes 3–5, washing fractions; and lanes 6–12, elution fractions. B, release of free fatty acids (FFA) was detected after incubation of 0.01 μg/ml (0.186 nm) purified rStep-PlaB with phospholipids for 60 min. The result represents the means ± S.D. of triplicate samples and is representative for at least three experiments. PG, dipalmitoylphosphatidylglycerol; PC, dipalmitoylphosphatidylcholine; LPG, 1-monopalmitoyl-lysophosphatidylglycerol; LPC, 1-monopalmitoyl-lysophosphatidylcholine; M, molecular weight standard.
Size Exclusion Chromatography Shows That Strep-PlaB Forms Homodimers
It has been described for the outer membrane PLA2 of E. coli that dimerization triggers activation of the enzyme (38). Therefore, we analyzed the oligomerization state of PlaB by means of several methods. As an initial experiment, SEC of 500 μg (1 mg/ml = 18.6 μm) rStrep-PlaB, generated from E. coli BL21 (pKK19), yielded an elution volume of 13.6 ml corresponding to an apparent molecular mass of about 110 kDa. Because the molecular mass of monomeric rStrep-PlaB as calculated from amino acid composition is 55.3 kDa, this suggested the occurrence of PlaB oligomers and especially dimers. Monomeric PlaB was not detected in substantial amounts (Fig. 2A).
FIGURE 2.
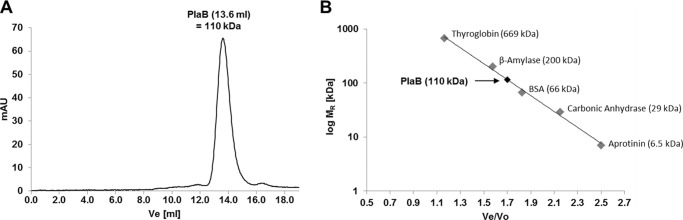
Size exclusion chromatography shows that PlaB forms homodimers. Size exclusion chromatogram of 500 μl, 1 mg/ml (18.6 μm), purified Strep-PlaB expressed from E. coli BL21 (pKK19) (A), and calibration curve (B) (Ve = elution volume of standard protein or PlaB; Vo = void volume of 8 ml) are shown. Results are representative of at least two independent experiments. mAU, milliabsorbance units.
Analytical Ultracentrifugation Shows That Strep-PlaB Forms Homodimers and Homotetramers
MALDI-TOF analyses of rStrep-PlaB preparations interestingly revealed the presence of a tetramer (222,343 Da/z) in addition to a dimer (110,680 Da/z) and a monomer (54,714 Da/z). A further peak corresponding to a trimer was considered as a laser-induced artifact (data not shown). Therefore, we applied further methods to analyze the presence of additional oligomeric states of PlaB. rStrep-PlaB was analyzed in SV experiments by means of analytical ultracentrifugation (AUC). At concentrations of 5.4 μm (0.3 mg/ml) and higher, c(s) analysis using the program SEDFIT (30) revealed that independent of protein concentration most of the protein sedimented with a sedimentation coefficient of 9.7 S (Fig. 3A). Taking diffusion broadening of the sedimenting boundary into account, c(s) analysis yielded a molecular mass of about 200 kDa. Therefore, in this concentration range PlaB forms most probably tetramers with a frictional ratio (f/f0) = 1.33 compared with an unhydrated spherical tetramer. The frictional ratio is a measure of macromolecule asymmetry and is expected to be in the range of 1.1 to 1.2 for a hydrated spherical protein (39). Accordingly, the shape of the rStrep-PlaB tetramer deviates somewhat from that of a sphere. In the c(s) distributions, aggregates with s values above 12 S were visible (Fig. 3A), and therefore the protein was further purified by SEC prior to sedimentation equilibrium centrifugation. At a protein concentration of 10.4 μm, analysis with a model of a single species yielded a molecular mass of 218 kDa (data not shown), confirming the tetrameric state of PlaB at protein concentrations above 5.4 μm. At lower protein concentrations, however, a second species was observed in SV, with an s20, w of 5.9 S (data not shown). Because the signal to noise ratio at these concentrations was rather low, SV centrifugation was repeated at 230 nm using SEC purified protein and lower protein concentrations. To allow for equilibration, the diluted samples were incubated for 7 h at 10 °C prior to SV. In addition to the tetrameric protein sedimenting with s20, w = 9.7 S, a second species sedimenting with s20, w = 5.9 S was observed at concentrations of 1.1 μm and below (Fig. 3B). As the fraction of the slower sedimenting species clearly increased with decreasing protein concentration, tetrameric PlaB seems to dissociate at lower protein concentrations. The second species is most probable a dimer, because its s value is higher than that of a spherical monomer, and c(s) analysis revealed a molecular mass of about 110 kDa. For dimeric PlaB, f/f0 = 1.38 was obtained, which is similar to the value for the tetrameric protein. Even at the lowest concentration used (0.13 μm), no significant amounts of monomeric PlaB were detected (Fig. 3B). Analytical ultracentrifugation therefore indicates that rStrep-PlaB exists in the lower micromolar concentration range in a dimer/tetramer equilibrium, with the protein being nearly completely dissociated into dimers at 0.13 μm and almost completely tetrameric above 5.4 μm.
