Background: Function and signaling mediated by SCUBE3 during development remain poorly understood.
Results: Loss and gain of function studies showed that SCUBE3 could modulate FGF8 signal activity for fast muscle development.
Conclusion: scube3 is critical for and acts at top of the regulatory hierarchy of fast fiber myogenesis by enhancing fgf8 signaling.
Significance: Scube3 is a key regulator for fast muscle development in vertebrates.
Keywords: Cell Differentiation, Fibroblast Growth Factor (FGF), Fibroblast Growth Factor Receptor (FGFR), Myogenesis, Zebrafish, Fast Muscle, scube3
Abstract
SCUBE3 (signal peptide CUB-EGF-like domain-containing protein 3) belongs to a newly identified secreted and cell membrane-associated SCUBE family, which is evolutionarily conserved in vertebrates. Scube3 is predominantly expressed in a variety of developing tissues in mice such as somites, neural tubes, and limb buds. However, its function during development remains unclear. In this study, we first showed that knockdown of SCUBE3 in C2C12 myoblasts inhibited FGF receptor 4 expression and FGF signaling, thus resulting in reduced myogenic differentiation. Furthermore, knockdown of zebrafish scube3 by antisense morpholino oligonucleotides specifically suppressed the expression of the myogenic marker myod1 within the lateral fast muscle precursors, whereas its expression in the adaxial slow muscle precursors was largely unaffected. Consistent with these findings, immunofluorescent staining of fast but not slow muscle myosin was markedly decreased in scube3 morphants. Further genetic studies identified fgf8 as a key regulator in scube3-mediated fast muscle differentiation in zebrafish. Biochemical and molecular analysis showed that SCUBE3 acts as a FGF co-receptor to augment FGF8 signaling. Scube3 may be a critical upstream regulator of fast fiber myogenesis by modulating fgf8 signaling during zebrafish embryogenesis.
Introduction
SCUBE3 [signal peptide-CUB (complement protein C1r/C1s, Uegf, and Bmp1)-EGF-like domain-containing protein 3] belongs to the secreted and membrane-associated SCUBE family composed of three members evolutionarily conserved from zebrafish to humans (1–8). Proteins encoded by these genes are organized in a modular fashion and share at least five protein domains: 1) an NH2-terminal signal peptide sequence, 2) nine copies of EGF-like repeats, 3) a spacer region, 4) three cysteine-rich motifs, and 5) one CUB domain at the COOH terminus. Scube gene expression is overlapping and/or complementary to each other in vertebrate embryos, with transcripts predominating in the somites, central nervous system, and notochord early during development (2, 3, 7). Thus, these Scube genes may have critical functions during development in these tissue sites, where delicate signaling of morphogens or growth factors are transmitted by cell surface receptors including receptor serine/threonine kinases and receptor tyrosine kinases (RTKs)2 (9, 10). We and others have recently shown that SCUBE proteins are involved in modulating the signal activity of hedgehog (Hh) (11) or bone morphogenetic protein and TGF-β (12–14), which bind and activate their corresponding G-protein coupled receptors or receptor serine/threonine kinases. However, whether SCUBE proteins can also regulate RTK signaling activity remains unknown.
Skeletal muscles are derived from somites that form by segmentation of the paraxial mesoderm in vertebrates (15, 16). Different fiber types within vertebrate muscles can be broadly classified as slow or fast muscle on the basis of their mechanical and metabolic properties (17). The slow muscle derives from the medially located adaxial cells and depends on Hh signals from the midline. The hh genes are expressed in the floor plate and notochord during myogenesis and are responsible for maintaining the early myogenic factors myf5 and myod1 in adaxial cells. Cells of the segmental plate located laterally to the adaxial cells develop into fast muscles (18). Studies in chick indicate that overexpression of fibroblast growth factor 8 (fgf8) promotes fibroblast growth factor receptor 4 (fgfr4, a type of RTK) and myod1 expression in somites, whereas inhibition of Fgfr4 signaling represses limb muscle differentiation (19). In zebrafish, lateral somatic cells require fgf8 signaling to initiate the expression of myod1 and, subsequently, to undergo terminal differentiation into fast muscles (20, 21). In addition, fgf8 and fgfr4 positively regulate their own expression through a feed-forward signaling loop (19, 22). However, how fgf8 signaling is regulated during myogenesis remains unclear.
In this study, we first demonstrated that SCUBE3 is involved in the modulation of FGF8 signaling and myogenic differentiation in C2C12 myoblasts. In addition, loss of function studies with an antisense morpholino oligonucleotide (MO) knockdown approach revealed that zebrafish scube3 plays an essential role in fast muscle development by acting as a co-receptor to augment Fgf8 signaling activity.
EXPERIMENTAL PROCEDURES
Ethics Statement
Animal handling protocols were reviewed and approved by the Institutional Animal Care and Utilization Committee of Academia Sinica (Protocol No. RMiIBMYR2010063).
Zebrafish
Wild-type AB strain zebrafish were maintained in a 14-h light/10-h dark cycle at 28.5 °C. Zebrafish embryos were collected by natural spawning and incubated in 0.3× Danieau's buffer (diluting by 1× Danieau's buffer: 58 mm NaCl, 0.7 mm KCl, 0.4 mm MgSO4, 0.6 mm Ca(NO3)2, and 5 mm HEPES (pH 7.6) with double distilled water) until observation or fixation. The definition of embryo stage was as described (23), and the stages are indicated as hours postfertilization. The 0.2 mm N-phenylthiourea in 0.3× Danieau's buffer was used to suppress melanin formation after the bud stage. Dechorionation of embryos was achieved by protease (0.01 g/ml) digestion.
Identification of Zebrafish scube3 Full-length cDNA
A TBLASTN search of the public database with mammalian SCUBE3 protein sequences identified one expressed sequence tag clone (GenBankTM accession number EB930285) containing the 5′ most coding sequences of zebrafish scube3. This cDNA clone was purchased from Open Biosystems (Lafayette, CO). After complete sequencing of the clone, we identified a full-length cDNA sequence of zebrafish scube3 (4,035 bp), which is composed of a 228-bp 5′ UTR, a 2,988-bp protein-coding sequence, and an 819-bp 3′ UTR. This sequence was deposited in GenBankTM (accession number KF730313).
Whole Mount in Situ Hybridization (ISH) and Immunofluorescent Staining
Whole mount ISH was performed essentially as described (24). An 801-bp cDNA in the 3′-UTR of zebrafish scube3 was used to synthesize antisense RNA riboprobe. All other probes were synthesized as described: scl (25), myod1 (26), and fgf8 (27). Whole mount immunofluorescent staining was performed with the following primary antibodies: anti-slow myosin heavy chain F59 (28) and anti-fast myosin light chain F310 (21). F59, F310, and MF20 antibodies were developed by F. E. Stockdale and D. A. Fischman and obtained from the Developmental Studies Hybridoma Bank, maintained by the University of Iowa. Staining was performed as previously described (29).
Morpholinos and mRNA Microinjection
Two scube3 antisense translation-blocking MOs (tMOs) were designed: tMO1, 5′-TGC ATG AAC ATC ATC GCA AAA CAC A-3′; and tMO2, 5′-TCC AAA ACC AGC GCG CAT CAG AAC A-3′ (Gene Tools, Philomath, OR). The scube3 tMO1 targets the 5′-UTR and scube3 tMO2 blocks the AUG translation start site of zebrafish scube3. The scube3 tMOs diluted in Danieau buffer (12) were injected at the one- or two-cell stage. Increasing doses of each tMO were injected to examine the phenotypic effects. As a control for nonspecific effects, each experiment was performed in parallel with a randomized control MO. For knockdown experiments, we usually injected 5 or 7.5 ng per embryo of scube3 tMO1 or tMO2. Sense mRNA encoding full-length scube1 was transcribed in vitro from pCS2+-scube3 plasmid by use of the mMESSAGE mMACHINE kit (Ambion).
Construction of MO Testing Plasmid and Expression Plasmids
To construct the scube3-GFP testing plasmid, we cloned cDNA containing the region targeted by scube3-tMOs (−34 to +20) into the EcoRI-XbaI sites of pCS2-EGFP vector. The scube3 cDNA fragment was obtained with the following complementary oligonucleotides: 5′-aat tcT TTG TGT TTT GCG ATG ATG TTC ATG CAT GTT CTG ATG CGC GCT GGT TTT GGA TTt-3′ and 5′-cta gaA ATC CAA AAC CAG CGC GCA TCA GAA CAT GCA TGA ACA TCA TCG CAA AAC ACA AAg-3′. The epitope-tagged version of Scube3, Fgf8, FGFR1, and FGFR4 expression plasmids were constructed as described (8).
Cell Culture, Transfection, and Generation of Stable Cells
Human embryonic kidney-293T (HEK-293T) cells and C2C12 myoblasts were maintained as described (5, 30) and transfected by use of Lipofectamine 2000 (Invitrogen) or FuGENE HD (Roche). For the RNAi-mediated silencing experiments, we used the vector-based shRNAs generated by The RNAi Consortium (31) to knock down the endogenous Scube3 (clones TRCN0000109645 and TRCN0000109648) in murine C2C12 myoblasts. A nontargeting shRNA for LacZ (clone TRCN0000072223) was used as a control. shRNA expression plasmid-transfected C2C12 myoblasts were maintained in normal growth medium for 24 h after transfection and supplemented with puromycin (selection antibiotics). Scube3 knockdown in a selected stable pool was confirmed by RT-PCR.
Immunoprecipitation and Western Blot Analysis
Two days after transfection, cell lysates were clarified by centrifugation at 10,000 × g for 20 min at 4 °C. Samples were incubated with 1 μg of the indicated antibody and 20 μl of 50% (v/v) protein A-agarose (Pierce) for 2 h with gentle rocking. After three washes with lysis buffer, precipitated complexes were solubilized by boiling in Laemmli sample buffer, fractionated by SDS-PAGE, and transferred to PVDF membranes, which were blocked with phosphate-buffered saline (pH 7.5) containing 0.1% gelatin and 0.05% Tween 20 and blotted with the indicated antibodies. After two washes, the blots were incubated with horseradish peroxidase-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) for 1 h. After washing the membranes, the reactive bands were visualized by use of the VisGlow chemiluminescent substrate, HRP system (Visual Protein).
Luciferase Reporter Assay
HEK-293T cells (2 × 105 cells/well) were seeded into 24-well plates and transfected with 0.3 μg of the FGF-responsive luciferase reporter I-FiRE-Luc, with 0.01 μg of the Renilla luciferase reporter used as an internal control, and together with 0.3 μg of empty vector or the zebrafish scube3 expression plasmid and 0.1 μg of FGFR1 or FGFR4 expression plasmid. Transfected cells were maintained in low serum medium (0.5% fetal bovine serum) for 18 h. Luciferase activity was measured after 24 h of treatment with rhFGF8 protein.
Confocal Immunofluorescence Analysis and Subcellular Fractionation
Confocal analysis was performed as previously described (32). Briefly, HeLa cells were fixed in 4% formaldehyde, blocked with 2% goat serum for 1 h, and incubated with mouse anti-FLAG antibody (Sigma) and rabbit anti-Myc antibody (Sigma) followed by Alexa Fluor 594-labeled anti-mouse IgG antibody and Alexa Fluor 488-labeled anti-rabbit IgG antibody (Molecular Probes). Fluorescence images were captured under a confocal microscope (model LSM 510; Carl Zeiss MicroImaging). Subcelluar fractionation was performed as described (8).
RESULTS
SCUBE3 Is Essential for Proper Myogenic Differentiation and FGF8 Signaling
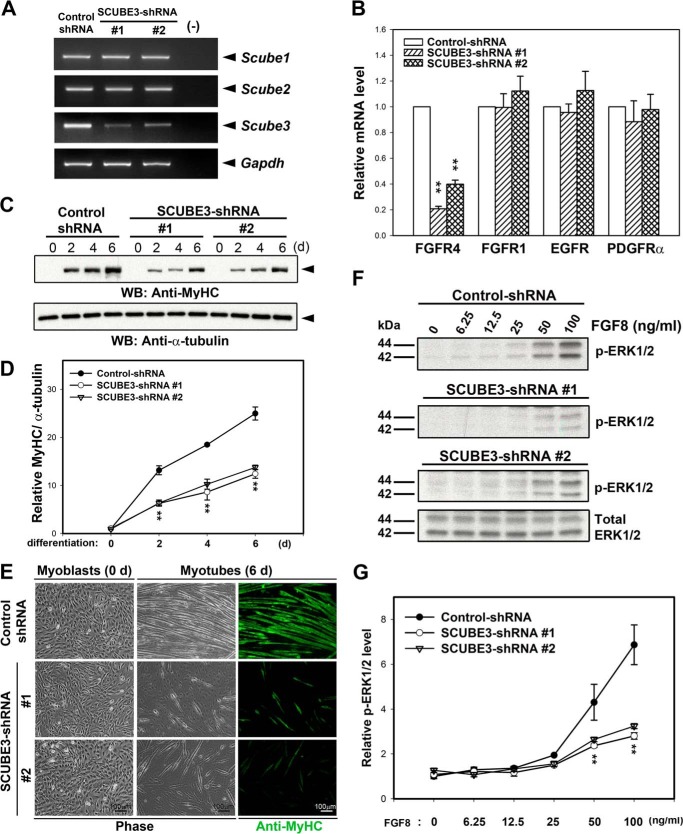
Because activation or suppression of RTK signaling usually leads to up- or down-regulation of its receptor expression via a feedback regulatory mechanism (19, 22), we first examined whether Scube3 knockdown could affect the expression of RTKs and downstream signaling. After transduction of recombinant lentivirus encoding two different SCUBE3-targeting shRNAs (shRNAs 1 and 2) or a control shRNA targeting bacterial β-galatosidase in C2C12 myoblasts, the efficiency and specificity of shRNA-mediated knockdown of Scube3 mRNA expression (but not Scube1 and Scube2) were confirmed by RT-PCR analysis because we lack an effective anti-mouse SCUBE3 antibody for Western blot analysis (Fig. 1A). The mRNA expression of FGFR4 but not several other RTKs, including FGFR1, EGF receptor, and PDGR-α, was concomitantly suppressed after SCUBE3 knockdown (Fig. 1B), which suggests a potential involvement of SCUBE3 in FGFR4-mediated signaling. Because FGFR4 can preferentially transmit FGF8 signaling and FGF8 has been implicated in skeletal muscle development (19, 20, 33), we further investigated whether SCUBE3 plays a role in myogenic differentiation in vitro. The control or Scube3 knockdown C2C12 myoblasts were induced for myogenic differentiation by culture under differentiation medium, and their differentiation traits were compared by Western blot analysis and immunofluorescence staining for myosin heavy chain (MyHC), a definitive marker for the activation of the myogenic program (30). Upon induction of differentiation, control C2C12 cells showed a significant up-regulation of MyHC by day 2 and subsequently transformed into multinucleated myofibers by day 6 (Fig. 1, C–E). However, knockdown of SCUBE3 markedly decreased MyHC expression and reduced the formation of multinucleated myofibers (14 ± 1 (shRNA 1) or 22 ± 2 (shRNA 2) versus 65 ± 4 (Control) myotubes/field; p < 0.01). Thus, SCUBE3 expression is required for an efficient myogenic differentiation in vitro.
FIGURE 1.
Scube3 knockdown causes defective myogenic differentiation and reduced FGF8 signaling. A, knockdown of Scube3 mRNA expression in murine C2C12 myoblasts. C2C12 myoblasts were infected with a lentiviral vector to generate a stable clone expressing an shRNA control or shRNA specific for mouse Scube3 (SCUBE3-shRNAs 1 and 2), respectively. These cells were maintained in normal growth medium for 24 h after transfection and supplemented with puromycin (selection antibiotics). The efficiency and specificity of Scube3 knockdown but not Scube1 and Scube2 in the selected stable pool was confirmed by RT-PCR analysis. Gapdh mRNA level was used as an internal control. B, specific down-regulation of FGFR4 expression in Scube3-silenced C2C12 myoblasts. The expression of FGFR4, FGFR1, EGF receptor, and PDGFRα mRNA was assessed by quantitative real time PCR normalized to GAPDH expression with individual primer sets. The data are means ± S.E. **, p < 0.01 (compared with the control). C, suppression of myosin heavy chain (MyHC) expression by SCUBE3 knockdown. Control and SCUBE3-shRNA 1 and 2 myoblasts were induced to differentiate for 0, 2, 4, and 6 days in differentiation medium. Cell lysates were analyzed for the expression of MyHC by Western blot analysis. Tubulin levels were used as a loading control. D, quantification of MyHC protein levels during myogenic differentiation. Relative MyHC levels were quantified by densitometric scanning and normalized by α-tubulin. The data are means ± S.E. of four experiments. **, p < 0.01 (compared with control). E, effect of SCUBE3 knockdown on morphological transformation and anti-MyHC immunostaining after myogenic differentiation. Control and SCUBE3-shRNA 1 and 2 myoblasts were analyzed by phase contrast microscopy or immunofluorescence staining for MyHC after 6 days of differentiation. F, effect of SCUBE3 knockdown on FGF8 signaling. Control myoblasts and Scube3 knockdown myoblasts (SCUBE3-shRNA 1 or 2) were treated with the indicated concentrations of FGF8 for 10 min. Cell lysates (20 μg) underwent Western blot analysis with antibody specific for phosphorylated ERK1/2 (p-ERK1/2; representative blots are shown in the top three panels) or total ERK1/2 protein (representative blots are shown in the bottom panel). G, quantification of relative phosphorylated ERK1/2 levels by densitometric scanning and normalized by total amount of ERK1/2. The data are means ± S.E. of two experiments. **, p < 0.01 (compared with control). d, days; WB, Western blot.
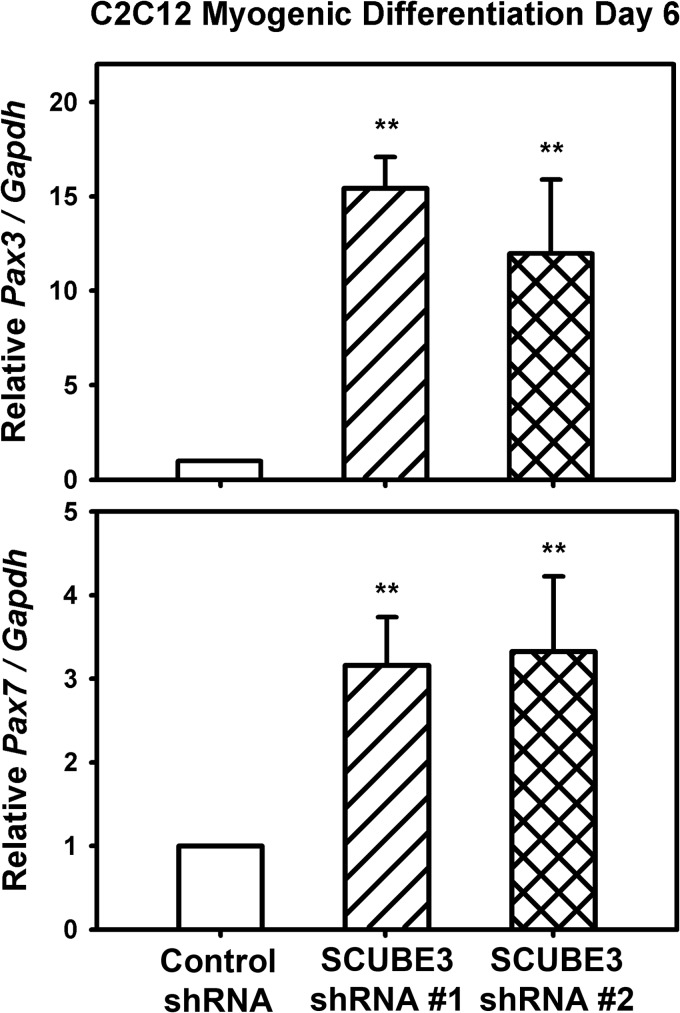
We then asked whether knockdown of SCUBE3 would result in decreased FGF8-dependent signaling by measuring the phosphorylation of ERKs in response to FGF8 (34). FGF8-induced ERK activation, as quantified by its phosphorylation, was significantly reduced in Scube3 knockdown C2C12 cells by two independent shRNAs (shRNAs 1 and 2) as compared with control cells (Fig. 1, F and G). In addition, it has been shown that Fgf signal pathway promotes myogenic differentiation by activating myod1 while it inhibits proliferation of myogenic precursors by repressing pax3 and pax7 expression (29, 36). In agreement with these reports, the Scube3 knockdown-induced reduction of myotube formation is accompanied by an increase of myogenic precursors caused by up-regulation of Pax3 and Pax7 expression in C2C12 cells (Fig. 2). Together, these results suggest that SCUBE3 may be involved in myogenic differentiation by modulating FGF8 signaling and downstream expression of Pax3/Pax7 in C2C12 myoblasts.
