Background: The wall-associated kinases (WAKs) serve as pectin receptors.
Results: A pectin methyl esterase and two transcription factor mutants suppress a dominant WAK allele.
Conclusion: De-esterification of pectin is required for WAK activation though EDS1 and PAD4.
Significance: The results provide a mechanism for the state of pectins to activate two different pathways.
Keywords: Arabidopsis, Cell Wall, Plant Defense, Receptor, Signaling, Receptors
Abstract
The wall-associated kinases (WAKs) have a cytoplasmic protein kinase domain that spans the plasma membrane and binds pectin in the extracellular matrix of plants. WAKs are required for cell expansion during Arabidopsis seedling development but are also an integral part of the response to pathogens and stress that present oligogalacturonides (OGs), which subsequently bind to WAKs and activate a MPK6 (mitogen-activated protein kinase)-dependent pathway. It was unclear how WAKs distinguish native pectin polymers and OGs to activate one or the other of these two pathways. A dominant allele of WAK2 constitutively activates the stress response, and we show here that the effect is dependent upon EDS1 and PAD4, transcriptional activators involved in the pathogen response. Moreover, the WAK2 dominant allele is suppressed by a null allele of a pectin methyl esterase (PME3) whose activity normally leads to cross-linking of pectins in the cell wall. Although OGs activate a transcriptional response in wild type, the response is enhanced in a pme3/pme3 null, consistent with a competition by OG and native polymers for activation of WAKs. This provides a plausible mechanism for WAKs to distinguish an expansion from a stress pathway.
Introduction
The cell wall of angiosperms is composed of a complex arrangement of cellulose, hemicellulose, and pectin. Pectins are synthesized in the Golgi as methyl esterified α1–4-d-galacturonic acid, and secreted into an extracellular matrix with cellulose, hemicellulose, and a variety of proteins to form the plant cell wall (1–6). Localized activity of pectin methyl esterases (PME)2 in the cell wall can reveal a charge on the pectins and can lead to a calcium-based cross-linking and a structural network that can have dramatic effects on cell enlargement (7, 8). Studies point to both a need for a cross-linking of the pectin to provide lateral structure and directionality of growth of root hairs and pollen tubes (9–11) and modification of a matrix to permit expansion of leaf cells (8, 12). Indeed there appear to be multiple roles for pectins in the cell wall, including the possibility that along with cellulose, pectins might also be load-bearing (8). Pectins are also the target of numerous pathogens that digest the wall as they approach the plant cell, thereby generating de-esterified pectin fragments or oligogalacturonides (OGs) (13, 14). Numerous studies demonstrate that OGs can activate a stress response by the plant, indicating that OGs signal pathogen presence (15).
The wall-associated kinases (WAKs) are known to bind both to long polymers of cross-linked pectin and to OGs (16–22). Notably, and in parallel to the two pectin types, WAKs have been assigned two distinct roles, one in cell expansion in seedlings (20) and another in a response to OGs generated by pathogens (21–23). During seedling growth, WAKs are required for cell expansion and have been shown to be involved in the pectin activation of MPK3 and a vacuolar invertase that can increase turgor-driven expansion (17, 20). The expression of all five WAKs, clustered on a 30-kb locus on chromosome 1 (16, 24), overlaps such that most tissues have some combination of these pectin receptors. Due to this overlapping expression and tight linkage, it has been hard to distinguish their respective contributions to pectin sensing (18). In vitro, WAK1 and WAK2 bind to long pectin chains reflective of a native pectin form (homogalacturonan) but have a preference for short OGs of degree of polymerization 9–15 (17, 25, 26). De-esterified pectins have a much higher binding to WAK1 than do esterified pectins (25).
However, WAKs are also required during the response to pathogen and help to mediate a stress response, which is coincident with the appearance in the cell wall of de-esterified OGs. The expression in a heterologous system of a fusion between the WAK1 extracellular domain and an unrelated kinase domain (ERF) leads to a response to OGs (22), suggesting that WAK1 is indeed a receptor for OG as well as for longer pectins. A dominant gain of function WAK2 allele, WAK2cTAP, constitutively activates a MPK6-dependent stress response in Arabidopsis, and this response also requires pectin binding by WAK (17, 21). Thus, a native pectin-based expansion response and the OG-activated stress response are distinguished by, on the one hand, the activation of MPK3 and invertase and, on the other, by MPK3 and MPK6 and stress-related proteins, respectively. We provide evidence that is consistent with a competition of newly generated OGs for WAKs that are bound to native longer polymers, thereby activating a stress response. Moreover, this response is dependent upon pectin de-esterification and the transcriptional regulators EDS1 and PAD4 (27).
EXPERIMENTAL PROCEDURES
Plant Growth
Arabidopsis thaliana Columbia was grown on soil or agar plates as described (21), at 22 °C, 16 h of light, 8 h of dark. For comparison within an experiment, triplicate samples grown at the same time were used. For treatment with OGs, seedlings were plated in a microtiter plate with 5 ml of 0.5× MS medium plus vitamins, vernalized for 3 days, and incubated at 22 °C with gentle shaking under 24-h light. After 7 days at 22 °C, OGs were added to 50 μg/ml unless otherwise noted and shaken for an additional 3 h, and then seedlings were frozen in liquid nitrogen. Experiments were done in biological triplicates.