FIGURE 3.
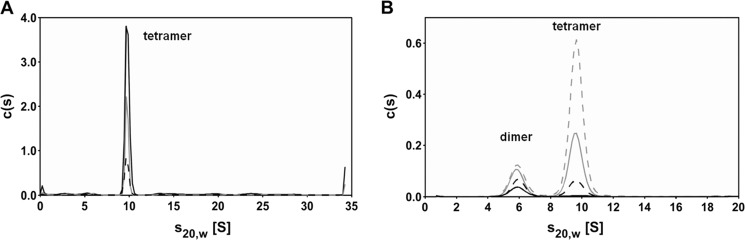
Analytical ultracentrifugation shows that PlaB forms homodimers and homotetramers. A, c(s) distributions of purified Strep-PlaB expressed from E. coli BL21(pKK19) at high concentrations (5.4 μm (black dashed line), 10.8 μm (gray line), and 24.4 μm (black solid line)) show that Strep-PlaB predominantly sediments as a tetramer with 9.7 S. SV experiments were performed in 100 mm Tris, pH 8.0, 100 mm NaCl, 1 mm EDTA, and absorbance was measured at 280 nm. For better comparison, all c(s) distributions have been converted to 12-mm path length. B, at lower protein concentrations (0.13 μm (black solid line), 0.27 μm (black dashed line), 0.54 μm (gray solid line), 1.1 μm (gray dashed line)), two protein species are visible in the c(s) distributions as follows: PlaB tetramers sedimenting with 9.7 S and PlaB dimers sedimenting with 5.9 S. Conditions are as in A, but rStrep-PlaB was subjected to SEC prior to SV, and absorbance was measured at 230 nm, and therefore EDTA was omitted from the buffer.
SAXS Analysis Reveals Shape Information of the PlaB Tetramers
As a further method to analyze the oligomeric state, SAXS was applied. The radius of gyration (Rg) determined from the SAXS curves for rStrep-PlaB was 5.16 nm. This value was in line with a hydrodynamic radius of 5.34 nm that correlates with the frictional ratio of 1.33 determined by analytical ultracentrifugation and indicated a molecular mass of about 200 kDa. This implied that PlaB was present as a tetramer at the concentration of 1 mg/ml (18.6 μm) used for SAXS experiments. The equilibrium between dimers and tetramers observed by analytical ultracentrifugation suggested that the PlaB tetramer assembles from the PlaB protomer in a two-step association via PlaB dimers that are predominant at lower concentrations. Hence, the PlaB tetramer must be classified as dimer of two dimers (D2 tetramer). An ab initio model of PlaB calculated from the SAXS curve under the assumption of D2 point group symmetry seems physically probable (Fig. 4), whereas models calculated assuming C2 or C4 point group symmetry exhibit shapes that seem unlikely for protein molecules (data not shown).
FIGURE 4.
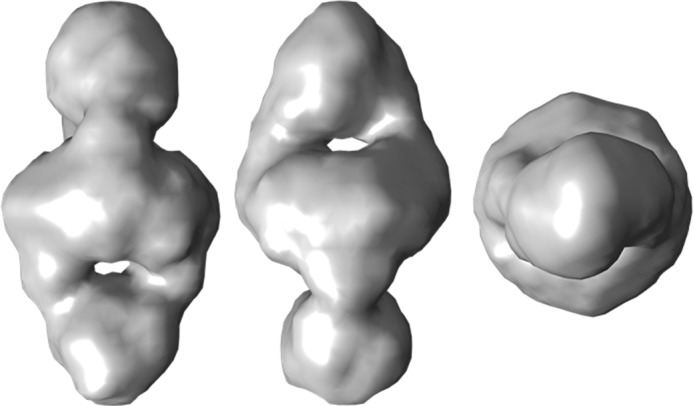
SAXS analysis confirms formation of PlaB tetramers. rStrep-PlaB was expressed from E. coli BL21(pKK19) and was purified. Low resolution ab initio model was calculated from the SAXS curve of Strep-PlaB under the assumption of D2 point group symmetry. From left to right: front, side, and top view.
