Background: Marijuana has been shown to have an immunomodulatory activity.
Results: ChIP-Seq results show genome-wide changes in histone methylation in immune cells treated with THC.
Conclusion: Histone modifications are associated with THC-mediated alterations in antigen-specific T cell response.
Significance: This study provides insights into the potential role of epigenetic changes induced by THC in gene regulation.
Keywords: Epigenetics, Histone Methylation, Immunosuppression, Lymphocyte, T-cell
Abstract
Marijuana is one of the most abused drugs due to its psychotropic effects. Interestingly, it is also used for medicinal purposes. The main psychotropic component in marijuana, Δ9-tetrahydrocannabinol (THC), has also been shown to mediate potent anti-inflammatory properties. Whether the immunomodulatory activity of THC is mediated by epigenetic regulation has not been investigated previously. In this study, we employed ChIP-Seq technology to examine the in vivo effect of THC on global histone methylation in lymph node cells of mice immunized with a superantigen, staphylococcal enterotoxin B. We compared genome-wide histone H3 Lys-4, Lys-27, Lys-9, and Lys-36 trimethylation and histone H3 Lys-9 acetylation patterns in such cells exposed to THC or vehicle. Our results showed that THC treatment leads to the association of active histone modification signals to Th2 cytokine genes and suppressive modification signals to Th1 cytokine genes, indicating that such a mechanism may play a critical role in the THC-mediated switch from Th1 to Th2. At the global level, a significant portion of histone methylation and acetylation regions were altered by THC. However, the overall distribution of these histone methylation signals among the genomic features was not altered significantly by THC, suggesting that THC activates the expression of a subset of genes while suppressing the expression of another subset of genes through histone modification. Functional classification of these histone marker-associated genes showed that these differentially associated genes were involved in various cellular functions, from cell cycle regulation to metabolism, suggesting that THC had a pleiotropic effect on gene expression in immune cells. Altogether, the current study demonstrates for the first time that THC may modulate immune response through epigenetic regulation involving histone modifications.
Introduction
Marijuana is the most frequently used illicit substance in the United States (1). In addition, many states in the United States have now legalized marijuana use, especially when authorized by a physician, for medical purposes, such as alleviation of nausea and vomiting from chemotherapy, wasting in AIDS patients, and chronic pain that is unresponsive to opioids (2, 3). Moreover, two states in the United States have legalized marijuana for recreational use. Thus, studies evaluating the risks and benefits of marijuana use are critical.
Δ9-Tetrahydrocannabinol (THC),2 the active psychotropic ingredient of marijuana, mediates its activity through cannabinoid receptors (CB1 and CB2). Cannabinoid receptors are typical transmembrane G protein-coupled receptors. Whereas CB1 is highly expressed in the brain, and to a lower extent in peripheral tissues (4), CB2 is predominant in immune cells (5). Therefore, besides its psychoactive effects, THC can suppress inflammation through activation of cannabinoid receptors on immune cells, using multiple pathways (6–8). THC has been shown to suppress Th1 while promoting Th2 cells (9, 10). In addition, THC induces CD11b+ Gr-1+ myeloid-derived suppressor cells (11–13) as well as Tregs (14), which have been shown to inhibit T cell proliferation. The induction of myeloid-derived suppressor cells by THC was associated with alterations in microRNA expression (15). Moreover, we also noted that prenatal exposure to THC causes T cell dysfunction in the offspring (16). Together, such data suggested that THC may trigger epigenetic modulations in immune cells.
Epigenetic modification has been implicated in the establishment and maintenance of differential gene expression in T cells (17). DNA methylation and histone modifications are common epigenetic pathways leading to alterations in gene expression. Epigenetic modifications have been shown to regulate T cell differentiation by modifying the chromatin at the related genes, such as Ifn-γ, Foxp3, and IL-4 (18). Genome-wide histone modification studies using the ChIP-Seq method in human T cells have linked histone methylation patterns to the specific gene activity in different T cell subtypes (17, 19–21). Histone methylation mainly occurs on the lysine and arginine residues, and lysines can be mono-, di-, or trimethylated. Histone H3 methylation on lysine 4, lysine 9, lysine 27, and lysine 36 are among the most extensively studied histone methylations (22). In general, histone H3 lysine 4 trimethylation (H3K4me3) in the promoter region is associated with transcription activation, whereas histone H3 lysine 27 trimethylation (H3K27me3) within the promoter region is associated with transcription repression. However, H3K4me3 and H3K27me3 that seem to be associated with opposite functions can co-exist in the same regions. This so-called “bivalent domain” has been found in embryonic stem cells and T cells and are proposed to lead to activation or suppression (23–25). Histone lysine 36 trimethylation (H3K36me3) has been linked to transcription elongation and is enriched in the body of active transcripts (26, 27). Histone lysine 9 trimethylation (H3K9me3) has been linked to the silencing of the gene. This mark is enriched in the telomeric region and terminal repeats (19, 27–29). However, it has been shown that H3K9me3 is also enriched in many promoters (30). Histone acetylation in general is associated with gene activation. One of the most well studied histone acetylation markers is histone H3 acetylation at lysine 9 (H3K9ac), which is enriched near the transcription start site (TSS) of highly expressed genes (31).
Staphylococcal enterotoxin B (SEB) is a bacterial superantigen that triggers a massive Th1-cytokine storm leading to lethal toxic shock syndrome (32). In this study, we investigated the effect of THC on SEB-induced T cell activation in vivo and determined whether THC modifies global histone methylation in activated immune cells. Using a ChIP-Seq approach, we compared genome-wide H3K4me3, H3K27me3, H3K36me3, H3K9me3, and H3K9ac patterns in SEB-activated popliteal lymph node (LN) cells in mice with or without THC pretreatment. Our data showed that a significant portion of histone methylation and acetylation regions are altered by THC treatment at the genomic level. However, the associated methylation markers, not the H3K9ac marker, in key Th1/Th2 cytokine genes are altered by THC treatment, which is consistent with the ability of THC to induce a shift in Th1-Th2 balance. Moreover, we identified many other genes whose expression may be regulated by THC through histone modification.
EXPERIMENTAL PROCEDURES
Mice and Cell Isolation
Female C57BL/6J mice were purchased from the National Institutes of Health (Frederick, MD). 6–7-Week-old mice received an intraperitoneal injection of THC (Sigma; 20 mg/kg of body weight) or the same amount of vehicle as described previously (33). Twenty-four h later, the mice received the same treatment again. Two h after the second treatment, 10 μg of SEB in 50 μl of PBS was injected in each foot pad (2 foot pads/mouse). Mice were euthanized 1, 3, or 5 days after SEB challenge. Popliteal LN were collected, and single cell suspension was prepared in RPMI1640 cell culture medium. We used a pretreatment regimen with THC because SEB triggers an acute cytokine storm, and moreover, such studies would indicate how marijuana abuse would alter the immune response when exposed to an infectious agent.
Staining and FACS Analysis for Intracellular Markers
LN cells were cultured in complete RPMI in the presence of 1 nm PMA (Sigma-Aldrich), 1 μm calcium ionophore (Sigma), and 2 μm protein transport inhibitor monensin (Biolegend, San Diego, CA) for 4 h. Cells were washed and resuspended in FACS buffer (PBS containing 2% FBS and 0.1% sodium azide). Fc receptors were blocked by adding anti-mouse CD16/CD32 (10 μg/ml) followed by surface staining for CD4. Intracellular staining for cytokines IL-4 and IFN-γ was performed using leukocyte activation mixture with BD Golgiplug (BD Biosciences) according to the manufacturer's instructions. Intranuclear staining for TBX21, GATA3, and Ki67 was done using the Fix/Perm reagent kit from Biolegend. Anti-mouse CD16/CD32 mAbs (Fc-block), PE-Cy7-conjugated anti-mouse CD4, and APC-conjugated anti-mouse IL-4 antibodies were purchased from BD Biosciences. Cells were analyzed in a BC FC 500 flow cytometer.