Preparation of OG
400 ml of 1% polygalacturonic acid (Sigma P3850, 85% de-esterified), pH 4.4 (NaOH), was autoclaved for 45 min, and then HCl was added dropwise to pH 2 while stirring. The preparation was centrifuged at 12,000 × g for 20 min, and the supernatant was adjusted to 50 mm NaOAc, 22.5% EtOH (pH 6 final). The sample was incubated at 4 °C for 12 h and centrifuged at 16,000 × g for 30 min. The pellet was resuspended in 50 ml of water and dialyzed versus five changes of water for 2 days at 4 °C using a 1000-kDa membrane. The solution was then lyophilized to powder. OGs were resuspended in water as needed and analyzed using Dionex chromatography to determine that the preparation had a degree of polymerization of predominately 9–15. From 4 g of material, 800 mg of OGs was recovered. Esterification was accomplished by adding 800 μl of MeOH and 40 μl of H2SO4 to 5 mg of OGs and incubation for 24 h. The OGs were pelleted in a microcentrifuge and resuspended in 1 ml of MeOH, 37.5 μl of H2SO4 for a further 24 h. The OGs were then washed three times in 1 ml of 80% ETOH, dried, and resuspended in water.
RNA
RNA was isolated from plant material using the RNeasy Plant Mini Kit (Qiagen). Quantitative PCR was as described (21, 28); 1 μg of RNA was used for a reverse transcription assay using oligo(dT) for first-strand synthesis in an Invitrogen Superscript III RT-PCR kit (Invitrogen catalog no. 18080-051). cDNA was then used for quantitative PCR using Power SYBR Green Master Mix (Applied Biosystems) and an Applied Biosystems StepOne system, version 2.1, which calculated the comparative CT (ΔΔCT) with the following cycles: 95 °C for 15 s, 56 °C for 1 min, repeated 38 times. Actin expression served as an internal standard, and wild-type untreated samples were set as the standard to which other samples were compared, in biological triplicate. Bar graphs in the figures show relative quantitation (RQ) maximum and minimum. Statistical analysis used ΔCT and ΔCTSE values in a two-tailed t test and ANOVA where indicated.
Genotyping
Plants were genotyped by PCR according to Ref. 21 and using primers listed in Table 1 and the following general T-DNA primers: p745, AACGTCCGCAATGTGTTATTAAGTTG; MLB1, GTGGACTCTTGTTCCAAACTG; LBb1.3, ATTTTGCCGATTTCGGAAC; LBa1, TGGTTCACGTAGTGGGCCATC.
TABLE 1.
Genes and mutant alleles tested for interaction with WAKs
When “cross only” is indicated, then the analysis was performed only by looking for phenotype segregations in the F1 and F2 of the cross. Oligonucleotides are listed in the order of forward and then reverse for WT allele and then with the indicated forward or reverse and the T-DNA primer.
| A. thaliana number | Gene name | Mutant | Forward/reverse primer for gene |
|---|---|---|---|
| At1g10210 | MPK1 | SALK_063897 | GGAACGTCGTTGGTCACTTAT/AGCAACTTTCTCGTTGGTGTC reverse + MLB1 |
| At3g45640 | MPK3 | SALK_151594 | AGCACCTGAGCTTCTGTTGAA/CCGTATGTTGGATTGAGTGCT forward + p745 |
| At4g01370 | MPK4 | SALK_056245 | CGGTGAAACAATGACACGAGA/CCGCTTCAACAGATGGTTACG reverse + MLB1 |
| At4g11330 | MPK5 | WiscDs/Lox430A12 | GTTAAGGAGCTACCTAAGTTCCCAAG/CATGAGATGAAGGAGAAACAGAGCT forward + p745 |
| At2g43790 | MPK6 | SALK_073907 | GGACTCTCCGTGAGATCAAGC/GAGTGGCTTACGGTCCATTAA forward + MLB1 |
| At1g18150 | MPK8 | SALK_037501 | TTCTTGGTACTCCACCTCCTGATCTTTCGGATCAAAGGCAAG forward + MLB1 |
| At3g18040 | MPK9 | SALK_064439 | CTGCAATCGACACACATTCAG/ATCGTTCGCCTTGATAACTTG reverse + p745 |
| At1g07880 | MPK13 | SALK_130193 | GACTCGGATCTCGAGTTCTTG/TGCTTCAATGCTTCATCCACT forward + p745 |
| At4g36450 | MPK14 | SALK_022928 | GCTTGCGAGAACTTATGAACAG/GTTGGATCAAACACAAGCATC reverse + p745 |
| At5g19010 | MPK16 | SALK_059737 | AACAGCATGCATTACCAAG/GCAGCAGCTGGATTTCTGAC reverse + p745 |
| At2g01450 | MPK17 | SALK_020801 | AACTCGTGACTGATCTGCTTG/ACTGGACAAACCAAGATTTCAG forward + p745 |
| At1g53510 | MPK18 | SALK_069399 | TAATCGCAATGCAGGACTATG/TCAAGTCATCCCTGAACAAGA forward + MLB1 |
| At2g42880 | MPK20 | SALK_090005 | AGCTCATGGAATCGGATCTT/GTTATATCGCGGCAACACAC forward + p745 |
| At2g23200 | Kinase | SALK_020561c | CTAGACGAGCACAATATAGCAAAAGTCGCAGG forward + LBb1.3 |
| CTCGAGTCTCCGATGAATCTGTAATCCAAATCT | |||
| At4g11900 | S locus kinase | WiscDsLoxHs215 | CTACGGAGTGTTGGTGTTCTTGTCTTTTCAAG cross only |
| _03H = CS920568 | GAACTGCCCCGAAACACCCATG | ||
| At3g45860 | CRK4 | SALK_063969.38.90 | GAGTATGCGATGTATGGCCAATTCTTCATG forward + LBa1 |
| GACGTATTGATAGATAGACGATCCACTAATCCTACTTGTTC | |||