Entire C-terminal 15-Amino Acid Region of PlaB Is Essential for Lipolytic Activities
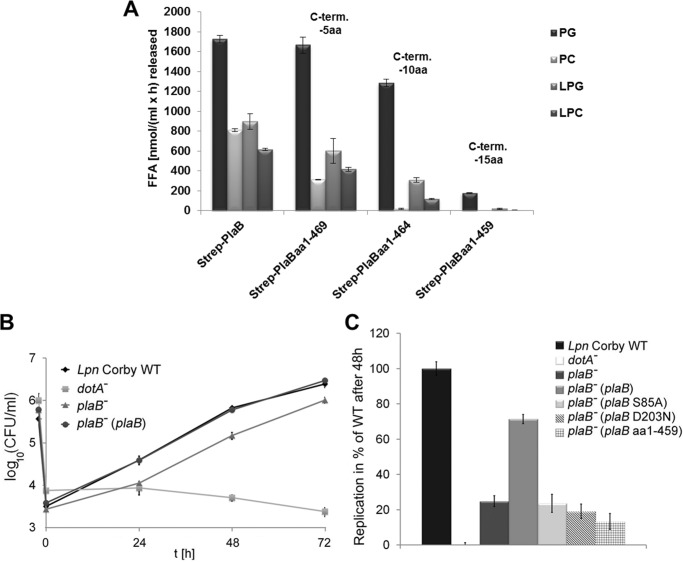
Earlier, we showed that the catalytically important amino acids (Ser-85, Asp-203, and His-251) of PlaB are contained in the N-terminal protein region of about 300 amino acids and that the C-terminal appendage of about 170 amino acids is nevertheless essential for PLA/LPLA activity. The removal of 15 C-terminal amino acids even abolished enzymatic activity (13). To narrow down which of the last 15 amino acids are essential, we performed further C-terminal protein truncations deleting 5 and 10 amino acids and analyzed purified rStrep-PlaB variants. As expected, the lack of 15 amino acids caused a severe loss of catalytic PLA and LPLA activities toward all substrates used in these experiments. The removal of 5 or 10 amino acids gradually decreased PLA activity, and the impact on PC-PLA was especially severe (Fig. 5A). In parallel with the shortage of the C terminus, a reduction of in vitro hemolysis was detected (data not shown).
FIGURE 5.
Entire C-terminal 15-amino acid region of PlaB is essential for lipolytic activity, and PlaB lipolytic activity is required for optimal intracellular replication. A, purified rStrep-PlaB and truncated variants were analyzed for enzymatic activities. Release of free fatty acid (FFA) was detected after incubation of 0.04 μg/ml (0.72 nm) purified rStep-PlaB and truncated variants with phospholipids for 45 min. C-term aa, C-terminal amino acid. RAW 264.7 macrophages were infected with L. pneumophila wild type, dotA−, and plaB− mutants and the complementing strain plaB−(plaB) expressing intact PlaB (B). C, RAW 264.7 macrophages were also infected with the plaB− mutant expressing active PlaB from pJB04, catalytic mutants expressing inactive PlaB from pJB06 and pJB12, or C-terminal truncated inactive PlaB from pKK50 at multiplicities of infection of 1. The dotA− strain was used as a virulence-attenuated control. The time point −2 h denotes the bacterial inoculum when the 2-h uptake period was started. At various time points before and after the uptake period, bacteria were quantified by plating aliquots on BCYE agar. The result represents the means ± S.D. of triplicate samples and are representative of at least two additional experiments. Replication of plaB− strains expressing plaB S85A, plaB D203N, or plaB aa1–459 and dotA− was significantly different from the wild type (*, p < 0.005, Student's t test, n = 3). PG, dipalmitoylphosphatidylglycerol; PC, dipalmitoylphosphatidylcholine; LPG, 1-monopalmitoyl-lysophosphatidylglycerol; LPC, 1-monopalmitoyllysophosphatidylcholine.
PlaB Catalytic Activity and the C-terminal 15 Amino Acids Contribute to Efficient Intracellular Replication of L. pneumophila in RAW 264.7 Mouse Macrophages
Previously, we showed that depletion of PlaB in L. pneumophila did not influence infection and replication in U937 macrophages and Acanthamoeba castellanii (14) but did attenuate infection and replication in an in vivo guinea pig model (22). Most interestingly, upon infection of RAW 264.7 mouse macrophages, we observed a replication defect of the plaB mutant (Fig. 5B). For example, within 48 h the wild type replicated about 200-fold, whereas the plaB mutant multiplied less than 60-fold. Provision of plaB in trans resulted in an increase of the replication rate to wild type levels (Fig. 5B). Defective replication of the plaB knock-out mutant in the model applied here gave us the opportunity to analyze whether loss of catalytic activity and not a PlaB-protein region with a different/unknown function is important for the observed defects. Indeed, we detected that provision of the plaB gene with respective mutations in amino acids of the catalytic triad, such as S85A and D203N, which were earlier shown to be essential for lipolytic activity (13), did not rescue replication efficiency in contrast to the wild type plaB-harboring strain (Fig. 5C). Furthermore, a strain harboring a C-terminal truncated plaB gene (PlaB −15 amino acids) exhibited strongly reduced PlaB activity (data not shown, for respective recombinant protein see Fig. 5A) and was not able to increase bacterial replication of the L. pneumophila plaB mutant (Fig. 5C). Our data show that PlaB catalytic activity and the C-terminal 15 amino acids contribute to efficient intracellular replication of L. pneumophila in RAW 264.7 mouse macrophages.