ChIP and ChIP-Seq
ChIP was performed using the Simple ChIP-enzymatic Chromatin IP Kit (Cell Signaling, catalog no. 9003). Briefly, cells were diluted to 5 × 106 cells/ml in the cell culture medium, and 37% formaldehyde was added to a final concentration of 1% to cross-link histone and DNA. After a 10-min incubation at room temperature, formaldehyde was quenched by adding glycine to a final concentration of 125 mm. Cells were then pelleted and washed with cold PBS twice. 5 × 106 cells were resuspended in 500 μl of micrococcal nuclease buffer and digested with 2000 units of the enzyme for 20 min at 37 °C. Nuclei were pelleted by centrifugation at 13,000 rpm for 1 min and resuspended in 1 ml of ChIP buffer. Nuclear membrane was disrupted by brief sonication (two sets of 10-s pulses), and lysates were clarified by centrifugation at 10,000 rpm for 10 min. The supernatant was used for chromatin immunoprecipitation. The ChIP antibodies were purchased from Abcam (Cambridge, MA). They were as follows: H3K4me3 (ab1012), H3K27me3 (ab6002), H3K9me3 (ab8898), H3K36me3 (ab9050), and H3K9ac (ab12179). Ten μg of antibody was used for each immunoprecipitation. Antibodies were incubated with the sample at 4 °C overnight with rotation. After the addition of beads, samples were incubated for another 2 h at 4 °C with rotation. After washing, the immunoprecipitated chromatin was eluted from the bead, and the cross-link was reversed by proteinase K digestion. DNA was then purified using spin columns and quantified. For ChIP-Seq, the library was constructed using Illumina's Chip Sequencing sample preparation kit (catalog no. 1003473) according to the manufacturer's instructions. Briefly, 10 ng of ChIP-enriched DNA was used for each library construction. First the DNA fragments were repaired to phosphorylated blunt ends using T4 DNA polymerase, Klenow polymerase, and T4 polynucleotide kinase. After DNA fragments were purified using the Qiagen PCR purification kit (Qiagen, catalog no. 28104), an “A” base was added to the 3′-end of the blunt DNA fragment by Klenow fragment (3′ to 5′ exo-) at 37 °C for 30 min. The product was purified by the MinElute purification kit (Qiagen, catalog no. 28004). Sequencing adapters were ligated to the ends of DNA fragments using DNA ligase at room temperature for 15 min, followed by purification with the MinElute PCR purification kit. The product was then separated in 2% agarose gel to remove excess adaptors and to select a size range of library. The fragments with a size range from 150 to 250 bp were excised and purified using the QIAquick gel extraction kit (Qiagen, catalog no. 28704). The library was then amplified by limited PCR (16 cycles) using primers provided by the kit. The concentration and distribution of the library were determined by Agilent Bioanalyzer 2100. The library was sequenced by an Illumina HiSeq2000 system at the Tufts University Genomic Core Facility.
Data Analysis
The HiSeq2000 platform generated single-end reads with a read length of 50 bp. Raw sequencing reads in the FASTQ format were mapped to mouse genome build mm9 using Bowtie software by allowing two mismatches in the read (34). The mapped reads (SAM file) were then filtered, and only uniquely mapped reads were used for the downstream analysis. SICER was used for the peak calling (35, 36). The peak calling parameters were 200-bp window size and 600-bp gap size except for H3K4me3 and H3K9ac, in which 200-bp window size and 200-bp gap size were used. The statistic threshold value (E value) was set as 0.01. The peaks (in WIG file format) were visualized in the UCSC genome browser (University of California, Santa Cruz, CA). SICER-generated scoreisland files were used to draw a circular overall methylation picture using the R program. The correlation heat map of these signals was generated using DiffBind software (37). Distribution of signal in various genomic features was calculated using CEAS software (38). The promoter region was defined as 3 kb upstream of the TSS. Annotation for peak associated genes was performed using the peak2gene program in Cistrome (39). Genes with the center of H3K4me3, H3K9me3, and H3K9ac located within 3 kb up- or downstream of their TSS were identified as H3K4me3- and H3K9me3-associated genes. Because H3K27me3 had broad peaks, its associated genes were identified as the peak center located within 5 kb up- and downstream of TSS. H3K36me3-associated genes were identified as the peak center located within 3 kb downstream of TSS. Biological functions of those genes were classified using the PANTHER classification system (Gene Ontology Consortium).
Quantitative Real-time PCR
Popliteal lymph nodes were collected and homogenized, and total RNA was isolated using TRIzol (Invitrogen). RNA was reverse transcribed into cDNA using random primer and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The relative abundance of gene expression was determined by real-time PCR using GAPDH as the internal reference.
RESULTS
THC-attenuated SEB-induced Cell Proliferation and Immune Response in Vivo
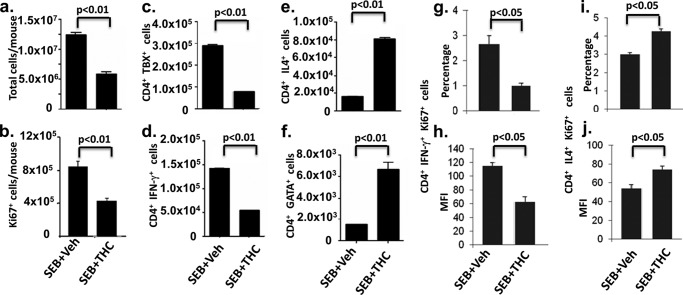
SEB is a superantigen that triggers robust T cell activation. Although THC is known to induce a switch in Th cell differentiation from Th1 to Th2 (9, 10), we attempted to corroborate these studies using SEB, so that we could use the same cells for epigenetic analysis. To this end, C57BL/6J mice were pretreated with THC or vehicle, as described previously (33), on day 0 and 1, and 2 h later, 10 μg of SEB was injected into each foot pad. Draining popliteal lymph nodes were harvested, and cells were analyzed 1, 3, and 5 days after SEB challenge. SEB exerted the most robust effect on CD4+ T cell proliferation 3 days after the treatment, as determined by total cell number (Fig. 1a) and Ki67 staining (Fig. 1b), and THC attenuated cell proliferation at all of those time points. Therefore, we chose the 3-day time point for the following study. In SEB + vehicle-treated mice, there was significant enlargement of draining popliteal LN with high cell yield (∼13 million cells) (Fig. 1a), when compared with LN from naive mice (∼1 million; data not shown), indicative of strong T cell proliferation caused by SEB. In contrast, in SEB + THC-treated mice, there was a significant decrease in total cell numbers as well as Ki67-positive cell number (Fig. 1b). We next determined the numbers of Th1 and Th2 cells by double-staining the cells for CD4 and IFN-γ/TBX21 to detect Th1 cells or for CD4 and IL-4/GATA3 to detect Th2 cells using flow cytometry. Based on the percentages of these cells, as detected by flow cytometry, we calculated the absolute numbers of Th1 or Th2 cells, respectively. In THC-pretreated mice, the numbers of CD4+ T cells TBX21-positive (Fig. 1c) and IFN-γ-positive (Fig. 1d) were significantly lower than those in vehicle-treated mice. On the other hand, IL-4-positive (Fig. 1e) and GATA-positive (Fig. 1f) CD4+ T cells were increased in THC-treated mice. The proliferation of Th1 and Th2 cells was further analyzed by FACS analysis for the expression of Ki67 on IFN-γ (Th1)- or IL-4 (Th2)-positive CD4+ T cells. Compared with vehicle-treated mice, cells from THC-treated mice had a decreased Ki67+ Th1 population (Fig. 1, g and h) and an increased Ki67+ Th2 population (Fig. 1, i and j). These results were consistent with what was shown in our previous studies and in those of others, that exposure to THC suppresses Th1 while enhancing Th2 response (10, 40, 41).
FIGURE 1.