| At1g35710 | LR kinase | SALK_143599.47.10 | GAATGGTTTTGCAAGAAGAATCTTTATGATTTTCG cross only |
| At5g60900 | RLK1 | SALK_146545.53.55 | CCGAGACTGCCTCAGAATCAGACATAAC reverse + LBb1.3 |
| GTTTGCGGTTTGGTTCTCTGGTCTACAAG | |||
| At4g39400 | BRI1 | ref × br1-5 | Cross only |
| At3g14310 | PME3 | ref × pme3-1 | CTAGTGTCGAACAATGGCACCATCAATGAAAG forward + TAG3 |
| GCCCCTTCAACAAGGCTTTACGAAC | |||
| At3g48090 | EDS1 | ref × eds1-2 | TCAGGTATCTGTTATTTATCATC WT, 1.3 kb; eds1-2, 0.4 kb |
| CCCTTTCTAGTTTCCTTGAGCTAAG | |||
| At3g52430 | PAD4 | ref × pad4-1 | TCGCATAAGACTAGGTAAGTCTT DdeI; WT, 100 bp; pad4-1, 80 bp |
| GCGTAAATCCATTTCTTTCCTA |
Western Blotting
Leaves were ground in 10 mm Tris, pH 7, 3% SDS, 100 mm DTT, 10% glycerol; centrifuged at 10,000 × g for 5 min; and measured for chlorophyll content by spectrophotometry at 660 nm, and adjusted for equal protein concentration. Bromphenol blue was added, and the sample was heated at 80 °C for 10 min and then separated by SDS-PAGE using 10% acrylamide gel and transferred to nitrocellulose membrane for 1500 mA h. Western blots were blocked with 5% (w/v) nonfat dry milk in Tris-buffered saline (TBS) supplemented with 3% Tween 20; incubated with peroxidase-antiperoxidase-soluble complex (Sigma) or the indicated antiserum and the appropriate secondary serum at 1:2500 dilution for 2 h each; and detected with chemiluminescence.
PME Activity
The Ruthenium Red agar diffusion assay was adapted from Bethke et al. (15). 0.1% pectin ≥85% esterified (Sigma P9561), 1% agarose, 12.5 mm citric acid, 50 mm Na2HPO4, pH 7.0, was microwaved, and 13 ml was poured per 10-cm Petri dish. The large end of a plastic pipette tip was used to create wells on the solid pectin agar plate for application of extracts. Extracts, in triplicate for each genotype, were prepared by homogenizing leaf tissue in 0.1 m sodium citrate, 0.2 Na2HPO4 buffer, 1.0 m NaCl (pH 5.0), centrifuging at 14,000 × g for 10 min at 4 °C, and standardized for concentration using a Nanodrop spectrophotometer. Equal amounts of protein extract were added to the wells and the plates and incubated at 37 °C for 16 h. The plates were then washed with 15 ml of water two times and then with 10 ml of 0.05% Ruthenium Red (MP Biochemicals, 0521810401) for 30 min while shaking slowly and destained with three washes of water. Plates were scanned, and stain intensity was quantified using ImageJ (National Institutes of Health).
Statistical Analysis
All pairwise analysis was performed using Prizm and R and a two-tailed t test, unpaired, or ANOVA as indicated. Curve fitting was performed using Prizm.
RESULTS
Results to date show that WAKs serve as pectin and OG receptors and can mediate either cell expansion or a response to stress. MPK3 and MPK6 are differentially involved in these two pathways. To identify possible co-receptors and additional components of the WAK signal transduction pathway, co-expression analysis using GeneInvestigator was employed. We focused on the genes encoding potential signaling components, and these are listed in Table 1. The goal was to make double mutants of these loci with the dominant hyperactive allele of WAK2cTAP to determine whether there was genetic interaction and hence evidence for involvement in pectin perception. For each of these genes, a mutant line was ordered from ABRC, and homozygous lines were identified by PCR with the appropriate primers (Table 1). The homozygous mutants were then crossed to a line homozygous for WAK2cTAP and, from the F2 individuals, were identified as homozygous for both WAK2cTAP and the indicated mutation. Phenotypes were scored on seedlings germinated on soil. Of nine lines tested, none had a visible effect on the stunted and necrotic growth of plants expressing WAK2cTAP (data not shown). The mutants were also crossed to plants homozygous for either wak1, wak2, or wak4, and F2 plants homozygous for both alleles were germinated on soil or on pond water medium that is known to reveal a weak root growth phenotype in wak2 nulls (20). No double mutant phenotypes were observed on either soil or agar. We have previously reported that a null allele of mpk6 suppresses the dominant effect of WAK2cTAP (21) and therefore also tested T-DNA insertion lines for 12 other Arabidopsis MPKs (courtesy of Patrick Krysan, University of Wisconsin). Again, none of these null lines had any visible effect on the WAK2cTAP phenotype, indicating the high degree of specificity of the mpk6 allele.