FIGURE 2.
Effects of Scube3 knockdown on the expression of Pax3 or Pax7. Control and SCUBE3-shRNA 1 and 2 myoblasts were induced to differentiate for 6 days in differentiation medium. Cells were collected to analyze the expression of Pax3 or Pax7 by quantitative RT-PCR. The data are means ± S.E. of three experiments. **, p < 0.01 (compared with control).
SCUBE3 Can Interact with FGF8 and FGFR4 and Enhances FGF8 Signaling
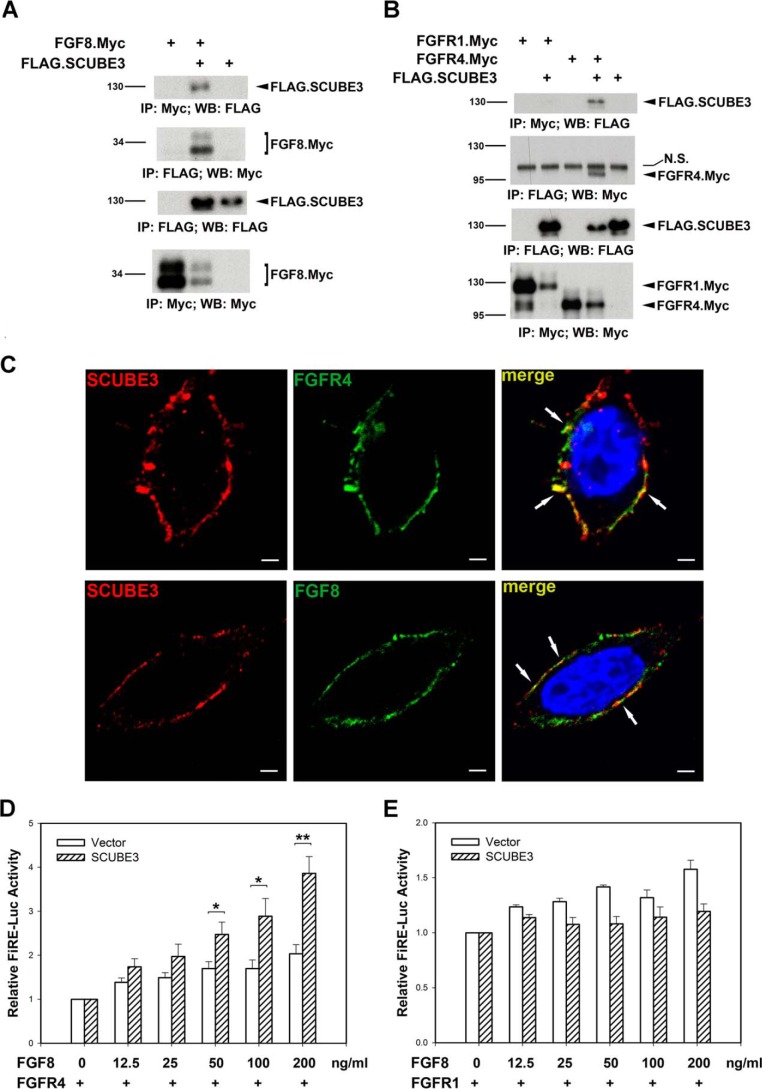
Because we observed a functional link between SCUBE3 and myogenic differentiation/FGF signaling, we then examined whether SCUBE3 could biochemically interact with FGF8 and its receptor, and if so, whether SCUBE3 could modulate FGF8-mediated signaling. HEK-293T cells were transfected with the expression plasmid encoding Myc-tagged FGF8 alone or with the FLAG-tagged SCUBE3 expression construct. Two days after transfection, cell lysates underwent immunoprecipitation with the anti-Myc monoclonal antibody, and precipitates were analyzed by immunoblotting with anti-FLAG monoclonal antibody to determine protein interaction. Immunoprecipitation with anti-Myc antibody resulted in specific co-immunoprecipitation of the SCUBE3 protein (Fig. 3A). Thus, SCUBE3 may indeed form a complex with FGF8.
FIGURE 3.
SCUBE3 forms a complex with FGF8 and FGFR4 and augments FGF signaling. A and B, SCUBE3 interacts with FGF8 and FGFR4. The expression plasmid encoding the Myc-tagged FGF8 (A) or Myc-tagged FGFR1 or FGFR4 (B) was transfected alone or with FLAG-tagged SCUBE3 in HEK-293T cells. After 2 days, cell lysates were prepared and underwent immunoprecipitation, followed by Western blot analysis with indicated antibodies to determine protein-protein interactions. Double bands seen for the FGF8.Myc were caused by differential glycosylation of this protein. N.S., nonspecific band; IP, immunoprecipitation; WB, Western blot. C, co-localization of SCUBE3 and FGF8 or FGFR4 was visualized by confocal immunofluorescence microscopy. SCUBE3 localization was detected by mouse anti-FLAG antibody and Alexa Fluor 594-conjugated goat anti-mouse IgG (red). FGF8 or FGFR4 was seen with rabbit anti-Myc antibody and Alexa Fluor 488-conjugated goat anti-rabbit IgG (green). The overlay demonstrates co-localization of SCUBE3 with FGF8 or FGFR4 on the plasma membrane (arrows). Scale bar, 2 μm. D and E, SCUBE3 augments FGF signaling through FGFR4. HEK-293T cells were transfected with FGF-responsive luciferase reporter (FiRE-Luc) and pRL-TK alone or with the expression plasmid encoding SCUBE3 combined with FGFR4 (D) or FGFR1 (E) expression plasmid. Transfected cells were incubated for 24 h with or without an increasing concentration of FGF8, and then luciferase activity was measured. Firefly luciferase values were normalized to that of Renilla activity for relative luciferase activity. The data are means ± S.E. *, p < 0.05; **, p < 0.01 (compared with the vector control).
FGF signaling is initiated when FGF protein binds to its receptor complex (37). Both FGFR1 and FGFR4 are expressed during skeletal development, but only FGFR4 shows high binding specificity with FGF8 (38, 39). Because SCUBE3 is also expressed on the cell surface (6), we next investigated whether SCUBE3 could interact with FGFR1 or FGFR4. HEK-293T cells were transfected with FGFR1 or FGFR4 expression plasmids alone (tagged with the Myc epitope for easy detection) or with FLAG-tagged SCUBE3. At 2 days after transfection, the FGFR1 or FGFR4 protein was immunoprecipitated with anti-Myc antibody, and individual immunoprecipitates were analyzed by immunoblotting for the presence of SCUBE3 (anti-FLAG) to determine their protein interactions. Interestingly, immunoprecipitation of FGFR4 but not FGFR1 pulled down SCUBE3 protein (Fig. 3B), so SCUBE3 could specifically interact with FGFR4 but not FGFR1 on the plasma membrane. Consistent with their biochemical interactions, co-localization of SCUBE3 with FGF8 or FGFR4 can be observed on the plasma membrane by using confocal immunofluorescence analysis (Fig. 3C).
Because of the biochemical interactions between SCUBE3 and FGF8 or FGFR4, we then evaluated whether SCUBE3 could affect FGF8 signaling. HEK-293T cells were transfected with an FGF-responsive luciferase reporter construct, FiRE-Luc (40), alone or with a plasmid encoding SCUBE3 combined with FGFR4 (Fig. 3D) or FGFR1 expression plasmids (Fig. 3E). Transfected cells were left untreated or were stimulated with increasing concentrations of FGF8 for 24 h, and then luciferase activity was measured. FGF8 could dose-dependently induce FGF-responsive luciferase activity in HEK-293T cells expressing FGFR4 or FGFR1 alone, and co-expression of SCUBE3 could further enhance FGF8-mediated signaling in the presence of FGFR4 (Fig. 3D) but not FGFR1 (Fig. 3E), which agrees with the specific interaction of SCUBE3 with FGFR4 but not FGFR1 (Fig. 3B). Together, these results suggest that SCUBE3 may act as a co-receptor by interacting with FGF8 and its receptor FGFR4 and subsequently augment FGF8 signal activity.