Effect of THC on lymph node cell proliferation and Th1 and Th2 subpopulations. C57BL/6J mice were treated with THC or vehicle as described under “Experimental Procedures” on days 0 and 1, and 2 h later, 10 μg of SEB was injected into each foot pad. Three days after SEB challenge, draining popliteal lymph nodes of SEB + vehicle- or SEB + THC-treated mice (n = 3) were harvested, and cells were analyzed. a, total cells in two popliteal lymph nodes in each mouse. b, cells were gated by CD4+ and analyzed by FACS for the expression of Ki67. c–f, based on flow cytometric analysis as described under “Experimental Procedures,” cell numbers of various CD4+ T cell subpopulations expressing IFN-γ, TBX21, IL-4, or GATA3 were depicted. g and h, overall frequency and mean fluorescence intensity (MFI) of Ki67, CD4, and IFN-γ triple-positive cells. i and j, overall frequency and MFI of Ki67, CD4, and IL-4 triple-positive cells. p values were determined by Student's t test. Error bars, S.E.
Genome-wide Histone H3 Methylation Profile in SEB-activated Lymph Node Cells
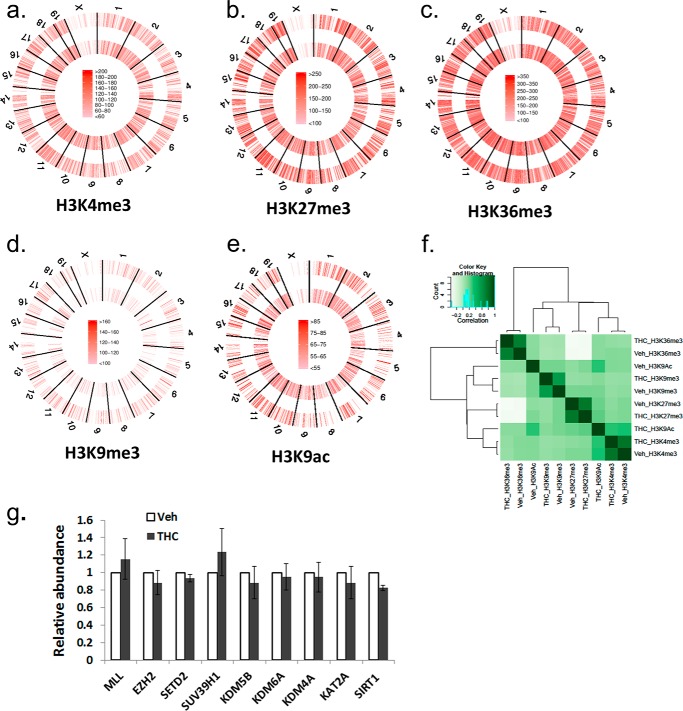
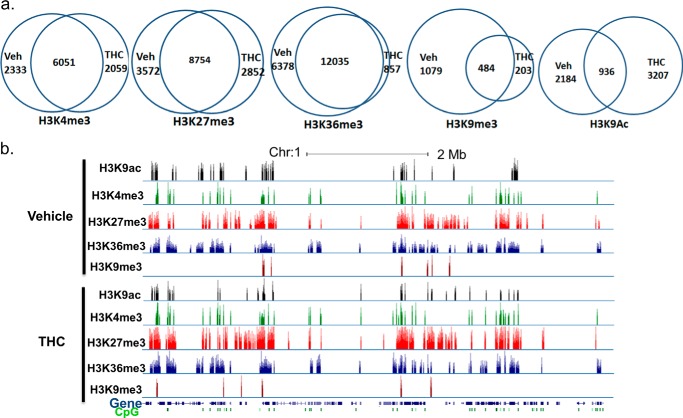
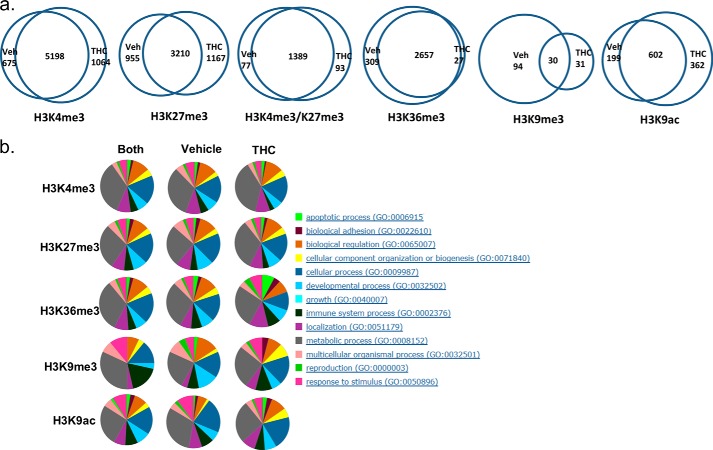
To determine whether THC exerts its immunosuppressive function through epigenetic modifications and to see if it has a global effect on histone modifications, we employed the ChIP-Seq method to examine the genome-wide histone H3 trimethylation pattern at Lys-4, Lys-9, Lys-27, and Lys-36 sites as well as acetylation at the Lys-9 site in draining popliteal lymph node cells from mice that received SEB + vehicle or SEB + THC. In our experiment, each ChIP library generated 150–210 million reads. Approximately, 60–70% of those reads were uniquely mapped to the mouse genome (mm9). A graphical display of H3K4me3, H3K27me3, H3K36me3, H3K9me3, and H3K9ac profiles across the whole genome is presented in Fig. 2, a–e. Although the overall signal level of each histone marker did not differ significantly between vehicle- and THC-treated cells, the distribution of the signal was altered, as demonstrated by correlation analysis (Fig. 2f). These results suggested that THC did not alter the overall activity of these histone modification enzymes, whereas genes associated with these histone markers were altered by THC treatment. We further examined the expression of major histone methyltranferase, demethylase, acetyltransferase, and deacetylase, which are known to control these histone modifications (42, 43). The expression of these enzymes did not differ significantly, as determined by real-time PCR (Fig. 2g). The unique and common genomic regions (intervals) containing these histone markers between vehicle- and THC-treated cells were further analyzed. Among these, whereas the occurrence of H3K36me3 markers was most abundant, H3K9me3 had the smallest number of signal regions (Fig. 3a). For H3K4me3, H3K27me3, and H3K36me3, there were more common regions than unique regions, whereas for H3K9me3 and H3K9ac, there were more unique regions. A representative histone methylation profile on a region of chromosome 1 is shown in Fig. 3b. In general, the H3K9ac, H3K4me3, and H3K9me3 had narrow signal peaks, whereas H3K27me3 and H3K36me3 had much broader peaks. These typical patterns were consistent with previous reports (19, 27). These results demonstrated that exposure to THC during an immune response to antigens such as SEB in vivo could alter histone modification, particularly H3K36me3, H3K9me3, and H3K9ac, thereby influencing global gene expression.
FIGURE 2.
Genome-wide histone H3 methylation level in lymph node cells. a–e, C57BL/6J mice were treated with SEB + THC or SEB + vehicle, as described in the legend to Fig. 1. The LN cells were studied for genome-wide histone H3 methylation and acetylation as described under “Experimental Procedures.” ChIP-Seq signal density is color-coded. Outer circle, SEB + vehicle-treated sample; inner circle, SEB + THC-treated sample. f, correlation of overall signal of these histone markers. The heat map was generated by DiffBind. g, relative mRNA abundance of histone-lysine N-methyltransferase Mll (H3K4me3), Ezh2 (H3K27me3), Setd2 (H3K36me3), Suv39h1 (H3K9me3), lysine-specific demethylase Kdm5b (H3K4me3), Kdm6a (H3K27me3), Kdm4a (H3K9me3 and H3K36me3), histone acetyltransferase Kat2a (H3K9ac), and NAD-dependent deacetylase Sirt1 (H3K9ac), as determined by real-time PCR. The amount in the vehicle-treated sample was set as 1. Error bars, S.E.
FIGURE 3.