EDS1 and PAD4
We next turned to candidate genes as possible members of the WAK pathway and chose EDS1 and PAD4 because these transcriptional modifiers are known to lie downstream of a variety of induced stress responses, in particular R-mediated innate immunity (27). Null alleles for both of these loci were crossed into the WAK2cTAP background, and plants homozygous for WAK2cTAP and either eds1-2 or pad4-1 were identified by Western blotting for WAK2cTAP and PCR, respectively. The results are shown in Fig. 1A and indicate that both eds1-2 and pad4-1 suppress the dwarf and necrotic WAK2cTAP-induced phenotype. The masses of each plant type were compared (Fig. 1B) and confirmed the visual differences seen in soil-grown plants. eds1-2/eds1-2 and pad4-1/pad4-1 each were not different from WT (t test, p > 0.01), but there was a significant difference between eds1-2/eds1-2 WAK2cTAP and WAK2cTAP and between pad4-1/pad4-1 WAK2cTAP and WAK2cTAP (t test, p < 0.01). The suppression by eds1-2 does not appear to be complete, based on plant mass, and pad4-1/pad4-1 WAKcTAP appeared to have a greater mass than WT (t test, p < 0.01). Each plant was also assayed for WAK2cTAP expression, and Fig. 1C shows that the levels of expressed protein are equivalent, relative to the actin standard. PCR with the appropriate primers shows that the individuals are homozygous mutant for the indicated locus (Fig. 1D). Thus, the stress response induced by WAK2cTAP is dependent upon both PAD4 and EDS1.
FIGURE 1.
eds1-2 and pad4-1 suppress WAK2cTAP. A, representative plants of the indicated genotype grown under the same conditions. B, wet mass of three plants of the indicated genotype. Shared colored asterisks between two bars indicate significance in the t test, p < 0.01. C, Western blot of equal total protein extracts from the indicated genotype, versus TAP tag to detect WAK2cTAP (cTAP) (top) and versus actin to indicate loading of equal protein amounts (bottom). D, genotypes, indicated above each lane, were determined using PCR and GelRed-stained agarose gels. PAD4 and pad4-1 alleles were distinguished by the absence or presence (respectively) of digestion with Dde1. EDS1 and eds1-2 were distinguished by smaller PCR product due to a deletion. Error bars, S.E.
PME3
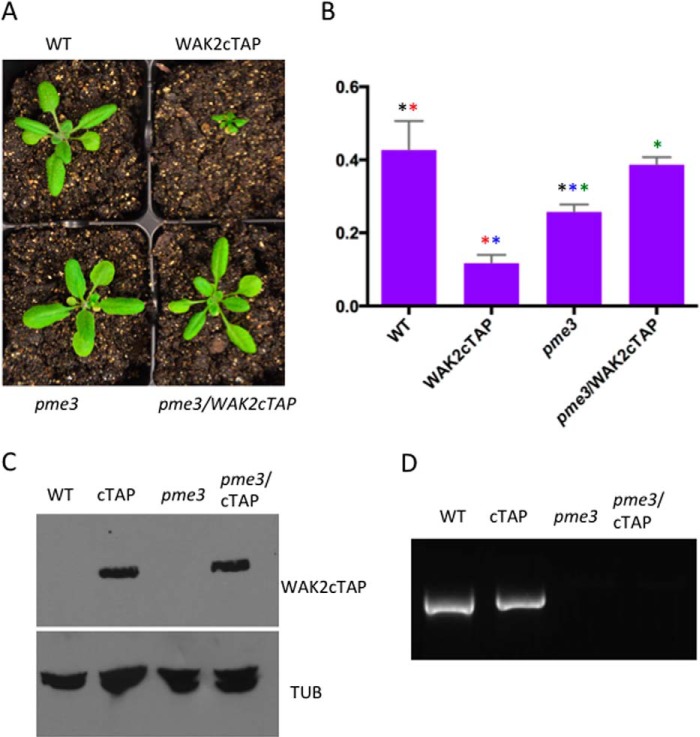
Much evidence indicates that WAKs bind to pectin both in vitro and in vivo and that pectins can activate a cellular response in a WAK-dependent fashion. In addition, WAKs appear to have a higher affinity for de-esterified than for esterified pectin in vitro. We therefore asked if the WAK2cTAP phenotype was affected by a mutation in the most abundantly expressed pectin methyl esterase, PME3 (29). Null alleles of this locus lead to altered branching and root growth, but little effect on leaf morphology and size has been reported (29). Plants at the seedling and rosette stage homozygous for pme3 and WAK2cTAP appear to have a wild type morphology (Fig. 2A) with a total plant mass not significantly different from wild type (t test, p > 0.01) but larger than WAK2cTAP (t test, p < 0.01; Fig. 2B). In the conditions used, pme3/pme3 mutants are slightly smaller than wild type (t test p < 0.01), but it is not clear why they are slightly smaller than pme3/pme3 WAK2cTAP (t test, p < 0.01). This difference disappears as the plants mature. The levels of WAK2cTAP expression were equivalent in lines expected to express the gene (Fig. 2C), and the pme3 genotypes were identified by PCR using the relevant primers (Fig. 2D). Thus, the de-esterification of pectin is required for the dominant effect of WAK2cTAP, and this is in agreement with previous results showing that WAK2cTAP requires an active kinase and pectin receptor domain (17, 21). This indicates that WAKs not only prefer to bind de-esterified pectins in vitro but also require this de-esterification for activation.