Mapping the Specific Domains of SCUBE3 That Interact with FGF8/FGFR4 for Enhancing FGF8 Signaling
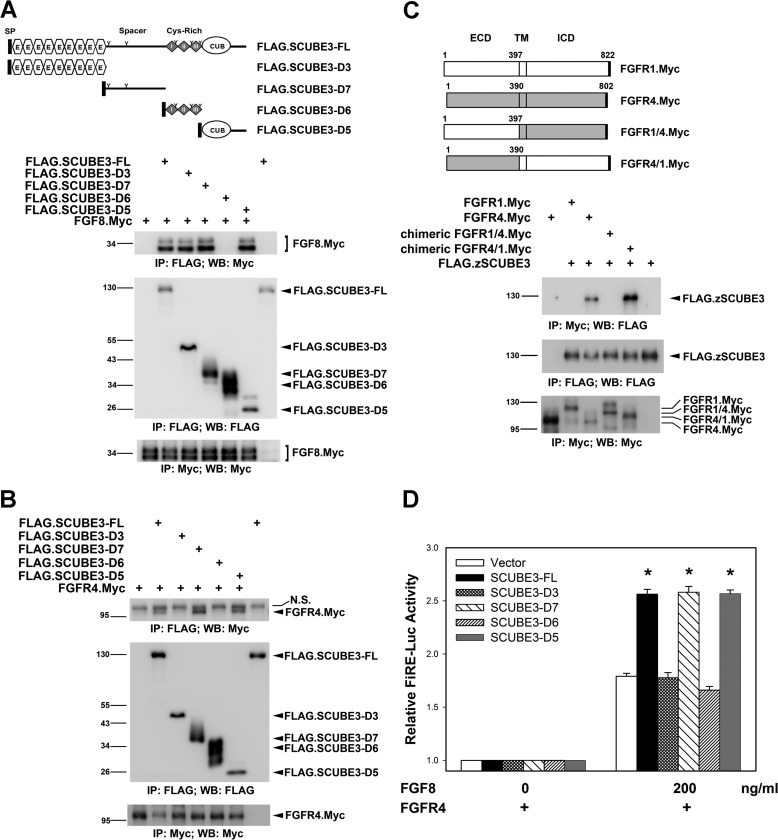
To further identify which SCUBE3 domains are responsible for its binding with FGF8 and FGFR4, we generated a series of SCUBE3 deletion mutants (Fig. 4A) and examined their interactions with FGF8 or FGFR4. HEK-293T cells were transfected with a Myc-tagged FGF8 or FGFR4 expression plasmid alone or with the FLAG-tagged SCUBE3 deletion constructs. Co-immunoprecipitation revealed showed that SCUBE3 could form a complex with FGF8 and FGFR4 commonly through its spacer region (D7 mutant) and CUB domain (D5 mutant), respectively (Fig. 4, A and B). Similarly, we generated chimeric receptor constructs between FGFR4 and FGFR1 to verify the specific domain involved in their interactions with SCUBE3. FGFR4 and chimeric FGFR4/1 containing the extracellular domain of FGFR4 fused with the transmembrane and intracellular domains of FGFR1 but not FGFR1 and chimeric FGFR1/4 containing the extracellular domain of FGFR1 fused with the transmembrane and intracellular domains of FGFR4 were co-immunoprecipitated with SCUBE3 proteins (Fig. 4C). Thus, the extracellular domain of FGFR4 may play an important role in forming a complex with SCUBE3.
FIGURE 4.
Molecular mapping for the functional domain of SCUBE3. A and B, the spacer and CUB domain of SCUBE3 can interact with FGF8 and FGFR4. The expression plasmid encoding Myc-tagged FGF8 (A) and Myc-tagged FGFR4 (B) was transfected alone or together with a series of FLAG-tagged SCUBE3 deletion constructs in HEK-293T cells as depicted on top of A. After 2 days, cell lysates were prepared and underwent immunoprecipitation, followed by Western blot analysis with indicated antibodies to determine the protein-protein interactions. IP, immunoprecipitation; N.S., nonspecific band; WB, Western blot. C, FGFR4 interacts with SCUBE3 through its extracellular domain. Myc-tagged FGFR1, FGFR4, and chimeric FGFR1/4 or FGFR4/1 expression plasmids were transfected alone or with the FLAG-tagged SCUBE3 in HEK-293T cells. Co-immunoprecipitation experiments were performed as described above to examine the protein-protein interactions. ECD, extracellular domain; TM, transmembrane domain; ICD, intracellular domain. D, SCUBE3 augments FGF signaling through its spacer region or CUB domain. HEK-293T cells were transfected with FiRE-Luc and pRL-TK alone (blank bars) or with the expression plasmid encoding FL or various SCUBE3 domain deletion constructs combined with FGFR4 expression plasmid. Transfected cells were incubated with or without FGF8 (200 ng/ml) for 24 h, and then luciferase activity was measured. Firefly luciferase values were normalized to Renilla activity to obtain relative luciferase activity. The data are means ± S.E. *, p < 0.05.
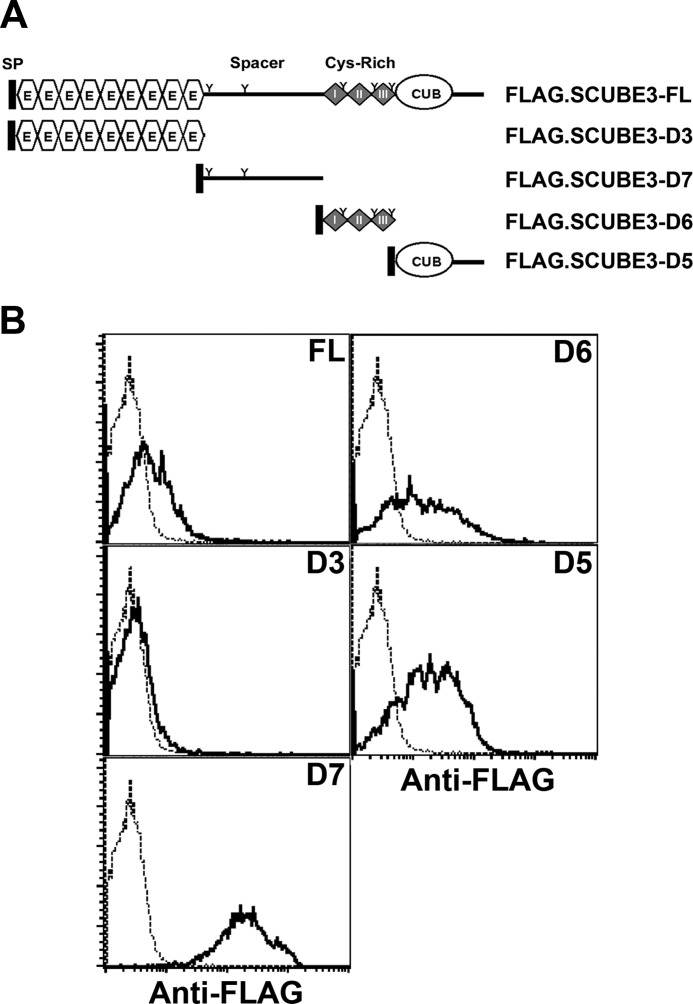
To further evaluate whether a minimal fragment of SCUBE3 interacting with FGF8/FGFR4 was sufficient to enhance FGF8 signaling, HEK-293T cells were transfected with an FGF-responsive luciferase reporter, alone or with a series of SCUBE3 domain deletion constructs in the presence of FGFR4. Transfected cells were then stimulated with FGF8 for 24 h, and luciferase activity was measured. Co-transfection of plasmids encoding the full-length (FL), spacer region (D7), or CUB domain (D5) but not EGF-like repeats (D3) or cysteine-rich motifs (D6) of SCUBE3 could specifically enhance FGF8-responsive luciferase activity (Fig. 4D). In agreement with its potential role as a co-receptor for FGFR4, SCUBE3-FL, the spacer region (D7) or the CUB domain (D5) could be expressed on the cell surface (Fig. 5). Therefore, the spacer region or CUB domain of SCUBE3 is a minimal fragment sufficient to augment FGF8 signaling.
FIGURE 5.
Flow cytometry of cell surface expression of SCUBE3. A, domain structure of SCUBE3-FL and its deletion constructs. A FLAG tag was added immediately after the signal peptide sequences, thus at the NH2 terminus, for easy detection. FL, amino acids 1–993; D3, amino acids 1–401; D6, amino acid 398–629; D7, amino acids 632–803; D5, amino acids 804–993. FL, full-length; SP, signal peptide; E, EGF-like repeats; Cys-Rich, cysteine-rich motifs; CUB, CUB domain. B, the spacer region, cysteine-rich motifs, or CUB domain of SCUBE3 is capable of tethering on cell surface. At 24 h after transfection of empty vector or these expression plasmids, cells were detached and stained with anti-FLAG monoclonal antibody to determine the cell surface expression by flow cytometry. Empty vector-transfected cells are presented as the basal expression level (dotted line), and SCUBE3-transfected cells are shown as a thick line.