C-terminal 15-Amino Acid Region Is Not Essential for Oligomerization
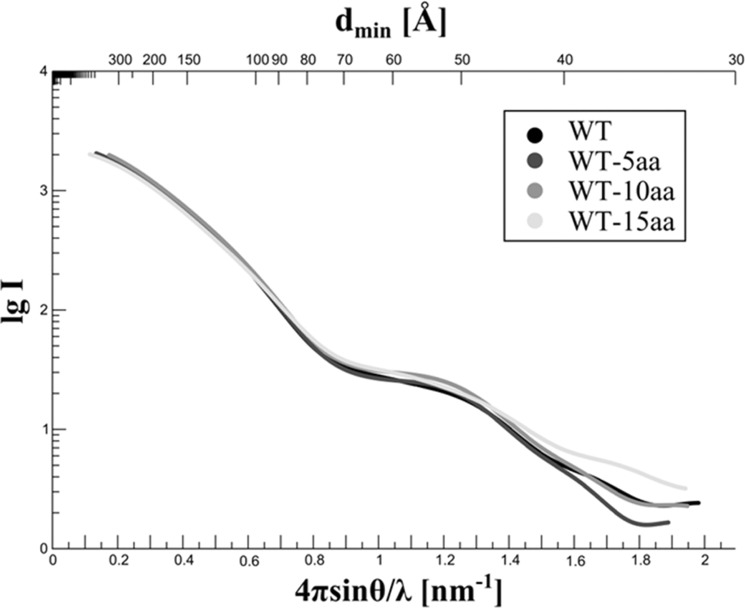
The loss of enzymatic activity upon deletion of the C-terminal 15 amino acids prompted us to speculate that those residues might regulate oligomerization and thereby enzymatic activity. We tested by means of SAXS experiments whether the gradual loss of activity resulting from deletion of the last 5, 10, or 15 amino acids correlates with a change of the oligomeric state of PlaB. Because wild type PlaB and the truncated variants showed similar CD spectra (data not shown), it was indicated that the C-terminal truncations are confined to unstructured regions and that the PlaB protomer does not undergo major conformational changes upon removal of the C terminus. SAXS curves of rStrep-PlaB and the three truncated variants coincided for low values of momentum transfer, which indicates a similar shape at low resolution for the wild type and variants and a comparable quaternary structure (Fig. 6). The Rg values determined from the SAXS curves for wild type and variants were similar within the procedural error margin with 5.16, 5.31, 5.25, and 5.10 nm, respectively. In conclusion, we observed that the entire C-terminal 15-amino acid region plays an important role for activity but is not essential for PlaB tetramerization.
FIGURE 6.
C-terminal 15-amino acid region of PlaB is not essential for oligomerization. Regularized SAXS curves determined for Strep-PlaB (WT) and the three C-terminally truncated variants lacking 5, 10, or 15 amino acids (WT-5aa, WT-10aa, and WT-15aa). The logarithm of scattering intensity is plotted versus the momentum transfer s = 4πsinθ/λ and the corresponding resolution dmin. The curves coincide well on the left-hand side of the diagram, which corresponds to low values of momentum transfer and indicates identical low- resolution structures of rStrep-PlaB and variants, i.e. the same oligomeric states. Deviations of the curves on the right-hand side result from minor structural differences apparent only at higher resolution.
PlaB Is Highly Active in the Nanomolar Concentration Range Only
Because we showed that PlaB exists in a dimer/tetramer equilibrium, we analyzed whether the two states differ in their lipolytic activity. We therefore tested rStrep-PlaB for activity at different concentrations. We observed an increase in activity from 0.2 to 70 nm PlaB but a decrease from concentrations of 0.14 to 18.6 μm (Fig. 7A). Accordingly, specific activity was decreasing in a sigmoidal pattern when the concentration was increased to 0.1 μm (Fig. 7B). Interestingly, our data showed that at concentrations above 5.4 μm, where the protein is exclusively in its tetrameric state, PlaB is inactive. Even at a concentration of 0.14 μm where the protein is expected to be dissociated almost completely into dimers, the specific activity is still very low (Fig. 3B). Definitely, the curve pattern, showing a strong increase in specific activity below 8.8 nm, implies that another PlaB species showing predominant activity is present in the lower nanomolar concentration range (Fig. 7B). Therefore, our data suggest that monomeric PlaB is actually the active form. The detection limits for SAXS (about 1 mg/ml = 18.6 μm) and AUC (about 0.13 μm) did not allow analysis at lower concentrations to check for the occurrence of PlaB monomers in the lower nanomolar concentration range. In summary, our results show that PlaB is highly active in the lower nanomolar concentration range and that the dimeric and tetrameric forms present at higher concentrations show very low specific activity or are inactive. Therefore, we propose that dissociation of the tetramer into monomers activates lipolytic activity.
FIGURE 7.
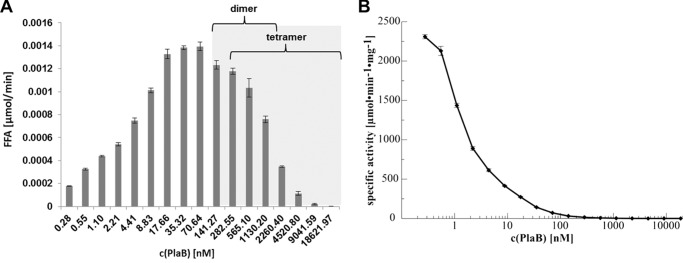
Lipid hydrolysis at different concentrations of PlaB. rStrep-PlaB was expressed from E. coli BL21 (pKK19) and was purified. Different concentrations of PlaB (18.6 μm (1 mg/ml) to 0.0003 μm (0.015 μg/ml)) were incubated with LPG for 15 min at 37 °C. To analyze the enzymatic activity, the release of free fatty acids was determined. The catalytic activity (amount of free fatty acids per time) is plotted versus concentration of rStrep-PlaB (A), and the specific activity (catalytic activity per mg of enzyme) is plotted versus concentration of rStrep-PlaB (B). The gray shaded area highlights the protein concentrations used in AUC experiments. The results represent the means ± S.D. of triplicate samples and are representative of at least two additional experiments.
DISCUSSION
We present here evidence that PlaB, in contrast to the above mentioned activation of phospholipases by oligomerization, forms inactive tetramers and that oligomer dissociation activates PLA (Figs. 7 and 8). We further showed that the C-terminal 15 amino acids important for enzymatic activity are not required for protein tetramerization (Fig. 6). Indeed, it is difficult to predict structure and oligomerization domains without a hint on the three-dimensional structure because PlaB is the first characterized homolog and the constituting member of a novel class of lipases (13). Structural analysis of PlaB may provide answers to questions we raised here, particularly concerning the nature of the interface for oligomer formation and enzyme inactivation by oligomerization.
FIGURE 8.

Model of the occurrence of PlaB-associated oligomerization states (monomer, dimer, and tetramer) and their relation to concentration and PLA activity.
Although many examples are known for enzyme activation upon oligomerization (40–48), only a few examples on enzyme activation upon dissociation have been described (49–52). One example is the purine nucleoside phosphorylase from bovine spleen. Purine nucleoside phosphorylase is a trimeric enzyme that dissociates into monomers upon dilution. Decreasing enzyme concentration from 20 to 0.02 μg/ml is accompanied by an approximate 50-fold increase in enzyme activity. It is therefore interpreted that the monomeric form is the fully active species. Further enzyme activity is influenced by phosphate concentration, which also determines oligomerization state and activity (50, 52). A second example, although the involved mechanism is different, is the Crotalus atrox venom PLA2. Dilution experiments show that this secreted PLA2 forms an extremely tight dimer. Fluorescence correlation spectroscopy data indicate that the addition of n-dodecylphosphocholine (C12-PN) results in dissociation of the dimer. On the basis of fluorescence correlation spectroscopy and tryptophan fluorescence results, it was postulated that an intermediate state is formed where the two monomers are in loose interaction within the protein-lipid comicelle. A complete dissociation of the dimer was observed with increasing concentrations of C12-PN. This indicates that the C. atrox PLA2 is dimeric in the absence of lipids and dissociates upon interaction with lipids and therefore supports the monomer hypothesis for PLA2 action (51).
What might be the reason for a down-regulation of PLA activity at higher protein concentrations, such as observed for PlaB? Phospholipases, such as PlaB, showing a high specific activity might be harmful for the producing bacterium when accumulating to significant concentrations. Therefore, from the perspective of L. pneumophila, it is of great importance that the protein contained in the bacterial cytoplasm is inactive to prevent disintegration of bacterial lipids. For example, PlaC, a PLA and acyltransferase of L. pneumophila, it has been shown that the zinc-metalloproteinase ProA cleaves a disulfide loop and thereby processes and activates PlaC after export to the external space (53). A proteolytic activation of PlaB seems unlikely because the protein confers activity as a recombinantly expressed and purified protein in the absence of additional factors. Therefore, other mechanisms must exist to bring the protein to the desired site and time into action. An effective way of activity up-regulation via oligomer dissociation and down-regulation via oligomer assembly is suggested by our present experiments. That this might be the case shows an approximate determination of the cellular PlaB concentration by means of Western blotting and subsequent band intensity analysis using rStrep-PlaB as the standard for quantification. When the size of an L. pneumophila bacterium of about 0.5 μm diameter and 1.5 μm length is assumed (54), about 64 monomers or 16 tetramers of PlaB are detected per cell and results in a cellular PlaB concentration of about 0.35 μm.3 At this concentration only a low specific activity of the protein was detected (see Fig. 7B), and considering molecular crowding in the cytoplasm, the occurrence of the tetrameric form would be even more promoted. Molecular crowding conditions refer to the notion that the cell cytoplasm contains high concentrations of high molecular weight components that occupy a substantial part of the volume. The effect of macromolecular crowding can influence biochemical processes in the cell such as assembly of macromolecular structures, protein folding, and protein-protein interactions (55–58). By means of analyzing enzymatic activity, it has been shown that PlaB is found in its active form at the surface of the bacteria and that the cell fractions with PlaB-derived lipolytic activity overlap with those containing the major outer membrane protein (22). Therefore, a fraction of formerly inactive PlaB is translocated to the bacterial surface and then may be present in an active dissociated form. So far, however, the mechanism of PlaB export and its distribution within the bacterial cell are unknown.