FIGURE 2.
pme3-1 suppresses WAK2cTAP. A, representative plants of the indicated genotype grown under the same conditions. B, wet mass of three plants of the indicated genotype. y axis, mass in g. Shared colored asterisks between two bars indicate significance in the t test, p < 0.01. C, Western blot of equal total protein extracts from the indicated genotype versus TAP tag to detect WAK2cTAP (top) and versus tubulin to indicate loading of equal protein amounts (bottom). D, genotypes, indicated above each lane, were determined using PCR, using primers to detect WT allele and GelRed-stained agarose gels. Error bars, S.E.
To determine whether the WAK2cTAP allele affects the levels of methyl esterification and if indeed pme3/pme3 has lower levels of de-esterified pectin, a Ruthenium Red assay that provides a relative measure of de-esterified pectin (Fig. 3A) was performed on leaf extracts from plants grown on soil. Fig. 3B shows that, relative to WT, pme3/pme3 plants have reduced levels (t test, p < 0.01) of de-esterified pectin as expected (30), WAK2cTAP has levels similar to that of wild type (t test, p > 0.01), and the double mutant is similar to the single pme3/pme3 line (t test, p > 0.01). Residual levels of PME activity in the pme3/pme3 mutants are probably due to the contribution of the remaining 66 PME genes (31).
FIGURE 3.
pme3-1/pme3-1 and pme3-1/pme3-1 WAK2cTAP have reduced PME activity. A, Ruthenium Red assay for relative levels of PME in plant extracts. A standard curve was generated by measuring in the pectin plate assay (see “Experimental Procedures”) dilutions of extract from WT leaves. Samples measured are shown in duplicate on plates and then measured after scanning and using ImageJ software. A larger range of concentrations was assayed before this experiment to focus on a level usable for subsequent assays. x axis, dilutions measured; y axis, relative activity. B, esterified pectin in dishes spotted with plant extracts (in triplicate vertical) from the indicated genotype and stained with Ruthenium Red to detect de-esterified pectin. There is a no extract spot at the top of each plate. Bar graph on right, quantitation of results from plates showing relative activity. Shared colored asterisks between two bars indicate significance in the t test, p < 0.01. Error bars, S.E.
The WAK2cTAP allele leads to a stress response, including the activation of a number of genes, including FADlox (FAD-linked oxidase) and CML41 (calmodulin-like protein) (17, 21, 28). To determine whether the pme3 allele suppresses this transcriptional response as well as the WAK2cTAP phenotype, RNA was isolated from each of the single and double mutants, the levels of FADlox and CML41 mRNA were measured by quantitative RT-PCR, and the results are shown in Fig. 4. WAK2cTAP plants express much higher levels of FADlox (6-fold, t test, p < 0.01) and CML41 (10-fold, t test, p < 0.01) RNA than WT plants, and this confirms our previous report (21). This high level is suppressed in the pme3/pme3 WAK2cTAP double mutant to amounts similar (t test p > 0.01) to both WT and the single pme3/pme3 line.
FIGURE 4.

A, pme3-1 suppresses the WAK2cTAP-induced transcriptional response. Relative expression of FADlox and CML41 mRNA, using actin as a standard, was determined by quantitative RT-PCR of RNA from the indicated genotype. B, eds1-2 and pad4-1 suppress the WAK2cTAP-induced transcriptional response. Relative expression of FADlox and CML41 mRNA, using actin as a standard, was determined by quantitative RT-PCR of RNA from the indicated genotype. Shared colored asterisks between two bars indicate significance in the t test, p < 0.01. Error bars, S.E.
A similar gene expression analysis was carried out for the eds1-2, pad4-1, and WAK2cTAP double mutants, and the results are shown in Fig. 4B. FADlox and CML41 gene expression were significantly higher in WAK2cTAP than WT plants (t test, p < 0.01), and this higher level was lowered in the double mutants because there was no significant difference (t test for each comparison, p < 0.01) between WT and eds1-2/eds1-2 WAKcTAP or pad4-1/pad4-1 WAK2cTAP. WT and eds1-2/eds1-2 FADlox levels were significantly different (t test, p < 0.01), but all other pairwise t tests of CML41 or FADlox levels between each gentoype showed no significant differences (except for WAK2cTAP). Thus, both eds1-2 and pad4-1 both suppressed the WAK2cTAP phenotype and elevated levels of gene expression.
pme3 Increases WAK Response
These results indicate that PME3 is required for the WAK2cTAP mediated stress response but also raise the question of whether PME3 activity or the protein itself is required. To test this, pme3/pme3 plants were treated with OGs that were >85% de-esterified, and the expression of FADlox was used as a measure of the WAK-activated pathway. The results, shown in Fig. 5A (x axis point 100) indicate that 100 μg/ml OGs provide a ∼700-fold activation of FADlox in both WT and pme3/pme3 plants. Thus, PME is not required for activation of the transcriptional response if the OGs are already de-esterified. The OG-activated transcription observed is over 100-fold higher than the steady state levels of FADlox expression in WAK2cTAP plants (Fig. 4) because the induction peaks at 3 h post-OG treatment and then decreases to steady state levels (21, 28). Because pme3/pme3 plants are still responsive, indeed more responsive, to de-esterified OG, it is unlikely that PME3 protein (versus activity) is also required as a cofactor in WAK induction. Rather, PME3 esterase activity is required in the absence of de-esterified pectins. In agreement with this finding is the absence of interaction of PME3 with the WAK extracellular domains in the yeast two-hybrid assay (data not shown).