Histone H3 methylation regions in activated lymph node cells. C57BL/6J mice were treated with SEB + THC or SEB + vehicle, as described in the legend to Fig. 1. The LN cells were studied for histone H3 methylation and acetylation regions. a, Venn diagrams of the overlap and unique regions of histone marker between the SEB + vehicle-treated (Veh) and SEB + THC-treated (THC) lymph node cells. b, representative ChIP-Seq result displayed in the UCSC genome browser.
Distribution of Histone H3 Methylation Signal in Genomic Features
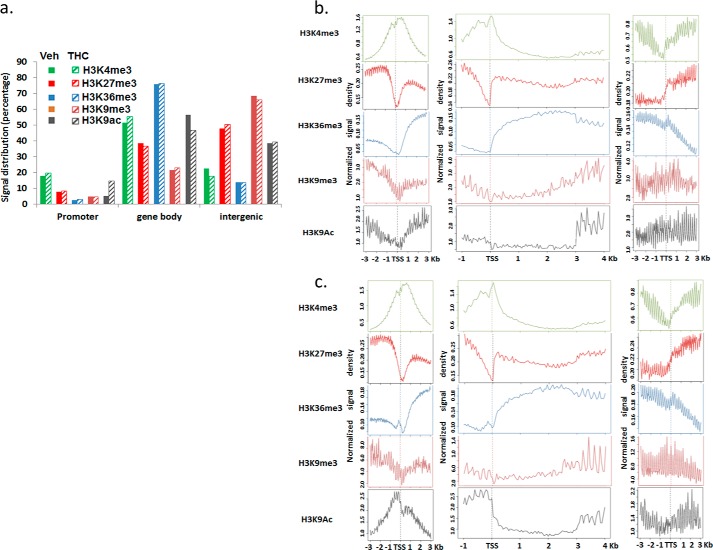
The distribution of histone markers was analyzed according to mouse genomic features. H3K4me3 was the most enriched in the promoter regions compared with others, whereas H3K27me3 was mostly located in the gene body and intergenic regions. H3K36me3 was mainly found within the gene body and H3K9me3 in the intergenic region (Fig. 4a). Furthermore, H3K4me3 was significantly increased near the TSS of genes, and its signal density decreased near the transcription termination site (TTS). This pattern was consistent with the notion that H3K4me3 is found in transcriptionally active promoters and is associated with gene activation. There was a dip of H3K4me3 signal density right before TSS (Fig. 4, b and c). A similar observation has been made by others, and this dip is thought to be due to the nucleosome loss in active genes (19). H3K27me3 level was lower near the TSS, and the signal was increased after the TTS (Fig. 4, b and c). This might be due to the reduced H3K27me3 modification in the active genes because H3K27me3 has been suggested to repress gene expression (44, 45). H3K36me3 signal was low before the TSS and after TTS but was enriched in the gene body, which was consistent with the indication that H3K36me3 associates with the transcription elongation (26). H3K9me3 has been implicated in gene repression. Its signal density decreased slightly near the TSS. Although the signal patterns of these histone methylation markers were similar in SEB + vehicle- and SEB + THC-treated cells, as sorted according to genomic features, many regions were differentially associated with these markers, suggesting that a different set of genes was expressed in these two samples. H3K9ac is associated with the promoter region of active genes. However, in SEB-activated lymph cells, its signal near the TSS was decreased, suggesting that SEB might affect histone acetylation or deacetylation enzymes. This pattern was reversed by THC treatment (Fig. 4, b and c), and H3K9ac was enriched near the TSS site as expected.
FIGURE 4.
Distribution of histone methylation signal among genomic features. C57BL/6J mice were treated with SEB + THC or SEB + vehicle, as described in the legend to Fig. 1. The LN cells were studied for histone markers as described under “Experimental Procedures.” a, the percentage of methylation signal located in the promoter regions (3 kb upstream of TSS), gene body (intron and exon), and intergenic region. b and c, the relative enrichment profile of each histone methylation near the TSS, within the transcript, and near the TTS in the SEB + vehicle-treated (b) and SEB + THC-treated (c) lymph node cells.
Genes Associated with Histone Methylation Markers
Because H3K4me3, H3K27me3, H3K9me3, and H3K9ac near the TSS are associated with gene activity, we identified genes that had these methylation signals near their TSS (Fig. 5a). Genes that had H3K36me3 signal in their transcript body were also identified. Overall, more genes were associated with H3K4me3, and fewer genes were associated with H3K36me3 and H3K9me3 signals in the THC-exposed cells relative to vehicle-treated cells. The number of genes that associated with H3K27me3 was similar between the SEB + vehicle-treated and SEB + THC-treated samples. Most genes with H3K4me3, H3K27me3, or H3K36me3 modification were common between the two samples. However, most of the H3K9me3-associated genes were unique to the vehicle treatment. Similarly, there were more H3K9ac-associated genes that were unique to the vehicle or THC treatment. Biological function classification showed that those genes were involved in a variety of pathways, from cell cycle to metabolism (Fig. 5b), suggesting that THC might have a much broader biological impact. The bivalent domains of H3K4me3 and H3K27me3 have been suggested to play a regulatory role in the differentiation in embryonic stem cells and T cells. There are a significant number of active genes that have both H3K4me3 and H3K27me3 in their promoters (25, 46). In this study, we also found that a significant number of genes had both H3K4me3 and H3K27me3 signal present near their TSS (Fig. 5a), suggesting that those genes might not be permanently activated or suppressed but rather more finely regulated. Although a large number of genes with this bivalent modification in their promoters were common to SEB + vehicle or SEB + THC treatment, a good number of such genes were unique to THC treatment. See the supplemental material for a list of genes that were differentially associated with these histone markers.
FIGURE 5.
Genes associated with histone methylation signal in lymph node cells. C57BL/6J mice were treated with SEB + THC or SEB + vehicle, as described in the legend to Fig. 1. The LN cells were studied for genes associated with histone markers. a, Venn diagrams of overlap and unique genes associated with each histone marker as well as H3K4me3/H3K27me3 bivalent modification in the SEB + vehicle-treated (Veh) and SEB + THC-treated (THC) samples. b, classification of these genes according to their cellular function.
Histone Methylation Pattern and Gene Expression
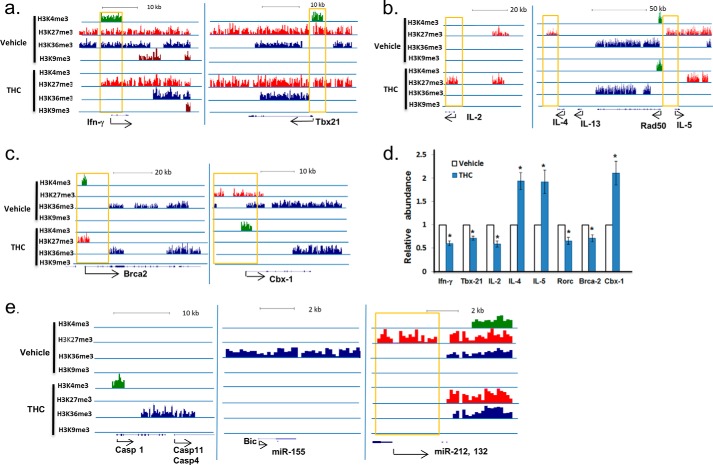
Because THC has been shown to shift the balance of Th1 and Th2, we examined these histone markers in the genomic regions of some of the Th1 and Th2 cytokines to determine their associated histone markers. IFN-γ is one of the most potent proinflammatory cytokines induced by SEB, and THC is known to suppress Ifn-γ expression (9, 14, 47), as was also seen in the current study (Fig. 1, b and c). In cells from SEB + vehicle-treated mice, the promoter of Ifn-γ was found to be associated with both active H3K4me3 and suppressive H3K27me3 signals. Its gene body also had active H3K36me3 signal. In cells from SEB + THC-treated mice, H3K4me3 and H3K36me3 diminished, indicating that the expression of Ifn-γ was suppressed (Fig. 6a). TBX21 is the transcription factor that controls the expression of Ifn-γ, which was decreased in SEB + THC-exposed cells (Fig. 1c). Correlating with this observation, we noted that the active signal H3K4me3 was present in the promoter region of Tbx21 in the SEB + vehicle-treated cells but absent in the SEB + THC-treated cells (Fig. 6a). On the other hand, IL-4 and IL-5, markers of Th2 cells, had H3K27me3 signal in their promoters in the SEB + vehicle-treated cells but lacked this signal in SEB + THC cells (Fig. 6b). Interestingly, H3K9ac marker was not found in the promoter regions of these genes in either vehicle- or THC-treated sample (data not shown). This result suggested that histone methylation, not histone H3 Lys-9 acetylation, correlated with THC-mediated Th1-Th2 shift in SEB-activated lymph cells. The mRNA expression of Ifn-γ, Tbx21, IL-4, and IL-5 was further validated by real-time PCR (Fig. 6d). We also noted that IL-2, involved in T cell proliferation, had suppressive H3K27me3 in its promoter region in THC-treated cells (Fig. 6b), which correlated with decreased mRNA expression (Fig. 6d).
