Background: Ubiquitin-modified TDP-43 protein aggregates characterize common neurodegenerative diseases.
Results: UBE2E ubiquitin-conjugating enzymes and ubiquitin isopeptidase Y (UBPY) are functional interactors of TDP-43 in cell culture and fly models.
Conclusion: Specific regulators of TDP-43 ubiquitination influence its aggregation and neurotoxic properties.
Significance: UBE2E and UBPY enzymes may modulate the course of TDP-43 diseases.
Keywords: Amyotrophic Lateral Sclerosis (ALS) (Lou Gehrig Disease), Drosophila, Proteasome, Protein Aggregation, Protein-Protein Interaction, TDP-43, Frontotemporal Dementia, Ubiquitin-conjugating Enzyme, Ubiquitin-specific Protease, Ubiquitination
Abstract
Trans-activation element DNA-binding protein of 43 kDa (TDP-43) characterizes insoluble protein aggregates in distinct subtypes of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. TDP-43 mediates many RNA processing steps within distinct protein complexes. Here we identify novel TDP-43 protein interactors found in a yeast two-hybrid screen using an adult human brain cDNA library. We confirmed the TDP-43 interaction of seven hits by co-immunoprecipitation and assessed their co-localization in HEK293E cells. As pathological TDP-43 is ubiquitinated, we focused on the ubiquitin-conjugating enzyme UBE2E3 and the ubiquitin isopeptidase Y (UBPY). When cells were treated with proteasome inhibitor, ubiquitinated and insoluble TDP-43 species accumulated. All three UBE2E family members could enhance the ubiquitination of TDP-43, whereas catalytically inactive UBE2E3C145S was much less efficient. Conversely, silencing of UBE2E3 reduced TDP-43 ubiquitination. We examined 15 of the 48 known disease-associated TDP-43 mutants and found that one was excessively ubiquitinated. This strong TDP-43K263E ubiquitination was further enhanced by proteasomal inhibition as well as UBE2E3 expression. Conversely, UBE2E3 silencing and expression of UBPY reduced TDP-43K263E ubiquitination. Moreover, wild-type but not active site mutant UBPY reduced ubiquitination of TDP-43 C-terminal fragments and of a nuclear import-impaired mutant. In Drosophila melanogaster, UBPY silencing enhanced neurodegenerative TDP-43 phenotypes and the accumulation of insoluble high molecular weight TDP-43 and ubiquitin species. Thus, UBE2E3 and UBPY participate in the regulation of TDP-43 ubiquitination, solubility, and neurodegeneration.
Introduction
Trans-activation element DNA-binding protein of 43 kDa (TDP-43)4 and fused-in-sarcoma (FUS) are neuropathologically and genetically linked to a neurological disease spectrum comprising frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS) (1, 2). These two disease-linked nucleic acid-binding proteins have structural and functional similarities (3). Specifically, TDP-43 binds to nucleic acids, including thousands of RNA species, thereby regulating transcription, splicing, and processing of mRNA and miRNAs as well as mRNA stability and transport. Moreover, TDP-43 can be incorporated into stress granules (SGs), pointing also to translational control functions. In human neurodegenerative diseases, TDP-43 shifts from predominantly nuclear to cytosolic cellular compartments. Characteristic intracellular inclusions are formed sometimes in the nucleus and conspicuously in the cytosol. Biochemically, TDP-43 in human diseases forms insoluble protein aggregates. Disease-associated TDP-43 modifications include phosphorylation, proteolytic formation of C-terminal fragments (CTFs), and ubiquitination (4). Candidate kinases and proteases have been proposed for the former, but the molecular mechanisms regulating TDP-43 ubiquitination and turnover are not well understood.
The accumulation and aggregation of neurotoxic proteins depends on the balance among synthesis, folding, and degradation pathways. The protein expression of TDP-43 is tightly regulated by binding to its own mRNA within the 3′-UTR, which leads to alternative splicing of polyadenylation sites and thus effective TDP-43 mRNA destabilization (5, 6). As for TDP-43 degradation, controversial reports in the literature point to proteasomal breakdown (7–10), autophagy (11, 12), or both (13–16). In a recent report, the ubiquitin ligase parkin was described to ubiquitinate TDP-43 in a complex manner, regulating subcellular TDP-43 transport (17).
TDP-43 exerts its diverse functions within multiple complexes. The first published interaction partner of TDP-43 was the survival of motoneuron (SMN) protein (18). This was corroborated by co-localization in distinct nuclear bodies (18–20). Outside the nucleus, there is a functional interaction of TDP-43 and SMN in axonal RNA transport granules in neuronal cell cultures (21). Here TDP-43 may participate in complexes containing SMN and other RNA-binding proteins such as fragile X mental retardation protein and Staufen (21, 22), pointing to functional roles in axonal mRNA transport and function.
Following a functional and structural rationale of TDP-43 being a heteronuclear ribonucleoprotein (hnRNP), Buratti et al. (23) found that TDP-43 interacts with hnRNP A/B. Such hnRNP interactions are mediated by sequences within the C-terminal glycine-rich domain of TDP-43 (23, 24). This initial finding was confirmed in subsequent proteomic screens of TDP-43 interactors (25, 26), establishing that TDP-43 interacts with many hnRNPs, thus contributing to hnRNP complexes involved in pre-mRNA splicing and other RNA processing steps (27).
In addition to the cluster of TDP-43-interacting proteins in hnRNP complexes, there is a cluster of TDP-43-interacting proteins involved in translation (25, 26). Beyond ribosomal proteins and core translation initiation factors, a number of proteins associated with SGs were found and confirmed to interact with TDP-43 (25, 28). In addition, TDP-43 interacts with FUS (26, 29), suggesting a functional interplay between TDP-43 and FUS, both of which are also recruited into SGs. Several components of the miRNA processing complexes Drosha and Dicer interacts with TDP-43 (26, 30), consistent with an influence of TDP-43 on the biogenesis of select miRNAs (30, 31).
Finally, TDP-43 interacts with itself either as a functional dimer (32) or potentially regulating self-aggregation via the C terminus (33). The TDP-43 C terminus harbors glutamine/asparagine-rich sequences with prion-like properties that confer self-interaction and co-aggregation with expanded poly(Q) proteins (34). Upon oxidative stress, TDP-43 may self-cross-link by disulfide bond formation (35). These latter processes might lead to TDP-43 aggregate sequestration and ultimately both loss of vital nuclear functions and/or proteotoxicity relevant to neurodegeneration.
To find novel protein interactors of TDP-43, we performed yeast two-hybrid (Y2H) screening with an adult brain cDNA library. We used as bait a CTF(193–414). This construct starts with the second RNA recognition motif and contains the glycine-rich domain as another presumed protein interaction domain. Synthetic CTF(193–414) partially mislocalized to the cytosol where it formed mildly insoluble aggregates (36). We discovered 10 potential TDP-43 protein-binding targets of which we could clone seven as complete cDNAs. Physical TDP-43 interactions were confirmed by co-immunoprecipitation and co-localization in human embryonic kidney HEK293E cells. Here we focus on the functional validation of enzymes involved in ubiquitin modifications: the ubiquitin-conjugating enzyme UBE2E3 (UbcH9) and the ubiquitin isopeptidase Y (UBPY) (USP8). UBE2E enzymes enhanced and UBPY decreased the ubiquitination of TDP-43. Proteasome inhibition also enhanced TDP-43 ubiquitination and shifted TDP-43 more into the insoluble protein fractions. In vivo functional interactions of TDP-43 with UBPY were confirmed in a Drosophila melanogaster model of TDP-43 neurotoxicity (37).
EXPERIMENTAL PROCEDURES
Constructs
FLAG-tagged, FL, and nuclear localization sequence (NLS)-mutated TDP-43 and polyglutamine-expanded ataxin-3 (Atx3-Q148-EGFP) constructs were described before (19). TDP-43 CTF (aa 193–414) was cloned into pcDNA3.1(−) with a 5′FLAG tag (NotI/EcoRI) via BamHI/HindIII. TDP-43 FL and CTF were subcloned into pcDNA3.1(−) with an mCherry tag (NotI/EcoRI) via BamHI/HindIII. CTF2 was subcloned into pEGFP-C1 via BamHI/HindIII. Furthermore, TDP-43 FL and CTF were inserted into pGADT7 (BamHI/NotI) and pGBKT7 (BamHI/XhoI) (Clontech). TDP-43K263E was generated by two-step site-directed mutagenesis of wild-type (WT) TDP-43 and cloned into pcDNA3.1(−) with a 5′-FLAG tag via BamHI/HindIII.
The LSM6, MED6, RBM45, RNF2, UBE2E3, and UBPY expression constructs were cloned from HEK293E cDNA with KpnI/BamHI forward and NotI backward primers into pcDNA3.1(+) (Invitrogen) and shuttled into pGADT7 (BamHI/XhoI), pCMV-Myc or -HA (BamHI/NotI), and pCMV-vector in which the Myc tag was changed to a triple FLAG tag (pCMV-FLAG) via Bsp120I/EcoRI. RACK1 was also amplified from HEK293E cDNA, cloned into pCMV-Myc (SfiI/SalI), and shuttled into pGADT7 via SfiI/SalI, and the vector was digested with SfiI/XhoI.
The E2 enzyme constructs UBE2E1, UBE2E2, UBE2N, and UBE2C were generated from HEK293E cDNA and inserted into pCMV-Myc via BglII/NotI. UBE2E3C145S and UBPYC786S were generated by two-step site-directed mutagenesis of UBE2E3 and UBPY constructs, respectively. UBE2E3 and UBPY point mutants as well as the truncated UBPY ΔC (aa 1–734) amplification products were cloned into pCMV-Myc (BglII/NotI). The pCMV-His6-ubiquitin expression construct was described before (38). Primer sequences are available on request.
Antibodies
The following antibodies were used for Western blot analysis (WB) or immunofluorescence staining (IF): mouse anti-α-tubulin (WB, 1:10,000; Sigma, clone B512), mouse anti-EEA1 (IF, 1:100; BD Transduction Laboratories, clone 14/EEA1), mouse anti-FLAG (IF, 1:500; Sigma, clone M2, affinity-purified), rabbit anti-FLAG (IF, 1:500; Cell Signaling Technology); peroxidase-conjugated anti-FLAG (WB, 1:1000–30,000; Sigma, clone M2), mouse anti-GAPDH (WB, 1:50,000; Biodesign International, clone 6C5), mouse anti-GM130 (IF, 1:500; BD Transduction Laboratories, clone 35/GM130), peroxidase-conjugated anti-HA (WB, 1:10,000; Roche Applied Science, clone 3F10), mouse anti-His6 (WB, 1:3000; Amersham Biosciences), rabbit anti-LC3B (WB, 1:1000; Cell Signaling Technology); rabbit anti-Living Colors DsRed (WB, 1:2000; Takara/Clontech); mouse anti-Myc (IF, 1:500; Sigma, clone 9E10), peroxidase-conjugated anti-Myc (WB, 1:10,000; Sigma, clone 9E10), rabbit anti-TDP-43 (WB, 1:2000; IF, 1:1000; ProteinTech Group), mouse anti-TDP-43 (WB, 1:2000; IF, 1:1000; Abnova, clone 2E2-D3), rabbit anti-UBE2E1 (WB, 1:4000; IF, 1:500; Abcam), rabbit anti-UBE2E2 (WB, 1:4000; IF, 1:500; Aviva Systems Biology), mouse anti-UBE2E3 (WB, 1:2000; Millipore, clone MABS17), mouse anti-UBE2E3 (IF, 1:150–500; OriGene, clone 7E8), mouse anti-ubiquitin (mono- and polyubiquitin; WB, 1:4000; Millipore MAB1510, clone Ubi-1), mouse anti-ubiquitin (mono- and polyubiquitin; WB, 1:500; Sigma UO508, clone 6C1), mouse anti-polyubiquitin (WB, 1:500; Biomol, clone FK1), and rabbit anti-UBPY (WB, 1:1000; IF, 1:500; Bethyl Laboratories). The secondary HRP-conjugated antibodies for WB were from Jackson ImmunoResearch Laboratories (1:15,000), and secondary Alexa Fluor 488- or 568-conjugated antibodies for IF were from Invitrogen (1:2000).
Y2H Screen
A human adult brain cDNA library (Mate & PlateTM Library) containing partial cDNAs was used for screening for TDP-43 CTF interactors with the Matchmaker Gold Yeast Two-Hybrid System (both Clontech). The cDNA library plasmids were already transformed into Saccharomyces cerevisiae strain Y187 and were mated with S. cerevisiae strain Y2HGold expressing TDP-43 CTF as bait. Mating efficiency was 5%, and ∼5.76 × 106 cDNA clones were screened. This represents 1.8-fold coverage of the library. The screen was performed on −LTHA + 2.5 mm 3-AT (−Leu/−Trp/−His/−Ade + 2.5 mm 3-amino-1,2,4-triazole (Sigma)) selective medium. Yeast colonies grown after 7–11 days (size, >1 mm) at 30 °C were seen as primary positive clones. These were selected for growth on −LT (−Leu/−Trp), −LTHA + 2.5 mm 3-AT, and −LTHA + X + AbA (α-X-Gal + 80 ng of aureobasidin) selective media. Additionally, yeast extracts of individual clones were prepared for WB to test for expression of HA-tagged prey proteins. Clones growing on all three selective media and expressing prey protein larger than 25 kDa (size of HA-tagged activation domain, approximately 22 kDa) were chosen for further study.
Individual clones were grown in liquid −Leu media to select for cDNA library plasmids, which were isolated as described (39). Plasmids were retransformed into Escherichia coli and reisolated to obtain pure cDNA library plasmids.
Purified library plasmids were co-transformed using a lithium acetate/single-stranded carrier DNA/polyethylene glycol method into yeast strain Y2HGold with bait vector pGBKT7-Ø, TDP-43, or CTF (retransformation). 6 × 104 yeast cells were spotted in duplicates on selective medium plates with increasing stringency. Yeast clones expressing prey protein and TDP-43 FL or CTF, not control vector, grown at least on −LTHA + 2.5 mm 3-AT were seen as true positive clones, and cDNA library plasmids were sequenced for identification.
Cell Culture, Transfections, and Proteasomal or Autophagosomal Inhibition
HEK293E cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FCS at 37 °C in 5% CO2. Transient transfections with DNA were performed with FuGENE 6 or X-tremeGENE 9 (Roche Applied Science) following the manufacturer's instructions. Transient silencing of UBE2E3 was performed with siGENOME siRNA (Thermo Scientific; target sequence, 5′-GCAUAGCCACUCAGUAUUU-3′). A scrambled siRNA was used as a control (Qiagen). HEK293E cells were transfected thrice in 72 h with 5 nm siRNA using HiPerfect (Qiagen) following the manufacturer's instructions. HEK293E cells were treated for the indicated time points with 10 μm MG-132 to inhibit proteasomal activity and/or with 20 nm bafilomycin A1 or 5 mm 3-methyladenine to inhibit basal autophagy (all Sigma-Aldrich).
Sequential Extraction of HEK293E Proteins
HEK293E cells were lysed in RIPA buffer (50 mm Tris/HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 10 mm NaPPi) and pelleted at 14,000 × g for 15 min at 4 °C. RIPA buffer-insoluble pellets were washed with RIPA buffer and lysed in 8 m urea buffer (10 mm Tris, pH 8.0, 100 mm NaH2PO4, 8 m urea). Total protein concentration of RIPA lysates was determined with the bicinchoninic acid (BCA) protein assay kit (Pierce). Total protein concentration of urea lysates was determined with the Bradford assay (Bio-Rad). 10 μg of protein was boiled in 1× Laemmli buffer at 95 °C for 10 min and subjected to WB.
Immunoprecipitation
HEK293E cells lysed in co-immunoprecipitation buffer (50 mm HEPES, pH 7.6, 10 mm KCl, 50 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 1.5 mm MgCl2, 10 mm NaPPi, 10% glycerol, 0.2% Nonidet P-40) containing 1× Complete protease inhibitors (Roche Applied Science) for 30 min on ice. Cell debris was removed by centrifugation at 14,000 × g at 4 °C for 15 min. Protein concentration in the supernatant was measured using the BCA protein assay kit, and 750 μg of such total protein lysate was incubated with EZviewTM Red anti-HA or anti-FLAG affinity gel (Sigma) for 3 h or over night at 4 °C. Beads were washed three times with co-immunoprecipitation buffer, and proteins were eluted in 2× Laemmli buffer at 95 °C. 10 μg of total protein lysates or one-fifth of eluated protein was subjected to WB.
Pulldown of Ubiquitinated Proteins
HEK293E cells were co-transfected with the indicated constructs and His6-ubiquitin for 24 or 48 h. Cells were washed with PBS and lysed with 8 m urea buffer containing 10 mm imidazole. The DNA was sheared by passing the lysate 20 times through a 23-gauge needle. Cell debris was pelleted at 14,000 × g for 15min at 4 °C, and protein concentration was determined with the Bradford assay (Bio-Rad protein assay). Total protein (750 μg) was incubated with Ni-NTA-agarose beads (Qiagen) for 4 h at room temperature. Beads were washed thrice with urea wash buffer (10 mm Tris, pH 6.3, 100 mm NaH2PO4, 8 m urea) containing 20 mm imidazole, and proteins were eluted in 3× Laemmli buffer with 500 mm imidazole at 95 °C for 10 min. Total protein and one-fifth of eluates were analyzed by WB.
Pulldown of Sequentially Extracted Ubiquitinated Proteins
HEK293E cells co-transfected with the indicated proteins and His6-ubiquitin were lysed under native conditions in Nonidet P-40 lysis buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 1% Nonidet P-40) containing 1× Complete protease inhibitors and 10 mm imidazole for 30 min on ice and pelleted at 14,000 × g for 15 min at 4 °C. Total protein concentration of the Nonidet P-40-soluble fraction was determined with the BCA protein assay kit. Nonidet P-40-insoluble pellets were washed twice with Nonidet P-40 buffer and lysed in 8 m urea buffer. The Bradford assay (Bio-Rad) was used to determine the protein concentration of the urea-soluble fraction. Pulldown with Ni-NTA-agarose beads was performed with 600 μg of Nonidet P-40 and 150 μg of urea lysates for 4 h at 4 °C (Nonidet P-40 lysates) or room temperature (urea lysates). Beads were washed thrice with either Nonidet P-40 buffer or urea wash buffer containing 20 mm imidazole, and proteins were eluted with 500 mm imidazole at 95 °C for 10 min. Total proteins or one-fifth of eluates was subjected to WB.
Western Blot Analysis
Denatured protein was separated on either 7.5, 10, 12.5, or 15% polyacrylamide gels or 4–12% Bis-Tris NuPAGE gradient gels (Invitrogen) and transferred onto Hybond-P polyvinylidene difluoride membrane (Millipore). Membranes were blocked in 5% skim-milk in TBS-Tween 20 or 5% BSA in TBS-Tween 20 and incubated with primary antibody in Western Blocking Reagent (Roche Applied Science) at 4 °C overnight. This was followed by incubation with HRP-conjugated secondary antibodies for 1 h at room temperature. Detection of proteins was performed with the Immobilon Western chemiluminescent HRP substrate (Millipore) on Amersham Biosciences HyperfilmTM ECL (GE Healthcare).
Immunofluorescence
HEK293E cells were seeded on poly-d-lysine- (Sigma) and collagen (Cohesion)-coated coverslips and fixed with 4% (w/v) paraformaldehyde in PBS for 20 min, permeabilized with 1% Triton X-100 in PBS for 5 min, and blocked for 1 h with 10% normal goat serum. Primary antibody incubation was performed in 1% BSA in PBS for 2 h at room temperature or overnight at 4 °C. Cells were incubated with secondary Alexa Fluor-conjugated antibodies in the dark for 1 h at room temperature. Nuclei were counterstained with Hoechst 33342 (2 μg/ml in PBS) for 10 min at room temperature, and coverslips were mounted in fluorescent mounting medium (Dako) onto microscope slides. Cells were analyzed with an ApoTome imaging system and processed with AxioVision software (Zeiss).
Fly Stocks
All D. melanogaster stocks were maintained on standard cornmeal-yeast agar-based fly food. Experiments were performed at 25 °C. The driver lines w[*]; P{w[+mC]= GAL4-ninaE.GMR}12 (BL1104) (GMR-Gal4 in text) and y[1] w[*]; P{w[+mC]=Act5C-GAL4}25FO1/CyO, y[+] (BL4414) (actin-Gal4 in text) were obtained from the Bloomington Drosophila Stock Center. The RNAi lines UBPY (v107623), UbcD1 (v26011), and UbcD2 (v31158) were obtained from the Vienna Drosophila RNAi Center (Austria). The random inserted UAS:TDP-43 transgenic line was generated by germ line transmission (BestGene) and was characterized before (37). Stable TDP-43 expression in the retina was achieved by recombination of the GMR-Gal4 driver with UAS:TDP-43 line 14 insertion.
RNA Extraction and RT-PCR
RNA was isolated from third instar larvae with the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. 600 ng of total RNA was reverse transcribed with anchored oligo(dT) primer using the Transcriptor High Fidelity cDNA Synthesis kit (Roche Applied Science). cDNA was used as template for transcription amplification in a 25-μl reaction with 5 μl of 5× GoTaq buffer, 0.1 μl of GoTaq polymerase (Promega), and 2 mm primer (sequences are available on request). Amplified PCR products were subjected to electrophoresis using a 2% agarose gel stained with ethidium bromide. Quantification was performed with ImageJ software.
Sequential Extraction of Fly Head Proteins
Flies were decapitated, and heads were homogenized in RIPA buffer containing 1× Complete protease inhibitors. Cell debris was pelleted at 14,000 × g for 15 min at 4 °C to obtain the RIPA buffer-soluble lysate. RIPA buffer-insoluble cell pellets were washed twice with RIPA buffer and lysed with 8 m urea buffer. An equivalent of one fly head (RIPA buffer-soluble) or five fly heads (urea-soluble) was loaded per lane in WB.
RESULTS
Y2H Screen for TDP-43 Interactors
To identify novel, (patho)physiologically relevant interactors of the neurodegeneration-associated protein TDP-43, we performed a Y2H screen using an adult human brain cDNA library. Initial attempts with human TDP-43 FL as bait failed to show any meaningful positive clones. However, a CTF comprising amino acids 193–414 yielded 10 novel primary positive clones that were confirmed upon retransformation. These 10 positive clones contained partial sequences that allowed the identification of homologous whole sequences (Table 1). We managed to clone seven of these as complete cDNAs, namely LSM6, MED6, RACK1, RBM45, RNF2, UBE2E3, and UBPY, and examined their TDP-43 co-immunoprecipitation and co-localization in HEK293E cells.
TABLE 1.