Identification and Embryonic Expression Pattern of Zebrafish scube3
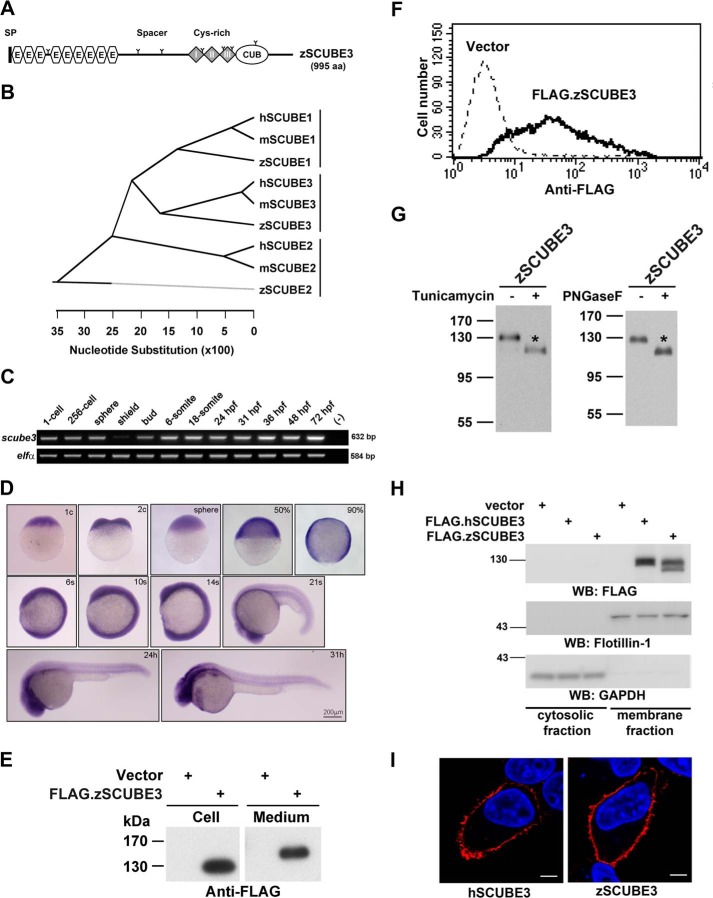
Next, we studied the role of Scube3 in regulating Fgf signaling and myogenesis in vivo with a zebrafish model. A homology search of the public database identified a full-length zebrafish scube3 cDNA clone coding a polypeptide of 995 amino acids that shares a conserved protein domain organization (Fig. 6A) and overall 72 or 73% sequence identity with its mouse or human orthologue, respectively. Furthermore, phylogenetic analysis showed that zebrafish Scube3 was clustered closely with mouse and human SCUBE3, separate from the groups of SCUBE1 or SCUBE2 protein sequences (Fig. 6B). Thus, zebrafish scube3 may be an orthologue of mammalian SCUBE3.
FIGURE 6.
Identification, embryonic expression, and characterization of zebrafish scube3. A, graphic illustration of the domain organization of zebrafish Scube3 protein. SP, signal peptide sequence; E, EGF-like domain; Cys-rich, cysteine-rich repeats. B, phylogenetic tree of the SCUBE protein family. Similarity of human (h), mouse (m), and zebrafish (z) SCUBE protein sequences was analyzed by use of Lasergene MEGALIGN program (Clustal W algorithm). Branch order indicates structural relative, and branch length reflects sequence identity. C and D, embryonic expression patterns of zebrafish scube3. Scube3 transcripts are maternally deposited in zebrafish embryos and distributed widely in the embryos throughout development. RT-PCR analysis of scube3 mRNA expression in 1-cell to 3-dpf embryos. C, the Ef-1α gene was used as the internal control. D, the expression pattern of scube3 was examined by whole mount in situ hybridization at indicated stages. Stages are indicated at the upper right of each micrograph. c, cell; %, % of epiboly; s, somite; h, hours postfertilization. E, secretion of zebrafish Scube3 into the conditioned medium. HEK-293T cells were transfected with empty vector (Vector) or expression plasmid encoding FLAG-tagged zebrafish Scube3 (FLAG.zSCUBE3). At 48 h post-transfection, the serum-free conditioned medium was collected, and cells were detached. Samples from cell lysate (Cell) or the serum-free conditioned culture medium (medium) were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Recombinant FLAG.zSCUBE3 proteins were detected by Western blot (WB) analysis with anti-FLAG antibody. F, cell surface expression of zebrafish Scube3 protein. HEK-293T cells were transfected with empty vector (Vector) or expression plasmid encoding FLAG.zSCUBE3 protein. At 48 h post-transfection, cells were detached and probed with anti-FLAG antibody and underwent flow cytometry. G, N-linked glycosylation of zebrafish SCUBE1. HEK-293T cells were transfected with expression vector encoding FLAG.zSCUBE1 protein. Transfected cells were cultured without (−) or with (+) tunicamycin (an inhibitor of N-glycosylation) for 24 h (left panel). Alternatively, cell lysates were left untreated (−) or were treated (+) with peptide N-glycosidase F (PNGaseF) to remove the N-linked oligosaccharide chain (right panel). Cell lysates derived from each treatment were examined by Western blot analysis with anti-FLAG antibody. H, SCUBE3 protein was targeted into the membrane. HEK-293T cells were transfected with the expression plasmids encoding FLAG-tagged SCUBE protein: human (h) or zebrafish (z) orthologue, respectively. Two days after transfection, cells were collected and homogenized. Samples of supernatant were centrifuged at 100,000 × g for 30 min to yield the supernatant (cytosolic fraction) and the pellet (membrane fraction). Each subcellular fraction was subjected to SDS-PAGE and immunoblot analysis using anti-FLAG, anti-flotillin-1 (a lipid raft membrane protein), or anti-GAPDH (a cytosolic protein) antibody. I, cell surface localization of SCUBE3 visualized by confocal immunofluorescent microscopy. After transfection of expression plasmid encoding FLAG-tagged SCUBE3, in HeLa cells, SCUBE3 expression was detected by mouse anti-FLAG antibody and Alexa Fluor 594-conjugated goat anti-mouse IgG (red). Scale bar, 5 μm.
We further examined the spatiotemporal expression pattern of scube3 during embryonic development by RT-PCR and whole mount ISH. Maternal scube3 transcripts are distributed evenly in blastomeres. Likewise, zygotic scube3 is globally expressed up to the 14-somite (s) stage. From 20 to 31 hours postfertilization, strong scube3 expression was detected in the brain and somitic region (Fig. 6, C and D). The maternal and ubiquitous expression patterns of scube3 during early embryonic stages implied that scube3 may play critical roles during early embryogenesis.
Because heterologous expression of human SCUBE3 manifested as secreted glycoproteins that could tether on the cell surface (6, 8), we examined whether the overexpressed zebrafish Scube3 protein might also behave in a similar manner. The zebrafish Scube3 protein was heterologously expressed by transient transfection in HEK-293T cells. Overexpressed zebrafish Scube3 (tagged with FLAG epitope) could be effectively secreted into the conditioned medium, targeted onto the cell surface, and post-translationally modified with N-linked oligosaccharides that could be inhibited by tunicamycin (an inhibitor of N-glycosylation) during biosynthesis or enzymatically removed by peptide-N-glycosidase F (Fig. 6, E–G). Furthermore, the expression of SCUBE3 on the cell surface was demonstrated by subcellular fractionation and confocal immunofluorescence analysis (Fig. 6, H and I).
Scube3 Regulates Fgf8 Signaling during Zebrafish Development
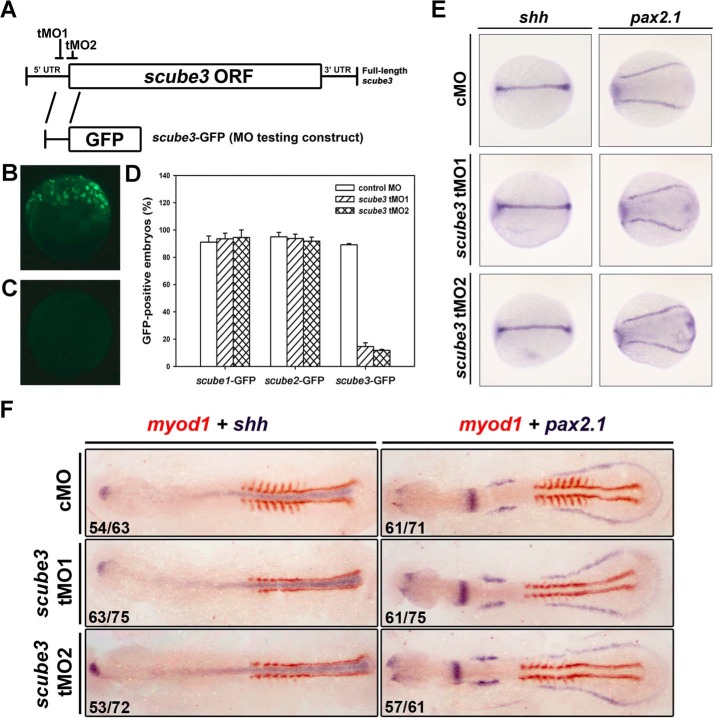
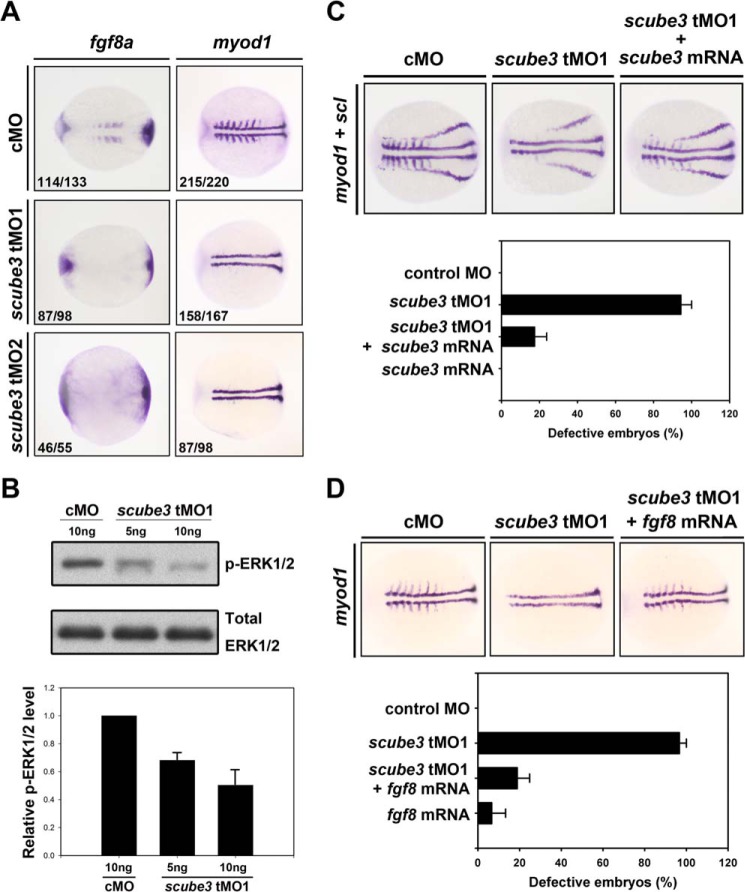
To further investigate the role of scube3 early during embryogenesis in zebrafish, we designed two nonoverlapping antisense tMOs targeting the 5′-UTR or at the start codon sequence of scube3 to block translation of scube3 (named scube3-tMO1 or tMO2; Fig. 7A). The efficiency of tMOs was analyzed by comparing their abilities to suppress in vivo the expression of GFP from ectopic mRNA containing tMO target sequence (scube3-GFP) with the control morpholino containing a random sequence that has a minimum effect on embryonic development. Both tMOs specifically and effectively blocked expression of scube3-GFP, whereas control MO did not (Fig. 7, B–D). We then evaluated the effect of scube3 knockdown (referred to as morphants) on the expression of 2 myogenesis-related FGF downstream target genes (fgf8 and myod1) by whole mount ISH. At 8 s, fgf8 expression in somites was significantly decreased in scube3 morphants (Fig. 8A, left panel). Interestingly, both scube3 morphants showed a specific loss of myod1 expression in the lateral fast muscle precursors but not in the adaxial slow muscle precursors (Fig. 8A, right panel). However, the expression of shh (an axial mesoderm marker) and pax2.1 (a lateral mesoderm marker) was preserved in scube3 morphants as well (Fig. 7, E and F). Importantly, loss of myod1 expression in the lateral somites could be rescued by co-injection of scube3 mRNA in the scube3 morphants (Fig. 8C), which confirms the specificity of scube3 tMO targeting. In addition, forced expression of fgf8 mRNA could restore myod1 expression in fast muscle precursors (Fig. 8D), suggesting that scube3 acts upstream of or in parallel to fgf8 in the initiation of myod1 expression in fast muscle precursors. Consistent with these genetic data, scube3 knockdown significantly inhibited Fgf8 downstream activation (phosphorylation) of ERK1/2 in a dose-dependent manner (Fig. 8B). To determine whether Scube3 functions through Hh signaling, we examined the expression of Hh target genes patched1 (ptc1) and engrailed (eng1a) in scube3 morphants. As shown in Fig. 9, scube3 knockdown barely affected expression of these Hh target genes, indicating that scube3 may not act through Hh signaling. Together, our data suggest that scube3 plays an important role in modulating Fgf signaling for early myod1 expression in fast muscle precursors.