FIGURE 6.
Histone methylation pattern and gene expression in lymph node cells. C57BL/6J mice were treated with SEB + THC or SEB + vehicle as described in the legend to Fig. 1. The LN cells were studied for histone methylation pattern and gene methylation, as described under “Experimental Procedures.” a and b, alteration of histone methylation in the promoter region of genes from cells exposed to SEB + vehicle (Vehicle) or SEB + THC (THC). c, example of genes with opposite histone methylation in SEB + vehicle-treated or SEB + THC-treated cells. d, relative mRNA abundance of selected genes, as determined by real-time PCR. The amount in the vehicle-treated sample was set as 1. e, example of potential genes and miRNAs whose expression might be regulated by THC. Error bars, S.E.
Besides these genes that are known to be regulated by THC, we also found other genes that were distinctively associated with active and suppressive methylation markers in vehicle- or THC-treated cells. For example, the promoter of Brca2, a tumor suppressor gene, had H3K4me3 and H3K27me3 signal in the SEB + vehicle- and SEB + THC-treated cells, respectively, suggesting that the expression of this gene might be suppressed by THC. On the contrary, CBX-1, a member of the heterochromatin protein family, had K3K27me3 signal in its promoter in the SEB + vehicle-treated cells but had K3K4me3 signal in the SEB + THC-treated cells (Fig. 6c). Real-time PCR results showed that Brca2 expression was indeed reduced, whereas Cbx-1 was increased with THC treatment (Fig. 6d). These validations indicated that histone methylation determinations in this study correlated well with expected gene expression changes. THC also induces apoptosis in immune cells. In macrophages and T cells, THC has been shown to act by inducing caspase-1 (48). Consistent with this, in SEB + THC-treated cells, Caspase-1 had H3K4me3 and H3K36me3 in its promoter and gene body, respectively (Fig. 6e).
A recent study showed that THC reduces Th17 (49). However, in this study, these histone methylation markers were not associated with Rorc, which regulates Th17 (data not shown). To determine whether Th17 is regulated by THC, we examined Rorc by real-time PCR. The expression of Rorc was decreased in THC-treated cells (Fig. 6), suggesting that THC modulates immune response by other histone modifications or by other mechanisms.
Besides protein coding genes, THC treatment also altered histone methylations in many noncoding RNAs. Long noncoding RNAs and miRNAs are important regulators of gene expression (50). For example, in the SEB + vehicle-treated mice, there was a strong H3K36me3 signal in the transcript of Bic/miR-155, whereas no signal was detected in SEB + THC-treated cells (Fig. 6e), suggesting that THC down-regulates Bic/miR-155 in the superantigen-activated LN cells. Another example is the miR-212 and miR-132 cluster. These two miRNAs are encoded from the intron of a non-coding transcript. Eighteen transcription start sites have been identified from 3 kb to 30 bp upstream of these miRNAs based on miRBase (Manchester, UK). The suppressive marker, H3K27me3, was present in all of these transcription start sites in the SEB + vehicle-treated cells but not in the SEB + THC-treated cells, suggesting that the suppressed expression of these miRNAs in SEB-activated lymph cells was reversed by THC treatment.
DISCUSSION
The immune response and the establishment of functionally specialized immune cell lineages are controlled by multiple transcription factors as well as epigenetic modifications, and these epigenetic modifications can be altered by various environmental factors or bioactive drug components. In this study, we examined the effect of THC on four histone methylation markers and one histone acetylation marker across the whole genome in SEB superantigen-activated lymph node cells in vivo. A significant amount of histone modification clusters were found to be unique to THC treatment. These results suggested that THC could specifically activate or suppress the expression of genes.
THC has been shown to have anti-inflammatory and immunosuppression properties and to induce apoptosis of immune cells (40). Indeed, the size of the popliteal lymph node was smaller, and the cell number was lower in SEB + THC-treated mice than that in the SEB + vehicle-treated mice. The histone methylation pattern in several proinflammatory and anti-inflammatory cytokines was consistent with data that indicated that THC suppressed proinflammatory cells such as Th1. H3K27me3, the suppression marker, was the only signal present in the promoter of Ifn-γ in the SEB + THC-treated sample in this study, and the expression of Ifn-γ was suppressed, although SEB is a potent agent to induce inflammation. In contrast, the Ifn-γ promoter in the SEB + vehicle-activated lymphocytes had both H3K4me3 and H3K27me3. The bivalent modification of H3K4me3 and H3K27me3 in the promoter of Ifn-γ suggested that the expression of Ifn-γ can be quickly modulated according to the external signal. Similarly, TBX21, a Th1-specific transcription factor that controls the expression of Ifn-γ, also had this bivalent modification in the SEB + vehicle-treated sample. This kind of modification might be critical for a balanced immune response because prolonged expression of proinflammatory cytokines can have adverse effects on the host. Despite a significant difference in overall H3K9ac pattern in vehicle- and THC-treated cells, we did not find a difference in the association of H3K9ac in these genes. The unexpected decrease of H3K9ac signal near the TSS of SEB + vehicle-treated cells may indicate that SEB affects the function of enzymes that regulate histone acetylation and deacetylation, and THC may partially relieve that effect. In the future, we will use other antigens to activate the immune cells to determine whether the H3K9ac pattern in this experiment is unique to SEB stimulation.
In this study, we identified many genes with bivalent modification. H3K4me3 and H3K27me3 bivalent modification has been proposed to explain the plasticity of T cell differentiation, and genes with bivalent modification can be either expressed or silenced (25). However, our study also demonstrated that some genes are oppositely modified in SEB + vehicle and SEB + THC samples. For example, the promoter of Brca2 had active H3K4me3 marker in the SEB + vehicle-treated sample but had suppressive H3K27me3 marker in the SEB + THC-treated sample. Whereas Cbx-1 had H3K27me3 in the SEB + vehicle-treated sample, it had H3K4me3 in the SEB + THC-treated sample. This suggested that the expression of these genes could be permanently altered by THC. Determination of whether this is the case, however, requires further investigation.
It is known that many histone modifications can independently regulate gene expression. For example, in human CD8+ T cells, some active genes are associated with high levels of H3K4me3, whereas others are associated with H3K9ac (17). That may explain the lack of histone methylation markers in Rorc while its expression is down-regulated by THC. It is possible that it is associated with other epigenetic modifications, such as other histone acetylation markers and DNA methylation.