Acknowledgments
We thank Lidia Litz (Hannover Medical School, Germany) for excellent technical assistance and Silke Tröller and Philipp Aurass (Robert Koch-Institut, Berlin and Wernigerode, Germany) for isolation and characterization of L. pneumophila Corby dotA::tn5 clone E01. We are indebted to Gottfried Wilharm and to the members of the division of Enteropathogenic Bacteria and Legionella (Robert Koch-Institut, Wernigerode, Germany) for helpful discussions and suggestions to the experiments. We further acknowledge Omar Metwally for critical reading of the manuscript and Manfred Nimtz (Helmholtz Centre for Infection Research, Braunschweig, Germany) for excellent MS analysis of PlaB.
This work was supported by the Robert Koch-Institute (to A. F.) and by Grant DFG FL359/4-2 from the German Research Foundation.
K. Kuhle and A. Flieger, unpublished observations.
- PLA
- phospholipase A
- LPLA
- lysophospholipase A
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- SEC
- size exclusion chromatography
- SAXS
- small angle x-ray scattering
- SV
- sedimentation velocity
- NSD
- normalized spatial discrepancy
- AUC
- analytical ultracentrifugation.
REFERENCES
- 1. Lang C., Flieger A. (2011) Characterisation of Legionella pneumophila phospholipases and their impact on host cells. Eur. J. Cell Biol. 90, 903–912 [DOI] [PubMed] [Google Scholar]
- 2. Kuhle K., Flieger A. (2014) Molecular Mechanisms in Legionella Pathogenesis, pp. 175–209, Springer Science and Media, New York [Google Scholar]
- 3. Newton H. J., Ang D. K., van Driel I. R., Hartland E. L. (2010) Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 23, 274–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aurass P., Schlegel M., Metwally O., Harding C. R., Schroeder G. N., Frankel G., Flieger A. (2013) The Legionella pneumophila Dot/Icm-secreted effector PlcC/CegC1 together with PlcA and PlcB promotes virulence and belongs to a novel zinc metallophospholipase C family present in bacteria and fungi. J. Biol. Chem. 288, 11080–11092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Viner R., Chetrit D., Ehrlich M., Segal G. (2012) Identification of two Legionella pneumophila effectors that manipulate host phospholipids biosynthesis. PLoS Pathog. 8, e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Upton C., Buckley J. T. (1995) A new family of lipolytic enzymes? Trends Biochem. Sci. 20, 178–179 [DOI] [PubMed] [Google Scholar]
- 7. Arpigny J. L., Jaeger K. E. (1999) Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343, 177–183 [PMC free article] [PubMed] [Google Scholar]
- 8. Wilhelm S., Tommassen J., Jaeger K. E. (1999) A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 181, 6977–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruiz-Albert J., Yu X. J., Beuzón C. R., Blakey A. N., Galyov E. E., Holden D. W. (2002) Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44, 645–661 [DOI] [PubMed] [Google Scholar]
- 10. Shinoda S., Matsuoka H., Tsuchie T., Miyoshi S., Yamamoto S., Taniguchi H., Mizuguchi Y. (1991) Purification and characterization of a lecithin-dependent haemolysin from Escherichia coli transformed by a Vibrio parahaemolyticus gene. J. Gen. Microbiol. 137, 2705–2711 [DOI] [PubMed] [Google Scholar]
- 11. Banerji S., Flieger A. (2004) Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology 150, 522–525 [DOI] [PubMed] [Google Scholar]
- 12. Blanc G., Renesto P., Raoult D. (2005) Phylogenic analysis of rickettsial patatin-like protein with conserved phospholipase A2 active sites. Ann. N.Y. Acad. Sci. 1063, 83–86 [DOI] [PubMed] [Google Scholar]
- 13. Bender J., Rydzewski K., Broich M., Schunder E., Heuner K., Flieger A. (2009) Phospholipase PlaB of Legionella pneumophila represents novel lipase family protein residues essential for lipolytic activity, substrate specificity, and Hemolysis. J. Biol. Chem. 284, 27185–27194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flieger A., Rydzewski K., Banerji S., Broich M., Heuner K. (2004) Cloning and characterization of the gene encoding the major cell-associated phospholipase A of Legionella pneumophila, plaB, exhibiting hemolytic activity. Infect. Immun. 72, 2648–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cazalet C., Rusniok C., Brüggemann H., Zidane N., Magnier A., Ma L., Tichit M., Jarraud S., Bouchier C., Vandenesch F. (2004) Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 16. Chien M., Morozova I., Shi S., Sheng H., Chen J., Gomez S. M., Asamani G., Hill K., Nuara J., Feder M. (2004) The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305, 1966–1968 [DOI] [PubMed] [Google Scholar]