FIGURE 7.
Specificity and effectiveness of scube3 tMOs and no effect of scube3 knockdown on the expression of other mesoderm genes. A, schematic presentation of the targeting sites of two scube3 tMOs and the scube3-GFP test construct. tMO1 targets the 5′-UTR, and tMO2 blocks at the AUG translation start site. B–D, efficiency of scube3 tMO. One-cell embryos were injected with 300 pg of tMO testing construct with 1 ng of control MO or scube3 tMO1 or tMO2, respectively. The expression of GFP was examined at 50–70% epiboly stage and photographed by epifluorescent microscopy. The images are representative of embryos injected with scube3-GFP plasmid in combination with control MO (B) or scube3-tMO1 (C). D, summary of translational blocking efficiency of scube3 tMOs. The lack of suppression effect on the scube1-GFP or scube2-GFP testing constructs confirms their specificity in blocking translation of scube3. The data are means ± S.E. E, whole mount in situ hybridization for shh and pax2.1 expression in the zebrafish embryos. Dorsal view of the eight-somite embryos. Anterior is to the left in all images. The expression of shh (the adaxial mesoderm marker) and pax2.1 (the intermediate mesoderm marker) was not affected in scube3 morphants. F, effect of scube3 knockdown on the expression of myod1. One-celled embryos were injected with control MO (cMO) or scube3 tMO1 or tMO2 (5 ng/embryo) and examined for expression of the indicated markers at the eight-somite stage by two-color RNA ISH. The embryos are shown in dorsal view with anterior to the left. Although the expression of shh (blue) and pax2.1 (blue) remained relatively unaltered, the expression level of myod1 (red) in the lateral somites were reduced in the scube3 morphants as compared with the control embryos. The numbers in the bottom left-hand corners of photographs are the numbers of affected embryos with phenotype similar to what is shown in the image (left) and the total number of observed embryos (right).
FIGURE 8.
Knockdown of scube3 suppressed FGF target genes and reduced FGF signaling in zebrafish embryos. A, effect of scube3 knockdown on the expression of FGF target genes (fgf8 and myod1) evaluated by whole mount in situ hybridization. The images are the dorsal view of the eight-somite embryos with anterior to the left. In scube3 morphants, fgf8 and myod1 expression was markedly reduced in the somites. Of note, scube3 knockdown specifically suppressed myod1 expression in lateral fast muscle precursors but not adaxial cells that will differentiate into slow muscles. The numbers in the bottom left-hand corners of photographs are the numbers of affected embryos with phenotype similar to what is shown in the image (left column) and the total number of observed embryos (right column). B, ERK activation in control or scube3 knockdown embryos. After Scube3 tMO1 injection, protein lysates (20 μg) derived from embryos (70% epiboly) were blotted with antibody specific to the phosphorylated form (p-ERK) or total ERK protein and quantified by densitometric scanning (mean ± S.E.) from three independent experiments (bottom panel). Note that scube3 knockdown significantly decreased p-ERK expression in a dose-dependent manner, which supports the role of scube3 in FGF signaling during zebrafish development. C and D, forced expression of scube3 or fgf8 mRNA restored myod1 expression in fast muscle precursors of scube3 morphants. C, co-injection of scube3 morphants with scube3 mRNA rescued myod1 expression. Scube3 morphants showed rescued expression of lateral myod1expression in somites when injected with 100 pg of synthetic scube3 mRNA at the one- or two-cell stage. The images are shown as dorsal views with anterior to the left. Quantification data are shown in the bottom panel. D, forced expression of fgf8 mRNA restored myod1 expression in lateral somites. Scube3 morphants showed rescued expression of lateral myod1 expression in somites when injected with 0.0625 pg of synthetic fgf8 mRNA at the one- or two-cell stage. The images are shown as dorsal views with anterior to the left. Quantification data are shown in the bottom panel. The data are means ± S.E. cMO, control MO.
FIGURE 9.
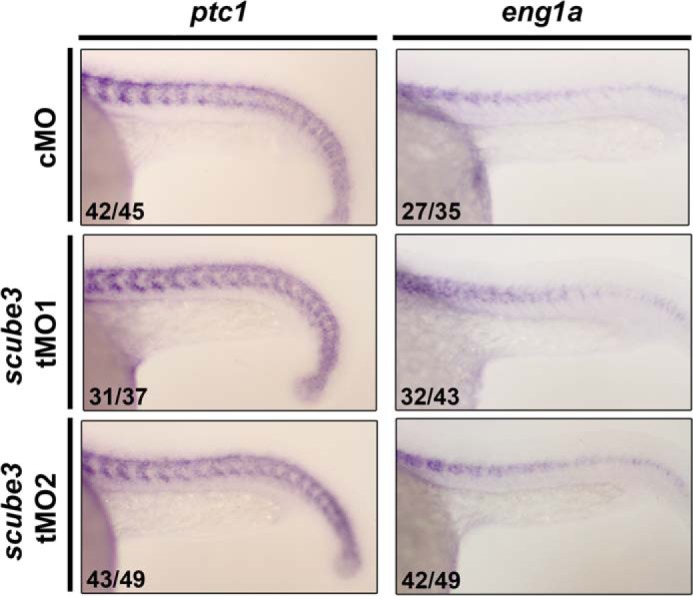
Knockdown of scube3 did not affect Hh target genes in zebrafish embryos. Effects of scube3 knockdown on the expression of Hh target genes (ptc1 and eng1a) were evaluated by whole mount in situ hybridization. The images are the lateral views of the 22-somite embryos with anterior to the left. No effect of scube3 knockdown on the expression of ptc1 and eng1a was observed. The numbers in the bottom left-hand corners of the photographs are the numbers of embryos with phenotype similar to what is shown in the image (left column) and the total number of observed embryos (right column). cMO, control MO.
Zebrafish scube3 Is Required for Efficient Fast Muscle Differentiation
To further validate the requirement for scube3 in the development of fast muscles in zebrafish, we examined the effect of scube3 knockdown on the expression of muscle type-specific MyHC by staining with specific antibodies to the slow and fast MyHC of zebrafish. Immunostaining with F59 monoclonal antibody, which specifically labels slow MyHC (28), was basically unaltered in control and scube3 morphants, whereas the F310 anti-fast MyHC staining signal (21) was significantly reduced in the scube3 morphants as compared with the control (Fig. 10). Therefore, scube3 expression was essential for proper fast muscle formation in zebrafish.
FIGURE 10.
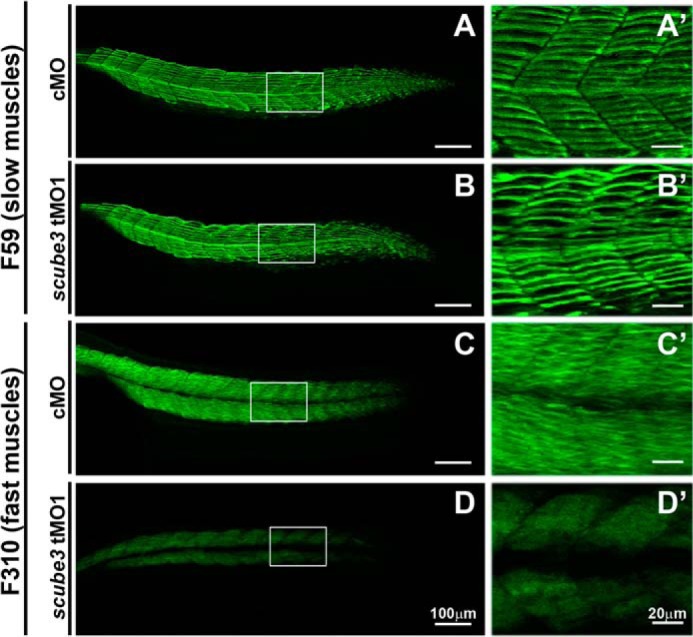
Scube3 is required for fast muscle differentiation. A, A′, B, and B′, immunofluorescent staining of slow muscle with F59 antibody in control (A and A′) and scube3 morphants (B and B′) at the 26-somite stage. The images are presented as lateral views with anterior to the left. C, C′, D, and D′, immunostaining of fast muscle with F310 antibody in control (C and C′) and scube3 morphants (D and D′) at the 26-somite stage. Corresponding higher magnification views are shown in A′, B′, C′, and D′. cMO, control MO.