Long noncoding RNAs and miRNAs are parts of the epigenetic regulation mechanism. Bic is a long noncoding RNA whose expression is elevated in the activated T cells (51, 52). Bic can be further processed into miR-155. It has been shown that Bic/miR-155 is essential for immune function, and mice with deficiency in Bic/miR-155 are immunodeficient (53). In a study of vulvar lichen sclerosus and lichen planus, autoimmune disorders that are characterized by a strong Th1 response, the expression of Bic/miR-155 was profoundly elevated (54). miR-155 has also been shown to be overexpressed in other autoimmune diseases and to enhance inflammatory T cell development (55). The altered histone methylation signal found in this study suggested that THC may also exert its function by regulating the expression of non-coding regulatory RNAs. Another example of histone methylation-mediated miRNA expression is miR-212 and miR-132. These miRNAs play important roles in immune response, apoptosis, and neuronal function. Expression of miR-212 enhances TRAIL-induced apoptosis, whereas inhibition of miR-212 renders cells resistant to TRAIL treatment (56). miR-132 has been indicated as an early response miRNA after viral infection and has been suggested as an innate immunity regulation miRNA (57). It has also been shown to potentiate anti-inflammatory signaling (58). Results from miR-212 and miR-132 knock-out mice indicated that these miRNAs regulate synaptic transmission and plasticity (59). Altered histone methylation signal in their transcription start sites after THC treatment suggested that THC could exert a broad biological effect by modulating miRNA expression.
In this study, we found that some genes have all four histone H3 methylations, whereas others only have one type of methylation signal. It is unclear whether the regulation of genes with more epigenetic modifications has greater complexity than those with fewer modification signals. It is also not clear whether genes with two active markers, such as H3K4me3 and H3K36me3 are more active than those with only one marker. It is also possible that the multiple modification signals may come from different types of cells found in the lymph node.
In summary, we demonstrate the association between THC-mediated histone modifications and a switch from Th1 to Th2 response against bacterial superantigen. The precise mechanisms through which THC regulates histone methylation remain to be further addressed. In the current study, we examined the expression of some major histone methyltranferase, demethylase, acetyltransferase, and deacetylase that are known to control these histone modifications (42, 43) and found that THC treatment failed to alter the expression of these enzymes, as determined by real-time PCR. However, it is possible that the expression of other enzymes might be altered by THC. In addition, THC could modulate the functional activity of these enzymes. Some studies suggested that THC could act directly on the epigenetic modification machinery. For example, anandamide, an endocannabinoid, has been shown to increase the DNA methylation level in human keratinocytes through p38 (60). As for histone modification, it has been shown that agonists of cannabinoid receptors can increase the number of H3K9me3-positive glioma stem-like cells, and this effect is blocked by CB antagonists (61). Interestingly, in four histone markers examined in this study, THC had the most profound effect on H3K9me3. Another example for the role of cannabinoids in histone modification is the association of increased overall histone H3 acetylation and decreased level of CB1 in Huntington disease (62), suggesting that cannabinoid signaling could affect histone acetylation enzymes. Furthermore, THC has been shown to alter histone deacetylase 3 in a dose-dependent manner (63). Histone deacetylase 3 is a member of the histone deacetylase family and, along with other histone deacetylases, is responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (64). Although we did not identify a significant change in the expression of Sirt1, the major deacetylase responsible for H3K9ac deacetylation in this study, we did observe a significant change in overall H3K9ac pattern after THC treatment (Fig. 4, b and c). Determination of whether the expression and activity of other histone acetylation enzymes are altered by THC requires further investigation. Another piece of evidence that suggests that cannabinoids may directly regulate epigenetic modification comes from cannabinoid receptor knock-out mice. In CB1 knock-out mice, it has been shown that CB1 regulates chromatin remodeling during spermiogenesis (65).
As for THC-mediated alteration in histone methylation, currently there is no study that indicates that THC directly regulates the expression or activity of histone methyltransferases or demethylases. However, THC could indirectly regulate the activity of enzymes involved in histone methylation. For example, cannabinoids have been shown to down-regulate the PI3K/AKT signaling pathway (66, 67), a pathway also known to cause global alterations of H3K27me3 (68). On the other hand, some studies showed that administration of THC increases phosphorylation of AKT in the mouse brain through CB1 (69). The discrepancy regarding the role of THC in AKT signaling may be due to the difference in cell type. Nonetheless, the effect of THC on AKT pathways may lead to regulation of histone methylation. AKT can phosphorylate EZH2 and suppress its methyltranferase activity, which results in a decrease of H3K27me3 (70). AKT also targets the association of histone with CBP, which regulates histone H3 acetylation (71). Additional studies are necessary to investigate whether the activity of EZH2 is altered by THC through the AKT pathway.
THC may also indirectly regulate histone methylation through other pathways, such as the estrogen receptor (ER) pathway. It has been shown that histone demethylases LSD1 and KDM2A are required for the induction of ER signaling after E2 stimulation (72). On the other hand, histone demethylase, KDM4B, is induced in an ER-α-dependent manner after E2 stimulation (73), indicating that activation of the ER pathway modulates histone methylation status. Many studies have shown that cannabinoid and estrogen pathways regulate each other. For example, some studies have suggested that both crude cannabis extract and THC inhibit the binding of estradiol to estradiol receptors in vivo (74, 75). Recent studies showed that some estrogen receptor modulators can bind to cannabinoid receptors (76, 77). These results have raised the possibility that THC could regulate histone methylation through ER signaling.
Thus, the current study opens new avenues to investigate the epigenetic pathways through which THC regulates the immune response. Because histone modifications can occur at many sites and at different levels, additional studies are necessary to address this because the current study focused on only certain histone markers. Second, the regulation of enzymes involved in histone modifications is very complex; thus, further investigations are necessary.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants P01AT003961, R01AT006888, R01ES019313, R01MH094755, and P20GM103641. This work was also supported by Veterans Affairs Merit Award BX001357.

This article contains supplemental material.
- THC
- Δ9-tetrahydrocannabinol
- H3K4me3
- histone H3 lysine 4 trimethylation
- H3K27me3
- histone H3 lysine 27 trimethylation
- H3K36me3
- histone H3 lysine 36 trimethylation
- H3K9me3
- histone H3 lysine 9 trimethylation
- H3K9ac
- histone H3 lysine 9 acetylation
- TSS
- transcription start site
- SEB
- staphylococcal enterotoxin B
- LN
- lymph node(s)
- TTS
- transcription termination site
- miRNA
- microRNA
- ER
- estrogen receptor.
REFERENCES
- 1. Substance Abuse and Mental Health Services Administration (2010) Results from the 2009 National Survey on Drug Use and Heath: Summary of National Findings, United States Department of Health and Human Services, Washington, D. C [Google Scholar]
- 2. Todaro B. (2012) Cannabinoids in the treatment of chemotherapy-induced nausea and vomiting. J. Natl. Compr. Canc. Netw. 10, 487–492 [DOI] [PubMed] [Google Scholar]
- 3. Cinti S. (2009) Medical marijuana in HIV-positive patients: what do we know? J. Int. Assoc. Physicians AIDS Care (Chic.) 8, 342–346 [DOI] [PubMed] [Google Scholar]
- 4. Galiègue S., Mary S., Marchand J., Dussossoy D., Carrière D., Carayon P., Bouaboula M., Shire D., Le Fur G., Casellas P. (1995) Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur. J. Biochem. 232, 54–61 [DOI] [PubMed] [Google Scholar]
- 5. Bouaboula M., Rinaldi M., Carayon P., Carillon C., Delpech B., Shire D., Le Fur G., Casellas P. (1993) Cannabinoid-receptor expression in human leukocytes. Eur. J. Biochem. 214, 173–180 [DOI] [PubMed] [Google Scholar]
- 6. Do Y., McKallip R. J., Nagarkatti M., Nagarkatti P. S. (2004) Activation through cannabinoid receptors 1 and 2 on dendritic cells triggers NF-κB-dependent apoptosis: novel role for endogenous and exogenous cannabinoids in immunoregulation. J. Immunol. 173, 2373–2382 [DOI] [PubMed] [Google Scholar]
- 7. Rao G. K., Zhang W., Kaminski N. E. (2004) Cannabinoid receptor-mediated regulation of intracellular calcium by Δ9-tetrahydrocannabinol in resting T cells. J. Leukoc. Biol. 75, 884–892 [DOI] [PubMed] [Google Scholar]
- 8. Newton C. A., Klein T. W., Friedman H. (1994) Secondary immunity to Legionella pneumophila and Th1 activity are suppressed by Δ-9-tetrahydrocannabinol injection. Infect. Immun. 62, 4015–4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klein T. W., Newton C. A., Nakachi N., Friedman H. (2000) Δ9-Tetrahydrocannabinol treatment suppresses immunity and early IFN-γ, IL-12, and IL-12 receptor β2 responses to Legionella pneumophila infection. J. Immunol. 164, 6461–6466 [DOI] [PubMed] [Google Scholar]
- 10. Yuan M., Kiertscher S. M., Cheng Q., Zoumalan R., Tashkin D. P., Roth M. D. (2002) Δ9-Tetrahydrocannabinol regulates Th1/Th2 cytokine balance in activated human T cells. J. Neuroimmunol. 133, 124–131 [DOI] [PubMed] [Google Scholar]
- 11. Hegde V. L., Nagarkatti M., Nagarkatti P. S. (2010) Cannabinoid receptor activation leads to massive mobilization of myeloid-derived suppressor cells with potent immunosuppressive properties. Eur. J. Immunol. 40, 3358–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kusmartsev S. A., Li Y., Chen S. H. (2000) Gr-1+ myeloid cells derived from tumor-bearing mice inhibit primary T cell activation induced through CD3/CD28 costimulation. J. Immunol. 165, 779–785 [DOI] [PubMed] [Google Scholar]
- 13. Bronte V., Apolloni E., Cabrelle A., Ronca R., Serafini P., Zamboni P., Restifo N. P., Zanovello P. (2000) Identification of a CD11b+/Gr-1+/CD31+ myeloid progenitor capable of activating or suppressing CD8+ T cells. Blood 96, 3838–3846 [PMC free article] [PubMed] [Google Scholar]
- 14. Hegde V. L., Hegde S., Cravatt B. F., Hofseth L. J., Nagarkatti M., Nagarkatti P. S. (2008) Attenuation of experimental autoimmune hepatitis by exogenous and endogenous cannabinoids: involvement of regulatory T cells. Mol. Pharmacol. 74, 20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hegde V. L., Tomar S., Jackson A., Rao R., Yang X., Singh U. P., Singh N. P., Nagarkatti P. S., Nagarkatti M. (2013) Distinct microRNA expression profile and targeted biological pathways in functional myeloid-derived suppressor cells induced by Δ9-tetrahydrocannabinol in vivo: regulation of CCAAT/enhancer binding protein α by microRNA-690. J. Biol. Chem. 288, 36810–36826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lombard C., Hegde V. L., Nagarkatti M., Nagarkatti P. S. (2011) Perinatal exposure to Δ9-tetrahydrocannabinol triggers profound defects in T cell differentiation and function in fetal and postnatal stages of life, including decreased responsiveness to HIV antigens. J. Pharmacol. Exp. Ther. 339, 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Araki Y., Wang Z., Zang C., Wood W. H., 3rd, Schones D., Cui K., Roh T. Y., Lhotsky B., Wersto R. P., Peng W., Becker K. G., Zhao K., Weng N. P. (2009) Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30, 912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morinobu A., Kanno Y., O'Shea J. J. (2004) Discrete roles for histone acetylation in human T helper 1 cell-specific gene expression. J. Biol. Chem. 279, 40640–40646 [DOI] [PubMed] [Google Scholar]
- 19. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 20. Roh T. Y., Cuddapah S., Cui K., Zhao K. (2006) The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. U.S.A. 103, 15782–15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lim P. S., Hardy K., Bunting K. L., Ma L., Peng K., Chen X., Shannon M. F. (2009) Defining the chromatin signature of inducible genes in T cells. Genome Biol. 10, R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Greer E. L., Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 24. Roh T. Y., Wei G., Farrell C. M., Zhao K. (2007) Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 17, 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T. Y., Watford W. T., Schones D. E., Peng W., Sun H. W., Paul W. E., O'Shea J. J., Zhao K. (2009) Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bannister A. J., Schneider R., Myers F. A., Thorne A. W., Crane-Robinson C., Kouzarides T. (2005) Spatial distribution of di- and trimethyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280, 17732–17736 [DOI] [PubMed] [Google Scholar]
- 27. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vakoc C. R., Mandat S. A., Olenchock B. A., Blobel G. A. (2005) Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19, 381–391 [DOI] [PubMed] [Google Scholar]
- 29. Wang Z., Zang C., Rosenfeld J. A., Schones D. E., Barski A., Cuddapah S., Cui K., Roh T. Y., Peng W., Zhang M. Q., Zhao K. (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Squazzo S. L., O'Geen H., Komashko V. M., Krig S. R., Jin V. X., Jang S. W., Margueron R., Reinberg D., Green R., Farnham P. J. (2006) Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16, 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karmodiya K., Krebs A. R., Oulad-Abdelghani M., Kimura H., Tora L. (2012) H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fraser J. D., Proft T. (2008) The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225, 226–243 [DOI] [PubMed] [Google Scholar]
- 33. Pandey R., Hegde V. L., Nagarkatti M., Nagarkatti P. S. (2011) Targeting cannabinoid receptors as a novel approach in the treatment of graft-versus-host disease: evidence from an experimental murine model. J. Pharmacol. Exp. Ther. 338, 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zang C., Schones D. E., Zeng C., Cui K., Zhao K., Peng W. (2009) A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blankenberg D., Von Kuster G., Coraor N., Ananda G., Lazarus R., Mangan M., Nekrutenko A., Taylor J. (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol., Chapter 19, Unit 19.10, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ross-Innes C. S., Stark R., Teschendorff A. E., Holmes K. A., Ali H. R., Dunning M. J., Brown G. D., Gojis O., Ellis I. O., Green A. R., Ali S., Chin S. F., Palmieri C., Caldas C., Carroll J. S. (2012) Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shin H., Liu T., Manrai A. K., Liu X. S. (2009) CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605–2606 [DOI] [PubMed] [Google Scholar]
- 39. Liu T., Ortiz J. A., Taing L., Meyer C. A., Lee B., Zhang Y., Shin H., Wong S. S., Ma J., Lei Y., Pape U. J., Poidinger M., Chen Y., Yeung K., Brown M., Turpaz Y., Liu X. S. (2011) Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12, R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagarkatti P., Pandey R., Rieder S. A., Hegde V. L., Nagarkatti M. (2009) Cannabinoids as novel anti-inflammatory drugs. Future Med. Chem. 1, 1333–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu L. X., Sharma S., Stolina M., Gardner B., Roth M. D., Tashkin D. P., Dubinett S. M. (2000) Δ-9-Tetrahydrocannabinol inhibits antitumor immunity by a CB2 receptor-mediated, cytokine-dependent pathway. J. Immunol. 165, 373–380 [DOI] [PubMed] [Google Scholar]
- 42. Shahbazian M. D., Grunstein M. (2007) Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 43. Klose R. J., Zhang Y. (2007) Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8, 307–318 [DOI] [PubMed] [Google Scholar]
- 44. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 45. Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vastenhouw N. L., Schier A. F. (2012) Bivalent histone modifications in early embryogenesis. Curr. Opin. Cell Biol. 24, 374–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karmaus P. W., Chen W., Crawford R., Kaplan B. L., Kaminski N. E. (2013) Δ9-Tetrahydrocannabinol impairs the inflammatory response to influenza infection: role of antigen-presenting cells and the cannabinoid receptors 1 and 2. Toxicol. Sci. 131, 419–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu W., Friedman H., Klein T. W. (1998) Δ9-Tetrahydrocannabinol induces apoptosis in macrophages and lymphocytes: involvement of Bcl-2 and caspase-1. J. Pharmacol. Exp. Ther. 286, 1103–1109 [PubMed] [Google Scholar]
- 49. Kozela E., Juknat A., Kaushansky N., Rimmerman N., Ben-Nun A., Vogel Z. (2013) Cannabinoids decrease the th17 inflammatory autoimmune phenotype. J. Neuroimmune Pharmacol. 8, 1265–1276 [DOI] [PubMed] [Google Scholar]
- 50. Mercer T. R., Mattick J. S. (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307 [DOI] [PubMed] [Google Scholar]
- 51. Tam W. (2001) Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene 274, 157–167 [DOI] [PubMed] [Google Scholar]
- 52. Haasch D., Chen Y. W., Reilly R. M., Chiou X. G., Koterski S., Smith M. L., Kroeger P., McWeeny K., Halbert D. N., Mollison K. W., Djuric S. W., Trevillyan J. M. (2002) T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol. 217, 78–86 [DOI] [PubMed] [Google Scholar]
- 53. Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A. (2007) Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Terlou A., Santegoets L. A., van der Meijden W. I., Heijmans-Antonissen C., Swagemakers S. M., van der Spek P. J., Ewing P. C., van Beurden M., Helmerhorst T. J., Blok L. J. (2012) An autoimmune phenotype in vulvar lichen sclerosus and lichen planus: a Th1 response and high levels of microRNA-155. J. Invest. Dermatol. 132, 658–666 [DOI] [PubMed] [Google Scholar]
- 55. O'Connell R. M., Kahn D., Gibson W. S., Round J. L., Scholz R. L., Chaudhuri A. A., Kahn M. E., Rao D. S., Baltimore D. (2010) MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Incoronato M., Garofalo M., Urso L., Romano G., Quintavalle C., Zanca C., Iaboni M., Nuovo G., Croce C. M., Condorelli G. (2010) miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 70, 3638–3646 [DOI] [PubMed] [Google Scholar]
- 57. Lagos D., Pollara G., Henderson S., Gratrix F., Fabani M., Milne R. S., Gotch F., Boshoff C. (2010) miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 12, 513–519 [DOI] [PubMed] [Google Scholar]
- 58. Shaked I., Meerson A., Wolf Y., Avni R., Greenberg D., Gilboa-Geffen A., Soreq H. (2009) MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31, 965–973 [DOI] [PubMed] [Google Scholar]
- 59. Remenyi J., van den Bosch M. W., Palygin O., Mistry R. B., McKenzie C., Macdonald A., Hutvagner G., Arthur J. S., Frenguelli B. G., Pankratov Y. (2013) miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS One 8, e62509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paradisi A., Pasquariello N., Barcaroli D., Maccarrone M. (2008) Anandamide regulates keratinocyte differentiation by inducing DNA methylation in a CB1 receptor-dependent manner. J. Biol. Chem. 283, 6005–6012 [DOI] [PubMed] [Google Scholar]
- 61. Aguado T., Carracedo A., Julien B., Velasco G., Milman G., Mechoulam R., Alvarez L., Guzmán M., Galve-Roperh I. (2007) Cannabinoids induce glioma stem-like cell differentiation and inhibit gliomagenesis. J. Biol. Chem. 282, 6854–6862 [DOI] [PubMed] [Google Scholar]
- 62. Sadri-Vakili G., Bouzou B., Benn C. L., Kim M. O., Chawla P., Overland R. P., Glajch K. E., Xia E., Qiu Z., Hersch S. M., Clark T. W., Yohrling G. J., Cha J. H. (2007) Histones associated with downregulated genes are hypo-acetylated in Huntington's disease models. Hum. Mol. Genet. 16, 1293–1306 [DOI] [PubMed] [Google Scholar]
- 63. Khare M., Taylor A. H., Konje J. C., Bell S. C. (2006) Δ9-Tetrahydrocannabinol inhibits cytotrophoblast cell proliferation and modulates gene transcription. Mol. Hum. Reprod. 12, 321–333 [DOI] [PubMed] [Google Scholar]
- 64. Guenther M. G., Lazar M. A. (2003) Biochemical isolation and analysis of a nuclear receptor corepressor complex. Methods Enzymol. 364, 246–257 [DOI] [PubMed] [Google Scholar]
- 65. Chioccarelli T., Cacciola G., Altucci L., Lewis S. E., Simon L., Ricci G., Ledent C., Meccariello R., Fasano S., Pierantoni R., Cobellis G. (2010) Cannabinoid receptor 1 influences chromatin remodeling in mouse spermatids by affecting content of transition protein 2 mRNA and histone displacement. Endocrinology 151, 5017–5029 [DOI] [PubMed] [Google Scholar]
- 66. Ellert-Miklaszewska A., Kaminska B., Konarska L. (2005) Cannabinoids down-regulate PI3K/Akt and Erk signalling pathways and activate proapoptotic function of Bad protein. Cell. Signal. 17, 25–37 [DOI] [PubMed] [Google Scholar]
- 67. Greenhough A., Patsos H. A., Williams A. C., Paraskeva C. (2007) The cannabinoid Δ9-tetrahydrocannabinol inhibits RAS-MAPK and PI3K-AKT survival signalling and induces BAD-mediated apoptosis in colorectal cancer cells. Int. J. Cancer 121, 2172–2180 [DOI] [PubMed] [Google Scholar]
- 68. Zuo T., Liu T. M., Lan X., Weng Y. I., Shen R., Gu F., Huang Y. W., Liyanarachchi S., Deatherage D. E., Hsu P. Y., Taslim C., Ramaswamy B., Shapiro C. L., Lin H. J., Cheng A. S., Jin V. X., Huang T. H. (2011) Epigenetic silencing mediated through activated PI3K/AKT signaling in breast cancer. Cancer Res. 71, 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ozaita A., Puighermanal E., Maldonado R. (2007) Regulation of PI3K/Akt/GSK-3 pathway by cannabinoids in the brain. J. Neurochem. 102, 1105–1114 [DOI] [PubMed] [Google Scholar]
- 70. Cha T. L., Zhou B. P., Xia W., Wu Y., Yang C. C., Chen C. T., Ping B., Otte A. P., Hung M. C. (2005) Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310, 306–310 [DOI] [PubMed] [Google Scholar]
- 71. Liu Y., Xing Z. B., Zhang J. H., Fang Y. (2013) Akt kinase targets the association of CBP with histone H3 to regulate the acetylation of lysine K18. FEBS Lett. 587, 847–853 [DOI] [PubMed] [Google Scholar]
- 72. Garcia-Bassets I., Kwon Y. S., Telese F., Prefontaine G. G., Hutt K. R., Cheng C. S., Ju B. G., Ohgi K. A., Wang J., Escoubet-Lozach L., Rose D. W., Glass C. K., Fu X. D., Rosenfeld M. G. (2007) Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kawazu M., Saso K., Tong K. I., McQuire T., Goto K., Son D. O., Wakeham A., Miyagishi M., Mak T. W., Okada H. (2011) Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One 6, e17830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sauer M. A., Rifka S. M., Hawks R. L., Cutler G. B., Jr., Loriaux D. L. (1983) Marijuana: interaction with the estrogen receptor. J. Pharmacol. Exp. Ther. 224, 404–407 [PubMed] [Google Scholar]
- 75. von Bueren A. O., Schlumpf M., Lichtensteiger W. (2008) Δ9-Tetrahydrocannabinol inhibits 17β-estradiol-induced proliferation and fails to activate androgen and estrogen receptors in MCF7 human breast cancer cells. Anticancer Res. 28, 85–89 [PubMed] [Google Scholar]
- 76. Kumar P., Song Z. H. (2014) CB2 cannabinoid receptor is a novel target for third-generation selective estrogen receptor modulators bazedoxifene and lasofoxifene. Biochem. Biophys. Res. Commun. 443, 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Prather P. L., FrancisDevaraj F., Dates C. R., Greer A. K., Bratton S. M., Ford B. M., Franks L. N., Radominska-Pandya A. (2013) CB1 and CB2 receptors are novel molecular targets for tamoxifen and 4OH-tamoxifen. Biochem. Biophys. Res. Commun. 441, 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.