FIGURE 5.

pme3-1/pme3-1 is more responsive to OGs than WT. A, relative expression (RQ) of FADlox mRNA using actin as a standard, determined by quantitative RT-PCR of RNA from the indicated plants (WT or pme3/pme3) treated with 0–100 μg/ml OG (x axis). B, same as A but for CML41 expression. C, Western blot of total cell extracts from the indicated genotype, probed with WAK or tubulin (TUB) antiserum. Error bars, S.E.
However, we also notice that the activation of FADlox in pme3/pme3 plants was consistently (and significantly, p < 0.01) higher than that of WT (Fig. 5). One possible explanation is that because pectins are more esterified in pme3/pme3 plants (Fig. 3), WAKs might be less tightly bound, and so, when presented with de-esterified OGs, the WAKs more readily bind the OGs than in WT. This model predicts a competition between OGs and native pectins. To test this, a concentration-dependent response curve was generated for both WT and pme3/pme3 plants, and we predicted that the pme3/pme3 plants would be more responsive because more WAK should be free of de-esterified pectin, and more should be available to bind OGs. Fig. 5A shows the results of treating plants with 0.1, 1, 10, and 100 μg/ml of OGs and measuring the induction of FADlox gene expression, where the relative quantitation levels were fitted to a curve. The pme3/pme3 plants were more responsive than WT at all concentrations of OG. At each concentration used, the levels of activation were significantly different between pme3/pme3 and WT (t test, p < 0.01 for each concentration of OG). Indeed, the WT 100-μg activation was similar to the pme3 10-μg activation (t test, p > 0.01). A two-way ANOVA between the two response curves also showed that the pme3/pme3 plants are different from WT in all three parameters (strain, OG, and strain/OG; p < 0.001). A similar analysis was performed with the CML41 gene, and although induction levels were lower, the differences remain significant (t test for each OG concentration, p < 0.01; two-way ANOVA, all pairwise comparisons, p < 0.001). These results are consistent with there being more WAKs available to bind to OGs in pme3/pme3 and also consistent with the idea that OGs are competing with native pectins for WAK binding. The amount of native WAK protein as assayed by Western in WT and pme3/pme3 plants is equivalent relative to a tubulin standard (Fig. 5C) and cannot account for the different response curves. Although preference by WAKs for de-esterified pectins and for OGs has been shown in vitro by competition assays, this is now apparent here in vivo.
The response to OGs was also tested in WAK2cTAP and pme3/pme3 WAK2cTAP, and the results are shown in Fig. 6. pme3/pme3 again shows higher induction than WT and levels similar to pme3/pme3 WAK2cTAP. WAK2cTAP is lower than WT, indicating that at steady state, this allele appears hyperactive, yet the total possible OG induction is less. Because there were four plant types tested, an ANOVA was used and showed that all pairwise comparisons were different (p < 0.001) with the exception of pme3/pme3 and pme3/pme3 WAK2cTAP that were similar, as expected. Individual t tests between pairs of plants at each concentration confirmed the significance of the results (p < 0.01). A similar analysis was performed with CML41, and the results are shown in Fig. 6B. Although pme3/pme3 plants do show higher induction levels for both FADlox and CML41, there are two notable difference between the FADlox and CML41 results. The first is that CML41 has higher initial basal levels in WAK2cTAP plants relative to WT (Fig. 6B, point 0). The second is that although WAK2cTAP, pme3/pme3 WAK2cTAP, and WT do have different responses (ANOVA, p < 0.001), their shapes are distinct from those of FADlox. At present, this is not understood but indicates a different saturation response and or feedback loop suggestive of the involvement of additional receptors.
FIGURE 6.

Response to OGs in pme3/pme3 and pme3/pme3 WAK2cTAP. Relative expression (RQ) of FADlox (A) and CML41 (B) mRNA using actin as a standard, determined by quantitative RT-PCR of RNA from the indicated plants treated with 0–100 μg/ml of OG (x axis).
DISCUSSION
Pectins have a major role in shaping the structure of developing plants cells but also serve as a primary protective barrier against invading pathogens. The WAKs bind to pectin polymers native to cell walls and to fragmented pectins or OGs generated by invading pathogens. These two types of pectins appear to activate through WAKs two very different responses. It is possible that part of the mechanism that distinguishes pectin types lies in the heterogeneity of the WAK family, and although the most abundant and ubiquitously expressed isoforms, WAK1 and WAK2, appear to have similar in vitro pectin binding activities (17, 20, 25), it has not been possible to distinguish the contribution of each gene using genetics. Here we show that a dominant WAK2 allele, WAK2cTAP, whose encoded protein requires a functional pectin binding domain and an active kinase (17, 21), is suppressed by a null allele of a pectin methyl esterase, pme3. Mutations in the WAK2cTAP extracellular domain that eliminate pectin binding also suppressed the phenotype (21), and the results reported here indicate that the activating pectin needs to be de-esterified. This is in agreement with the in vitro binding activities of WAK1 and -2, which have a higher binding of de-esterified over esterified pectins in vitro (25, 26).
The results point to a need for de-esterification of pectins for WAK activation. WAK2cTAP is dominant, hyperactive, and pectin-inducible, but care must be taken in interpretation because dominant alleles can affect pathways not normally activated by endogenous receptors. This possibility cannot be completely discounted at this time, but we think it unlikely for several reasons. First, de-esterified OGs activate a similar stress response and do so through WAKs. Second, mutations that affect pectin binding also affect WAK2cTAP, and kinase activity is required. Last, the results concerning the effect of OGs on wild type and pme3/pme3 mutants are consistent with a need for de-esterification for activity and WAK2cTAP activating a relevant pathway. It remains possible that PME3 is required not only for its esterase activity but also in some unknown physical capacity. Future studies exploring the localization and regulation of PME3 and its physical partners will be of interest.
The pme3/pme3 mutant is indeed more responsive to OGs than WT plants, and one possible interpretation is that there is more available WAK to receive incoming OGs when WAK is bound less tightly to esterified pectin. This model implies that OGs are competing with WAKs for native pectin, and indeed in vitro studies see this very event (25, 26). At this point, we cannot discount the possibility that pme3 eliminates a negative feedback loop that, as a result, leads to increased FADlox expression, relative to WT. Exploration of this more complicated explanation awaits analysis of the pathway components. Alternative interpretations of the increased response to OGs by pme3/pme3 are also possible, including an increased porosity of the wall in a pme3/pme3 mutant such that OGs have more access to membrane receptors. This is less likely because, first, few structural differences in the walls were detected in the pme3/pme3 mutant (29), and second, treatment of WT seedlings with fluorescent OGs results in a rapid (minutes) and apparent ubiquitous coating of the plasma membrane (data not shown), and the OG treatment given here to detect the transcriptional response was 3 h. It was also observed that methyl esterified OGs had no ability to induce the stress response as assayed by the induction of FADlox transcription.3 Indeed, most pectinases expressed by pathogens prefer as targets de-esterified pectins (13), and subsequently, the predominant OGs generated upon infection are de-esterified. Our results also indicate that the native PME activity in wild type has insufficient time or activity to de-esterify the added OGs of degree of polymerization 9–15.
To identify components of the WAK signaling mechanism, we also tested the genetic interaction between WAK alleles and mutants of co-expressed and other logical candidate genes. MPK6 had been identified previously in this manner (21), and we show here that eds1 and pad4 also suppress the WAK2cTAP phenotype and hence are involved in WAK signaling. We tentatively place WAK, MPK6, EDS1, PAD4, and FADLox activation in one sequential pathway, with obvious gaps at each step. The remaining 25 loci tested included receptor-like kinases and most of the Arabidopsis MPKs (32), and these had no visible effect on either the WAK2cTAP phenotype or on plants homozygous for the wak1, wak2, or wak4 null alleles. We also tested for genetic interaction between pme3 and wak1, wak2 or wak4 but failed to detect alterations in phenotype, and it is possible that a redundancy in the WAK gene family masks any potential interactions. The analysis of the OG induction of CML41 in WAKcTAP, pme3/pme3, and combined mutants, also revealed an additional layer of complexity in that the response curves were distinct from those seen for FADlox. Given that both genes are induced by multiple biotic and abiotic events, one would expect a complex interaction that may involve alternate receptor interactions that remain undefined. We hope in the future to identify these components that relate to the WAKs.
Taken together, the results suggest that WAKs distinguish the state of pectin in the cell wall on the basis of methyl esterification and perhaps size. We suggest that WAKs bound to native polymers are released to bind OGs of higher affinity and thereby activate a distinct response pathway. The mechanism by which a different downstream signaling path is initiated remains to be determined, but it may indeed require additional receptors, either WAKs or other members of the large Arabidopsis receptor-like kinase (RLK) family.
Acknowledgments
We thank Chris and Shauna Somerville, Nadav Sorek, Clarice Souze, Bill Underwood, Heidi Szemenyei, and Jack Bateman for helpful discussions; Dave Carlon and John Lichter for help with statistical analysis; and Stephan Bauer for use of the Dionex.
This work was supported by the National Science Foundation Grant IOS-1146245 (to B. D. K.). Students were also supported by grants from the National Center for Research Resources (5P20RR016463-12) and the NIGMS (8 P20 GM103423-12) from the National Institutes of Health.
B. D. Kohorn, unpublished results.
- PME
- pectin methyl esterase(s)
- WAK
- wall-associated kinase
- OG
- oligogalacturonide
- ANOVA
- analysis of variance.
REFERENCES
- 1. Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H. (2004) Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211 [DOI] [PubMed] [Google Scholar]
- 2. Anderson C. T., Carroll A., Akhmetova L., Somerville C. (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152, 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kohorn B. D. (2000) Plasma membrane-cell wall contacts. Plant Physiol. 124, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christensen U., Alonso-Simon A., Scheller H. V., Willats W. G., Harholt J. (2010) Characterization of the primary cell walls of seedlings of Brachypodium distachyon: a potential model plant for temperate grasses. Phytochemistry 71, 62–69 [DOI] [PubMed] [Google Scholar]
- 5. Caffall K. H., Mohnen D. (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900 [DOI] [PubMed] [Google Scholar]
- 6. Mohnen D. (2008) Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11, 266–277 [DOI] [PubMed] [Google Scholar]
- 7. Wolf S., Mouille G., Pelloux J. (2009) Homogalacturonan methyl-esterification and plant development. Mol. Plant 2, 851–860 [DOI] [PubMed] [Google Scholar]
- 8. Peaucelle A., Braybrook S., Höfte H. (2012) Cell wall mechanics and growth control in plants: the role of pectins revisited. Front. Plant Sci. 3, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rojas E. R., Hotton S., Dumais J. (2011) Chemically mediated mechanical expansion of the pollen tube cell wall. Biophys. J. 101, 1844–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bosch M., Hepler P. K. (2005) Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17, 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winship L. J., Obermeyer G., Geitmann A., Hepler P. K. (2010) Under pressure, cell walls set the pace. Trends Plant Sci. 15, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peaucelle A., Braybrook S. A., Le Guillou L., Bron E., Kuhlemeier C., Höfte H. (2011) Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21, 1720–1726 [DOI] [PubMed] [Google Scholar]
- 13. Ferrari S., Savatin D. V., Sicilia F., Gramegna G., Cervone F., Lorenzo G. D. (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Espino J. J., Gutiérrez-Sánchez G., Brito N., Shah P., Orlando R., González C. (2010) The Botrytis cinerea early secretome. Proteomics 10, 3020–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bethke G., Grundman R. E., Sreekanta S., Truman W., Katagiri F., Glazebrook J. (2014) Arabidopsis pectin methylesterases contribute to immunity against Pseudomonas syringae. Plant Physiol. 164, 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wagner T. A., Kohorn B. D. (2001) Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13, 303–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohorn B. D., Johansen S., Shishido A., Todorova T., Martinez R., Defeo E., Obregon P. (2009) Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 60, 974–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kohorn B. D., Kohorn S. L. (2012) The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohorn B. D., Kobayashi M., Johansen S., Friedman H. P., Fischer A., Byers N. (2006) Wall-associated kinase 1 (WAK1) is crosslinked in endomembranes, and transport to the cell surface requires correct cell-wall synthesis. J. Cell Sci. 119, 2282–2290 [DOI] [PubMed] [Google Scholar]
- 20. Kohorn B. D., Kobayashi M., Johansen S., Riese J., Huang L. F., Koch K., Fu S., Dotson A., Byers N. (2006) An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 46, 307–316 [DOI] [PubMed] [Google Scholar]
- 21. Kohorn B. D., Kohorn S. L., Todorova T., Baptiste G., Stansky K., McCullough M. (2012) A dominant allele of Arabidopsis pectin-binding wall-associated kinase induces a stress response suppressed by MPK6 but not MPK3 mutations. Mol. Plant 5, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G. (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He Z. H., He D., Kohorn B. D. (1998) Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 14, 55–63 [DOI] [PubMed] [Google Scholar]
- 24. He Z. H., Cheeseman I., He D., Kohorn B. D. (1999) A cluster of five cell wall-associated receptor kinase genes, Wak1–5, are expressed in specific organs of Arabidopsis. Plant Mol. Biol. 39, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 25. Decreux A., Messiaen J. (2005) Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 46, 268–278 [DOI] [PubMed] [Google Scholar]
- 26. Decreux A., Thomas A., Spies B., Brasseur R., Van Cutsem P., Messiaen J. (2006) In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry 67, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 27. Wagner S., Stuttmann J., Rietz S., Guerois R., Brunstein E., Bautor J., Niefind K., Parker J. E. (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14, 619–630 [DOI] [PubMed] [Google Scholar]
- 28. Denoux C., Galletti R., Mammarella N., Gopalan S., Werck D., De Lorenzo G., Ferrari S., Ausubel F. M., Dewdney J. (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1, 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guénin S., Mareck A., Rayon C., Lamour R., Assoumou Ndong Y., Domon J. M., Sénéchal F., Fournet F., Jamet E., Canut H., Percoco G., Mouille G., Rolland A., Rustérucci C., Guerineau F., Van Wuytswinkel O., Gillet F., Driouich A., Lerouge P., Gutierrez L., Pelloux J. (2011) Identification of pectin methylesterase 3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis thaliana. New Phytol. 192, 114–126 [DOI] [PubMed] [Google Scholar]
- 30. Raiola A., Lionetti V., Elmaghraby I., Immerzeel P., Mellerowicz E. J., Salvi G., Cervone F., Bellincampi D. (2011) Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol. Plant Microbe Interact. 24, 432–440 [DOI] [PubMed] [Google Scholar]
- 31. Harholt J., Suttangkakul A., Vibe Scheller H. (2010) Biosynthesis of pectin. Plant Physiol. 153, 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andreasson E., Ellis B. (2010) Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15, 106–113 [DOI] [PubMed] [Google Scholar]