- 17. D'Auria G., Jiménez-Hernández N., Peris-Bondia F., Moya A., Latorre A. (2010) Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glöckner G., Albert-Weissenberger C., Weinmann E., Jacobi S., Schunder E., Steinert M., Hacker J., Heuner K. (2008) Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298, 411–428 [DOI] [PubMed] [Google Scholar]
- 19. Kozak N. A., Buss M., Lucas C. E., Frace M., Govil D., Travis T., Olsen-Rasmussen M., Benson R. F., Fields B. S. (2010) Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J. Bacteriol. 192, 1030–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moliner C., Raoult D., Fournier P.-E. (2009) Evidence that the intra-amoebal Legionella drancourtii acquired a sterol reductase gene from eukaryotes. BMC Res. Notes 2, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qin T., Cui Y., Cen Z., Liang T., Ren H., Yang X., Zhao X., Liu Z., Xu L., Li D. (2012) Draft genome sequences of two Legionella dumoffii strains, TEX-KL and NY-23. J. Bacteriol. 194, 1251–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schunder E., Adam P., Higa F., Remer K. A., Lorenz U., Bender J., Schulz T., Flieger A., Steinert M., Heuner K. (2010) Phospholipase PlaB is a new virulence factor of Legionella pneumophila. Int. J. Med. Microbiol. 300, 313–323 [DOI] [PubMed] [Google Scholar]
- 23. Fang G.-D., Yu V. L., Vickers R. M. (1989) Disease due to the Legionellaceae (other than Legionella pneumophila): historical, microbiological, clinical, and epidemiological review. Medicine 68, 116–132 [DOI] [PubMed] [Google Scholar]
- 24. Muder R. R., Yu V. L. (2002) Infection due to Legionella species other than L. pneumophila. Clin. Infect. Dis. 35, 990–998 [DOI] [PubMed] [Google Scholar]
- 25. Jepras R. I., Fitzgeorge R. B., Baskerville A. (1985) A comparison of virulence of two strains of Legionella pneumophila based on experimental aerosol infection of guinea-pigs. J. Hyg. 95, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Banerji S., Bewersdorff M., Hermes B., Cianciotto N. P., Flieger A. (2005) Characterization of the major secreted zinc metalloprotease-dependent glycerophospholipid: cholesterol acyltransferase, PlaC, of Legionella pneumophila. Infect. Immun. 73, 2899–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bertani G. (1951) Studies on lysogenesis I: the mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Edelstein P. H. (1981) Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14, 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Merzbacher M., Detsch C., Hillen W., Stülke J. (2004) Mycoplasma pneumoniae HPr kinase/phosphorylase. Eur. J. Biochem. 271, 367–374 [DOI] [PubMed] [Google Scholar]
- 30. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laue M. T., Shah B. D., Rigdeway T. M., Pelletier S. L. (1992) in Biochemistry and Polymer Science (Harding S. E., ed) pp. 90–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 32. Wyszomirski K. H., Curth U., Alves J., Mackeldanz P., Möncke-Buchner E., Schutkowski M., Krüger D. H., Reuter M. (2012) Type III restriction endonuclease EcoP15I is a heterotrimeric complex containing one Res subunit with several DNA-binding regions and ATPase activity. Nucleic Acids Res. 40, 3610–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Franke D., Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Svergun D. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. St-Denis A., Caouras V., Gervais F., Descoteaux A. (1999) Role of protein kinase C-α in the control of infection by intracellular pathogens in macrophages. J. Immunol. 163, 5505–5511 [PubMed] [Google Scholar]
- 37. Weber S. S., Ragaz C., Reus K., Nyfeler Y., Hilbi H. (2006) Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Snijder H. J., Dijkstra B. W. (2000) Bacterial phospholipase A: structure and function of an integral membrane phospholipase. Biochim. Biophys. Acta 1488, 91–101 [DOI] [PubMed] [Google Scholar]
- 39. Lebowitz J., Lewis M. S., Schuck P. (2002) Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng J., Goldstein R., Stec B., Gershenson A., Roberts M. F. (2012) Competition between anion binding and dimerization modulates Staphylococcus aureus phosphatidylinositol-specific phospholipase C enzymatic activity. J. Biol. Chem. 287, 40317–40327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Oliveira A. H., Giglio J. R., Andrião-Escarso S. H., Ito A. S., Ward R. J. (2001) A pH-induced dissociation of the dimeric form of a lysine 49-phospholipase A2 abolishes Ca2+-independent membrane damaging activity. Biochemistry 40, 6912–6920 [DOI] [PubMed] [Google Scholar]
- 42. Dekker N., Tommassen J., Lustig A., Rosenbusch J. P., Verheij H. M. (1997) Dimerization regulates the enzymatic activity of Escherichia coli outer membrane phospholipase A. J. Biol. Chem. 272, 3179–3184 [DOI] [PubMed] [Google Scholar]
- 43. Griffon N., Jin W., Petty T. J., Millar J., Badellino K. O., Saven J. G., Marchadier D. H., Kempner E. S., Billheimer J., Glick J. M. (2009) Identification of the active form of endothelial lipase, a homodimer in a head-to-tail conformation. J. Biol. Chem. 284, 23322–23330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kosk-Kosicka D., Bzdega T. (1988) Activation of the erythrocyte Ca2+-ATPase by either self-association or interaction with calmodulin. J. Biol. Chem. 263, 18184–18189 [PubMed] [Google Scholar]
- 45. Kosk-Kosicka D., Bzdega T., Wawrzynow A. (1989) Fluorescence energy transfer studies of purified erythrocyte Ca2+-ATPase. Ca2+-regulated activation by oligomerization. J. Biol. Chem. 264, 19495–19499 [PubMed] [Google Scholar]
- 46. Magro A. J., Soares A. M., Giglio J. R., Fontes M. R. (2003) Crystal structures of BnSP-7 and BnSP-6, two Lys49-phospholipases A 2: quaternary structure and inhibition mechanism insights. Biochem. Biophys. Res. Commun. 311, 713–720 [DOI] [PubMed] [Google Scholar]
- 47. Marchi-Salvador D. P., Corrêa L. C., Magro A. J., Oliveira C. Z., Soares A. M., Fontes M. R. (2008) Insights into the role of oligomeric state on the biological activities of crotoxin: crystal structure of a tetrameric phospholipase A2 formed by two isoforms of crotoxin B from Crotalus durissus terrificus venom. Proteins Struct. Funct. Bioinformatics 72, 883–891 [DOI] [PubMed] [Google Scholar]
- 48. Snijder H. J., Ubarretxena-Belandia I., Blaauw M., Kalk K. H., Verheij H. M., Egmond M. R., Dekker N., Dijkstra B. W. (1999) Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401, 717–721 [DOI] [PubMed] [Google Scholar]
- 49. Beernink P. T., Tolan D. R. (1996) Disruption of the aldolase A tetramer into catalytically active monomers. Proc. Natl. Acad. Sci. 93, 5374–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ropp P. A., Traut T. W. (1991) Purine nucleoside phosphorylase. Allosteric regulation of a dissociating enzyme. J. Biol. Chem. 266, 7682–7687 [PubMed] [Google Scholar]
- 51. Sanchez S. A., Chen Y., Müller J. D., Gratton E., Hazlett T. L. (2001) Solution and interface aggregation states of Crotalus atrox venom phospholipase A2 by two-photon excitation fluorescence correlation spectroscopy. Biochemistry 40, 6903–6911 [DOI] [PubMed] [Google Scholar]
- 52. Traut T. W. (1994) Dissociation of enzyme oligomers: a mechanism for allosteric regulation. Crit. Rev. Biochem. Mol. Biol. 29, 125–163 [DOI] [PubMed] [Google Scholar]
- 53. Lang C., Rastew E., Hermes B., Siegbrecht E., Ahrends R., Banerji S., Flieger A. (2012) Zinc metalloproteinase ProA directly activates Legionella pneumophila PlaC glycerophospholipid: cholesterol acyltransferase. J. Biol. Chem. 287, 23464–23478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Faulkner G., Garduño R. A. (2013) Electron microscopy of legionella and legionella-infected cells. Methods Mol. Biol. 954, 279–307 [DOI] [PubMed] [Google Scholar]
- 55. Chebotareva N. A., Kurganov B. I., Livanova N. B. (2004) Biochemical effects of molecular crowding. Biochemistry (Mosc.) 69, 1239–1251 [DOI] [PubMed] [Google Scholar]
- 56. Ellis R. J. (2001) Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 11, 114–119 [DOI] [PubMed] [Google Scholar]
- 57. Zhou H.-X., Rivas G., Minton A. P. (2008) Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 37, 375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou Y.-L., Liao J.-M., Chen J., Liang Y. (2006) Macromolecular crowding enhances the binding of superoxide dismutase to xanthine oxidase: implications for protein-protein interactions in intracellular environments. Int. J. Biochem. Cell Biol. 38, 1986–1994 [DOI] [PubMed] [Google Scholar]