Overview of positive Y2H hits with UniProt accession number, similarity of cDNA library with homologous sequence, size of proteins, and strength of interaction with TDP-43 FL and CTF upon retransformation
Targets cloned FL and confirmed here are shown in bold. Strength of interaction: −, none; +, weak; ++, moderate; +++, strong.
| Symbol | Name | Accession number (translated amino acids in Y2H) | Identities | Protein size | Homologous hit | Strength of interaction in Y2H with TDP-43 |
|
|---|---|---|---|---|---|---|---|
| FL | CTF | ||||||
| aa | |||||||
| BEX2 | Brain-expressed X-linked protein 2 | Q9BXY8 (aa 1–128) | 118/128 (92%) | 128 | − | +++ | |
| CTAGE5 | Cutaneous T-cell lymphoma-associated antigen 5 | O15320 (aa 442–615)a | 132/174 (75%) | 804 | − | +++ | |
| GPR137B | Integral membrane protein GPR137B | O60478 (aa 292–399) | 108/108 (100%) | 399 | + | +++ | |
| LSM6 | U6 snRNA-associated Sm-like protein LSm6 | P62312 (aa 62–80) | 19/19 (100%) | 80 | − | ++ | |
| cDNA FLJ57086 | cDNA FLJ57086, highly similar to RNA polymerase transcriptional regulation mediator, subunit 6 homolog | B4DTY4 (aa 1–128) | 128/128 (100%) | 128 | MED6 | − | +++ |
| MED6 | Mediator of RNA polymerase II transcription subunit 6 | O75586 (aa 1–121)b | 119/121 (98%) | 246 | |||
| RACK1 (GNB2L1) | Receptor for activated C kinase 1 | P63244 (aa 78–290)c | 213/213 (100%) | 317 | − | +++ | |
| RBM45 | RNA-binding protein 45 | Q8IUH3 (aa 59–284)c | 214/226 (94%) | 476 | − | +++ | |
| RNF2 | E3 ubiquitin-protein ligase RING2 | Q99496 (aa 276–336) | 61/61 (100%) | 336 | − | ++ | |
| UBE2E3 | Ubiquitin-conjugating enzyme E2 E3 | Q969T4 (aa 126–207) | 82/82 (100%) | 207 | UBE2E1, UBE2E2 | − | ++ |
| UBPY (USP8) | Ubiquitin isopeptidase Y | P40818 (aa 877–1088)b | 212/212 (100%) | 1118 | + | +++ | |
a cDNA sequence ends with stop codon, although reference sequence has later stop codon.
b MED6 was assumed to be a positive interactor because its first 121 amino acids share 98% homology with cDNA FLJ57086.
c Partial cDNA sequences from Y2H hits were not sequenced through to the stop codon.
Co-immunoprecipitation of TDP-43 Interactors
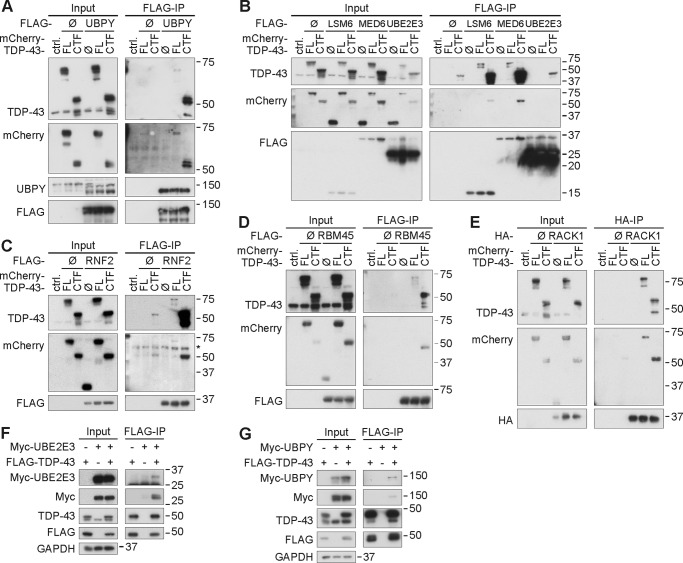
HEK293E cells were co-transfected with mCherry-tagged TDP-43 FL or CTF constructs along with each of the seven cloned Y2H interactor cDNAs. Most immunoprecipitates of the tagged interactor proteins were also immunoreactive for the mCherry-TDP-43 fusion proteins (Fig. 1, A–E). FLAG-RACK1 co-immunoprecipitated mCherry-tagged TDP-43 FL and CTF with similar strength (Fig. 1E). In the other cases, co-immunoprecipitation efficiency was higher for CTF, but some TDP-43 FL co-immunoprecipitation was detected for most targets, suggesting physiological interactions. Furthermore, co-immunoprecipitation of endogenous TDP-43 and 35-kDa fragment with FLAG-UBPY was also detected (see Fig. 6F). Compared with the other interactors, the co-immunoprecipitation strength of UBE2E3 was weaker (Fig. 1B). FLAG-UBE2E3 co-immunoprecipitation with mCherry-tagged TDP-43 FL could not be detected, and co-immunoprecipitation with CTF was only slightly above background. However, FLAG-tagged TDP-43 FL did co-immunoprecipitate Myc-UBE2E3 (Fig. 1F). In this same converse co-immunoprecipitation setup, the interaction of TDP-43 with UBPY was confirmed (Fig. 1G). As the demonstration of a somewhat unusual direct physical interaction of the E2 ubiquitin-conjugating enzyme UBE2E3 with its putative substrate protein TDP-43 requires special experimental conditions, binding could be weak or transient as a dynamic enzyme/substrate pair and/or require stabilization by cellular E3 ubiquitin ligases. The interaction is further complicated by the solubility shifts of TDP-43 in the presence of UBE2E3 (see below) that likely account for the reduced TDP-43 input levels in Fig. 1B. Despite these issues, the interaction of TDP-43 with UBE2E3 could be functionally meaningful (see below).
FIGURE 1.
Co-immunoprecipitation of TDP-43 with Y2H interactors. A–E, HEK293E cells were co-transfected with mCherry-TDP-43 FL, CTF, or control vector (ctrl.) along with control vectors (Ø), FLAG-tagged UBPY (A), LSM6, MED6, UBE2E3 (B), RNF2 (C), RBM45 (D), or HA-tagged RACK1 (E). Then cells were lysed and subjected to immunoprecipitation with anti-FLAG (A–D) or anti-HA (E). Western blotting was performed with total cell lysates (Input) and immunoprecipitates (FLAG-IP and HA-IP, respectively). Proteins were detected with antibodies against TDP-43; DsRed (mCherry), FLAG, or HA; and UBPY as indicated. The endogenous TDP-43 bands confirm even input loading. F and G, immunoprecipitation with anti-FLAG beads from HEK293E cell lysates co-expressing FLAG-TDP-43 FL and either Myc-UBE2E3 (F) or Myc-UBPY (G). Total protein (Input) and immunoprecipitated proteins (FLAG-IP) were subjected to WB with antibodies against UBE2E3, UBPY, Myc, TDP-43, FLAG, and GAPDH as a loading control.
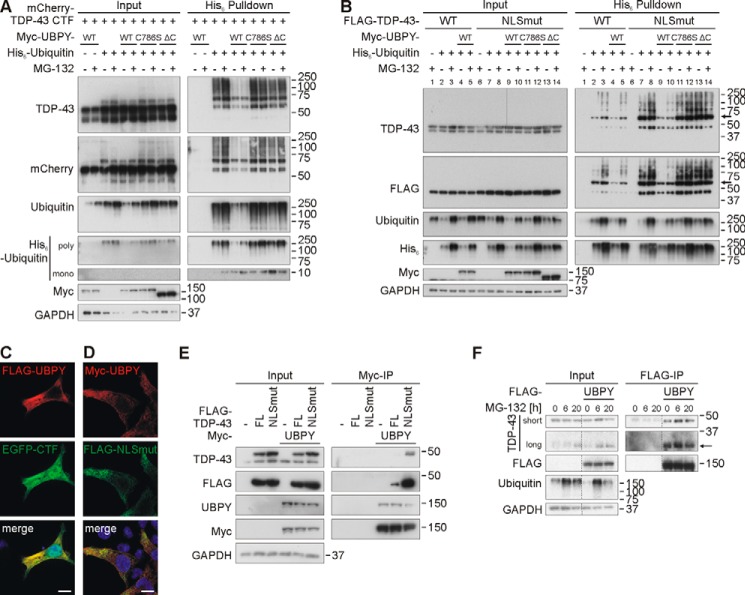
FIGURE 6.
Transfected UBPY deubiquitinates TDP-43 CTF and NLSmut. A, UBPY deubiquitinates mCherry-TDP-43 CTF independently of proteasomal status. mCherry-tagged TDP-43 was overexpressed with His6-ubiquitin and Myc-UBPY WT, Myc-UBPYC786S mutant, UPBY lacking the ubiquitin C-terminal hydrolase domain (ΔC), or vector controls in HEK293E cells for 48 h. After proteasomal inhibition with MG-132 (10 μm) for 2 h, cells were lysed, and Ni-NTA purification of His6-ubiquitin-conjugated proteins was performed. Total cell lysates (Input) and pulldown eluates (His6 Pulldown) were Western blotted and stained with antibodies against TDP-43, mCherry, ubiquitin, His6, Myc, and GAPDH. B, UBPY deubiquitinates cytosolically localized, NLS-mutated TDP-43. The experiment was performed as in A, but instead of mCherry-TDP-43 CTF, FLAG-TDP-43 WT or NLS-mutated TDP-43 (NLSmut) was overexpressed. Western blots were stained with antibodies against the indicated proteins. Monoubiquitinated TDP-43 is indicated by an arrow. C and D, immunostainings of HEK293E cells overexpressing FLAG-UBPY (red) and EGFP-TDP-43 CTF (green) (C) or Myc-UBPY (red) and cytosolic FLAG-TDP-43 NLSmut (green) (D). Nuclei were stained with Hoechst 33342 (blue). Merged images are on the right. Scale bars apply to all images and correspond to 10 μm. E, co-immunoprecipitation of overexpressed WT TDP-43 or NLS-mutated TDP-43 with UBPY. HEK293E cells were transfected with FLAG-TDP-43 (WT) or TDP-43 with a mutated NLS and Myc-UBPY. Cell lysates were subjected to immunoprecipitation for Myc-UBPY. Total cell lysates (Input) and (co-)immunoprecipitated proteins were subjected to Western blotting. Blots were sequentially stained with antibodies against TDP-43, FLAG, UBPY, Myc, and GAPDH. F, co-immunoprecipitation of endogenous TDP-43 FL and CTF with UBPY. HEK293E cells were transfected with FLAG-UBPY, and proteasomal inhibition was performed for the indicated time points starting 30–44 h post-transfection. Cell lysates were immunoprecipitated for FLAG-UBPY. Total cell lysates (Input) and (co-)immunoprecipitated proteins were analyzed by WB. Proteins were stained with antibodies against TDP-43, FLAG, mono- and polyubiquitin, and GAPDH. A thin arrow points to a 35-kDa TDP-43 fragment. IP, immunoprecipitate.
Co-localization of TDP-43 with Its Interactors
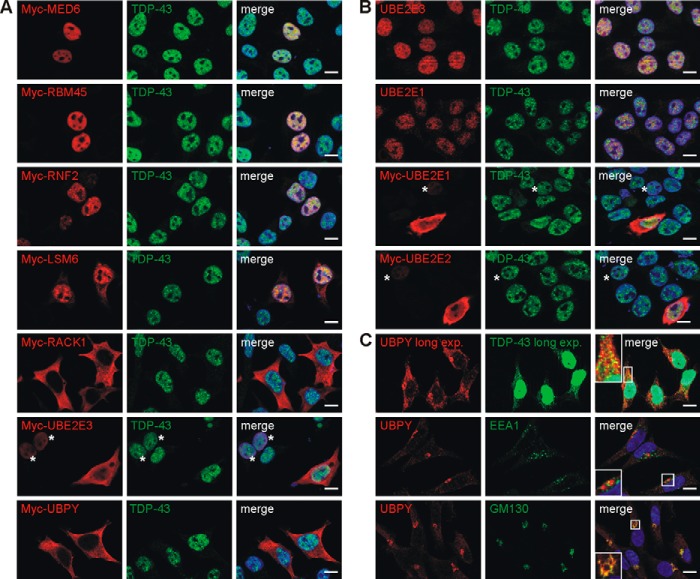
Under normal conditions, TDP-43 is quantitatively most abundant in the nucleus but can shuttle to the cytosol (40). To see where the novel interactions could take place, Myc-tagged constructs were transfected into HEK293E cells, and dual label immunofluorescence microscopy was performed with antibodies against the endogenous TDP-43 (Fig. 2A). As expected, most of the endogenous TDP-43 was found in the nucleus, but granular TDP-43-immunoreactive material was observed in the cytosol upon longer exposure (see Fig. 2C). MED6, RBM45, and RNF2 were mostly nuclear very similar to TDP-43. LSM6 partitioned to the nucleus and cytosol with strong overlap in the nucleus. RACK1 was distributed to cytosolic and membrane compartments with apparently little co-immunostaining. Finally, overexpressed UBPY was exclusively localized to the cytoplasm, whereas UBE2E3 localization depended on the extent of overexpression. In moderately expressing cells, Myc-UBE2E3 signal was always nuclear (Fig. 2A, UBE2E3, *) where it co-localized with TDP-43. Strongly overexpressing cells showed in addition to the “normal” nuclear staining additional Myc-UBE2E3 signal in the cytoplasm (Fig. 2A). Co-expression of FLAG- or EGFP-tagged TDP-43 FL with the interactor proteins yielded the same results, and the subcellular localization of the targets did not appreciably change upon co-expression of EGFP-CTF (data not shown).
FIGURE 2.
TDP-43 co-localization with its interactors. A, HEK293E cells were dual labeled with rabbit antibody against endogenous TDP-43 (green) and the indicated Myc-tagged interactors (mouse anti-Myc, red). B, non-transfected HEK293E cells or those transfected with each of the indicated Myc-tagged UBE2E enzyme were stained with mouse anti-TDP-43 (green) and rabbit anti-UBE2E1 or rabbit anti-UBE2E2 (red) and with rabbit anti-TDP-43 (green) and mouse anti-UBE2E3 (red) as indicated. C, non-transfected HEK293E cells were stained with mouse anti-TDP-43 (green) and rabbit anti-UBPY (red). TDP-43 signal was overexposed in TDP-43/UBPY co-staining to visualize cytoplasmic TDP-43 (inset). Lower panels, HEK293E were stained for endogenous UBPY with rabbit anti-UBPY (red) and for early endosomal marker EEA1 or cis-Golgi marker GM130 (green), respectively. A–C, merged images, including nuclear counterstaining with Hoechst 33342 (blue), are to the right. Asterisks (*) label moderately Myc-UBE2E-expressing cells. Scale bars correspond to 10 μm. exp., exposure.
We detected the endogenous proteins UBE2E3 and UBPY with selective antibodies. Similar to TDP-43, endogenous UBE2E3 was predominantly detected in the nucleus as was the related UBE2E1 (Fig. 2B). Antibody against the third family member, UBE2E2, did not immunostain the endogenous protein. When overexpressed, all three Myc-tagged UBE2E proteins showed in addition to the normal nuclear staining a large proportion of cytosolic localization (Fig. 2B). Endogenous UBPY was found in the cytosol as was the transfected Myc-UBPY (Fig. 2C). The cytosolic endogenous UBPY in HEK293E cells was enriched in the Golgi apparatus where it co-localized with the marker GM130 (Fig. 2C). In addition, UBPY was present in vesicular compartments (Fig. 2C), consistent with its role in endocytic sorting (41). Although separate UPBY and TDP-43 vesicular and granular structures were abundant (after long exposure to visualize cytosolic TDP-43), a considerable fraction showed co-localization of UBPY and TDP-43 (Fig. 2C). The cytosolic interactions of TDP-43 may not be quantitatively abundant and/or may be transient but could reflect functionally relevant interactions in cells.
Potential Functional Implications of the Novel TDP-43 Interactors
Some of the hits (Table 1) are noteworthy. For example, the transmembrane protein GPR137B transcript was divergently regulated after TDP-43 knockdown (27). Several interactors point to functions in RNA processing. The interaction of TDP-43 with LSM6 could indicate a role in LSm complexes, a group of RNA processing particles, both in the nucleus and cytosol (42). The developmentally regulated RNA-binding protein RBM45 was very recently reported to co-aggregate with TDP-43 in ALS/FTLD (43). Of particular interest is RACK1 as it has roles in neurite outgrowth and translational control in SGs (44, 45), two cellular functions potentially affected by TDP-43. RACK1 has been found in a proteomic TDP-43 interactor screen (25) and associated with TDP-43 in a global protein interaction map of D. melanogaster (46). In addition, the interaction with UBPY is apparent in that interaction map (46). Moreover, RACK1 was found in a co-aggregation proteome along with several RNA-binding proteins implicated with SGs and translation control in a cell culture model of TDP-43 proteinopathy (47).
UBPY is a deubiquitinating enzyme (DUB) best known for its regulation of receptor endocytic sorting (41). TDP-43 could be a novel substrate for UBPY, potentially regulating TDP-43 turnover and cellular functions by controlling its ubiquitination state. Conversely, UBE2E3 is an E2 ubiquitin-conjugating enzyme, and RNF2 is an E3 ubiquitin ligase. Thus, these three enzymes could regulate TDP-43 ubiquitinations in both directions. Here we specifically validated the influence of UBE2E3 and UBPY on TDP-43 ubiquitination. For this purpose, we first established a cellular TDP-43 ubiquitination assay.
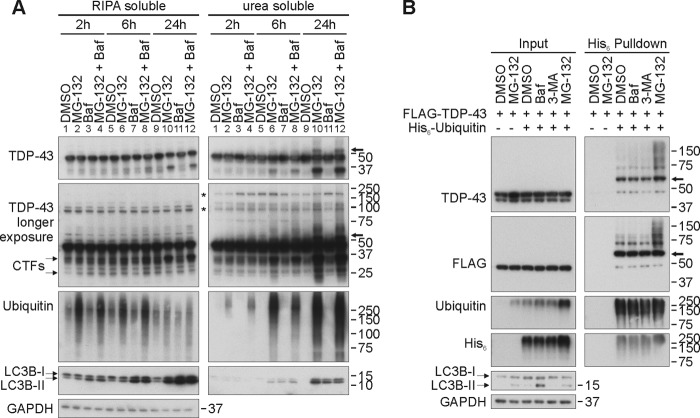
When assessing the contribution of proteasomal and autophagic degradation pathways for TDP-43 using pharmacological inhibitors in HEK293E cells, we found hardly any accumulation of TDP-43 within a 1-day observation period by either treatment with the proteasome inhibitor MG-132, the lysosomal inhibitor bafilomycin A1, or the autophagy inhibitor 3-methyladenine (Fig. 3). Thus, TDP-43 is a relatively long lived protein with apparently slow turnover. Eventually TDP-43 may be a target of microautophagy and in larger aggregates subject to macroautophagy, but this is not occurring within the time frame of the present experimental setup. In marked contrast, the distinct 35- and 25-kDa endogenous CTFs dramatically accumulated in an insoluble fraction upon proteasome inhibition for 6–24 h but not after lysosomal inhibition (Fig. 3A). Inhibiting both degradation pathways had no synergistic effect. Thus, CTFs are rapidly turned over predominantly by the proteasome at least in this cell culture model.
FIGURE 3.
TDP-43 is ubiquitinated upon proteasomal inhibition. A, HEK293E cells were treated with lysosomal inhibitor bafilomycin A1 (Baf; 20 nm), proteasomal inhibitor MG-132 (10 μm), both, or DMSO for 2, 6, or 24 h as depicted. Cells were subjected to sequential extraction with RIPA buffer and urea buffer. Lysates were Western blotted and probed with antibodies against TDP-43, ubiquitin, LC3, and GAPDH as a loading control. Asterisks (*) mark nonspecific bands. B, to confirm that the observed higher molecular smear is ubiquitinated TDP-43, HEK293E cells overexpressing FLAG-TDP-43 and His6-tagged ubiquitin or vector control (−) were treated for 3 h with MG-132, bafilomycin A1, autophagy inhibitor 3-methyladenine (3-MA; 5 mm), or DMSO. Ni-NTA purification for His6-tagged ubiquitin was performed with urea buffer-soluble protein lysates. Western blots of total protein lysates (Input) and purified Ni-NTA-agarose eluates (His6 Pulldown) were probed with antibodies against TDP-43, FLAG, ubiquitin, LC3, or GAPDH as indicated. Arrows point to putative monoubiquitinated TDP-43 (see text for details).
Interestingly, the accumulation of ubiquitinated TDP-43 species in proteasome-inhibited HEK293E cells was accompanied by a shift into more insoluble fractions (Fig. 3A). Thus, proteasome inhibition and the accompanying accumulation of insoluble ubiquitinated TDP-43 species provide a cellular assay for the validation of ubiquitinating enzymes and DUBs for TDP-43. For this purpose, HEK293E cells were transiently transfected with FLAG-TDP-43 and His6-ubiquitin. After treatment with inhibitors, cells were lysed harshly using 8 m urea, and His6-ubiquitin conjugates were pulled down with nickel beads. Specifically after treatment with the proteasome inhibitor MG-132, higher molecular mass smears of FLAG-TDP-43 appeared (Fig. 3B), suggesting TDP-43 ubiquitination. Note that ∼55-kDa TDP-43 bands were seen after MG-132 treatment in the insoluble fraction (Fig. 3A). Similar bands were observed in His6-ubiquitin pulldowns even without proteasomal inhibition (Fig. 3B). Their molecular mass is compatible with monoubiquitination (see below).
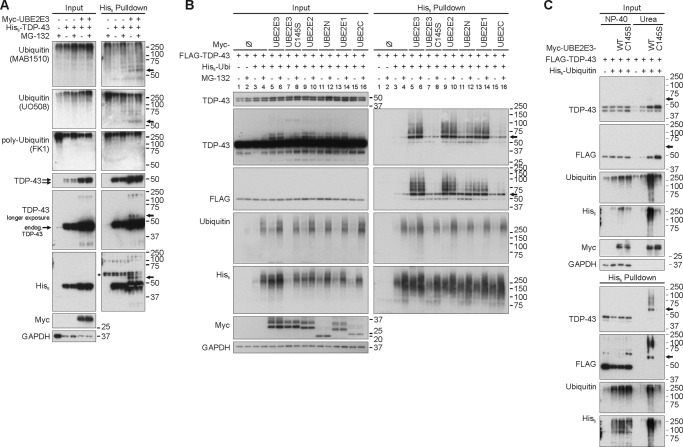
Regulation of TDP-43 Ubiquitination by UBE2E Enzymes
First we measured whether UBE2E3 could enhance TDP-43 ubiquitination. HEK293E cells were transiently transfected with His6-tagged TDP-43 and Myc-UBE2E3. Pulldowns from lysates obtained using harsh lysis conditions revealed ∼55-kDa TDP-43 bands specifically when UBE2E3 was co-transfected (Fig. 4A). The nature of such ∼55-kDa bands as monoubiquitinated TDP-43 was evidenced by immunoreactivity of the endogenous ubiquitin modifications with antibodies that recognize both mono- and polyubiquitin (MAB1510 and UO508) but not with an antibody that recognizes only polyubiquitin (FK1). Thus, ∼55-kDa TDP-43 bands are annotated by arrows as monoubiquitinated TDP-43 in the figures.
FIGURE 4.
UBE2E enzymes enhance TDP-43 ubiquitination and mediate insolubility. A, His6-TDP-43 (+) was co-expressed with Myc-UBE2E3 (+) or vector controls (−) in HEK293E cells in the presence of endogenous (endog.) ubiquitin. After proteasomal inhibition with MG-132 (10 μm) for 2 h, cells were lysed harshly, and Ni-NTA purification of His6-TDP-43-conjugated proteins was performed. Total cell lysates (Input) and pulldown eluates (His6 Pulldown) were Western blotted and probed for TDP-43, His6, Myc, and GAPDH. Mono- and polyubiquitin were detected with mouse anti-ubiquitin (MAB1510 and UO508), and polyubiquitinated proteins were detected with mouse anti-ubiquitin (clone FK1). B, HEK293E cells were triple transfected for 48 h with FLAG-TDP-43, His6-vector control (−), or His6-ubiquitin (His6-Ubi) (+) and Myc-tagged E2 enzyme UBE2E3, catalytically inactive UBE2E3C145S, UBE2E2, UBE2N, UBE2E1, or UBE2C or Myc-vector control (−). Prior to lysis with urea buffer, cells were treated with MG-132 (10 μm) or DMSO for 2 h. His6-ubiquitin-conjugated proteins were pulled down from cell lysates. Total protein lysates and Ni-NTA-agarose eluates were Western blotted and stained for TDP-43, FLAG, ubiquitin, His6, Myc-tagged E2 enzymes, and GAPDH. C, FLAG-TDP-43 was overexpressed with His6-vector control (−) or His6-ubiquitin (+) and Myc-vector control (−) or Myc-UBE2E3 WT or C145S in HEK293E cells. Proteasome was inhibited for 2 h, and sequential extraction for Nonidet P-40 (NP-40)-soluble and urea-soluble proteins was performed. His6-ubiquitin-conjugated proteins were isolated from both fractions, and Western blots of total cell lysates and pulldown eluates were stained for TPD-43, FLAG, ubiquitin, His6, Myc, and GAPDH. The amount of analyzed insoluble protein corresponded to approximately one-twentieth (5%) of soluble protein concentration. Arrows point to putative monoubiquitinated TDP-43. Ø, control vectors.
Effects on the formation of higher molecular mass (multiple mono- or poly-)ubiquitinated TDP-43 species were detected by pulldown of co-transfected His6-ubiquitin in HEK293E cells. Overexpression of Myc-UBE2E3 enhanced FLAG-TDP-43 ubiquitination (Fig. 4B, lanes 5 and 6). This effect depended on UBE2E3 catalytic activity as the active site mutant C145S failed to promote such TDP-43 ubiquitination (Fig. 4B, lanes 7 and 8). The closely related UBE2E1 and UBE2E2 had effects similar to UBE2E3 (Fig. 4B, lanes 13 and 14 and lanes 9 and 10, respectively). In contrast, the more distantly related ubiquitin-conjugating enzymes UBE2N and UBE2C promoted TDP-43 ubiquitination to a much lesser extent than did the UBE2E enzymes despite similar transfection levels (Fig. 4B, lanes 11 and 12 and lanes 15 and 16, respectively). Although MG-132 slightly enhanced the amounts of polyubiquitinated TDP-43 in the absence of exogenous E2 or in the presence of the weakly reactive UBE2N and UBE2C, the strong UBE2E-enhanced TDP-43 ubiquitinations were hardly stabilized by MG-132 treatment (Fig. 4). Thus, UBE2E-mediated TDP-43 ubiquitinations are not straightforward proteasome-targeting signals. Rather, these ubiquitinated TDP-43 species were shifted into insoluble fractions (Fig. 4C). Furthermore, the co-expression of UBE2E3 WT and C145S increased amounts of insoluble TDP-43.
Conversely, silencing of UBE2E3 by siRNA treatment reduced TDP-43 ubiquitinations stabilized by proteasome inhibition (Fig. 5A). Transfection of WT UBE2E3 rescued TDP-43 ubiquitination in siUBE2E3-treated HEK293E cells (Fig. 5B, compare lanes 8 and 10) in contrast to the inactive mutant UBE2E3C145S (Fig. 5B, lanes 14). This control experiment rules out off-target effects and emphasizes the importance of catalytic UBE2E3 activity for TDP-43 ubiquitination.
FIGURE 5.
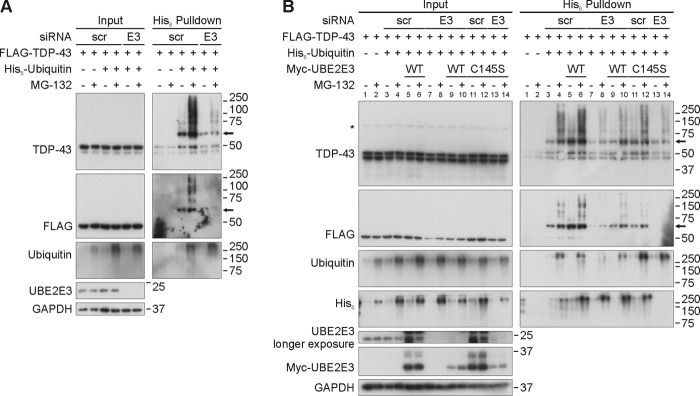
UBE2E3 silencing decreases ubiquitinated TDP-43 and can be rescued with UBE2E3 overexpression. A, silencing of UBE2E3 reduces ubiquitinated TDP-43 level. HEK293E cells were silenced three times with scrambled (scr) or UBE2E3-directed (E3) siRNA over 72 h. 4 h after the third silencing, cells were transfected with FLAG-TDP-43 and His6-ubiquitin or vector control (−). After 24 h of protein overexpression, proteasome was inhibited with MG-132 (10 μm) for 2 h, and urea-soluble cell lysates were prepared. Pulldown of His6-ubiquitin-conjugated proteins was performed, and total cell lysates (Input) and pulldown eluates (His6 Pulldown) were Western blotted and stained with antibodies against TDP-43, ubiquitin, His6, UBE2E3, and GAPDH as indicated. B, rescue of the effect of UBE2E3 silencing on TDP-43 ubiquitination with UBE2E3 overexpression. Silencing of UBE2E3 was performed as in A. 4 h after the third silencing, HEK293E cells were triple transfected with FLAG-TDP-43, His6-ubiquitin, or vector control and Myc-tagged UBE2E3 WT, catalytically inactive variant (C145S), or Myc-vector control. After 2 h of MG-132 treatment, cell lysates were prepared as in A, and His6-ubiquitin-conjugated proteins were purified with Ni-NTA-agarose. Western blots of total cell lysates and pulldown eluates were stained with antibodies against TDP-43, FLAG, ubiquitin, His6, UBE2E3, or GAPDH as indicated. An asterisk (*) labels an unspecific band, and an arrow points to monoubiquitinated FLAG-TDP-43.
Regulation of TDP-43 Ubiquitination by UBPY
To determine TDP-43 DUB activity of UBPY, we used two other TDP-43 constructs: mCherry-CTF that showed robust ubiquitination even in the absence of MG-132 (Fig. 6A) and a FLAG-TDP-43 with mutated nuclear localization sequence (NLSmut). In both cases, co-transfection with Myc-UBPY strongly reduced His6-ubiquitin pulldown of mCherry-CTF (Fig. 6A) and FLAG-TDP-43(NLSmut) (Fig. 6B, compare lanes 7 and 8 with lanes 9 and 10). In contrast, the catalytically inactive UBPYC786S point mutant (48) and a mutant in which we deleted the C terminus (ΔC) containing the ubiquitin C-terminal hydrolase domain of UBPY (49) both failed to decrease ubiquitination of mCherry-CTF (Fig. 6A) and FLAG-TDP-43(NLSmut) (Fig. 6B, lanes 11–14).
Note that UBPY is almost exclusively localized in the cytosol (see Fig. 2). It is likely that UBPY has better access to the cytosolically localized NLSmut TDP-43 and CTF proteins lacking the NLS. This view is supported by the extensive cytosolic co-localization of transiently co-transfected UBPY with EGFP-CTF and FLAG-TDP-43(NLSmut) in HEK293E cells (Fig. 6, C and D). Consistently, FLAG-TDP-43(NLSmut) showed much greater co-immunoprecipitation than WT FLAG-TDP-43 with Myc-UBPY (Fig. 6E). Likewise, the interaction with mCherry-CTF was stronger than with TDP-43 FL (see above). In addition, we analyzed whether Myc-UBPY binds endogenous CTFs that are stabilized by proteasomal inhibition. Indeed, we observed co-immunoprecipitation of endogenous CTF and full-length TDP-43 with overexpressed Myc-UBPY (Fig. 6F). The strength of co-immunoprecipitation was independent of the duration of MG-132 treatment and was even detected in the absence of inhibitor.
Ubiquitination of Pathogenic TDP-43 Mutants
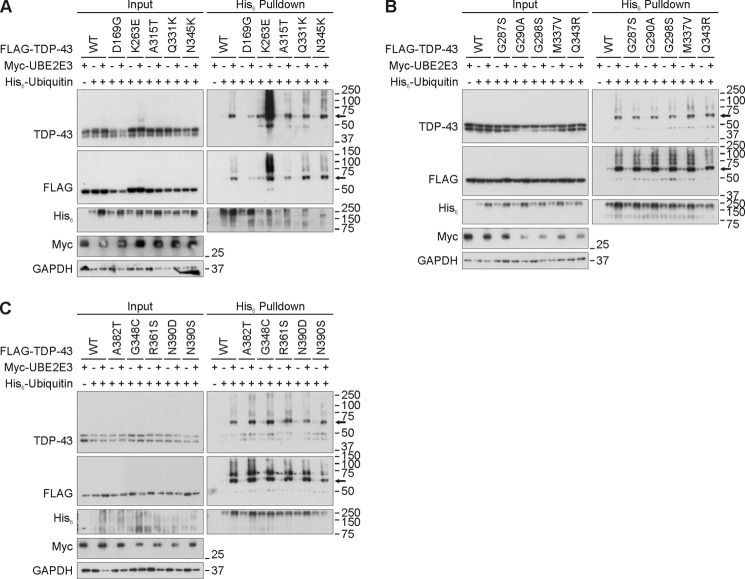
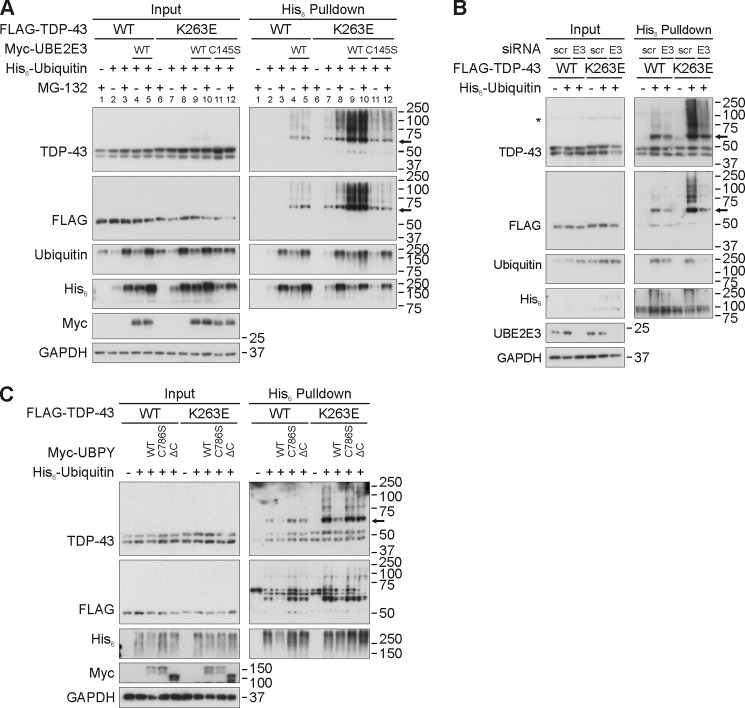
Ubiquitinated TDP-43 inclusions are the neuropathological hallmarks of human disease. To date, 48 mutations in the human gene encoding TDP-43 (TARDBP) have been linked mostly to ALS and FTLD (50). We first analyzed 15 of these TDP-43 mutants with regard to ubiquitination and UBE2E3 responsiveness in transiently transfected HEK293E cells. Almost all of the mutant proteins behaved like WT TDP-43 in that they showed increased His6-ubiquitin pulldown after co-transfection with Myc-UBE2E3 (Fig. 7). However, one mutant, K263E, was different from the others. This extremely non-conservative substitution was found in a Hungarian man who developed FTLD as well as supranuclear palsy and chorea but no signs of motor neuron disease (51). Unlike the other TDP-43 proteins studied, TDP-43K263E was ubiquitinated already under basal conditions, and the amount of ubiquitinated K263E mutant was dramatically increased upon UBE2E3 overexpression (Fig. 7A). Thus, we examined the ubiquitination-prone point mutant TDP-43K263E systematically for the modifying effects of the enzymes UBE2E3 and UBPY (Fig. 8). Overexpression of UBE2E3 dramatically increased the amounts of ubiquitinated TDP-43K263E in pulldown experiments, whereas the catalytically inactive UBE2E3C145S was much less effective (Fig. 8A). Treatment with the proteasomal inhibitor MG-132 enhanced the ubiquitination of TDP-43K263E without UBE2E3 co-transfection, but with UBE2E3, the extremely high level of TDP-43K263E ubiquitination seemed to become saturated (Fig. 8A). Conversely, UBE2E3 silencing reduced the moderate pulldown of His6-ubiquitinated WT TDP-43 as well as the strong ubiquitination of TDP-43K263E (Fig. 8B). Finally, co-expression of Myc-UBPY, but not the catalytically inactive C786S and ΔC UBPY mutants, reduced both the moderate ubiquitination of WT TDP-43 and the strong ubiquitination of TDP-43K263E (Fig. 8C). Thus, UBE2E3 and UPBY are capable of regulating the ubiquitination of WT TDP-43 and the ubiquitination-prone mutant K263E.
FIGURE 7.
Effect of UBE2E3 overexpression on ubiquitination of pathogenic TDP-43 mutants. A–C, HEK293E cells were triple transfected with His6-ubiquitin (+) or vector control (−), Myc-UBE2E3 (+) or vector control (−), and FLAG-TDP-43 WT or mutants as indicated. Urea-soluble cell lysates were prepared, and His6-ubiquitin-conjugated proteins were purified with Ni-NTA-agarose. Total cell lysates (Input) and pulldown eluates were subjected to WB and stained for TDP-43, FLAG, His6, Myc, and GAPDH. An arrow points to monoubiquitinated FLAG-TDP-43.
FIGURE 8.
Regulation of TDP-43K263E ubiquitination by proteasomal inhibition, UBE2E3, and UBPY. A, HEK293E cells were triple transfected with FLAG-TDP-43 WT or K263E, His6-vector control (−), or His6-ubiquitin (+) and Myc-vector control, Myc-UBE2E3, or catalytically inactive UBE2E3C145S. Cells were treated with MG-132 (10 μm) or DMSO for 2 h and lysed with urea buffer, and His6-ubiquitin-conjugated proteins were pulled down from cell lysates. Total protein lysates and Ni-NTA-agarose eluates were subjected to WB with antibodies detecting TDP-43, FLAG, ubiquitin, His6, Myc, and GAPDH. B, knockdown of UBE2E3 decreases the level of ubiquitinated K263E. HEK293E cells were silenced three times with scrambled (scr) or siUBE2E3 (E3). 4 h after the third silencing, cells were double transfected with FLAG-TDP-43 WT or K263E and His6-vector control (−) or His6-ubiquitin. 24 h after transfection, urea lysates were prepared, and a pulldown of His6-ubiquitin-conjugated proteins was performed. Total cell lysates (Input) and eluates (His6 Pulldown) were Western blotted and stained with antibodies against TDP-43, FLAG, ubiquitin, His6, UBE2E3, and GAPDH as indicated. An asterisk (*) labels an unspecific band. C, UBPY can deubiquitinate K263E. FLAG-tagged TDP-43 WT or K263E was overexpressed with His6-ubiquitin and Myc-UBPY, catalytically inactive mutants C786S, ΔC, or vector controls in HEK293E cells. Urea lysates were prepared, and Ni-NTA purification of His6-tagged proteins was performed. Total cell lysates and pulldown eluates were Western blotted and stained with antibodies against TDP-43, FLAG, ubiquitin, His6, Myc, and GAPDH. In A–C, monoubiquitinated TDP-43 is indicated with an arrow.
Specificity of UBE2E3- and UBPY-regulated Ubiquitinations
To check whether UBE2E3 and UBPY were reacting specifically with TDP-43 proteins or generally with aggregating proteins, we determined the ubiquitinations of very long poly(Q)-expanded Atx3. No ubiquitination was detected for the EGFP fusion protein even in the presence of Myc-UBE2E3. In contrast, His6-ubiquitination was detected for Atx3-Q148-EGFP. Myc-UBE2E3 co-transfection hardly enhanced the His6-ubiquitination of Atx3-Q148-EGFP, while Myc-UBPY slightly reduced the EGFP signal in His6-ubiquitin pulldowns (Fig. 9). Thus, there seems to be some specificity of the TDP-43 interaction with UBE2E3, whereas UBPY also could act on aggregating poly(Q) proteins. It is noteworthy that RNAi of a Caenorhabditis elegans ortholog of UBPY (E01B7.1) enhances the formation of poly(Q) protein aggregates (52).
FIGURE 9.
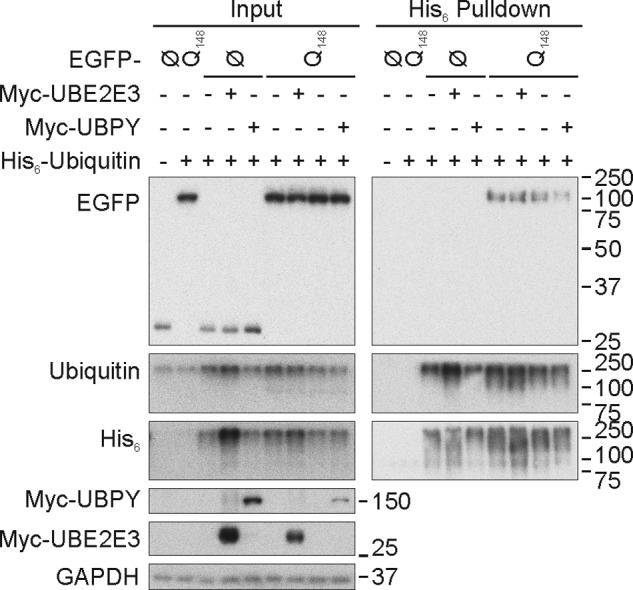
Effect of UBE2E3 and UBPY overexpression on ubiquitination of Atx3-Q148-EGFP. HEK293E cells were triple transfected with His6-ubiquitin (+) or vector control (−), Myc-UBE2E3 (+) or vector-control (−), and EGFP-N1-Atx3-Q148 (Q148) or vector control (Ø) as indicated, and urea-soluble cell lysates were prepared. His6-ubiquitin-conjugated proteins were purified with Ni-NTA-agarose. Western blotting was performed with total cell lysates (Input) and pulldown eluates, and blots were stained for GFP (Living Colors), ubiquitin, His6, Myc, and GAPDH.
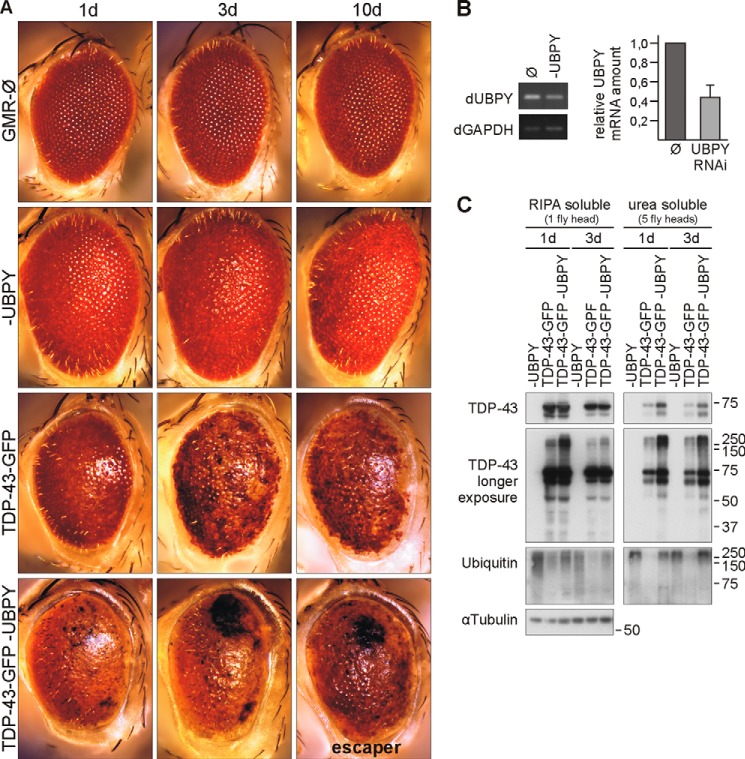
UBPY Deficiency Enhances TDP-43 Neurotoxicity in Flies
To determine the effects of UBPY deficiency on TDP-43 and to provide in vivo evidence for a functional interaction, we turned to D. melanogaster. Specifically, we used an established model for TDP-43 neurodegeneration (37). We first expressed human TDP-43 fused to GFP under the control of an eye-specific driver (GMR-Gal4). As reported before (37), the high expresser line 14 developed a progressive rough eye phenotype and depigmentation (Fig. 10A). UBPY RNAi (Fig. 10B) alone caused a very mild rough eye phenotype but no depigmentation (Fig. 10A), whereas the GMR driver line alone only presented some ommatidial disorganization (Fig. 10A). UBPY silencing enhanced the phenotype of line 14 TDP-43-GFP transgenic flies. Black lesions developed in the eyes, and after 10 days, most of these flies were dead (Fig. 10A). These very strong phenotypes were observed only in combination but never with transgenic TDP-43-GFP or UBPY RNAi alone.
FIGURE 10.
UBPY deficiency enhances TDP-43 neurotoxicity in D. melanogaster. A, light microscope eye images of 1-, 3-, and 10-day-old male flies expressing TDP-43-GFP (line 14 high expression), UBPY RNAi, or both under the control of the eye-specific driver GMR-Gal4. The driver line is shown for comparison (top row). B, UBPY silencing under control of actin-Gal4 was lethal. Silencing efficiency was confirmed in larvae expressing UBPY RNAi. C, analysis of TDP-43 protein level in flies expressing human TDP-43-GFP (line 14), UBPY RNAi, or both for 1 and 3 days (d). Fly heads were collected and subjected to sequential extraction. RIPA and urea lysate volumes equivalent to one and five fly heads, respectively, were Western probed as indicated with antibodies against TDP-43, ubiquitin, and α-tubulin as a loading control. Ø, vector control; dGAPDH, Drosophila GAPDH; dUBPY, Drosophila UBPY.
Biochemical extractions showed increased amounts of insoluble TDP-43 in the heads of line 14 transgenic flies with silenced UBPY (Fig. 10C). Moreover, UBPY-silenced fly heads contained more higher molecular mass TDP-43 and ubiquitin smears especially in the insoluble fractions of TDP-43 transgenic flies, likely reflecting pathological TDP-43 ubiquitination (Fig. 10C). UBPY silencing increased the amount of apparently ubiquitinated transgenic TDP-43-GFP species throughout the time course and importantly led to an accumulation of TDP-43-GFP in the insoluble fraction. Thus, the accumulation of insoluble, likely ubiquitinated TDP-43-GFP accompanies neurodegeneration in the D. melanogaster eye and is enhanced by UBPY knockdown.
DISCUSSION
The linkage of the nucleic acid-binding proteins TDP-43 and FUS to distinct subtypes of FTLD and ALS implicated dysfunctions of RNA processing with these neurodegenerative diseases. Both TDP-43 and FUS are involved in several RNA processing steps, including RNA splicing, miRNA processing, mRNA transport, and regulation (3). Consequently, TDP-43 has been incidentally found in experimental systems in a number of protein complexes, including heteronuclear ribonucleoprotein particles, miRNA processing complexes, and RNA transport and stress granules (see the Introduction). Here we screened for TDP-43 interactors by Y2H for the first time using a library from adult human brain, which is the tissue affected by TDPopathies. We identified 10 novel potential interactors of which seven were confirmed by co-immunoprecipitation and co-localization in HEK293E cells. Among these are the RNA-binding proteins LSM6, MED6, and RBM45, further emphasizing the role of TDP-43 in RNA processing. Specifically, our findings suggest a role of TDP-43 in nuclear and/or cytosolic LSm RNA processing complexes (42) as well as in the nuclear mediator complex that regulates transcriptional activation (53). TDP-43 has been found previously in mediator complex preparations (54). Moreover, the potential interaction of TDP-43 with RACK1 could point to a regulatory role in SG formation and/or to additional roles of the extremely multifunctional RACK1 protein (55).
As TDP-43 belongs to the large and highly diverse group of neurodegeneration-associated misfolding proteins, which are found deposited in diseased brain as ubiquitinated aggregates, we focused on this aspect of the cluster of novel TDP-43 interactors found here. RBM45 has been reported to accumulate within ubiquitin-positive TDP-43 inclusions in human patients (43). Likewise, RACK1 has been found in a co-aggregating proteome in a cell culture model of TDPopathy (47). More specifically, we identified three enzymes directly involved in ubiquitination reactions: the ubiquitin-conjugating enzyme UBE2E3, the ubiquitin ligase RNF2, and the DUB UBPY. We provide direct evidence that the UBE2E class of ubiquitin-conjugating enzymes promotes TDP-43 ubiquitination, whereas UBPY reduces it. Catalytically inactive UBE2E3 and UPBY were much less reactive toward TDP-43. UBPY knockdown enhanced the formation of insoluble and likely ubiquitinated TDP-43 species as well as neurotoxicity in transgenic D. melanogaster, providing in vivo support for a role of UBPY as a potentially neuroprotective DUB for TDP-43. We could not unequivocally assess the modifier effects of RNAi for the two D. melanogaster orthologs of UBE2E enzymes. GMR-driven silencing of UbcD1 was lethal, and UbcD2 RNAi led to eye depigmentation on its own. For the third enzyme, RNF2, we could not observe strong effects on TDP-43 ubiquitination. Perhaps this reflects technical problems with transfection of functional, catalytically active RNF2; a lack of appropriate cellular context and activation in the model system used; or that RNF2 is a functionally false positive interactor of TDP-43. The cognate ubiquitin ligase(s) of the UBE2E class of ubiquitination-promoting enzymes remains to be shown. One other candidate ubiquitin ligase for TDP-43 was recently reported, namely parkin (17). Parkin and UBE2E2 (UbcH8) were shown to interact (56, 57), and for the polycomb protein RNF2 (RING1b), UBE2E1 (UbcH6) promoted histone H2A monoubiquitination (58). Whether UBE2E3 functionally interacts with additional, distinct ubiquitin ligases remains to be explored. In other words, TDP-43 turnover might also be regulated by an additional ubiquitin ligase(s) that remains to be identified.
Ubiquitin is a universal modifier regulating trafficking and turnover of proteins and organelles (38, 59, 60). Ubiquitin linkage to target proteins is mediated by a three-step catalytic process: first ubiquitin forms an energy-rich thioester with the E1 ubiquitin-activating enzyme, and then activated ubiquitin is transferred to one of ≈35 E2 ubiquitin-conjugating enzymes (61), which in turn transfers ubiquitin to a multitude of E3 ubiquitin ligases that contribute to the final isopeptide bond formation with ubiquitinated substrate proteins. DUBs regulate protein ubiquitination states or recycle ubiquitin for example when proteins polyubiquitinated with chains linked via Lys-48 are targeted to proteasomal degradation. This is the main catabolic pathway of short lived proteins and provides a defense system against misfolded and aggregating proteins such as those that occur during aging and particularly in patients with neurodegenerative proteinopathies (62). It is widely believed that the protein refolding and degradation capacity declines in age-related neurodegenerative proteinopathies. All neuropathological protein aggregates, including TDP-43 lesions in FTLD-TDP and ALS, contain ubiquitin (4, 63).
Here we found that UBE2E enzymes may participate in TDP-43 ubiquitination. The physical interaction of an E2 ubiquitin-conjugating enzyme with a putative substrate protein is unusual. However, RING-type ubiquitin ligases stay bound to their cognate E2s and transfer ubiquitin to the substrate protein directly from the E2 (64). In such E2-E3 complexes, many E2s are believed to influence the type of ubiquitin linkage and even substrate specificity. Perhaps there is a TDP-43 interaction interface on UBE2E3 that allowed reaction in the yeast two-hybrid screen. In transfected cells, optimized conditions for co-immunoprecipitations between TDP-43 and UBE2E3 might argue for the requirement for a complex-stabilizing E3. Parkin is a candidate for such a RING-type ubiquitin ligase for TDP-43 (17). Alternatively, UBE2E3 could be a novel example of a highly dynamic and/or weak physical interaction of an E2 directly interacting with its target protein, TDP-43.
Functionally, endogenous ubiquitin tended to monoubiquitinate His6-TDP-43 in the presence of UBE2E3 (Fig. 4A), although the experimental setup suffered from high background for polyubiquitinated proteins. When His6-ubiquitin was used as a probe, higher molecular mass smears of (multiple mono- or polyubiquitinated) FLAG-TDP-43 were pulled down after co-transfection with UBE2E family members (Fig. 4B). Proteasome inhibition with MG-132 hardly stabilized UBE2E-mediated TDP-43 ubiquitination. Moreover, UBE2E overexpression did not lead to reduction of TDP-43 steady-state levels within the observation period as would be expected for proteasome-targeting ubiquitinations. Note that TDP-43 expression is not only regulated by protein turnover but also via self-regulation of its mRNA, keeping cellular TDP-43 levels within carefully controlled margins (5, 6). Instead, forced TDP-43 ubiquitination by active UBE2E3 causes a shift into insoluble fractions (Fig. 4C). Perhaps TDP-43 ubiquitination subtly affects its tertiary or quaternary structure and thus incorporation into proper protein complexes in appropriate subcellular localizations. Despite the detectable shift of a portion of ubiquitinated TDP-43 into the insoluble fraction, UBE2E co-transfection even followed by 6-h proteasome inhibition did not lead to overt depletion of functional TDP-43 in the acute cell culture system as evidenced by unaffected expression of histone deacetylase 6 (19) and splicing of the ribosomal S6 kinase 1 Aly/REF-like target (65) both at the mRNA and protein levels (data not shown). However, long term accumulation of ubiquitinated TDP-43 in insoluble fractions in aging human patients could eventually lead to pathological proteasome overload, preventing the effective degradation of ubiquitinated TDP-43 that eventually becomes too insoluble to be degraded (see Fig. 11). UBE2E1 (UbcH6) was reported previously to bind and ubiquitinate ataxin-1, in this case successfully promoting ataxin-1 degradation (66). Thus, the overall effects of aggregating protein ubiquitinations by UBE2E enzymes may differ according to solubility of the target protein and/or the degradative capacity of the cellular system. RNAi of the TDP-43 DUB UBPY also led to the accumulation of insoluble, apparently ubiquitinated TDP-43 in Drosophila and enhanced neurotoxicity. Interestingly, pharmacological DUB inhibition was similarly shown to cause aggregation of ubiquitinated tau in an oligodendroglial cell culture model (67).
FIGURE 11.
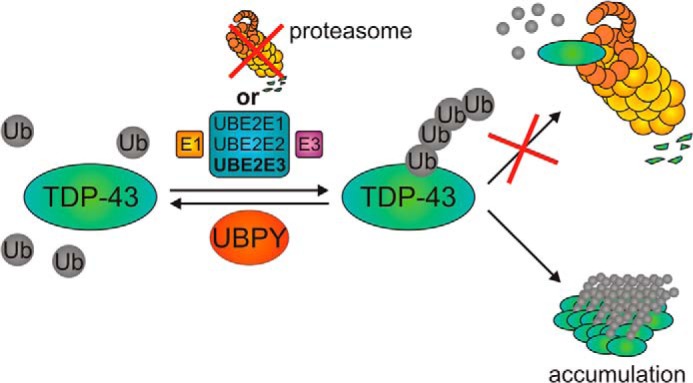
Schematic overview of regulation of TDP-43 ubiquitination by UBE2E ubiquitin-conjugating enzymes, UBPY, and proteasomal inhibition. Ub, ubiquitin.
The exact molecular mechanisms of how disease-linked mutations confer TDPopathy are poorly understood. We examined 15 of the 48 known pathogenic TDP-43 mutations. Almost all of them were ubiquitinated by UBE2E3 like WT TDP-43 at least within the short observation period in transiently transfected HEK293E cells. However, one unusually not ALS- but FTLD-TDP-linked mutation (51) caused excessive TDP-43K263E ubiquitination even under basal conditions that could still be regulated by UBE2E3 and UBPY. The K263E substitution replaces a positively charged lysine residue with a negatively charged aspartic acid side chain. Such altered charge distribution and/or severe structural distortion could account for the observed reduced electrophoretic motility of TDP-43K263E (see Figs. 7 and 8). The K263E mutation resides in the second RNA recognition motif, where the Lys-263 residue may contact bound RNA (68). The K263E mutation could disrupt appropriate TDP-43 incorporation into physiological complexes and/or intramolecular stabilization, ultimately giving rise to a misfolded protein that evidently provides a challenge for cellular handling. Aberrant folding and abortive proteasomal degradation could be a straightforward explanation for the pathogenic effect of the K263E mutation.
Although the tight regulation of TDP-43 synthesis is well studied (5, 6), the mechanisms of proteolytic breakdown are less well understood. Conflicting results suggesting TDP-43 breakdown by the proteasome, autophagy machinery, or both (see the Introduction) have been reported. In mouse motor neurons, impairment of proteasome activity but not autophagy leads to the formation of cytosolic TDP-43 inclusions and other ALS-like phenotypes (69). In transiently transfected HEK293E cells, we found that TDP-43 FL (both WT and mutant) was turned over in >24 h, whereas the commonly observed 35- and 25-kDa CTFs were rapidly turned over by the proteasome but not autophagy.
Taken together, the regulation of TDP-43 ubiquitination may control TDP-43 neurotoxicity, which appears to be enhanced when TDP-43 accumulates in ubiquitinated and insoluble forms. This could be achieved by inefficient deubiquitination upon UBPY deficiency. The exact relationships among TDP-43 fragmentation and ubiquitination, turnover, protein solubility, and the mechanisms of cell death in flies and mammals remain to be further elucidated. Meanwhile, our study identified UBPY as a potential suppressor of TDP-43 neurotoxicity that might become useful in future (pre)clinical studies.
This work was supported by the German Center for Neurodegenerative Diseases (DZNE) within the Helmholtz Association, specifically a DZNE Ph.D. stipend (to F. H.) and the DZNE Virtual Institute “RNA Dysmetabolism in ALS”; German Research Foundation (Deutsche Forschungsgemeinschaft) Grants KA1675/3-1 and KA1675/3-2; the German Competence Network “Degenerative Dementias”; and the Hertie Foundation.
- TDP-43
- trans-activation element DNA-binding protein of 43 kDa
- ALS
- amyotrophic lateral sclerosis
- CTF
- C-terminal fragment
- DUB
- deubiquitinating enzyme
- FL
- full-length
- FTLD
- frontotemporal lobar degeneration
- FUS
- fused-in-sarcoma
- hnRNP
- heteronuclear ribonucleoprotein
- SG
- stress granule
- SMN
- survival of motoneuron
- UBE2
- E2 ubiquitin-conjugating enzyme
- UBPY
- ubiquitin isopeptidase Y
- Y2H
- yeast two-hybrid
- EGFP
- enhanced GFP
- WB
- Western blot analysis
- IF
- immunofluorescence staining
- RIPA
- radioimmune precipitation assay
- Atx3
- ataxin-3
- Ni-NTA
- nickel-nitrilotriacetic acid
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- NLS
- nuclear localization sequence
- NLSmut
- mutated nuclear localization sequence
- aa
- amino acids.
REFERENCES
- 1. Mackenzie I. R., Rademakers R., Neumann M. (2010) TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 9, 995–1007 [DOI] [PubMed] [Google Scholar]
- 2. Van Langenhove T., van der Zee J., Van Broeckhoven C. (2012) The molecular basis of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum. Ann. Med. 44, 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fiesel F. C., Kahle P. J. (2011) TDP-43 and FUS/TLS: cellular functions and implications for neurodegeneration. FEBS J. 278, 3550–3568 [DOI] [PubMed] [Google Scholar]
- 4. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 5. Avendaño-Vázquez S. E., Dhir A., Bembich S., Buratti E., Proudfoot N., Baralle F. E. (2012) Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev. 26, 1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ayala Y. M., De Conti L., Avendaño-Vázquez S. E., Dhir A., Romano M., D'Ambrogio A., Tollervey J., Ule J., Baralle M., Buratti E., Baralle F. E. (2011) TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 30, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J.-P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P.-F., Camu W., Meininger V., Dupre N., Rouleau G. A. (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40, 572–574 [DOI] [PubMed] [Google Scholar]
- 8. Rutherford N. J., Zhang Y.-J., Baker M., Gass J. M., Finch N. A., Xu Y.-F., Stewart H., Kelley B. J., Kuntz K., Crook R. J., Sreedharan J., Vance C., Sorenson E., Lippa C., Bigio E. H., Geschwind D. H., Knopman D. S., Mitsumoto H., Petersen R. C., Cashman N. R., Hutton M., Shaw C. E., Boylan K. B., Boeve B., Graff-Radford N. R., Wszolek Z. K., Caselli R. J., Dickson D. W., Mackenzie I. R., Petrucelli L., Rademakers R. (2008) Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 4, e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Y.-J., Gendron T. F., Xu Y.-F., Ko L.-W., Yen S.-H., Petrucelli L. (2010) Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA binding protein-43 C-terminal fragments. Mol. Neurodegener. 5, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Eersel J., Ke Y. D., Gladbach A., Bi M., Götz J., Kril J. J., Ittner L. M. (2011) Cytoplasmic accumulation and aggregation of TDP-43 upon proteasome inhibition in cultured neurons. PLoS One 6, e22850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caccamo A., Majumder S., Deng J. J., Bai Y., Thornton F. B., Oddo S. (2009) Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability. J. Biol. Chem. 284, 27416–27424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E. M., Isaacs A., Brech A., Stenmark H., Simonsen A. (2007) Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179, 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brady O. A., Meng P., Zheng Y., Mao Y., Hu F. (2011) Regulation of TDP-43 aggregation by phosphorylation and p62/SQSTM1. J. Neurochem. 116, 248–259 [DOI] [PubMed] [Google Scholar]
- 14. Scotter E. L., Vance C., Nishimura A. L., Lee Y.-B., Chen H.-J., Urwin H., Sardone V., Mitchell J. C., Rogelj B., Rubinsztein D. C., Shaw C. E. (2014) Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J. Cell Sci. 127, 1263–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Urushitani M., Sato T., Bamba H., Hisa Y., Tooyama I. (2010) Synergistic effect between proteasome and autophagosome in the clearance of polyubiquitinated TDP-43. J. Neurosci. Res. 88, 784–797 [DOI] [PubMed] [Google Scholar]
- 16. Wang X., Fan H., Ying Z., Li B., Wang H., Wang G. (2010) Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci. Lett. 469, 112–116 [DOI] [PubMed] [Google Scholar]
- 17. Hebron M. L., Lonskaya I., Sharpe K., Weerasinghe P. P., Algarzae N. K., Shekoyan A. R., Moussa C. E. (2013) Parkin ubiquitinates Tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6). J. Biol. Chem. 288, 4103–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang I.-F., Reddy N. M., Shen C.-K. (2002) Higher order arrangement of the eukaryotic nuclear bodies. Proc. Natl. Acad. Sci. U.S.A. 99, 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fiesel F. C., Voigt A., Weber S. S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M. L., Kern J. V., Rasse T. M., Schmidt T., Springer W., Kirchner R., Bonin M., Neumann M., Baekelandt V., Alunni-Fabbroni M., Schulz J. B., Kahle P. J. (2010) Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 29, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsuiji H., Iguchi Y., Furuya A., Kataoka A., Hatsuta H., Atsuta N., Tanaka F., Hashizume Y., Akatsu H., Murayama S., Sobue G., Yamanaka K. (2013) Spliceosome integrity is defective in the motor neuron diseases ALS and SMA. EMBO Mol. Med. 5, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fallini C., Bassell G. J., Rossoll W. (2012) The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum. Mol. Genet. 21, 3703–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Z., Fan D., Gui B., Shi L., Xuan C., Shan L., Wang Q., Shang Y., Wang Y. (2012) Neurodegeneration-associated TDP-43 interacts with fragile X mental retardation protein (FMRP)/Staufen (STAU1) and regulates SIRT1 expression in neuronal cells. J. Biol. Chem. 287, 22560–22572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y. M., Baralle F. E. (2005) TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 280, 37572–37584 [DOI] [PubMed] [Google Scholar]
- 24. D'Ambrogio A., Buratti E., Stuani C., Guarnaccia C., Romano M., Ayala Y. M., Baralle F. E. (2009) Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 37, 4116–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Freibaum B. D., Chitta R. K., High A. A., Taylor J. P. (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 9, 1104–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ling S.-C., Albuquerque C. P., Han J. S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D. W. (2010) ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. U.S.A. 107, 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buratti E., Baralle F. E. (2010) The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol. 7, 420–429 [DOI] [PubMed] [Google Scholar]
- 28. Liu-Yesucevitz L., Bilgutay A., Zhang Y.-J., Vanderweyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., Petrucelli L., Wolozin B. (2010) Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 5, e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S. H., Shanware N. P., Bowler M. J., Tibbetts R. S. (2010) Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J. Biol. Chem. 285, 34097–34105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawahara Y., Mieda-Sato A. (2012) TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U.S.A. 109, 3347–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buratti E., De Conti L., Stuani C., Romano M., Baralle M., Baralle F. (2010) Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 277, 2268–2281 [DOI] [PubMed] [Google Scholar]
- 32. Shiina Y., Arima K., Tabunoki H., Satoh J. (2010) TDP-43 dimerizes in human cells in culture. Cell. Mol. Neurobiol. 30, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang I.-F., Chang H.-Y., Hou S.-C., Liou G.-G., Way T.-D., Shen C.-K. (2012) The self-interaction of native TDP-43 C terminus inhibits its degradation and contributes to early proteinopathies. Nat. Commun. 3, 766. [DOI] [PubMed] [Google Scholar]
- 34. Fuentealba R. A., Udan M., Bell S., Wegorzewska I., Shao J., Diamond M. I., Weihl C. C., Baloh R. H. (2010) Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J. Biol. Chem. 285, 26304–26314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cohen T. J., Hwang A. W., Unger T., Trojanowski J. Q., Lee V. M. (2012) Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. EMBO J. 31, 1241–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Furukawa Y., Kaneko K., Nukina N. (2011) Molecular properties of TAR DNA binding protein-43 fragments are dependent upon its cleavage site. Biochim. Biophys. Acta 1812, 1577–1583 [DOI] [PubMed] [Google Scholar]
- 37. Voigt A., Herholz D., Fiesel F. C., Kaur K., Müller D., Karsten P., Weber S. S., Kahle P. J., Marquardt T., Schulz J. B. (2010) TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS One 5, e12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 39. Hoffman C. S., Winston F. (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57, 267–272 [DOI] [PubMed] [Google Scholar]
- 40. Ayala Y. M., Zago P., D'Ambrogio A., Xu Y.-F., Petrucelli L., Buratti E., Baralle F. E. (2008) Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 121, 3778–3785 [DOI] [PubMed] [Google Scholar]
- 41. Clague M. J., Urbé S. (2006) Endocytosis: the DUB version. Trends Cell Biol. 16, 551–559 [DOI] [PubMed] [Google Scholar]
- 42. Tharun S. (2009) Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int. Rev. Cell Mol. Biol. 272, 149–189 [DOI] [PubMed] [Google Scholar]
- 43. Collins M., Riascos D., Kovalik T., An J., Krupa K., Krupa K., Hood B. L., Conrads T. P., Renton A. E., Traynor B. J., Bowser R. (2012) The RNA-binding motif 45 (RBM45) protein accumulates in inclusion bodies in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) patients. Acta Neuropathol. 124, 717–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. (2008) A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. (2008) Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 46. Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., Zerhusen B., Malcolm R., Varrone Z., Collis A., Minto M., Burgess S., McDaniel L., Stimpson E., Spriggs F., Williams J., Neurath K., Ioime N., Agee M., Voss E., Furtak K., Renzulli R., Aanensen N., Carrolla S., Bickelhaupt E., Lazovatsky Y., DaSilva A., Zhong J., Stanyon C. A., Finley R. L., Jr., White K. P., Braverman M., Jarvie T., Gold S., Leach M., Knight J., Shimkets R. A., McKenna M. P., Chant J., Rothberg J. M. (2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 47. Dammer E. B., Fallini C., Gozal Y. M., Duong D. M., Rossoll W., Xu P., Lah J. J., Levey A. I., Peng J., Bassell G. J., Seyfried N. T. (2012) Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PLoS One 7, e38658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Row P. E., Prior I. A., McCullough J., Clague M. J., Urbé S. (2006) The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 281, 12618–12624 [DOI] [PubMed] [Google Scholar]
- 49. Avvakumov G. V., Walker J. R., Xue S., Finerty P. J., Jr., Mackenzie F., Newman E. M., Dhe-Paganon S. (2006) Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8). J. Biol. Chem. 281, 38061–38070 [DOI] [PubMed] [Google Scholar]
- 50. Lattante S., Rouleau G. A., Kabashi E. (2013) TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum. Mutat. 34, 812–826 [DOI] [PubMed] [Google Scholar]
- 51. Kovacs G. G., Murrell J. R., Horvath S., Haraszti L., Majtenyi K., Molnar M. J., Budka H., Ghetti B., Spina S. (2009) TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov. Disord. 24, 1843–1847 [DOI] [PubMed] [Google Scholar]
- 52. Nollen E. A., Garcia S. M., van Haaften G., Kim S., Chavez A., Morimoto R. I., Plasterk R. H. (2004) Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. U.S.A. 101, 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taatjes D. J. (2010) The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 35, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sato S., Tomomori-Sato C., Parmely T. J., Florens L., Zybailov B., Swanson S. K., Banks C. A., Jin J., Cai Y., Washburn M. P., Conaway J. W., Conaway R. C. (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 [DOI] [PubMed] [Google Scholar]
- 55. Gibson T. J. (2012) RACK1 research—ships passing in the night? FEBS Lett. 586, 2787–2789 [DOI] [PubMed] [Google Scholar]
- 56. Imai Y., Soda M., Takahashi R. (2000) Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275, 35661–35664 [DOI] [PubMed] [Google Scholar]
- 57. Zhang Y., Gao J., Chung K. K., Huang H., Dawson V. L., Dawson T. M. (2000) Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. U.S.A. 97, 13354–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006) Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Clague M. J., Urbé S. (2010) Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685 [DOI] [PubMed] [Google Scholar]
- 60. Komander D., Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 61. van Wijk S. J., Timmers H. T. (2010) The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24, 981–993 [DOI] [PubMed] [Google Scholar]
- 62. Tai H.-C., Schuman E. M. (2008) Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9, 826–838 [DOI] [PubMed] [Google Scholar]
- 63. Lehman N. L. (2009) The ubiquitin proteasome system in neuropathology. Acta Neuropathol. 118, 329–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Metzger M. B., Pruneda J. N., Klevit R. E., Weissman A. M. (2014) RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fiesel F. C., Weber S. S., Supper J., Zell A., Kahle P. J. (2012) TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 40, 2668–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hong S., Lee S., Cho S.-G., Kang S. (2008) UbcH6 interacts with and ubiquitinates the SCA1 gene product ataxin-1. Biochem. Biophys. Res. Commun. 371, 256–260 [DOI] [PubMed] [Google Scholar]
- 67. Seiberlich V., Goldbaum O., Zhukareva V., Richter-Landsberg C. (2012) The small molecule inhibitor PR-619 of deubiquitinating enzymes affects the microtubule network and causes protein aggregate formation in neural cells: implications for neurodegenerative diseases. Biochim. Biophys. Acta 1823, 2057–2068 [DOI] [PubMed] [Google Scholar]
- 68. Lukavsky P. J., Daujotyte D., Tollervey J. R., Ule J., Stuani C., Buratti E., Baralle F. E., Damberger F. F., Allain F. H. (2013) Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat. Struct. Mol. Biol. 20, 1443–1449 [DOI] [PubMed] [Google Scholar]
- 69. Tashiro Y., Urushitani M., Inoue H., Koike M., Uchiyama Y., Komatsu M., Tanaka K., Yamazaki M., Abe M., Misawa H., Sakimura K., Ito H., Takahashi R. (2012) Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J. Biol. Chem. 287, 42984–42994 [DOI] [PMC free article] [PubMed] [Google Scholar]