Abstract
Precise control of the balance between protein phosphorylation, catalyzed by protein kinases, and protein dephosphorylation, catalyzed by protein phosphatases, is essential for cellular homeostasis. Deregulation of this balance leads to pathophysiological states, driving diseases such as cancer, heart disease, and diabetes, among many others. Aberrant phosphorylation of components of the pathways that control cell growth cell survival are particularly prevalent in cancer. One of the most studied tumor suppressors in these pathways is the lipid phosphatase, PTEN, which dephosphorylates the lipid second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3), thus preventing activation of the oncogenic kinase AKT. In 2005, the discovery of a family of protein phosphatases whose members directly dephosphorylate and inactivate AKT introduced a new negative regulator of the phosphatidylinositol-3-kinase (PI3K) oncogenic pathway. PH domain Leucine-rich repeat Protein Phosphatase (PHLPP) isozymes comprise a novel tumor suppressor family whose two members, PHLPP1 and PHLPP2, are deleted as frequently as PTEN in cancers such as those of the prostate. PHLPP is thus a novel therapeutic target to suppress oncogenic pathways and is a potential candidate biomarker to stratify patients for the appropriate targeted therapeutics. This review discusses the role of PHLPP in terminating AKT signaling and how pharmacological intervention would impact this pathway.
Keywords: PHLPP, AKT, PI3K, PTEN, p53
1. INTRODUCTION
Poised as a central transducer of growth and proliferative signaling, the phosphatidylinositol-3 kinase (PI3K) signaling pathway plays an essential role in maintaining cellular homeostasis. Binding of growth factors to cell surface receptors triggers the activation of this lipid kinase, a requisite event in turning on the protein kinase AKT; activated AKT initiates a cascade of phosphorylation events that ultimately control survival and proliferation. This pathway is opposed by both lipid and protein phosphatases and dysregulation of the precise balance between phosphorylation, and dephosphorylation results in pathogenic states. Thus, it is not surprising that this pathway is one of the most commonly mutated pathways in human cancer. In prostate cancer, the PI3 kinase pathway has been reported to be aberrantly regulated in 42% of primary tumors and a staggering 100% of metastatic tumors (1). This high rate of dysregulation is a consistent theme in many cancers, including uterine, brain, breast, colon and lung (2). Thus, targeting players in the pathway that either propagate signaling (oncogenes) or terminate signaling (tumor suppressors) constitutes a major therapeutic effort.
A. Propagating growth factor signaling
Binding of growth factors to receptor tyrosine kinases (RTKs) results in recruitment of the lipid kinase PI3 kinase to the plasma membrane, where it phosphorylates phosphatidylinositol-4,5-trisphosphate (PIP2) to generate the rare lipid second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3) (3) (Figure 1). This recruitment can be through direct interaction of the class I type PI3K with the RTK or via scaffold proteins such as IRS-1. Generation of PIP3 recruits the Ser/Thr protein kinase AKT by engaging its amino-terminal PH domain, a membrane-targeting module that, in the case of AKT, specifically binds 3’-phosphorylated inositol lipids such as PIP3 (4; 5). Membrane binding exposes the carboxy-terminal kinase domain of AKT for phosphorylation by the upstream kinase, phosphoionsitol dependent kinase 1 (PDK-1), an event that triggers a second phosphorylation at the C-terminus (see Side Bar for discussion of mechanisms of phosphorylation of AKT). Once fully phosphorylated, AKT is locked in an active conformation and diffuses through the cytosol to distinct subcellular compartments, including the nucleus (6). The advent of genetically-encoded reporters to visualize the spatio-temporal dynamics of AKT activation reveal that the kinase is activated rapidly at the plasma membrane, with kinetics mirroring the rate of formation of PIP3, that the activity then propagates through the cytosol with slightly slower kinetics, and is eventually detected in the nucleus (7).
Figure 1. AKT is a central mediator of PI3K signaling.
Cartoon showing details of the regulation of AKT: AKT is co-translationally phosphorylated on the turn motif (orange circle, Thr450 in AKT1) by ribosome and ER-localized mTORC2 (orange oval). This phosphorylation stabilizes AKT and protects it from proteosome-mediated degradation. Signals that generate PIP3 engage AKT’s PH domain and thus recruit it to the plasma membrane (middle panel; membrane-engaged species). Membrane-binding unmasks the activation loop, resulting in phosphorylation by PDK-1 (pink circle, Thr308 in AKT1) and a subsequent tightly-coupled phosphorylation on the hydrophobic motif (green circle, Ser473 in AKT1). Although mTORC2 has been proposed to phosphorylate this site too, mechanisms that displace the PH domain (e.g. tethering AKT to the membrane via myristoylation) bypass the requirement for mTORC2. This latter phosphorylation depends on the intrinsic catalytic activity of AKT, suggesting that, similar to PKC, this site is autophosphorylated following structuring of the active site by the PDK-1 phosphorylation of Thr308. The fully-phosphorylated species of AKT is locked in an active conformation and diffuses throughout the cell to mediate down stream signaling (bottom right species). Signaling is suppressed by dephosphorylation of PIP3 by the tumor suppressor PTEN and acutely terminated by direct dephosphorylation of AKT to regenerate the mono-phosphorylated (phospho-Thr 450), auto-inhibited species. Dephosphorylation is catalyzed in part by the recently discovered PHLPP phosphatases, which directly dephosphorylate the hydrophobic motif, and by okadaic acid-sensitive phosphatases, such as PP2A, which dephosphorylate the activation loop.
SIDE BAR 1.
Regulation of AKT by Phosphorylation
AKT comprises a family of three structurally-related kinases that have an N-terminal PIP3-sensing PH domain and a C-terminal kinase domain (Figure 1). In its inactive state, AKT is constitutively phosphorylated at a C-terminal residue termed the turn motif (Thr450 in AKT1); this phosphorylation is dependent on mTORC2, a kinase complex that colocalizes on the ER with ribosomal protein S6 (113), and occurs co-translationally (114). Upon PIP3-dependent recruitment to the membrane, all three AKT isoforms are phosphorylated by PDK-1 at a site near the entrance to the active site termed the activation loop (115; 116); this is Thr308 on AKT1. This triggers a tightly-coupled phosphorylation at a second C-terminal site termed the hydrophobic motif; this is Ser473 on AKT1. The mechanism of phosphorylation at this second C-terminal site has been highly contested. Autophosphorylation, as occurs for protein kinase C at the analogous site (117), as well as phosphorylation by a parade of upstream kinases, including DNA-PK and mTORC2, has been proposed (118–120).
Many of AKT’s substrates promote cell growth, proliferation, and cell survival (8; 9). Cell growth effects are mediated in part by phosphorylation of the tumor suppressor TSC2, a GTPase-activating protein (GAP) that, upon phosphorylation by AKT, relieves its suppression of the GTPase Rheb, in turn activating the Ser/Thr kinase mTOR in the rapamycin-sensitive mTORC1 complex (10). mTORC1, in turn, phosphorylates a number of proteins, including S6 kinase and 4EBP1 to promote protein translation and lipid biogenesis (11). Cell proliferation is promoted by phosphorylation of cell cycle regulators such as p27 and the E3 ubiquitin ligase HDM2 (human homolog to murine double minute 2), and pro-survival responses are regulated by phosphorylation of substrates such as GSK3β and members of Forkhead Box O (FoxO) family of transcription factors (see (12)).
B. Terminating growth factor signaling
Whereas lipid and protein kinases promote PI3K signaling, lipid and protein phosphatases suppress PI3K signaling. First, the lipid phosphatase PTEN dephosphorylates PIP3 to remove the activating lipid second messenger (13–15). However, AKT continues to signal, even in the absence of PIP3, until it itself becomes dephosphorylated by protein phosphatases (Figure 1). Dephosphorylation at the activation loop (Thr308 in AKT1) is sensitive to okadaic acid and is likely catalyzed by PP2A-type phosphatases (16). In contrast, dephosphorylation of the hydrophobic motif (Ser473) is catalyzed by the okadaic acid-insensitive PP2C-type family of phosphatases, the recently discovered PH domain Leucine-rich repeat Protein Phosphatases (16). Loss of both phosphates inactivates AKT. It is noteworthy that loss of phosphate at only the hydrophobic motif impairs the intrinsic catalytic activity of AKT by 90% based on biochemical studies (17). However, a subset of AKT substrates (namely TSC2 and GSK3β, but not FOXO3) continues to be phosphorylated in cells in which AKT is only phosphorylated on the activation loop in cells (18; 19). One possibility is that localized AKT signaling on protein scaffolds increases the local concentration of tethered substrates such as TSC2 sufficiently to allow effective phosphorylation despite the 10-fold reduction in catalytic activity compared to fully-phosphorylated AKT. It is also possible that AKT phosphorylated at Thr308 vs both Thr308 and Ser473 has altered substrate specificity. Alternatively, other kinases may compensate for AKT in situations where hydrophobic motif phosphorylation is prevented (18).
The AKT pathway is exquisitely sensitive to feedback regulation: the signaling output is under strong control by a negative feedback loop wherein activated S6K phosphorylates IRS-1, an event that leads to the degradation of IRS-1, thus reducing PI3K activation and consequently AKT activation (20; 21). This loop is effectively suppressed by inhibition of mTORC1 with rapamycin or by deletion of S6K (22). This feedback loop dampens AKT signaling in the face of high agonist input. Targeting signaling molecules within this feedback loop is particularly challenging, as discussed below (Section 4C).
2. PHLPP primer
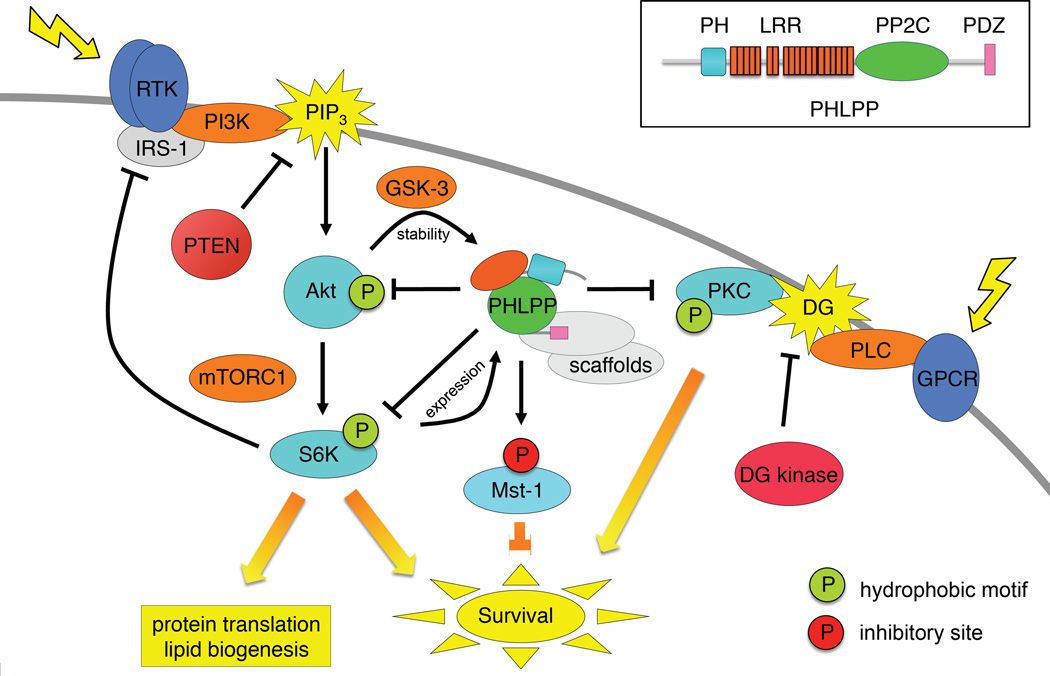
The PHLPP family of Ser/Thr phosphatases is encoded by two genes: PHLPP1, which has two predominant splice variants, PHLPP1α (1205 amino acids), and PHLPP1β (1717 amino acids; also known as suprachiasmatic nucleus oscillatory protein, SCOP (23)), and PHLPP2 (1323 amino acids) (16; 24). The discovery of this family of phosphatases exemplifies Pasteur’s aphorism that ‘chance favours the prepared mind’: reasoning that a phosphatase involved in terminating AKT signalling may have a membrane-targeting module like many of the players involved in lipid second messenger signalling (notably a PH domain such as that of AKT and its upstream kinase PDK-1), a query of the NCBI database for genes predicted to encode a PH domain and a phosphatase domain revealed the existence of precisely one such gene family. The protein family was named after its domain composition: PHLPP (pronounced ‘flip’) for PH domain Leucine-rich repeat Protein Phosphatase (Figure 2, inset) (16). A previous study had identified an mRNA that oscillated in a circadian rhythm-dependent manner in the suprachiasmatic nucleus (SCN); the predicted protein was named SCOP for SCN circadian oscillatory protein and corresponds to one of the splice variants of PHLPP1, PHLPP1β (23) (see below).
Figure 2. PHLPP suppresses lipid second messenger signaling.
PHLPP (center) directly dephosphorylates and inactivates AKT, PKC, and S6K, at their hydrophobic phosphorylation motif (green circle) thus opposing cell survival and growth signals, and directly dephosphorylates and activates Mst1 at an inhibitory site (red circle), thus promoting pro-apoptotic signals. Indicated are two negative feedback loops which control the steady-state levels of PHLPP: high AKT activity up regulates PHLPP levels by suppressing GSK-3-dependent degradation, and high S6K activity up regulates PHLPP expression. Note that targeting the PI3 kinase pathway within these feedback loops will have the consequence of decreasing PHLPP levels, thus promoting pro-survival pathways mediated by PKC and inhibiting pro-apoptotic pathways mediated by Mst1. The lipid phosphatase PTEN opposes the lipid kinase phosphatidylinositol 3 kinase (PI3K) by dephosphorylating PIP3 and the lipid kinase DG kinase opposes phospholipase C by phosphorylating DG. Scaffolds play an essential role in controlling substrate specificity of PHLPP and are indicated in grey. Also shown is the negative feedback loop from S6K to the scaffold IRS-1, which dampens PI3K activation and hence AKT activation (see Figure 4 also). Inset shows conserved domain structure of mammalian PHLPP family members showing PH domain (cyan), Leucine-rich repeats (LRR; orange), PP2C domain (green), and C-terminal PDZ-binding motif (pink); PHLPP1β and PHLPP2 also have a predicted RA domain preceding the PH domain (not shown).
Both PHLPP genes reside on chromosomal regions that are commonly lost in diverse cancers, consistent with their tumor suppressive function. PHLPP1 is on chromosome 18q21.33, a region that undergoes loss of heterozygosity (LOH) in a high percentage of colon cancers (25; 26). Similarly, LOH is common at the PHLPP2 locus, on chromosome 16q22.3, in breast and ovarian cancers, prostate cancer, and hepatocellular carcinomas (27–31); in addition to PHLPP2, the 16q22.3-16q23.1 region also harbors potential tumor suppressors such as Breast cancer anti-estrogen resistance 1 (BCAR1) (27).
A. Domain Structure of PHLPP
The PHLPP family is conserved all the way to yeast; interestingly, the yeast homologue, CYR1 (NCBI, HomoloGene database), is fused at the C-terminus to an additional enzyme, adenylate cyclase (32) and contains a putative Ras Association (RA) domain, Leu Rich Repeat (LRR) segment, and PP2C domain, but lacks the PH domain and PDZ ligand found in human PHLPP. The C-terminal PDZ ligand first appeared in the Bilateria phylum (animals having bilateral symmetry). Drosophila have a single PHLPP isozyme that contains all domains except the PH domain. The PH domain was acquired late in evolution and is present in all mammalian PHLPP isozymes. PHLPP1 and PHLPP2 diverged in the Euteleostomi clade (which includes 90% of living vertebrates); human PHLPP1 and PHLPP2 share 58% and 63% amino identity in the PP2C domain and PH domain, respectively (16; 24).
The integration of regulatory/scaffolding modules in the same polypeptide as the phosphatase domain set PHLPP aside from most Ser/Thr phosphatases, which typically consist of a separate phosphatase domain and separate regulatory proteins that drive localization and specificity (33). In this regard, PHLPP is more like the protein tyrosine phosphatases, which usually have targeting domains (e.g. PDZ or SH2 domains) present in the same polypeptide as the catalytic domain (34; 35). This built-in targeting likely plays a determining role in restricting the substrate specificity of PHLPP in cells.
Amino Terminus
The amino terminus of PHLPP1β and PHLPP2 contain a predicted Ras Association (RA) domain, an approximately 90 kDa globular domain that binds Ras (36). It remains to be determined whether this region in PHLPP binds to Ras, but PHLPP1β (SCOP) was reported to bind to Ras in lipid rafts via the LRR domain (37). Additionally, the amino terminus of these isozymes contains a poly-Thr and poly-Ser low complexity region. The function of these determinants remains to be examined.
PH domain
PH domains are β-strand rich globular structures of approximately 120 amino acids of which a small subset serve as membrane-targeting modules that specifically recognize phosphoinositides with varying affinity (38–40). The PH domain of PHLPP has been classified as a ‘weak’ binder of phosphoinositides based on an algorithm developed by Teruel and coworkers for predicting phosphoinositide binding of these domains (41). Consistent with this, only the middle Arg of an R-x-R-x-F consensus sequence for phosphoinositide binding is present in the PH domains of PHLPP1 and PHLPP2 (42). Although the isolated domain localizes to plasma membrane under some conditions (41), its role in regulating the membrane localization of PHLPP remains to be determined.
LRR
PHLPP1 and PHLPP2 contain approximately 20 LRR segments, a structural motif consisting of 20 – 30 residues enriched in Leu and other hydrophobic residues (43). These repeating segments typically fold into a horseshoe structure, with a curved parallel β sheet on the inner surface of the shoe and α helices splayed on the outer surface. These segments play key roles in protein;protein interactions and are likely to be key regulators of PHLPP.
Phosphatase domain
The phosphatase domain of PHLPP belongs to the PP2C subclass of the PPM (Protein Phosphatase Mg2+-activated) family of Ser/Thr phosphatases. This subclass contains 11 other known PP2C members: PP2C α, β, γ, δ, ε, ζ, and η, Wip1, CaMKP (also referred to as hFEM2 or POPX2), CaMKP-N (POPX1), and NERPP-2C (33; 44; 45). The PP2C domain of PHLPP1 and 2 form a distinct subclass within the PP2C family (46). PP2C members, including PHLPP, are notorious for their resistance to phosphatase inhibitors such as okadaic acid and calyculin A.
The crystal structure of PP2Cα with bound phosphate identified a group of invariant carboxylate-containing residues that coordinate two metal ions (47): these residues are conserved in the PHLPP phosphatase domain, with Asp698, Gly699, Lys872, and Aps901 in PHLPP1α corresponding to Asp60, Gly61, Asp239, and Asp282 in PP2Cα. Similarly, an invariant basic residue that anchors the phosphate in the active site of PP2Cα (Arg33) is present in both PHLPP isoforms (Lys673 in PHLPP1 and Arg795 in PHLPP2).
The PP2C domain of PHLPP has intrinsic catalytic activity towards protein substrates and synthetic substrates such as p-nitrophenylphosphate (16; 24). This activity depends on Mn2+. Recently, coupling of biochemical and virtual screens identified a number of first-generation PHLPP inhibitors, which, if optimized for higher affinity binding, may hold promise as pharmacological tools to assess PHLPP function in cells (48).
PDZ Ligand
The last 3 residues of both PHLPP1 and PHLPP2 conform to the consensus sequence for a PDZ (post synaptic density protein (PSD95), Drosophila disc large tumor suppressor (Dlg1), and zonula occludens-1 protein (zo-1)) ligand. Specifically, they encode a Type 1 PDZ ligand, TPL for PHLPP1 and TAL for PHLPP2 (24). PDZ ligands tether their parent protein to scaffolds that serve as platforms to coordinate signalling (49). As discussed below, a number of scaffolds for the PDZ ligand of PHLPP have been identified. Additionally, the integrity of the PDZ ligand is required for PHLPP1 to recognize AKT in cells (16).
B. Substrates of PHLPP
PHLPP was originally discovered in a rational search for the phosphatase that dephosphorylates the hydrophobic motif of AKT and PKC; it was subsequently also shown to dephosphorylate the hydrophobic motif of S6K (Figure 2). This function, as the hydrophobic motif phosphatase, remains the best characterized function of PHLPP to date. However, given that there are over 100,000 phospho-Ser/Thr sequences in cells (50), and fewer than 40 Ser/Thr phosphatases, it is likely that PHLPP has an abundance of other substrates.
Hydrophobic motif phosphatase
PHLPP selectively dephosphorylates the hydrophobic motif of AKT, PKC, and S6 kinase (16; 24; 51; 52). In the case of AKT, dephosphorylation of this site (Ser473 of AKT1) reduces its intrinsic catalytic activity, leading to increased apoptosis and decreased proliferation (16; 53). In the case of PKC, dephosphorylation at this site (Ser660 of PKCβII) destabilizes PKC and shunts it to degradation pathways. In the case of S6 kinase, dephosphorylation of the hydrophobic motif (Thr389) reduces its activity, with the functional consequence of reducing overall protein translation (54). This selectivity for the hydrophobic motif may reflect unique interactions of the motif with the substrate binding cavity of the phosphatase domain; molecular docking reveals that the signature Phe at the P-1 position of the motif is accommodated in a hydrophobic pocket (Figure 3B) (48). Thus, PHLPP terminates signaling of three AGC kinases by selective dephosphorylation of their hydrophobic motif.
Figure 3. PHLPP phosphatase domain structure and model with hydrophobic motif peptide substrate bound.
A. Homology model of PP2C domain of PHLPP2, based on crystal structure of PP2Cα (47), showing two of the active site acidic residues (Asp 806, Asp 1024; pink) that coordinate two Mn2+ (yellow spheres) (48); Leu 1016 which is present as Ser in 30% of the population is highlighted in orange. B. Surface rendition of the active site of PHLPP2 docked with a phosphorylated hydrophobic motif peptide from AKT (HFPQFpSYSAS; phosphorylated Ser (Ser473) in magenta; active site residues and Mn2+ colored as in A.
Mst1
Mst-1 (the human homologue of Hippo) is a pro-apoptotic kinase that is regulated by an inhibitory phosphorylation on Thr387 catalyzed by AKT. Upon dephosphorylation of this site, Mst1 becomes activated. Pardee and co-workers recently identified PHLPP as the phosphatase of this inhibitory site (55). Specifically, they showed that PHLPP binds Mst1 and catalyzes the direct dephosphorylation of Thr387, activating the enzyme to phosphorylate its downstream targets p38 and JNK to induce apoptosis and growth arrest. This activation of Mst1 by PHLPP is further enhanced because PHLPP inactivates AKT, thus impairing the phosphorylation of Thr387. Thus, this double regulation of Mst1 at its inhibitory site by direct dephosphorylation of PHLPP and, indirectly, by inactivation of AKT provides a highly-sensitive mechanism by which PHLPP promotes apoptosis, consistent with its major role as a tumor suppressor (55).
C. Localization
Protein scaffolds play key roles in directing downstream signaling by poising kinases and phosphatases near relevant substrates, thus coordinating efficient signal propagation and signal termination (56). Nowhere is this better understood than in the signaling mediated by the archetypal kinase, PKA; here, scaffolds known as A Kinase Anchoring Proteins (AKAPs) position PKA, phosphatases, and substrates on signaling platforms to ensure high specificity and fidelity in downstream signaling (57). Although much less is known about the role of scaffolds in AKT signaling, compartmentalization is likely to be as critical for specificity and fidelity in AKT signaling as it is for PKA signaling. Certainly examples of compartmentalization exist, but they are few. Notably, β-arrestin has been shown to coordinate AKT and PP2A on the same signaling platform (58). The scaffold CNK1, which contains multiple protein interaction domains, including a PDZ domain, scaffolds AKT to enhance FOXO phosphorylation (59). Interestingly, this scaffold also coordinates Mst-1, another PHLPP substrate. No partner for the PDZ domain of CNK1 has been identified (60), and it will be particularly interesting to determine whether PHLPP is scaffolded next to these two substrates. Additionally, lipid rafts have been shown to concentrate AKT signaling (61). Scaffold interactions likely play an essential role in driving spatial and temporal specificity in the ability of PHLPP to suppress AKT signaling. As noted above, loss of the PDZ ligand of PHLPP1 removes its ability to recognize AKT in cells.
Compartmentalization in PHLPP signaling is supported by studies showing specificity in the regulation of AKT isoforms by PHLPP isoforms (24; 62). Specifically, knockdown and co-immunoprecipitation studies have revealed that PHLPP1 binds and dephosphorylates AKT2 and AKT3, but not AKT1 in cells, whereas PHLPP2 binds and dephosphorylates AKT1 and AKT3, but not AKT2, in cells (24). Furthermore, knockdown of specific PHLPP isoforms differentially regulates AKT substrates, suggesting that a scaffold may coordinate signaling between specific PHLPP isoforms, AKT isoforms, and a select set of AKT substrates. Thus, knockdown in H157 cells of PHLPP1 or AKT2 affects the phosphorylation of HDM2 and GSK3α, but not substrates such as p27, whose phosphorylation is affected upon knockdown of either PHLPP2 or AKT3. This wiring in signaling has also been demonstrated in pancreatic cells, where PHLPP1 regulates AKT2 and PHLPP2 regulates AKT1 (63). AKT2, but not AKT1, is up regulated in human pancreatic ductal adenocarcinomas. Importantly, high levels of PHLPP1 (but not PHLPP2) have been reported to correlate with longer survival times of patients with human pancreatic ductal adenocarcinomas, presumably by suppressing AKT2 activity (63). These data strongly support specific wiring in signaling to and from AKT. Given that deletion of the last 3 amino acids of PHLPP1 no longer allows PHLPP1 to recognize AKT in cells, PDZ scaffolds likely are critical determinants in this wiring.
PDZ Domain Scaffolds
A number of PDZ domains have been identified as binding partners for PHLPP. The PDZ domain protein NHERF1, previously shown to bind PTEN via its PDZ ligand (64), has been recently shown to also bind PHLPP1 and PHLPP2 via their PDZ ligands (65; 66). NHERF is a signaling platform for a variety of signaling molecules (67). Relevant to AKT signaling, it binds the EGF and PDGF receptors through its PDZ ligand (68; 69). PHLPP1 also binds the scaffold Scribble via a PDZ domain interaction (70).
Other scaffolds
Levels of the tumor suppressor scaffold FKBP5 (FK506 binding protein 5) have been shown to be a predictor of the response to chemotherapeutics, with higher levels promoting greater sensitivity to chemotherapeutics (71). This scaffold was recently shown to coordinate PHLPP and AKT in pancreatic cancer cells, thus facilitating the PHLPP-mediated dephosphorylation of AKT (62). This led to the proposal that the scaffold facilitates termination of AKT signaling by PHLPP, allowing greater sensitivity to chemotherapeutics (62; 72). This hypothesis is strongly supported by studies using a pancreatic mouse xenograft model that show that down-regulation of FKBP5 promotes tumor growth and resistance to the chemotherapeutic gemcitabine (73). Thus, levels of FKBP5 serve as a biomarker for the chemotherapeutic responsiveness in pancreatic cancer.
3. PHLPP regulation and how this is disrupted in disease
Protein:protein interactions critically regulate the access of PHLPP to its substrates. Whether post-translational modifications alter these interactions, or whether these binding partners alter the intrinsic catalytic activity of PHLPP, remains to be determined. An abundance of putative phosphorylation, acetylation, and ubiquitination sites are predicted for PHLPP (74). Additionally, several phosphorylated sequences have been reported, including Y1712 directly preceding the PDZ ligand in PHLPP1 (75). PHLPP activity and expression levels are likely to be under exquisite control by multiple inputs. To date, two feedback pathways have been identified that control the steady state levels of PHLPP: one that controls the rate of degradation of PHLPP and one that controls the rate of biosynthesis.
A. Feedback loop from AKT controls PHLPP degradation
Curiously, high levels of AKT activity correlate with high levels of both PHLPP1 and PHLPP2 in a variety of cancer cell lines (76; 77). One mechanism that accounts for this is a feedback loop in which high AKT activity increases the activity of its negative regulator, PHLPP, by promoting the stability of the phosphatase (Figure 2). Specifically, Gao and coworkers showed that GSK3β, which is inhibited upon phosphorylation by AKT, phosphorylates a site on PHLPP1 to create a phosphodegron (76). This phosphodegron is recognized by β-TrCP (β-transducin repeats-containing protein), an E3 ligase that serves as the substrate recognition subunit in the SCF (Skp1-Cullin 1-F-box protein) protein complex, causing the ubiquitination and subsequent degradation of PHLPP. Levels of PHLPP1 and β-TrCP are inversely correlated in a number of colon cancer cell lines, suggesting that high levels of this E3-ligase may be responsible, in part, for the low level of PHLPP1 often associated with tumors.
A screen of the NCI60 panel of tumor cell lines identified a subset of tumors in which the negative feedback loop between AKT and PHLPP1 is lost (77). Specifically, the cellular levels of PHLPP1 were found to be insensitive to manipulation of AKT activity in high-grade glioblastoma (GBM) cell lines. Cellular fractionation revealed that, in astrocytoma cell lines and normal brain tissue, β-TrCP1 is predominantly cytoplasmic, whereas in GBM cell lines and patient-derived tumor neurospheres, β-TrCP1 is confined to the nucleus and thus spatially separated from PHLPP1, which is predominantly cytoplasmic. As a result, in GBM, although PHLPP1 is properly phosphorylated by GSK3β, it can no longer interact with β-TrCP1, and PHLPP1 levels are no longer sensitive to AKT activity. Consistent with this, reintroduction of β-TrCP1 to the cytosol of GBM cells was sufficient to restore the ability of AKT to control PHLPP1 levels. This novel mechanism for the dysregulation of PHLPP1 levels by mislocalization of β-TrCP may be relevant to the many other SCF β-TrCP substrates whose function is dysregulated in cancer. Interestingly, PHLPP1 mRNA levels were consistently reduced in a majority of the GBM cell lines tested compared to low grade astrocytomas, suggesting that dysregulation of PHLPP at the transcriptional level may be responsible for promoting AKT signaling in this disease.
B. Feedback loop from mTOR and S6 kinase controls PHLPP biosynthesis
The rate of biosynthesis of PHLPP is also controlled by a feedback loop. Specifically, the rate of translation of PHLPP is controlled by the kinase complex mTORC1 via its substrates S6 kinase and 4EBP1 (Figure 2). Thus, treating cells with the mTORC1 inhibitor rapamycin or genetically interfering with the mTORC1 complex decreases PHLPP1 and PHLPP2 protein levels without affecting protein degradation or mRNA expression; this rapamycin-induced down regulation of PHLPP levels is blocked by expression of a constitutively-active, mTORC1-independent, construct of S6K (T389E has a phosphomimetic at the hydrophobic motif, the site phosphorylated by mTORC1) (78). