Background: Mechanisms for sterol- and geranylgeraniol-induced dislocation of reductase from ER membranes into cytosol for degradation are unclear.
Results: VCP/p97 mediates extraction of reductase across ER membranes, whereas the proteasome 19 S regulatory particle (RP) dislodges extracted reductase into cytosol.
Conclusion: VCP/p97 and proteasome 19 S RP mediate sequential steps in reductase degradation.
Significance: These results define a new step in the reductase degradative pathway.
Keywords: Cholesterol Metabolism, Endoplasmic Reticulum (ER), Endoplasmic Reticulum-associated Protein Degradation (ERAD), Membrane Trafficking, Proteasome, ER-associated Degradation, Cytosolic Dislocation, Membrane Extraction
Abstract
Accelerated endoplasmic reticulum (ER)-associated degradation (ERAD) of the cholesterol biosynthetic enzyme 3-hydroxy-3-methylglutaryl-coenzyme A reductase results from its sterol-induced binding to ER membrane proteins called Insig-1 and Insig-2. This binding allows for subsequent ubiquitination of reductase by Insig-associated ubiquitin ligases. Once ubiquitinated, reductase becomes dislocated from ER membranes into the cytosol for degradation by 26 S proteasomes through poorly defined reactions mediated by the AAA-ATPase valosin-containing protein (VCP)/p97 and augmented by the nonsterol isoprenoid geranylgeraniol. Here, we report that the oxysterol 25-hydroxycholesterol and geranylgeraniol combine to trigger extraction of reductase across ER membranes prior to its cytosolic release. This conclusion was drawn from studies utilizing a novel assay that measures membrane extraction of reductase by determining susceptibility of a lumenal epitope in the enzyme to in vitro protease digestion. Susceptibility of the lumenal epitope to protease digestion and thus membrane extraction of reductase were tightly regulated by 25-hydroxycholesterol and geranylgeraniol. The reaction was inhibited by RNA interference-mediated knockdown of either Insigs or VCP/p97. In contrast, reductase continued to become membrane-extracted, but not cytosolically dislocated, in cells deficient for AAA-ATPases of the proteasome 19 S regulatory particle. These findings establish sequential roles for VCP/p97 and the 19 S regulatory particle in the sterol-accelerated ERAD of reductase that may be applicable to the ERAD of other substrates.
Introduction
Endoplasmic reticulum (ER)2-associated degradation (ERAD) is a highly conserved cellular process through which misfolded or unassembled secretory and membrane proteins are selectively degraded by cytosolic 26 S proteasomes (1). In addition to its central role in maintenance of ER homeostasis, the ERAD pathway contributes to the feedback control of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase (2, 3). In animal cells, the ER-localized reductase catalyzes conversion of HMG-CoA to mevalonate, which constitutes a rate-controlling step in synthesis of cholesterol and nonsterol isoprenoids including heme A, ubiquinone, dolichol, and the farnesyl and geranylgeranyl groups found attached to many cellular proteins (4). The ERAD of reductase is triggered by its sterol-induced binding to the ER membrane proteins Insig-1 and Insig-2 (5, 6). This binding is mediated entirely by the N-terminal membrane domain of reductase, which contains eight membrane-spanning segments and precedes a cytosolic C-terminal domain that exerts catalytic activity (7, 8). A pair of Insig-associated ubiquitin ligases, gp78 and Trc8, combine to initiate ubiquitination of reductase on two cytosolic lysine residues in the membrane domain of the enzyme (6, 9, 10). Ubiquitination targets reductase for recognition by VCP/p97, an AAA (ATPases associated with diverse cellular activities)-ATPase that has been implicated in the extraction of ERAD substrates from ER membranes and their subsequent delivery to cytosolic 26 S proteasomes for degradation (11). The 20-carbon nonsterol isoprenoid geranylgeraniol augments sterol-accelerated ERAD of reductase but does not appreciably affect its sterol-induced ubiquitination (6, 12). Thus, geranylgeraniol appears to augment sterol-accelerated ERAD of reductase by modulating postubiquitination steps in the reaction.
A key event in the ERAD pathway is dislocation of substrates from ER membranes into the cytosol where they become fully accessible to proteasomes for degradation. Although cytosolic dislocation has been demonstrated for both soluble substrates sequestered within the ER lumen and membrane-bound substrates with one or more membrane-spanning segments (1, 13), underlying mechanisms for the reaction remain to be completely elucidated. Previous studies showed that sterols cause intact, full-length reductase to become dislocated from ER membranes into the cytosol (14, 15). This sterol-induced cytosolic dislocation of reductase was augmented by nonsterol isoprenoids, and the reaction required ubiquitination of the enzyme as well as the actions of Insigs and VCP/p97 (15). More recently, we found that the in vitro addition of sterols triggered dislocation of ubiquitinated reductase from membranes of permeabilized cells through an Insig-dependent reaction that was enhanced by geranylgeraniol (12). Despite these lines of evidence supporting the relevance of cytosolic dislocation to ERAD of reductase, only a small fraction of the protein (∼10%) was recovered from the cytosol of intact or permeabilized cells treated with sterols (12, 15). This apparent inefficiency, which hampers efforts to further elucidate mechanisms for cytosolic dislocation and to determine how geranylgeraniol modulates the reaction, may result from a yet to be identified step in reductase ERAD that precedes dislocation. However, identification of this putative intermediate step in cytosolic dislocation of reductase requires development of a robust assay for the reaction.
In the current study, we developed an assay that measures the sterol-induced extraction of reductase across ER membranes. Intact membranes isolated from cells expressing a form of reductase containing T7 epitopes in the N-glycosylated lumenal loop between transmembrane domains 7 and 8 were subjected to in vitro digestion with the protease trypsin. Trypsinolysis produced protected fragments of reductase that were observed in anti-T7 immunoblots. A substantial fraction of the lumenal T7 epitopes in reductase became susceptible to trypsin digestion when cells were treated with the oxysterol 25-hydroxycholesterol (25-HC) and geranylgeraniol prior to harvest and subcellular fractionation. This result indicates that sterols together with geranylgeraniol trigger extraction of reductase across the ER membrane, resulting in exposure of the lumenal loop between transmembrane domains 7 and 8 to the cytosol. The sterol-induced membrane extraction of reductase as determined by susceptibility of the lumenal T7 epitope to trypsinolysis was inhibited by RNA interference (RNAi)-mediated knockdown of Insigs or VCP/p97. In contrast, the extraction of reductase across membranes continued in cells subjected to RNAi-mediated knockdown of AAA-ATPases of the proteasome 19 S regulatory particle (RP) (16, 17). Interestingly, knockdown of the 19 S RP inhibited not only the proteasome-mediated ERAD of reductase, but the treatment also blunted its sterol-induced cytosolic dislocation. These observations considered together with our previous studies (15) provide biochemical evidence that VCP/p97 mediates the sterol-induced extraction of ubiquitinated reductase across ER membranes, whereas the 19 S RP mediates release of membrane-extracted reductase into the cytosol for proteasomal degradation.
EXPERIMENTAL PROCEDURES
Materials
We obtained MG-132 from Boston Biochem (Cambridge, MA); trypsin and trypsin inhibitor from Sigma; horseradish peroxidase-conjugated donkey anti-mouse, anti-rabbit, and anti-biotin IgGs (affinity-purified) as well as biotin-conjugated anti-mouse IgGs (affinity-purified) from Jackson ImmunoResearch Laboratories (West Grove, PA); geranylgeraniol from Santa Cruz Biotechnology (Dallas, TX); and 25-hydroxycholesterol from Steraloids (Newport, RI). Apomine was synthesized by the Core Medicinal Chemistry laboratory at the University of Texas Southwestern Medical Center. Other reagents including lipoprotein-deficient serum (LPDS; d > 1.215 g/ml), sodium compactin, and sodium mevalonate were prepared or obtained from previously described sources (18, 19).
Cell Culture
Monolayers of CHO-K1 cells were maintained in tissue culture at 37 °C in 8–9% CO2. Stock cultures were maintained in medium A (1:1 mixture of Ham's F-12 medium and Dulbecco's modified Eagle's medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 5% FCS. UT-2 cells, a clone of reductase-deficient CHO-K1 cells (20), were maintained in medium B (medium A supplemented with 5% FCS and 0.2 mm mevalonate). UT-2/pHMG-Red-T7 cells were generated as follows. On day 0, UT-2 cells were set up at a density of 4 × 105 cells/100-mm dish in medium B. On day 1, the cells were transfected with 2 μg/dish pCMV-HMG-Red-T7, which encodes full-length hamster reductase with two copies of the T7 epitope inserted in the lumenal loop between transmembrane helices 7 and 8 of reductase (5), using FuGENE 6 transfection reagent (Roche Applied Science) as described previously (21). Following incubation for 16 h at 37 °C, the cells were switched to medium C (medium B supplemented with 700 μg/ml G418). Fresh medium was added every 2–3 days until colonies formed after about 2 weeks. Individual colonies were isolated using cloning cylinders, and expression of T7-tagged reductase was determined by immunoblot analysis. Cells from a single colony of cells expressing a moderate level of transfected reductase (as determined by immunoblot analysis with monoclonal anti-T7 IgG) were selected, cloned by limiting dilution, and maintained in medium C.
Isolation of Cell Membranes
Protocol 1
Triplicate dishes of cells were harvested into culture medium and washed with PBS. The cell pellets were resuspended in 500 μl of Buffer A (10 mm HEPES-KOH (pH 7.4), 10 mm KCl, 1.5 mm MgCl2, 5 mm sodium EDTA, 5 mm sodium EGTA, 250 mm sucrose, 5 mm dithiothreitol, and 0.1 mm leupeptin), passed through a 22.5-gauge needle 30 times, and centrifuged at 1000 × g for 7 min at 4 °C. The resulting postnuclear supernatants were then subjected to an additional round of centrifugation at 16,000 × g for 15 min at 4 °C. The resulting pellets were designated as the membrane fraction.
Protocol 2
Triplicate dishes of cells were harvested into culture medium, washed with PBS, resuspended in Buffer A, and lysed for preparation of postnuclear supernatants as described above. The postnuclear supernatants were then subjected to centrifugation at 100,000 × g for 45 min at 4 °C. The resulting pellet and supernatant fractions were designated membrane and cytosol, respectively.
Trypsin Proteolysis
Cells were harvested, and the membrane fractions were prepared using Protocol 1 or 2 as indicated in the figure legends. The membrane pellets were resuspended in 100 μl of Buffer B (Buffer A without dithiothreitol and leupeptin supplemented with 100 mm NaCl). The resuspended membranes were pooled together when appropriate and divided into 56-μl aliquots for trypsin digestion. Some of the aliquots of membranes were treated with 1% Nonidet P-40 (final concentration) prior to trypsin digestion as indicated in the figure legends. Varying amounts of trypsin were added to the resuspended membranes in a volume of 2 μl, and samples were incubated at 30 °C for the times indicated in the figure legends. Trypsin proteolysis was terminated by the addition of Buffer C (62.5 mm Tris-HCl (pH 6.8), 15% (w/v) SDS, 8 m urea, 10% (v/v) glycerol, and 100 mm dithiothreitol) containing 40 μg of soybean trypsin inhibitor and 4× SDS loading buffer (1× final concentration). Samples were then heated at 37 °C for 20 min and subjected to SDS-PAGE followed by immunoblot analysis as described below.
Immunoblot Analysis
The protein concentration of membrane and cytosol fractions was determined using the Coomassie Bradford and BCA protein assay kits (Thermo Scientific, Rockford, IL). Aliquots of the fractions were mixed with an equal volume of Buffer C and 4× SDS loading buffer added to a final concentration of 1×. For anti-Scap immunoblots, fractions were directly mixed with 4× SDS loading buffer to a final concentration of 1×. Samples were fractionated on SDS-polyacrylamide gels calibrated with prestained molecular mass markers (Bio-Rad) after which the proteins were transferred to nitrocellulose membranes and subjected to immunoblot analysis. Primary antibodies used for immunoblotting were as follows: IgG-A9, a mouse monoclonal antibody against hamster HMG-CoA reductase (22); monoclonal anti-T7 tag IgG (EMD Biosciences, Darmstadt, Germany); IgG-9D5, a mouse monoclonal antibody against hamster Scap (23); IgG-18, a mouse monoclonal antibody against VCP/p97 (BD Transduction Laboratories); rabbit polyclonal anti-calnexin IgG (Novus Biologicals, Littleton, CO); MLO7, rabbit polyclonal antiserum against rat GM130 (24); IgG-TBP1-19, a mouse monoclonal antibody against human Rpt5 (Enzo Life Sciences, Farmingdale, NY); IgG-p42-23, a mouse monoclonal antibody against human Rpt4 (Biomol International/Enzo Life Sciences, Ann Arbor, MI), and rabbit polyclonal anti-Rpt2 IgG (Enzo Life Sciences). Bound antibodies were visualized with peroxidase-conjugated donkey anti-mouse or anti-rabbit IgG using the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) according to the manufacturer's instructions. Filters were exposed to film at room temperature.
RNAi
RNAi was carried out as described previously with minor modifications (15). Duplexes of small interfering RNA (siRNA) were designed and synthesized by Dharmacon/Thermo Fisher Scientific. Sequences of the siRNAs used in the current study are listed in Table 1; the siRNA duplex against the control gene, green fluorescent protein (GFP), was described previously (25).
TABLE 1.
siRNAs used in this study
| Name | GenBankTM accession number | Sequence (5′–3′) |
|---|---|---|
| GFP | CAGCCACAACGUCUAUAUCUU | |
| Insig-1 | NM_001244079.1 | AAACAUAGGACGACAGUUAUU |
| Insig-2 | NM_001244078.1 | UGACAGGCAUCUAGGAGAAUU |
| VCP/p97 | NM_009503.4 | GAAUAGAGUUGUUCGGAAUUU |
| Rpt1-1 | NM_011188.3 | GGGAUUUGGCUGCAGAUAAUU |
| Rpt1-2 | GCUCACUGGUAUUAAAGAAUU | |
| Rpt2-1 | NM_008947.3 | GCCACAAACCGAAUAGAAAUU |
| Rpt2-2 | AGAAGGAUGACAAGGACAAUU | |
| Rpt3-1 | NM_011874.2 | GGCCAAGGACUUCGAGAAAUU |
| Rpt3-2 | AGGUAAUCAUGGCCACAAAUU | |
| Rpt4-1 | NM_025959.3 | CAGGAAGAUUAGAUAGAAAUU |
| Rpt4-2 | sGCUACAAACAGACCAGAUAUU | |
| Rpt4-3 | GAUCAUGGCUACAAACAGAUU | |
| Rpt5-1 | NM_008948.2 | GAAUAUGACUCUCGGGUGAUU |
| Rpt5-2 | UCAAGAUCAUGAAGAGUGAUU | |
| Rpt5-3 | CGGCAGGAGAAGAUGGCGAUU | |
| Rpt6-1 | NM_008950.1 | CUGAACUGGUACAGAAAUUUU |
| Rpt6-2 | CCAAGAACAUCAAGGUUAUUU |
UT-2/pHMG-Red-T7 and CHO-K1 cells were set up for experiments in 60- or 100-mm dishes as described in the figure legends. Unless indicated otherwise, the cells were transfected on days 1 and 2 with duplexes of siRNA using a ratio of 2 μl of LipofectamineTM RNAiMAX (Invitrogen) to 40–50 pmol of siRNA duplexes/dish. Cells were washed with 2 ml of medium A (without antibiotics) and refed 1.6 ml of medium A (without antibiotics)/dish. Duplexes of siRNAs were diluted in Opti-MEM I reduced serum medium (Invitrogen) to a final volume of 370 μl/dish. Lipofectamine RNAiMAX was diluted with Opti-MEM I to a final volume of 30 μl/dish and incubated at room temperature for 5 min. The siRNA and Lipofectamine RNAiMAX mixtures were combined and further incubated for 10–20 min at room temperature. The combined siRNA and Lipofectamine RNAiMAX mixtures (400 μl) were then added to each dish. Incubation conditions are described in the figure legends. Following incubations, triplicate dishes of cells were harvested and pooled for analysis. When 100-mm dishes were used, cells were transfected on day 1 with duplexes of siRNA using a ratio of 6 μl of Lipofectamine RNAiMAX to 120 pmol of siRNA duplexes/dish. Cells were washed with 5 ml of medium A (without antibiotics) and refed 4.8 ml of medium A (without antibiotics)/dish. Duplexes of siRNAs were diluted in Opti-MEM I reduced serum medium (Invitrogen) to a final volume of 1110 μl/dish. Lipofectamine RNAi MAX was diluted with Opti-MEM I to a final volume of 90 μl/dish and incubated at room temperature for 5 min. The siRNA and Lipofectamine RNAiMAX mixtures were combined and further incubated for 10–20 min at room temperature. The combined siRNA and Lipofectamine RNAiMAX mixtures (1.2 ml) were then added to each dish. Incubation conditions are described in the figure legends. Following incubations, triplicate dishes of cells were harvested and pooled for analysis.
RESULTS
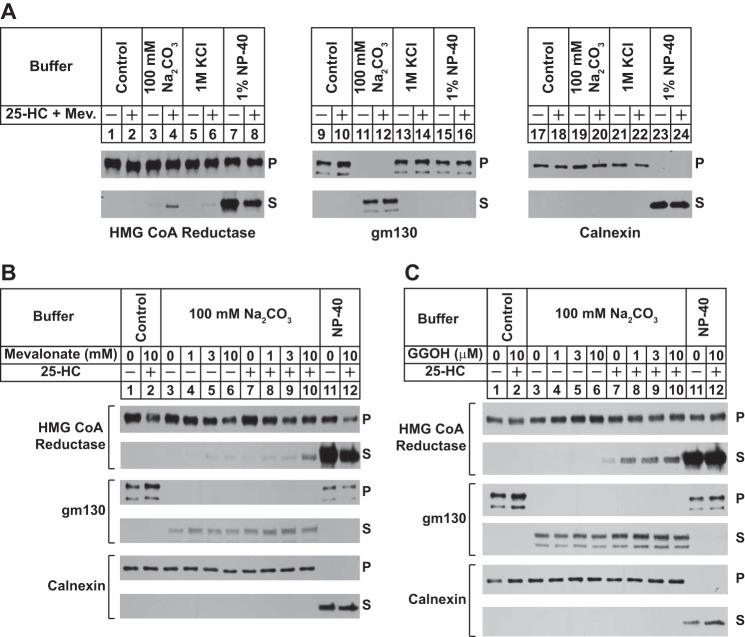
A plausible explanation for the inefficiency of reductase dislocation from ER membranes of intact or permeabilized cells is the existence of additional postubiquitination steps that occur prior to cytosolic release. For example, ubiquitinated reductase may become extracted across the ER membrane and subsequently dislocated into the cytosol for degradation. To measure this putative extraction step, we examined the membrane association of reductase in sterol-treated cells. Reductase-deficient UT-2 cells (20) were transfected with an expression plasmid encoding a previously described version of full-length reductase, designated HMG-Red-T7, which contains two copies of the T7 epitope in the lumenal loop between transmembrane helices 7 and 8 (5). A stable line expressing HMG-Red-T7 was isolated and designated UT-2/pHMG-Red-T7. In the experiment of Fig. 1A, sterol-depleted UT-2/pHMG-Red-T7 cells were treated with the proteasome inhibitor MG-132 (to block reductase ERAD) in the absence or presence of 25-HC plus mevalonate (to provide nonsterol isoprenoids). Following treatments, cells were harvested for subcellular fractionation. Resulting membranes were then washed in control Tris-buffered saline (TBS), 100 mm Na2CO3, 1 m KCl, or 1% Nonidet P-40 buffers and subsequently separated into pellet and supernatant fractions by centrifugation. Immunoblot analysis of these fractions revealed that reductase remained associated with the pellet when membranes were washed with the control TBS buffer (Fig. 1A, top and bottom panels, lanes 1 and 2). However, 25-HC plus mevalonate caused a small fraction of reductase to become released from membranes that were treated with the high pH buffer containing 100 mm Na2CO3 (bottom panel, lane 4). Treatment of the membranes with the nonionic detergent Nonidet P-40 released reductase into the supernatant regardless of the absence or presence of 25-HC plus mevalonate (lanes 7 and 8). The samples were also immunoblotted for two control proteins, GM130 and calnexin. The Golgi-localized GM130 lacks transmembrane helices and is associated with membranes via hydrophobic interactions (26). Consistent with this, the protein was fully released from membranes by high pH (bottom panel, lanes 11 and 12). Calnexin, a molecular chaperone that is integrated in ER membranes by a single membrane-spanning segment (27), was only released into the supernatant when membranes were dissolved in Nonidet P-40 (bottom panel, lanes 23 and 24).
FIGURE 1.
Peripheral association of HMG-CoA reductase with membranes of sterol-treated cells. UT-2/pHMG-Red-T7 cells were set up for experiments on day 0 in medium C at a density of 2 × 105 (A) or 5 × 105 (B and C) cells/100-mm dish. On day 3, the cells were switched to medium A supplemented with 5% LPDS, 10 μm compactin, 50 μm mevalonate, and 1 μm MG-132 in the absence or presence of 1 μg/ml 25-HC; 10 mm mevalonate (Mev.; A); 1, 3, or 10 mm mevalonate (B); and 1, 3, or 10 μm geranylgeraniol (GGOH; C) as indicated. Following incubation for 16 h at 37 °C, cells were harvested, resuspended in buffer containing 20 mm Tris-HCl (pH 7.4) and 250 mm sucrose, and lysed. Resulting lysates were then subjected to centrifugation at 1000 × g to generate postnuclear supernatants that were subjected to an additional round of centrifugation at 20,000 × g. Membrane pellets were resuspended in an equal volume of one of the following buffers: 20 mm Tris-HCl (pH 7.4) and 140 mm NaCl (Control), 20 mm Tris-HCl (pH 7.4) and 1 m KCl, 100 mm Na2CO3, and 20 mm Tris-HCl (pH 7.4) and 1% Nonidet P-40 (NP-40). Following rotation for 2 h at 4 °C, samples were layered on buffer containing 20 mm Tris-HCl (pH 7.4) and 500 mm sucrose and subjected to centrifugation for 30 min at 100,000 × g. Equal proportions of the resulting pellet (P) and supernatant (S) fractions were subjected to SDS-PAGE followed by immunoblot analysis with anti-T7 IgG (against reductase), MLO7 (against GM130), and anti-calnexin IgG.
The experiment shown in Fig. 1B examines the individual and combined effects of sterol and nonsterol isoprenoids on the membrane association of reductase. The results show that when added to cells alone high concentrations of mevalonate (3 and 10 mm) or 25-HC caused a slight increase in the amount of reductase released from membranes by the high pH wash (Fig. 1B, second panel, compare lane 3 with lanes 4–7). The effect of high concentrations of mevalonate can be attributed to its conversion to sterol as well as nonsterol isoprenoids in UT-2/pHMG-Red-T7 cells. The combination of 10 mm mevalonate and 25-HC enhanced the high pH-mediated release of reductase from isolated membranes (lane 10). GM130 was only released by high pH wash (fourth panel, lanes 3–10), whereas Nonidet P-40 treatment of membranes caused the complete release of calnexin from membranes (sixth panel, lanes 11 and 12). To more clearly demonstrate the effect of nonsterol isoprenoids on the membrane association of reductase, we conducted an experiment using geranylgeraniol, which, unlike mevalonate, cannot become incorporated into sterols. The results show that reductase remained associated with the membrane pellet when cells were treated with geranylgeraniol alone (Fig. 1C, second panel, lanes 4–6), but the nonsterol isoprenoid combined with 25-HC to cause release of reductase from membranes (Fig. 1C, second panel, lanes 8–10).
Results of Fig. 1 show that sterol and nonsterol isoprenoids combine to trigger the extraction of reductase across ER membranes. However, the reaction appeared to be inefficient as determined by membrane association of reductase. Thus, we next sought to develop a more robust assay for sterol-induced membrane extraction of reductase that exploits the lumenal orientation of the T7 epitope in HMG-Red-T7 (Fig. 2A). This orientation was confirmed by subjecting intact membranes from sterol-deprived UT-2/pHMG-Red-T7 cells to in vitro digestion with trypsin, which produced protected fragments of reductase that were detected by anti-T7 immunoblot analysis; these fragments were completely destroyed by trypsinolysis when membrane were first dissolved in Nonidet P-40 detergent (data not shown). The lumenal orientation of the T7 epitope in HMG-Red-T7 is further supported by the observation that the trypsin-protected fragments were sensitive to digestion by either peptide-N-glycosidase F or endoglycosidase H, endoglycosidases that removed the single N-glycan in reductase (data not shown).
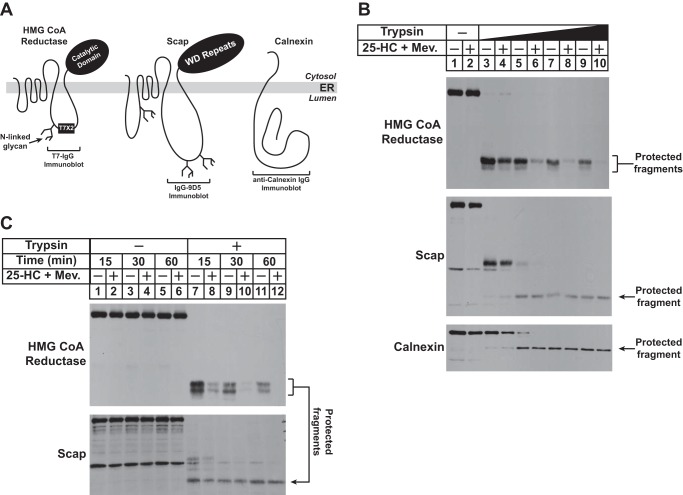
FIGURE 2.
Sterol and nonsterol isoprenoids enhance susceptibility of lumenal T7 epitope in HMG-Red-T7 to trypsinolysis. A, topology of HMG-CoA reductase, Scap, and calnexin denoting the location of two T7 epitope tags in reductase encoded by pCMV-HMG-Red-T7, the epitope in Scap recognized by monoclonal IgG-9D5, and the region in calnexin recognized by anti-calnexin IgG. Sites for N-linked glycosylation in HMG-CoA reductase and Scap are indicated. B and C, UT-2/pHMG-Red-T7 cells were set up for experiments on day 0 at a density of 5 × 105 cells/100-mm dish in medium C. On day 2, the cells were refed medium B supplemented with 5% LPDS, 10 μm compactin, 50 μm mevalonate (Mev.), and 1 μm MG-132 in the absence or presence of 1 μg/ml 25-HC plus 10 mm mevalonate as indicated. Following incubation at 37 °C for 16 h, the cells were harvested for subcellular fractionation using Protocol 1 as described under “Experimental Procedures.” The resulting membrane fractions were resuspended in Buffer B and digested with 2–20 μg of trypsin for 30 min at 30 °C (B) or with 20 μg of trypsin for the indicated amount of time at 30 °C (C). Following treatments, reactions were terminated, and the samples were subjected to SDS-PAGE followed by immunoblot analysis with anti-T7 IgG (against reductase), IgG-9D5 (against Scap), and anti-calnexin IgG.