Human SCUBE3 mRNA Can Rescue the myod1-defected Phenotype in scube3 Morphants
To examine whether the function of SCUBE3 in regulating fast muscle development is conserved between zebrafish and mammals, we further determined the effect of overexpression of human SCUBE3 on rescuing myod1 expression defects in scube3 morphants. Forced expression of human SCUBE3 by mRNA microinjection rescued the expression of myod1 in the lateral somites in scube3 morphants (Fig. 11), which suggests that human SCUBE3 may share a preserved function with the zebrafish orthologue, at least for the development of fast muscle.
FIGURE 11.
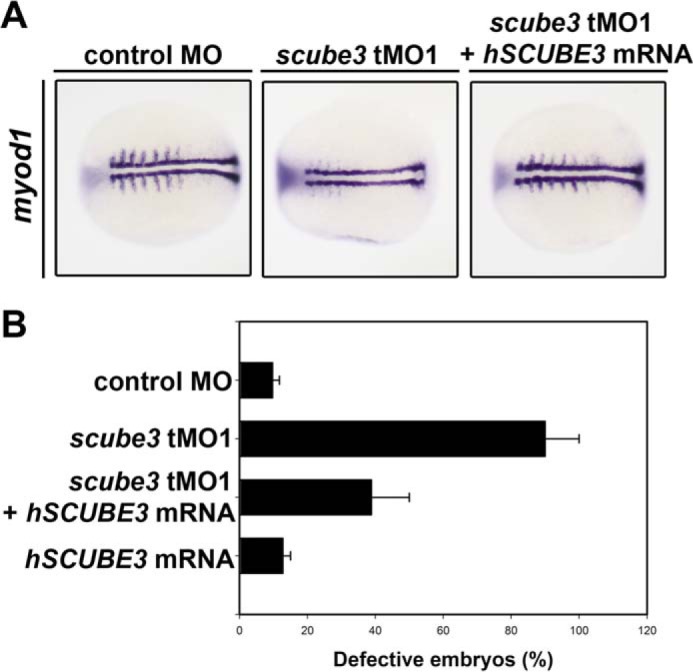
Human SCUBE3 mRNA restores myod1 expression in the lateral somites in the scube3 morphants. A, visualization of the rescue experiments using myod1 expression. The images are shown as dorsal views with anterior to the left. Scube3 morphants showed rescued expression of lateral myod1 expression in somites when injected with 5 pg of synthetic human SCUBE3 mRNA at the one- to two-cell stage. B, corresponding quantified data are shown. The data are means ± S.E.
DISCUSSION
SCUBE3 is the last member identified in the SCUBE family (6), and its biological function has remained largely unexplored. In the present study, we used a combination of molecular, biochemical, and genetic approaches to elucidate a novel biological role of SCUBE3 as a key regulator, possibly through acting as a co-receptor, in promoting Fgf8 signaling during fast muscle development in zebrafish.
Despite a conserved protein domain composition with high homology among these three Scube members, knockdown of two other scube genes (i.e. scube1 and scube2) produced no apparent defect on myod1 expression in the lateral somites (4, 12). These observations can be due simply to a distinctive, nonoverlapping spatiotemporal expression pattern and thus nonredundant functions for each scube gene during early zebrafish embryogenesis. For example, zebrafish scube2 selectively expressed in the developing midbrain and hindbrain and in posterior paraxial stripes has been implicated in the development of slow muscle fibers and the ventral neural tube by modulating the hedgehog signal transduction pathway (4). Alternatively, because of their secretion nature, cellular distribution or functional activation of each Scube protein may be determined by as yet unidentified factor(s) expressed in a lineage-specific manner. This hypothesis is further supported by our observations that gain of function (forced expression) of scube3 mRNA did not affect fast muscle differentiation in zebrafish embryos (data not shown).
Because the genetic program that drives myogenesis is evolutionarily conserved among vertebrates (15, 16), SCUBE3 might also be involved in skeletal muscle development in mammals; our studies demonstrated that mouse Scube3 could modulate FGF signaling and myogenic differentiation in myoblasts in vitro (Fig. 1) and that human SCUBE3 mRNA could rescue myod1 expression in fast muscle precursors in scube3 morphants (Fig. 11). However, targeted inactivation of mouse Scube3 showed normal embryonic development and survival (41). Although the precise functional differences of SCUBE3 in different species remain to be further investigated, Scube1 and/or Scube2 may compensate for the loss of Scube3, given that these mouse genes, as opposed to their zebrafish orthologues with distinctive embryonic expression profiles, seem to be expressed in overlapping tissues such as in the dermomyotome of the somites for Scube2 and Scube3 (2, 3, 7). In addition, the functional redundancy could be due to the cooperation between members of the fgf/fgfr gene family, a well known phenomenon among highly homologous mammalian genes. For example, despite the well documented importance of fgfr4 in muscle development in the chick (19), fgfr4 null mice develop normally, with no evident muscle defects (42). Therefore, a double or triple knock-out of these Scube genes may be required to further define their roles in muscle or other developmental processes.
Muscles of the vertebrate consist of two distinct fibers with different contractile and metabolic properties. In zebrafish, two distinct cell populations occupying separate regions give rise to slow and fast muscle fibers (15). Fgf8 signaling is essential for the initiation and maintenance of myod1 expression and proper myogenic differentiation of fast fibers in zebrafish (20, 21). Thus, its signaling must be finely controlled in a spatial and temporal fashion. Previous studies showed that FGFs including FGF8 are involved in increasing the expression of their genes via a positive feedback loop of FGF signaling (22). Our biochemical and molecular studies suggest that SCUBE3 may modulate FGF8 signal transduction during receipt of the FGF8 signal at the plasma membrane, possibly through its action as a FGF8 co-receptor by enhancing cellular responses to FGF8 (Figs. 3 and 4), thus constituting a critical component of this FGF8 feed-forward signaling for the differentiation of fast muscle precursors (Fig. 8A). Overall, these findings agree with our genetic studies that position scube3 at the top of the fgf8 regulatory hierarchy of fast myogenesis and an ordered epistatic relationship of scube3-fgf8-myod1 pathway during zebrafish fast muscle development (Fig. 8). In addition, it is noteworthy that a recent report showed that scube3 could cooperate with scube1 and scube2 to participate in Hh signaling for slow muscle development in scube triple MO embryos (11), indicating that Scube3 is a multifunctional protein that acts either alone or together with other Scube proteins in a context-dependent manner.
One interesting finding is that we demonstrated a specific interaction of SCUBE3 with the extracellular domain of FGFR4 (Fig. 4C) that consists of three Ig modules; Ig2 and Ig3 determine the affinity and specificity of FGFR4 for FGF and heparin binding (35, 43). However, whether Ig2 and Ig3 of FGFR4 are also involved in its specific interaction with SCUBE3 remains to be investigated.
In summary, our findings reveal that zebrafish scube3 plays an important role in fast fiber development by modulating Fgf signaling. Given the wide spectrum expression of scube3 in a variety of developing tissues (3), further study is needed to fully elucidate the involvement of SCUBE3 in FGF-mediated stage and tissue-specific function during embryonic development or even in FGF-related pathological processes.
Acknowledgments
We thank the Taiwan Zebrafish Core Facility at Academia Sinica for providing zebrafish and some ISH probes used in this study. RNAi reagents were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center of Academia Sinica. The F59, F310, and MF20 monoclonal antibodies developed by F. E. Stockdale and D. A. Fischman were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the United States NICHD, National Institutes of Health and maintained by the Department of Biology of the University of Iowa (Iowa City, IA).
This work was supported by Ministry of Science and Technology Grants NSC 102-2320-B-001-015-MY3 and MOST 103-2325-B-001-004 (to R.-B. Y.) and NSC 98-2311-B-002-006-MY3 (to S.-J. L.) and National Taiwan University Grants NTU CESRP-10R70602A5 and NTU ERP-10R80600. The Taiwan Zebrafish Core Facility at Academia Sinica is supported by Ministry of Science and Technology Grant 102-2321-B-001-038, and the National RNAi Core Facility Platform located at the Institute of Molecular Biology/Genomic Research Center of Academia Sinica is supported by the National Core Facility Program for Biotechnology Grant NSC 100-2319-B-001-002 from the Ministry of Science and Technology.
- RTK
- receptor tyrosine kinase
- FGFR
- FGF receptor
- Hh
- hedgehog
- MO
- morpholino oligonucleotide
- tMO
- translation-blocking MO
- ISH
- in situ hybridization
- MyHC
- myosin heavy chain
- FL
- full-length.
REFERENCES
- 1. Cheng C. J., Lin Y. C., Tsai M. T., Chen C. S., Hsieh M. C., Chen C. L., Yang R. B. (2009) SCUBE2 suppresses breast tumor cell proliferation and confers a favorable prognosis in invasive breast cancer. Cancer Res. 69, 3634–3641 [DOI] [PubMed] [Google Scholar]
- 2. Grimmond S., Larder R., Van Hateren N., Siggers P., Hulsebos T. J., Arkell R., Greenfield A. (2000) Cloning, mapping, and expression analysis of a gene encoding a novel mammalian EGF-related protein (SCUBE1). Genomics 70, 74–81 [DOI] [PubMed] [Google Scholar]
- 3. Haworth K., Smith F., Zoupa M., Seppala M., Sharpe P. T., Cobourne M. T. (2007) Expression of the Scube3 epidermal growth factor-related gene during early embryonic development in the mouse. Gene Expr. Patterns 7, 630–634 [DOI] [PubMed] [Google Scholar]
- 4. Kawakami A., Nojima Y., Toyoda A., Takahoko M., Satoh M., Tanaka H., Wada H., Masai I., Terasaki H., Sakaki Y., Takeda H., Okamoto H. (2005) The zebrafish-secreted matrix protein you/scube2 is implicated in long-range regulation of hedgehog signaling. Curr. Biol. 15, 480–488 [DOI] [PubMed] [Google Scholar]
- 5. Tsai M. T., Cheng C. J., Lin Y. C., Chen C. C., Wu A. R., Wu M. T., Hsu C. C., Yang R. B. (2009) Isolation and characterization of a secreted, cell-surface glycoprotein SCUBE2 from humans. Biochem. J. 422, 119–128 [DOI] [PubMed] [Google Scholar]
- 6. Wu B. T., Su Y. H., Tsai M. T., Wasserman S. M., Topper J. N., Yang R. B. (2004) A novel secreted, cell-surface glycoprotein containing multiple epidermal growth factor-like repeats and one CUB domain is highly expressed in primary osteoblasts and bones. J. Biol. Chem. 279, 37485–37490 [DOI] [PubMed] [Google Scholar]
- 7. Xavier G. M., Cobourne M. T. (2011) Scube2 expression extends beyond the central nervous system during mouse development. J. Mol. Histol. 42, 383–391 [DOI] [PubMed] [Google Scholar]
- 8. Yang R. B., Ng C. K., Wasserman S. M., Colman S. D., Shenoy S., Mehraban F., Komuves L. G., Tomlinson J. E., Topper J. N. (2002) Identification of a novel family of cell-surface proteins expressed in human vascular endothelium. J. Biol. Chem. 277, 46364–46373 [DOI] [PubMed] [Google Scholar]
- 9. Cadena D. L., Gill G. N. (1992) Receptor tyrosine kinases. FASEB J. 6, 2332–2337 [DOI] [PubMed] [Google Scholar]
- 10. Thisse B., Thisse C. (2005) Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev. Biol. 287, 390–402 [DOI] [PubMed] [Google Scholar]
- 11. Johnson J. L., Hall T. E., Dyson J. M., Sonntag C., Ayers K., Berger S., Gautier P., Mitchell C., Hollway G. E., Currie P. D. (2012) Scube activity is necessary for Hedgehog signal transduction in vivo. Dev. Biol. 368, 193–202 [DOI] [PubMed] [Google Scholar]
- 12. Tsao K.-C., Tu C.-F., Lee S.-J., Yang R.-B. (2013) Zebrafish scube1 (signal peptide-CUB (complement protein C1r/C1s, Uegf, and Bmp1)-EGF (epidermal growth factor) domain-containing protein 1) is involved in primitive hematopoiesis. J. Biol. Chem. 288, 5017–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu Y. Y., Peck K., Chang Y. L., Pan S. H., Cheng Y. F., Lin J. C., Yang R. B., Hong T. M., Yang P. C. (2011) SCUBE3 is an endogenous TGF-β receptor ligand and regulates the epithelial-mesenchymal transition in lung cancer. Oncogene 30, 3682–3693 [DOI] [PubMed] [Google Scholar]
- 14. Yang H.-Y., Cheng C.-F., Djoko B., Lian W.-S., Tu C.-F., Tsai M.-T., Chen Y.-H., Chen C.-C., Cheng C.-J., Yang R.-B. (2007) Transgenic overexpression of the secreted, extracellular EGF-CUB domain-containing protein SCUBE3 induces cardiac hypertrophy in mice. Cardiovasc. Res. 75, 139–147 [DOI] [PubMed] [Google Scholar]
- 15. Stickney H. L., Barresi M. J., Devoto S. H. (2000) Somite development in zebrafish. Dev. Dyn. 219, 287–303 [DOI] [PubMed] [Google Scholar]
- 16. Bentzinger C. F., Wang Y. X., Rudnicki M. A. (2012) Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 4, a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hughes S. M., Salinas P. C. (1999) Control of muscle fibre and motoneuron diversification. Curr. Opin. Neurobiol. 9, 54–64 [DOI] [PubMed] [Google Scholar]
- 18. Ochi H., Westerfield M. (2007) Signaling networks that regulate muscle development: lessons from zebrafish. Dev. Growth Differ. 49, 1–11 [DOI] [PubMed] [Google Scholar]
- 19. Marics I., Padilla F., Guillemot J.-F., Scaal M., Marcelle C. (2002) FGFR4 signaling is a necessary step in limb muscle differentiation. Development 129, 4559–4569 [DOI] [PubMed] [Google Scholar]
- 20. Groves J. A., Hammond C. L., Hughes S. M. (2005) Fgf8 drives myogenic progression of a novel lateral fast muscle fibre population in zebrafish. Development 132, 4211–4222 [DOI] [PubMed] [Google Scholar]
- 21. Hamade A., Deries M., Begemann G., Bally-Cuif L., Genêt C., Sabatier F., Bonnieu A., Cousin X. (2006) Retinoic acid activates myogenesis in vivo through Fgf8 signalling. Dev. Biol. 289, 127–140 [DOI] [PubMed] [Google Scholar]
- 22. Kim G.-Y., Kim H.-Y., Kim H.-T., Moon J.-M., Kim C.-H., Kang S., Rhim H. (2012) HtrA1 is a novel antagonist controlling fibroblast growth factor (FGF) signaling via cleavage of FGF8. Mol. Cell Biol. 32, 4482–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995) Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- 24. Thisse C., Thisse B. (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- 25. Liao E. C., Paw B. H., Oates A. C., Pratt S. J., Postlethwait J. H., Zon L. I. (1998) SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 12, 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weinberg E. S., Allende M. L., Kelly C. S., Abdelhamid A., Murakami T., Andermann P., Doerre O. G., Grunwald D. J., Riggleman B. (1996) Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development 122, 271–280 [DOI] [PubMed] [Google Scholar]
- 27. Reifers F., Böhli H., Walsh E. C., Crossley P. H., Stainier D. Y., Brand M. (1998) Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125, 2381–2395 [DOI] [PubMed] [Google Scholar]
- 28. Devoto S. H., Melançon E., Eisen J. S., Westerfield M. (1996) Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 122, 3371–3380 [DOI] [PubMed] [Google Scholar]
- 29. Feng X., Adiarte E. G., Devoto S. H. (2006) Hedgehog acts directly on the zebrafish dermomyotome to promote myogenic differentiation. Dev. Biol. 300, 736–746 [DOI] [PubMed] [Google Scholar]
- 30. Gutiérrez J., Brandan E. (2010) A novel mechanism of sequestering fibroblast growth factor 2 by glypican in lipid rafts, allowing skeletal muscle differentiation. Mol. Cell Biol. 30, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Root D. E., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M. (2006) Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods 3, 715–719 [DOI] [PubMed] [Google Scholar]
- 32. Lin Y. C., Chen C. C., Cheng C. J., Yang R. B. (2011) Domain and functional analysis of a novel breast tumor suppressor protein, SCUBE2. J. Biol. Chem. 286, 27039–27047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scata K. A., Bernard D. W., Fox J., Swain J. L. (1999) FGF receptor availability regulates skeletal myogenesis. Exp. Cell Res. 250, 10–21 [DOI] [PubMed] [Google Scholar]
- 34. Wu Z., Woodring P. J., Bhakta K. S., Tamura K., Wen F., Feramisco J. R., Karin M., Wang J. Y., Puri P. L. (2000) p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell Biol. 20, 3951–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. (2000) Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750 [DOI] [PubMed] [Google Scholar]
- 36. Hammond C. L., Hinits Y., Osborn D. P., Minchin J. E., Tettamanti G., Hughes S. M. (2007) Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev. Biol. 302, 504–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Böttcher R. T., Niehrs C. (2005) Fibroblast growth factor signaling during early vertebrate development. Endocr. Rev. 26, 63–77 [DOI] [PubMed] [Google Scholar]
- 38. Itoh N., Ornitz D. M. (2004) Evolution of the Fgf and Fgfr gene families. Trends Genet. 20, 563–569 [DOI] [PubMed] [Google Scholar]
- 39. Kwiatkowski B. A., Kirillova I., Richard R. E., Israeli D., Yablonka-Reuveni Z. (2008) FGFR4 and its novel splice form in myogenic cells: interplay of glycosylation and tyrosine phosphorylation. J. Cell Physiol. 215, 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang R.-B., Ng C. K., Wasserman S. M., Kömüves L. G., Gerritsen M. E., Topper J. N. (2003) A novel interleukin-17 receptor-like protein identified in human umbilical vein endothelial cells antagonizes basic fibroblast growth factor-induced signaling. J. Biol. Chem. 278, 33232–33238 [DOI] [PubMed] [Google Scholar]
- 41. Xavier G. M., Panousopoulos L., Cobourne M. T. (2013) Scube3 is expressed in multiple tissues during development but is dispensable for embryonic survival in the mouse. PLoS One 8, e55274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao P., Caretti G., Mitchell S., McKeehan W. L., Boskey A. L., Pachman L. M., Sartorelli V., Hoffman E. P. (2006) Fgfr4 is required for effective muscle regeneration in vivo: delineation of a MyoD-Tead2-Fgfr4 transcriptional pathway. J. Biol. Chem. 281, 429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pellegrini L., Burke D. F., von Delft F., Mulloy B., Blundell T. L. (2000) Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407, 1029–1034 [DOI] [PubMed] [Google Scholar]