Having established the membrane orientation of the T7 epitope in HMG-Red-T7, we next set out to determine whether in vitro trypsin digestion could be used to monitor sterol- and geranylgeraniol-induced extraction of the protein across ER membranes. In the experiment of Fig. 2B, UT-2/pHMG-Red-T7 cells were subjected to treatment with MG-132 in the absence or presence of 25-HC plus mevalonate prior to harvest and subcellular fractionation. Aliquots of the resulting membrane fractions were then incubated with increasing amounts of trypsin and subsequently analyzed by anti-T7 immunoblotting. In the absence of trypsin, equivalent amounts of full-length reductase were detected in membranes isolated from cells incubated with or without 25-HC plus mevalonate (Fig. 2B, top panel, lanes 1 and 2). The lowest concentration of trypsin produced the expected protected fragments in both the absence and presence of 25-HC plus mevalonate (lanes 3 and 4). At higher concentrations of trypsin, 25-HC plus mevalonate enhanced the susceptibility of the protected fragments of reductase to digestion (top panel, compare lanes 5, 7, and 9 with lanes 6, 8, and 10). To control for membrane integrity, samples were immunoblotted for the cholesterol-sensing escort protein Scap and calnexin. Scap is integrated in ER membranes with a topology similar to that of reductase: the N-terminal domain of Scap contains eight membrane-spanning helices and is followed by a large cytosolic domain that contains WD repeats (Fig. 2A) (23, 28). The monoclonal antibody IgG-9D5 recognizes residues 540–707 of Scap that corresponds to the large, N-glycosylated lumenal loop between transmembrane helices 7 and 8 of the protein; this antibody was used previously to define the membrane orientation of Scap (28). Trypsinolysis of membranes led to the appearance of a protected fragment of Scap that was detected in IgG-9D5 immunoblots, and importantly, the susceptibility of this fragment to trypsinolysis was not influenced by the absence or presence of 25-HC plus mevalonate (Fig. 2B, middle panel, lanes 5–10). Calnexin is a type II ER membrane protein that consists of a large N-terminal lumenal domain, a single transmembrane helix, and a short cytoplasmic tail (Fig. 2A) (27). Immunoblot analysis with polyclonal antibodies against the lumenal domain of calnexin revealed generation of a protected fragment of the protein in the presence of trypsin; the amount of this protected fragment did not change when cells were treated with the combination of 25-HC and mevalonate (bottom panel, lanes 5-10). Fig. 2C shows an experiment in which membranes from cells treated with 25-HC and mevalonate were incubated with a fixed concentration of trypsin for various periods of time. Protected fragments of reductase were observed in immunoblots of membranes from sterol-deprived cells treated with trypsin for the shortest period of time, 15 min (Fig. 2C, top panel, lane 7); the amount of these fragments was reduced in membranes from 25-HC plus mevalonate-treated cells (lane 8). Longer incubations with trypsin led to a slight reduction of the protected fragments of reductase in membranes from sterol-deprived cells (lanes 9 and 11), a likely reflection of the basal membrane extraction and degradation of reductase. The susceptibility of these fragments to trypsin was enhanced when cells were treated with 25-HC plus mevalonate prior to subcellular fractionation (lanes 10 and 12). The expected protease-protected fragment of Scap was observed in the IgG-9D5 immunoblot, and importantly, the susceptibility of this fragment to trypsinolysis was not influenced by the absence or presence of 25-HC plus mevalonate (Fig. 2C, bottom panel, lanes 7–12).
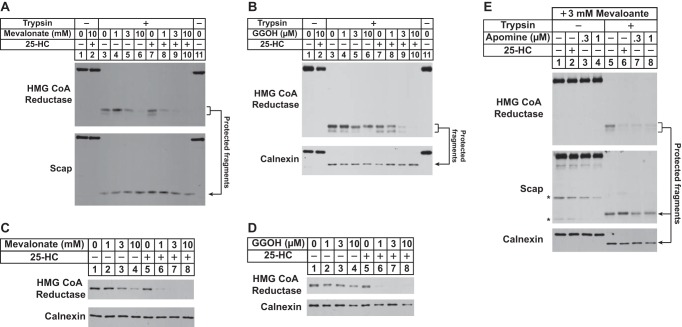
The contribution of sterol and nonsterol isoprenoids to the membrane extraction of reductase as determined by sensitivity to trypsin digestion was examined by treating UT-2/pHMG-Red-T7 cells with 25-HC, mevalonate, and geranylgeraniol individually or in combination prior to harvest and subcellular fractionation. Immunoblot analysis shows that in the absence of 25-HC a high concentration (10 mm) of mevalonate caused a significant fraction of the protected fragment of reductase to become fully digested by trypsin in vitro (Fig. 3A, top panel, lanes 3–6). This is consistent with conversion of mevalonate to both sterol and nonsterol isoprenoids that maximally stimulate the degradation (2, 6) as well as the membrane extraction of reductase. Treatment of the cells with 25-HC alone did not render the protected fragment susceptible to trypsinolysis (lane 7); however, 25-HC together with low concentrations of mevalonate (1 and 3 mm) caused the fragment to disappear upon trypsin digestion (lanes 8 and 9). Levels of the protected fragment of Scap remained constant regardless of whether or not cells were treated with 25-HC and/or mevalonate (bottom panel, lanes 3–10). Similar results were obtained when we measured the effect of geranylgeraniol on the membrane extraction of reductase (Fig. 3B). The trypsin-protected fragment of reductase persisted in membranes from cells treated individually with geranylgeraniol (Fig. 3B, top panel, lanes 4–6) or 25-HC (lane 7). However, geranylgeraniol combined with 25-HC to cause the protected fragments to become susceptible to trypsinolysis (lanes 8–10). These results obtained for membrane extraction of HMG-Red-T7 are consistent with those that demonstrate the requirement of sterol and nonsterol isoprenoids for accelerated ERAD of the protein (Fig. 3, C and D).
FIGURE 3.
Sterol and nonsterol requirements for extraction of HMG-CoA reductase across ER membranes as determined by protease protection. UT-2/pHMG-Red-T7 cells were set up for experiments on day 0 as described in the legend for Fig. 2B. On day 2, cells were refed medium A supplemented with 5% LPDS, 10 μm compactin, 50 μm mevalonate, and 1 μm MG-132 in the absence or presence of 1 μg/ml 25-HC plus the indicated concentration of mevalonate (A and C), 0–10 μm geranylgeraniol (GGOH; B and D), or 3 mm mevalonate plus either 1 μg/ml 25-HC or the indicated concentration of apomine (E). A, B, and E, following incubation at 37 °C for 16 h, the cells were harvested for subcellular fractionation using Protocol 1. The resulting membrane fractions were resuspended in Buffer B and pooled where appropriate, and equal amounts of membrane suspensions were digested with 10–20 μg of trypsin for 30 min at 30 °C. Reactions were then terminated, and samples were subjected to SDS-PAGE followed by immunoblot analysis with anti-T7 IgG (against reductase), IgG-9D5 (against Scap), and anti-calnexin IgG. C and D, following incubation for 16 h at 37 °C, cells were harvested for subcellular fractionation. Aliquots of resulting membrane fractions (10 μg of protein/lane) were subjected to SDS-PAGE followed by immunoblot analysis with anti-T7 IgG (against reductase) and anti-calnexin IgG. Asterisks denote non-specific bands.
In cultured cells, the 1,1-bisphosphonate ester apomine and its structurally related analog SR-12813 mimic 25-HC in stimulating Insig-dependent ubiquitination and subsequent ERAD of reductase (29, 30). Thus, we next designed an experiment to evaluate apomine in triggering the extraction of reductase across ER membranes. In membranes from UT-2/pHMG-Red-T7 cells treated with 25-HC plus mevalonate (3 mm), the protected fragments of reductase were digested by trypsin as expected (Fig. 3E, top panel, compare lanes 5 and 6). Treatment of the cells with apomine and 3 mm mevalonate rendered the protected fragments susceptible to trypsinolysis (lanes 7 and 8), indicating that, like 25-HC, apomine induces extraction of reductase across ER membranes en route to degradation.
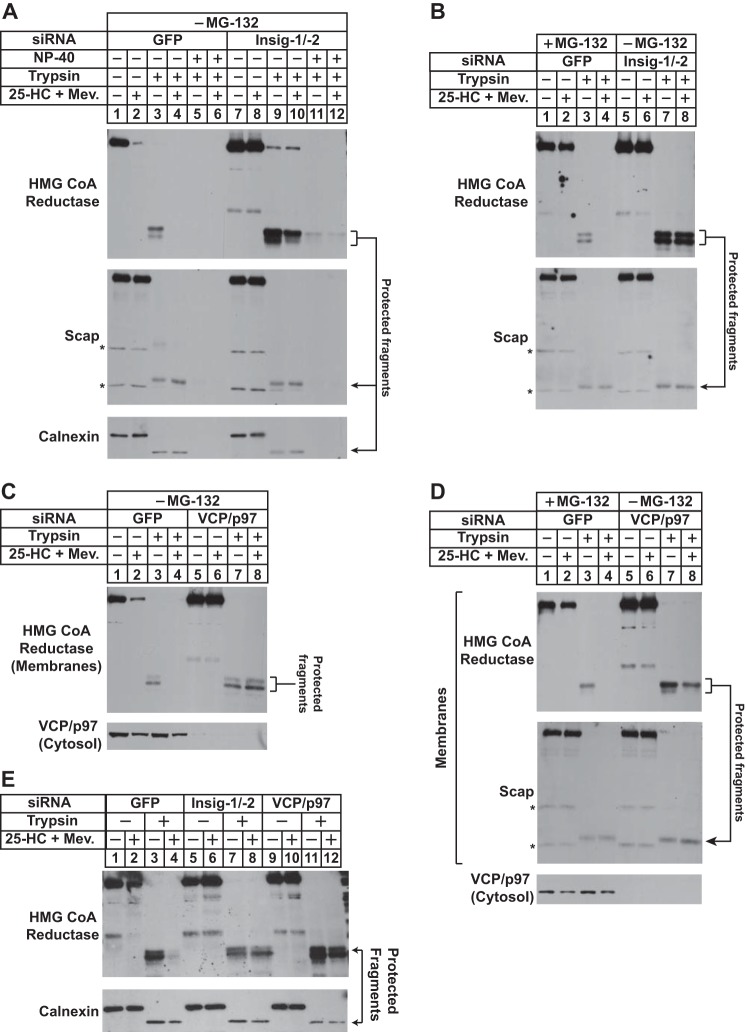
To further establish the physiologic relevance of the membrane extraction of reductase, we next examined a role for Insigs in the reaction using RNAi. UT-2/pHMG-Red-T7 cells were transfected with duplexes of siRNAs targeting a control mRNA encoding GFP or mRNAs encoding Insig-1 and Insig-2. Following siRNA transfection, cells were treated in the absence or presence of 25-HC plus mevalonate and MG-132 prior to harvest, subcellular fractionation, and treatment of membranes with or without trypsin. The results show that in the presence of 25-HC plus mevalonate and absence of MG-132 reductase was appropriately degraded from membranes of cells transfected with GFP siRNA (Fig. 4A, top panel, lanes 1 and 2). Trypsin digestion of sterol-deprived membranes led to the appearance of the expected protected fragments of reductase (lane 3). These protected fragments were absent in membranes from sterol-treated cells (lane 4). However, this absence was due to accelerated ERAD of reductase rather than from extraction of the protein across ER membranes. Treatment with Nonidet P-40 disrupted membrane integrity and rendered the protected fragments of reductase susceptible to trypsinolysis (lane 5). As expected, RNAi-mediated knockdown of Insig-1 and Insig-2 blocked sterol-accelerated degradation of reductase (Fig. 4A, top panel, compare lanes 1 and 2 with lanes 7 and 8). The 25-HC plus mevalonate-dependent susceptibility of the protected fragments to trypsinolysis was blunted in Insig-1 and -2 knockdown cells (lanes 9 and 10). The fragments were fully digested by trypsin in the presence of Nonidet P-40 (lanes 11 and 12), indicating their sequestration in the ER lumen. Levels of trypsin-protected fragments of Scap and calnexin were not affected by the absence or presence of 25-HC plus mevalonate in control or Insig-1/-2 knockdown cells (middle and bottom panels, lanes 3, 4, 9, and 10).
FIGURE 4.
RNAi-mediated knockdown of Insigs or VCP/p97 blunts sterol-induced extraction of HMG-CoA reductase across ER membranes. UT-2/pHMG-Red-T7 cells were set up on day 0 at a density of 1.5–2 × 105 cells/60-mm dish in medium C. On days 1 and 2, the cells were transfected with the indicated siRNA duplexes as described under “Experimental Procedures.” Following transfection on day 2, cells received a direct addition of medium A containing 5% LPDS, 10 μm compactin, and 50 μm mevalonate (Mev.) in the absence or presence of 1 μg/ml 25-HC plus 10 mm mevalonate (final concentrations). In B and D, some of the cells (lanes 1–4) were also treated with 1 μm MG-132; all of the cells in E were treated with 1 μm MG-132. Following incubation at 37 °C for 16 h, cells were harvested for subcellular fractionation using Protocol 1(A and B) or Protocol 2 (C and D). Aliquots of the resulting membrane fractions were resuspended in Buffer B and pooled where appropriate, and equal amounts of membrane suspensions were digested with 10 μg of trypsin. Some of the samples (lanes 5, 6, 11, and 12 in A) were treated with 1% Nonidet P-40 (NP-40) prior to proteolysis. After 30 min at 30 °C, the reactions were terminated, and samples were subjected to SDS-PAGE followed by immunoblot analysis with anti-T7 IgG (against reductase), IgG-9D5 (against Scap), IgG-18 (against VCP/p97), and anti-calnexin IgG. Asterisks denote non-specific bands.
In the experiment of Fig. 4B, the membrane extraction assay was conducted with membranes from cells in which reductase degradation was blocked by either proteasome inhibition or RNAi-mediated knockdown of Insigs. In control cells transfected with the GFP siRNA and treated with MG-132, sterol-accelerated degradation of reductase was blocked (Fig. 4B, top panel, lanes 1 and 2). Trypsin digestion led to the appearance of the protected fragment of reductase in membranes isolated from cells incubated in the absence (lane 3), but not in the presence, of 25-HC plus mevalonate (lane 4). Knockdown of Insig-1 and Insig-2 resulted in resistance of the protected fragments of reductase to trypsinolysis (lanes 7 and 8), demonstrating their requirement in the membrane extraction of reductase.
Considering its role in cytosolic dislocation of reductase (15), we next conducted RNAi experiments to determine whether VCP/p97 is required for membrane extraction of the enzyme. As expected, knockdown of VCP/p97 blunted sterol-accelerated reductase ERAD (Fig. 4C, top panel, compare lanes 1 and 2 with lanes 5 and 6). In the absence of MG-132, the trypsin-protected fragments of reductase were observed in membranes from control-transfected cells deprived of sterols (lane 3) but not in membranes isolated from cells treated with 25-HC plus mevalonate (lane 4) because of accelerated reductase degradation. The protected fragments of reductase in membranes from VCP/p97 knockdown cells resisted trypsin digestion (lanes 7 and 8) regardless of whether the cells had been treated with or without 25-HC plus mevalonate before harvest. In the presence of MG-132, 25-HC plus mevalonate rendered the protected fragments of reductase susceptible to trypsinolysis in GFP knockdown cells (Fig. 4D, top panel, lanes 3 and 4). However, these fragments were again resistant to digestion in VCP/p97 knockdown cells (lanes 7 and 8), consistent with a key role for VCP/p97 in sterol-induced extraction of reductase to the cytosolic face of the ER. It should be noted that RNAi-mediated knockdown of VCP/p97 or Insig-1 and Insig-2 prevented membrane extraction of reductase in MG-132-treated cells (Fig. 4E).
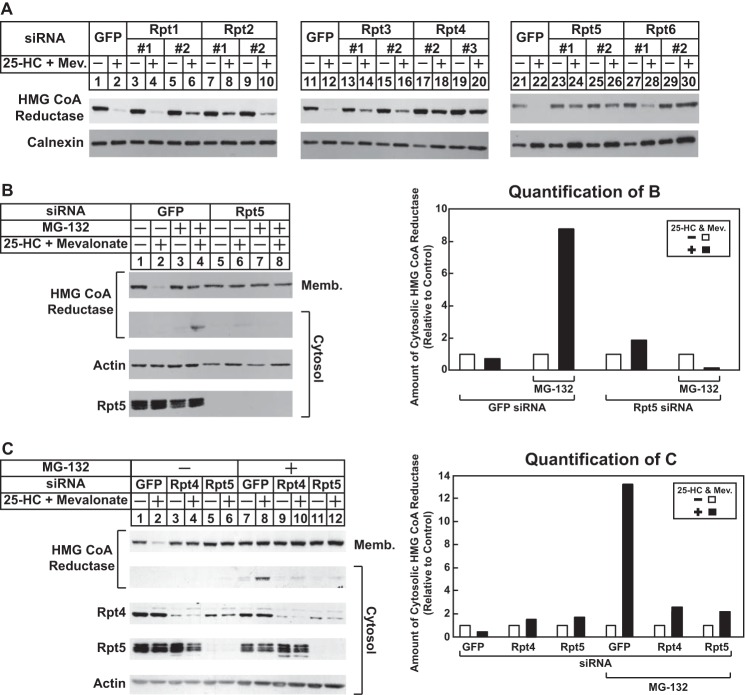
In previous studies, we reconstituted Insig-mediated, sterol-accelerated ERAD of mammalian reductase in Drosophila S2 cells (30). Building on this observation, we recently conducted a high throughput genome-wide RNAi screen in S2 cells. Several genes encoding components of the 26 S proteasome were identified among those required for sterol-accelerated ERAD of reductase in S2 cells.3 Some of these genes encode Drosophila homologs of the AAA-ATPase subunits of the proteasome 19 S RP (16, 17). The 19 S RP uses energy from ATP hydrolysis to unfold substrates, trigger the activation of the 20 S core particle (CP) of the proteasome, and translocate the substrate into the proteolytic chamber of the 20 S CP. Although the requirement for the 19 S RP in reductase degradation in Drosophila cells was not surprising, it led us to the realization that the regulatory particle has been implicated in cytosolic dislocation of several ERAD substrates (17). To determine whether the 19 S RP is required for cytosolic dislocation of reductase, we began by screening siRNAs that target the AAA-ATPases of the regulatory particle for the ability to block reductase degradation. The results show that knockdown of these ATPases blunted reductase degradation to various degrees (Fig. 5A), which is likely due to the efficiency of their RNAi-mediated knockdown. Thus, we focused on three AAA-ATPases, Rpt2, Rpt5, and Rtp4, whose knockdown consistently inhibited reductase degradation and for which specific antibodies are commercially available.
FIGURE 5.
RNAi-mediated knockdown of AAA-ATPases of the proteasome 19 S regulatory particle blunts sterol-accelerated degradation and cytosolic dislocation of HMG-CoA reductase. CHO-K1 cells were set up on day 0 at a density of 5 × 105 cells/100-mm dish (A and C) or 1.5 × 105 cells/60-mm dish (B) in medium A containing 5% FCS. On day 1, cells were transfected with the indicated siRNA duplexes in A, Rpt5-1 in B, and Rpt4-3 or Rpt5-1 in C as described in the legend for Fig. 4. Four hours following transfection, cells were depleted of sterols for 16 h by the direct addition of medium A containing 5% LPDS, 10 μm compactin, and 50 μm mevalonate (Mev.) (final concentrations). A, sterol-depleted cells were switched to identical medium in the absence or presence of 1 μg/ml 25-HC plus 10 mm mevalonate as indicated. After 4–5 h at 37 °C, cells were harvested and subjected to cellular fractionation. Aliquots of resulting membrane fractions (15–20 μg of protein/lane) were subjected to SDS-PAGE and immunoblot analysis with IgG-A9 (against reductase) and anti-calnexin IgG. B and C, sterol-depleted cells were pretreated for 1 h with medium A supplemented with 5% LPDS, 10 μm compactin, and 50 μm mevalonate in the absence or presence of 10 μm MG-132. Cells then received medium A containing 5% LPDS, 10 μm compactin, and 50 μm mevalonate in the absence or presence of 10 μm MG-132 and 1 μg/ml 25-HC plus 10 mm mevalonate. After 4 h at 37 °C, cells were harvested, and postnuclear supernatants were subjected to centrifugation at 1 × 106 g for 30 min at 4 °C. Aliquots of the resulting membrane (Memb.) pellet (15–20 μg of protein/lane) and cytosolic supernatant (10–40 μg of protein/lane) fractions were subjected to SDS-PAGE and immunoblot analysis with IgG-A9 (against reductase), IgG-TBP1-19 (against Rpt5), IgG-p42-23 (against Rpt4), and anti-actin IgG. Bands corresponding to cytosolically dislocated reductase were quantified using ImageJ software. The intensities of these bands in the absence of 25-HC plus mevalonate were arbitrarily set as 1 for each of the RNAi-mediated knockdowns. Results are representative of at least three independent experiments.
In the experiment of Fig. 5B, CHO-K1 cells transfected with control GFP or Rpt5 siRNA duplexes were treated in the absence or presence of 25-HC plus mevalonate and MG-132. The cells were subsequently harvested, and postnuclear supernatants were separated into membrane and cytosol fractions by 100,000 × g centrifugation. The results show that 25-HC plus mevalonate stimulated ERAD of reductase from membranes as expected (Fig. 5B, top panel, compare lanes 1 and 2). This degradation was blocked by treatment of the cells with MG-132 (lane 4) or by the RNAi-mediated knockdown of Rpt5 (compare lanes 5 and 6). In the presence of MG-132, full-length reductase appeared in the cytosol but only when cells were also subjected to treatment with 25-HC plus mevalonate (second panel, lane 4). In contrast, reductase was not present in the cytosol of Rpt5 knockdown cells regardless of the absence or presence of MG-132 (second panel, lanes 5–8). In unpublished studies,4 we found that Rpt5 co-precipitated with cytosolic reductase in 25-HC plus mevalonate-treated cells, consistent with a role of the 19 S RP in reductase dislocation. The sterol-accelerated ERAD (Fig. 5C, top panel, lanes 3 and 4) and the cytosolic dislocation of reductase (bottom panel, lanes 9 and 10) were also blunted by RNAi-mediated knockdown of the Rpt4 AAA-ATPase subunit of the 19 S RP. Knockdown of the other AAA-ATPases of the 19 S RP similarly blunted the sterol-induced cytosolic dislocation of reductase (data not shown). Moreover, ubiquitinated proteins accumulated in Rpt4 and Rpt5 knockdown cells to levels similar to those observed in MG-132-treated cells (data not shown). This indicates that the effect of Rpt4 and Rpt5 knockdown on reductase dislocation results from disruption of the entire 19 S RP rather than a specific effect of the individual ATPase subunits.
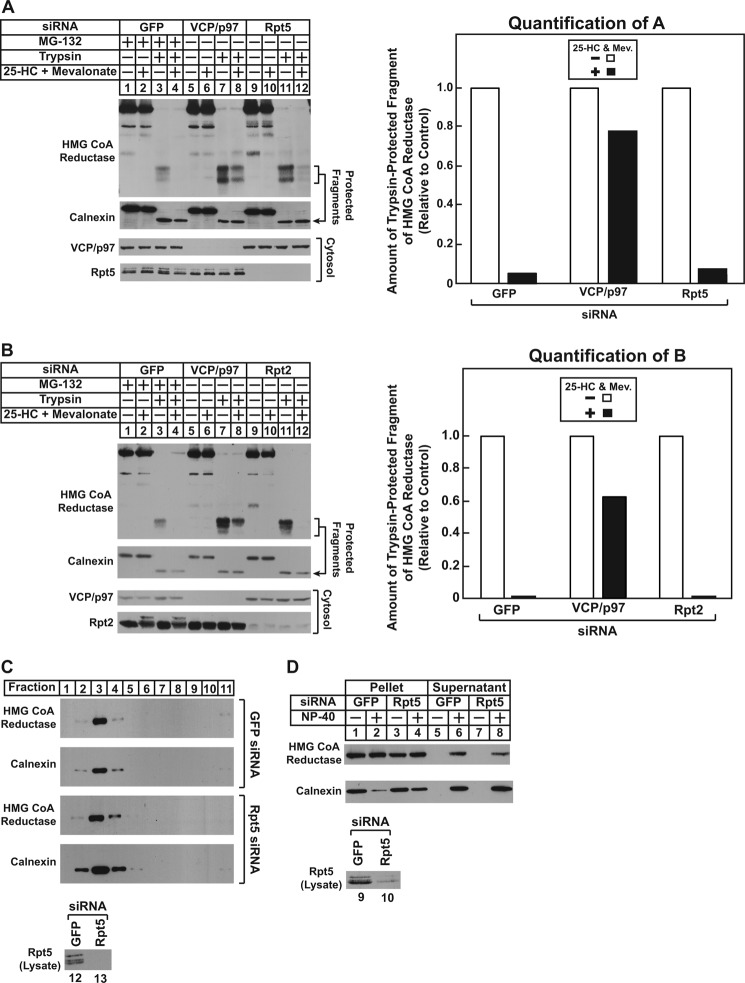
We next examined whether ATPases of the 19 S RP are required for sterol-induced membrane extraction of reductase using the trypsin protection assay. As shown in Fig. 6A, reductase was extracted across membranes of UT-2/pHMG-Red-T7 cells transfected with control GFP siRNA and treated with 25-HC plus mevalonate in the presence of MG-132 (top panel, compare lanes 3 and 4). The RNAi-mediated knockdown of VCP/p97 blunted sterol-induced membrane extraction of reductase as expected (lanes 7 and 8); however, the enzyme continued to become extracted in 25-HC plus mevalonate-treated Rpt5 knockdown cells (lanes 11 and 12). At least three replicate experiments confirm the continued membrane extraction of reductase in Rpt5 knockdown cells (data not shown). Similar results were obtained in cells lacking the Rpt2 AAA-ATPase subunit of the 19 S RP. Knockdown of VCP/p97, but not Rpt2, blunted membrane extraction of reductase (Fig. 6B, top panel, compare lanes 3, 4, 7, 8, 11, and 12).
FIGURE 6.
Sterol and nonsterol isoprenoids trigger membrane extraction of HMG-CoA reductase in cells deficient for AAA-ATPases of the proteasome 19 S regulatory particle. UT-2/pHMG-Red-T7 (A and B) and CHO-K1 (C and D) cells were set up for experiments, transfected with the Rpt5-1 or Rpt2-1 siRNA duplex as indicated, and treated in the absence or presence of 1 μm MG-132, 1 μg/ml 25-HC, and 10 mm mevalonate (Mev.) as described in the legends for Figs. 4 and 5. A and B, following treatments, cells were harvested for subcellular fractionation using Protocol 1, and aliquots of resulting membranes were subjected to trypsinolysis as described in the legends for Figs. 4 and 5. After trypsinolysis, samples together with aliquots of the cytosol were subjected to SDS-PAGE, and immunoblot analysis was carried out with anti-T7 IgG (against reductase), IgG-18 (against VCP/p97), IgG-TBP1-19 (against Rpt5), anti-Rpt2 IgG, and anti-calnexin IgG. Bands corresponding to the protected fragments of reductase in the anti-T7 immunoblots were quantified using ImageJ software. The intensities of the protected fragments of reductase in the absence of 25-HC plus mevalonate were arbitrarily set as 1 for each of the RNAi-mediated knockdowns. C, membranes from control and Rpt5 knockdown cells treated with 25-HC plus 10 mm mevalonate and 10 μm MG-132 were resuspended in TBS containing 71.5% sucrose, and the samples were then overlaid with TBS containing 65 and 10% sucrose. After centrifugation at 100,000 × g for 16 h at 4 °C, aliquots were removed from the top to the bottom of the gradient and analyzed by SDS-PAGE and immunoblot analysis with anti-T7 IgG (against reductase), anti-calnexin IgG, and IgG-TBP1-19 (against Rpt5). D, membranes from control and Rpt5 knockdown cells treated with 10 μm MG-132, 1 μg/ml 25-HC, and 10 mm mevalonate were resuspended in TBS with or without 0.1% Nonidet P-40 (NP-40). Following rotation at 4 °C for 1 h, the samples were separated into soluble supernatant and insoluble pellet fractions by centrifugation. Aliquots of the fractions were then analyzed by immunoblotting with IgG-A9 (against reductase) and anti-calnexin IgG.
The results of Fig. 6, A and B, indicate that VCP/p97 and the 19 S RP mediate distinct steps (membrane extraction and cytosolic dislocation) in the reductase ERAD pathway. However, an alternative explanation for these results is that VCP/p97 solely mediates membrane extraction and cytosolic dislocation of reductase. The 19 S RP has been shown to exhibit chaperone activity in vitro (31) and to prevent aggregation of dislocated catalytic A chains of the toxin ricin (32). In the absence of the 19 S RP, membrane-extracted and/or cytosolic reductase may become aggregated and pellet with membranes upon centrifugation. To rule out this possibility, membranes isolated from control and Rpt5 knockdown cells were resuspended in buffer containing a high concentration of sucrose; the samples were then overlaid with buffer of decreasing sucrose concentration. Following centrifugation overnight, aliquots taken from the top to bottom of the gradient were analyzed by immunoblot. The results show that the majority of reductase as well as calnexin floated to the lower sucrose gradient in control knockdown cells (Fig. 6C, top two panels, lanes 2–4). A similar pattern was observed in Rpt5 knockdown cells (bottom two panels, lanes 2–4), indicating that reductase remained membrane-associated regardless of the absence or presence of Rpt5. The cytosolic protein Bag6 has been identified in a ubiquitin ligase-associated multiprotein complex that chaperones dislocated ERAD substrates to proteasomes for degradation (33). In Bag6 knockdown cells, degradation of these substrates is blocked, their solubility in the detergent Nonidet P-40 is reduced, and the proteins accumulate in detergent-insoluble aggregates. Another line of evidence indicating that the 19 S RP does not perform a chaperone function in reductase degradation is provided by the detergent solubility experiment shown in Fig. 6D. The results of this experiment show that the solubility of reductase in Nonidet P-40 remained unchanged in control and Rpt5 knockdown cells (top panel, compare lanes 6 and 8).
DISCUSSION
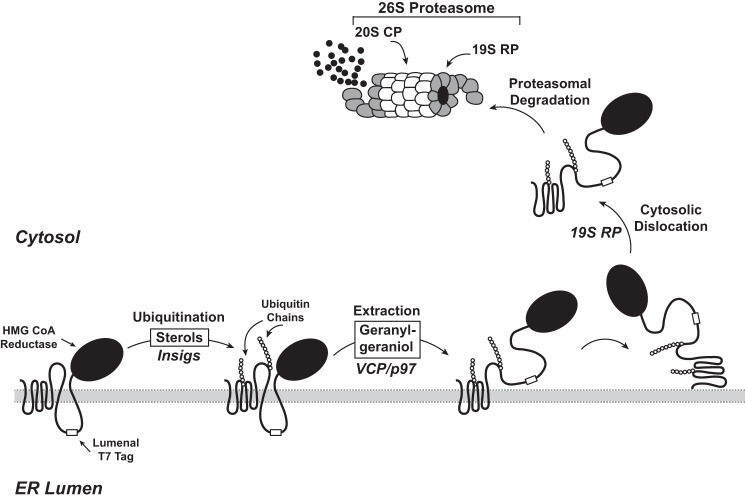
Fig. 7 shows a model for the sterol-accelerated ERAD of reductase based on data presented in the current study. Similar to previously proposed models (10), the reaction is initiated by sterol-induced binding of reductase to Insigs, resulting in gp78/Trc8-mediated ubiquitination of the protein. The current results indicate that once ubiquitinated reductase becomes extracted across the ER membrane through a reaction mediated by the AAA-ATPase VCP/p97. Membrane-associated, extracted reductase then becomes dislodged from membranes and dislocated into the cytosol where the protein is degraded by proteasomes. The novel aspect of this revised model is the proposed membrane extraction step that is distinct from and precedes cytosolic dislocation of reductase. Initial evidence for this putative intermediate step is provided by experiments examining the membrane association of reductase (Fig. 1A). When proteasomes were inhibited by MG-132, the oxysterol 25-HC and the nonsterol isoprenoid geranylgeraniol caused a fraction of reductase to become peripherally associated with membranes as indicated by its release by high pH wash (Fig. 1, B and C). These results indicate that sterol and nonsterol isoprenoids trigger reductase to become fully extracted across the ER membrane prior to degradation. These results taken together with our previous observation that geranylgeraniol also combines with 25-HC to maximally stimulate reductase ERAD but does not appreciably affect sterol-induced ubiquitination of the protein (6, 12) form the basis for the conclusion that geranylgeraniol modulates postubiquitination steps in reductase ERAD (see Fig. 7).
FIGURE 7.
Model for the Insig-mediated, sterol-accelerated ER-associated degradation of HMG-CoA reductase. The diagram shows a schematic representation of the sterol-accelerated ERAD of HMG-CoA reductase. Sterol-induced binding to Insigs results in ubiquitination of HMG-CoA reductase on two cytosolic lysine residues in the membrane domain. Ubiquitination of reductase, which is mediated by two Insig-associated ubiquitin ligases called gp78 and Trc8, marks the enzyme for VCP/p97-mediated extraction across ER membranes through a reaction that is enhanced by geranylgeraniol. Partially extracted reductase with the T7-tagged lumenal loop between transmembrane helices 7 and 8 exposed to the cytosol is indicated as an intermediate in the complete extraction of reductase. Once extracted, reductase is released from membranes into the cytosol through a reaction mediated by the proteasome 19 S RP and finally delivered into the 20 S proteasome CP for degradation.
Considering that reductase is embedded in ER membranes, it is logical to postulate that its extraction across ER membranes must precede cytosolic dislocation as depicted in Fig. 7. However, it remained to be determined whether membrane extraction and cytosolic dislocation of reductase are distinct and separable or coupled reactions. To distinguish between these possibilities, we sought to develop an assay for membrane extraction of reductase that is more robust than measuring its association with membranes. For this purpose, we exploited a previously described version of reductase containing T7 epitopes in the lumenal loop between transmembrane domains 7 and 8 (Fig. 2A) (6). In vitro digestion of intact membranes isolated from sterol-deprived cells with trypsin generated fragments of reductase detected in anti-T7 immunoblots that were protected from proteolysis. Trypsinolysis of membranes from sterol-treated cells led to a marked reduction in the amount of protected fragments in a manner that was proportional to the amount of trypsin in the assay (Fig. 2B) and duration of incubation (Fig. 2C). The extraction of reductase across the ER membrane as determined by susceptibility of its lumenal T7 epitopes to trypsin digestion required the additions of both 25-HC and mevalonate or geranylgeraniol to cells prior to harvest and subcellular fractionation (Fig. 3, A and B). In addition, membrane extraction of reductase was triggered by apomine (Fig. 3E), a 1,1-bisphosphonate ester that mimics sterols in accelerating reductase ERAD (29, 30). RNAi-mediated knockdown of Insig-1 and Insig-2 not only blunted sterol-accelerated ERAD of reductase, but the treatment also blocked its membrane extraction (Fig. 4, A, B, and E). Knockdown of VCP/p97 similarly abolished both the sterol-induced membrane extraction and ERAD of reductase (Fig. 4, C, D, and E). These observations indicate that sterol- and geranylgeraniol-regulated susceptibility of the lumenal T7 epitopes in reductase to trypsinolysis results from its Insig-dependent, VCP/p97-mediated extraction across membranes, exposing the lumenal epitopes to the cytosolic face of the ER. It is important to note that a significant fraction of reductase was observed to be extracted in the trypsin protection assay, whereas only a small fraction of the protein was found to be extracted in membrane association studies. A plausible explanation for this difference is that membrane association studies using the high pH wash measure reductase that has become completely membrane-extracted, whereas the trypsin protection assay measures both completely and partially extracted populations of reductase (see Fig. 7). Partially extracted reductase may result from a feedback mechanism that prevents accumulation of ERAD substrates in the cytosol when proteasomes are impaired or from the saturation of proteins required for extraction of reductase across membranes.
The 26 S proteasome consists of two large subcomplexes designated the 20 S CP and the 19 S RP (16, 17, 34). The 20 S CP comprises four stacked heptameric rings organized in a barrel structure with openings at each end. The two inner rings are composed of β-subunits that exhibit proteolytic activity, whereas the two outer rings consist of structural α-subunits. Both ends of the 20 S CP are associated with the 19 S RP, which is composed of a base that contains six AAA-ATPases and a lid containing non-ATPase subunits. Factors within the lid of the 19 S RP mediate recruitment of ubiquitinated substrates and their subsequent deubiquitination. Binding of the base to the 20 S CP activates proteolysis, and the AAA-ATPases of the base facilitate substrate unfolding and delivery into the proteolytic chamber for degradation. In addition, the 19 S RP has been implicated in cytosolic dislocation of ERAD substrates (17, 35–37); however, the majority of these studies focused on soluble substrates sequestered within the ER lumen. In some cases, dislocation of these substrates was independent of ubiquitination and the action of VCP/p97. In the few studies in which a role for the 19 S RP in the ERAD of membrane-bound substrates was examined, degradation appeared to be coupled to retrotranslocation and extraction; proteasome inhibition did not cause the appearance of these substrates in the cytosol (38, 39). This contrasts with our previous and current findings that MG-132 uncoupled sterol-induced cytosolic dislocation of reductase from its sterol-accelerated proteasomal degradation. Treatment of cells with the inhibitor caused reductase to appear in the cytosol of sterol-treated cells (15). This appearance, which required reductase ubiquitination as well as the presence of Insigs and VCP/p97, indicated that reductase may become dislocated from ER membranes before proteasomal degradation commences. Notably, AAA-ATPases of the 19 S RP have been shown to be required for the ERAD of Hmg2p, a homolog of reductase in the yeast Saccharomyces cerevisiae (37). However, a role for the 19 S RP in the cytosolic dislocation of Hmg2p was not appraised in the study.
In the current study, we appraised a role for the 19 S RP in postubiquitination steps of sterol-accelerated reductase degradation. Consistent with a role for proteasomes in sterol-accelerated reductase ERAD, the reaction was blunted by the proteasome inhibitor MG-132 or by the knockdown of AAA-ATPases of the 19 S RP (16, 17) (Fig. 5A). In surprising contrast to results obtained when proteolytic activity of proteasomes was inhibited, knockdown of the 19 S RP ATPases prevented sterol-induced dislocation of reductase into the cytosol (Fig. 5, B and C). Experiments utilizing the trypsin protection assay revealed that although VCP/p97 knockdown inhibited sterol-induced extraction of reductase across membranes, the reaction continued in cells deficient for the 19 S RP (Fig. 6, A and B). Knockdown of the 19 S RP did not appear to cause membrane-extracted reductase to become aggregated as indicated by its association with membranes in knockdown cells (Fig. 6C) and its detergent solubility (Fig. 6D). These findings not only demonstrate that the 19 S RP and 20 S CP mediate, respectively, the cytosolic dislocation and proteolytic degradation of reductase, they also support our proposal that membrane extraction (mediated by VCP/p97 and enhanced by geranylgeraniol) and cytosolic dislocation (mediated by the 19 S RP) are sequential, separable steps in the reductase ERAD pathway (see Fig. 7). It is noteworthy that dislocation of at least one lumenal ERAD substrate, the μs heavy chain of secretory IgM, involves consecutive membrane extraction and cytosolic dislocation steps mediated by VCP/p97 and proteasomes, respectively (40). However, a major difference exists in the mechanism for membrane extraction and cytosolic dislocation of reductase and μs: proteolytic activity of proteasomes is required for dislocation of μs, whereas reductase dislocation can only be observed upon proteasome inhibition. Proteasome inhibition is known to cause several membrane-bound ERAD substrates in addition to reductase to accumulate in the cytosol of cells (13, 41–47). It will thus be important to determine in future studies whether membrane extraction and release into the cytosol through sequential actions of VCP/p97 and the proteasome 19 S RP are a general ERAD mechanism or a peculiarity in the sterol-accelerated ERAD of reductase. Finally, the current findings together with our previous studies (6, 12) pinpoint the action of geranylgeraniol in reductase ERAD to reactions that underlie VCP/p97-mediated membrane extraction of ubiquitinated reductase. The establishment of a robust assay for membrane extraction of reductase provides a means to further characterize the mechanism for geranylgeraniol-mediated augmentation of reductase ERAD and to identify the target of the nonsterol isoprenoid, which may be an unknown membrane protein or ubiquitinated reductase itself.
Acknowledgments
We thank Drs. Michael S. Brown and Joseph L. Goldstein for continued encouragement and advice. We also thank Muleya Kapaale, Hue Dao, and Shomanike Head for help with tissue culture and Rebecca Faulkner and Marc Schumacher for critical evaluation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL20948, GM090216, and T32GM008203, a Division of Cell and Molecular Biology Training Program grant (to L. L. M.).
D.-J. Jun and R. A. DeBose-Boyd, unpublished observations.
L. L. Morris, I. Z. Hartman, D.-J. Jun, J. Seemann, and R. A. DeBose-Boyd, unpublished studies.
- ER
- endoplasmic reticulum
- RP
- regulatory particle
- CP
- core particle
- ERAD
- ER-associated degradation
- 25-HC
- 25-hydroxycholesterol
- LPDS
- lipoprotein-deficient serum
- VCP
- valosin-containing protein.
REFERENCES
- 1. Brodsky J. L. (2012) Cleaning up: ER-associated degradation to the rescue. Cell 151, 1163–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown M. S., Goldstein J. L. (1980) Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J. Lipid Res. 21, 505–517 [PubMed] [Google Scholar]
- 3. Hampton R. Y. (2002) ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14, 476–482 [DOI] [PubMed] [Google Scholar]
- 4. Goldstein J. L., Brown M. S. (1990) Regulation of the mevalonate pathway. Nature 343, 425–430 [DOI] [PubMed] [Google Scholar]
- 5. Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. (2003) Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell 11, 25–33 [DOI] [PubMed] [Google Scholar]
- 6. Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., DeBose-Boyd R. A. (2003) Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J. Biol. Chem. 278, 52479–52490 [DOI] [PubMed] [Google Scholar]
- 7. Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., Goldstein J. L. (1985) Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J. Biol. Chem. 260, 522–530 [PubMed] [Google Scholar]
- 8. Roitelman J., Olender E. H., Bar-Nun S., Dunn W. A., Jr., Simoni R. D. (1992) Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J. Cell Biol. 117, 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song B. L., Sever N., DeBose-Boyd R. A. (2005) Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 10. Jo Y., Lee P. C., Sguigna P. V., DeBose-Boyd R. A. (2011) Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc. Natl. Acad. Sci. U.S.A. 108, 20503–20508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meyer H., Bug M., Bremer S. (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 [DOI] [PubMed] [Google Scholar]
- 12. Elsabrouty R., Jo Y., Dinh T. T., DeBose-Boyd R. A. (2013) Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from membranes of permeabilized cells. Mol. Biol. Cell 24, 3300–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bagola K., Mehnert M., Jarosch E., Sommer T. (2011) Protein dislocation from the ER. Biochim. Biophys. Acta 1808, 925–936 [DOI] [PubMed] [Google Scholar]
- 14. Leichner G. S., Avner R., Harats D., Roitelman J. (2009) Dislocation of HMG-CoA reductase and Insig-1, two polytopic endoplasmic reticulum proteins, en route to proteasomal degradation. Mol. Biol. Cell 20, 3330–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hartman I. Z., Liu P., Zehmer J. K., Luby-Phelps K., Jo Y., Anderson R. G., DeBose-Boyd R. A. (2010) Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J. Biol. Chem. 285, 19288–19298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ehlinger A., Walters K. J. (2013) Structural insights into proteasome activation by the 19S regulatory particle. Biochemistry 52, 3618–3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bar-Nun S., Glickman M. H. (2012) Proteasomal AAA-ATPases: structure and function. Biochim. Biophys. Acta 1823, 67–82 [DOI] [PubMed] [Google Scholar]
- 18. Goldstein J. L., Basu S. K., Brown M. S. (1983) Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98, 241–260 [DOI] [PubMed] [Google Scholar]
- 19. DeBose-Boyd R. A., Brown M. S., Li W. P., Nohturfft A., Goldstein J. L., Espenshade P. J. (1999) Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99, 703–712 [DOI] [PubMed] [Google Scholar]
- 20. Mosley S. T., Brown M. S., Anderson R. G., Goldstein J. L. (1983) Mutant clone of Chinese hamster ovary cells lacking 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem. 258, 13875–13881 [PubMed] [Google Scholar]
- 21. Rawson R. B., DeBose-Boyd R., Goldstein J. L., Brown M. S. (1999) Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 274, 28549–28556 [DOI] [PubMed] [Google Scholar]
- 22. Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., Brown M. S. (1983) Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase and its mRNA in rat liver as studied with a monoclonal antibody and a cDNA probe. J. Biol. Chem. 258, 8450–8455 [PubMed] [Google Scholar]
- 23. Sakai J., Nohturfft A., Cheng D., Ho Y. K., Brown M. S., Goldstein J. L. (1997) Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 272, 20213–20221 [DOI] [PubMed] [Google Scholar]
- 24. Diao A., Rahman D., Pappin D. J., Lucocq J., Lowe M. (2003) The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 160, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jo Y., Hartman I. Z., DeBose-Boyd R. A. (2013) Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol. Biol. Cell 24, 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. (1995) Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131, 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ou W. J., Bergeron J. J., Li Y., Kang C. Y., Thomas D. Y. (1995) Conformational changes induced in the endoplasmic reticulum luminal domain of calnexin by Mg-ATP and Ca2+. J. Biol. Chem. 270, 18051–18059 [DOI] [PubMed] [Google Scholar]
- 28. Nohturfft A., Brown M. S., Goldstein J. L. (1998) Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. J. Biol. Chem. 273, 17243–17250 [DOI] [PubMed] [Google Scholar]
- 29. Roitelman J., Masson D., Avner R., Ammon-Zufferey C., Perez A., Guyon-Gellin Y., Bentzen C. L., Niesor E. J. (2004) Apomine, a novel hypocholesterolemic agent, accelerates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase and stimulates low density lipoprotein receptor activity. J. Biol. Chem. 279, 6465–6473 [DOI] [PubMed] [Google Scholar]
- 30. Nguyen A. D., Lee S. H., DeBose-Boyd R. A. (2009) Insig-mediated, sterol-accelerated degradation of the membrane domain of hamster 3-hydroxy-3-methylglutaryl-coenzyme A reductase in insect cells. J. Biol. Chem. 284, 26778–26788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Braun B. C., Glickman M., Kraft R., Dahlmann B., Kloetzel P. M., Finley D., Schmidt M. (1999) The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1, 221–226 [DOI] [PubMed] [Google Scholar]
- 32. Pietroni P., Vasisht N., Cook J. P., Roberts D. M., Lord J. M., Hartmann-Petersen R., Roberts L. M., Spooner R. A. (2013) The proteasome cap RPT5/Rpt5p subunit prevents aggregation of unfolded ricin A chain. Biochem. J. 453, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Q., Liu Y., Soetandyo N., Baek K., Hegde R., Ye Y. (2011) A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell 42, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tomko R. J., Jr., Hochstrasser M. (2013) Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 82, 415–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee R. J., Liu C. W., Harty C., McCracken A. A., Latterich M., Römisch K., DeMartino G. N., Thomas P. J., Brodsky J. L. (2004) Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. EMBO J. 23, 2206–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wahlman J., DeMartino G. N., Skach W. R., Bulleid N. J., Brodsky J. L., Johnson A. E. (2007) Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell 129, 943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lipson C., Alalouf G., Bajorek M., Rabinovich E., Atir-Lande A., Glickman M., Bar-Nun S. (2008) A proteasomal ATPase contributes to dislocation of endoplasmic reticulum-associated degradation (ERAD) substrates. J. Biol. Chem. 283, 7166–7175 [DOI] [PubMed] [Google Scholar]
- 38. Mayer T. U., Braun T., Jentsch S. (1998) Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J. 17, 3251–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walter J., Urban J., Volkwein C., Sommer T. (2001) Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 20, 3124–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elkabetz Y., Shapira I., Rabinovich E., Bar-Nun S. (2004) Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates: roles of endoplasmic reticulum-bound p97/Cdc48p and proteasome. J. Biol. Chem. 279, 3980–3989 [DOI] [PubMed] [Google Scholar]
- 41. Huppa J. B., Ploegh H. L. (1997) The α chain of the T cell antigen receptor is degraded in the cytosol. Immunity 7, 113–122 [DOI] [PubMed] [Google Scholar]
- 42. Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. (1996) The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84, 769–779 [DOI] [PubMed] [Google Scholar]
- 43. Wiertz E. J., Tortorella D., Bogyo M., Yu J., Mothes W., Jones T. R., Rapoport T. A., Ploegh H. L. (1996) Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432–438 [DOI] [PubMed] [Google Scholar]
- 44. VanSlyke J. K., Deschenes S. M., Musil L. S. (2000) Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol. Biol. Cell 11, 1933–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garza R. M., Sato B. K., Hampton R. Y. (2009) In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J. Biol. Chem. 284, 14710–14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiong X., Chong E., Skach W. R. (1999) Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J. Biol. Chem. 274, 2616–2624 [DOI] [PubMed] [Google Scholar]
- 47. Faulkner R. A., Nguyen A. D., Jo Y., DeBose-Boyd R. A. (2013) Lipid-regulated degradation of HMG-CoA reductase and Insig-1 through distinct mechanisms in insect cells. J. Lipid Res. 54, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]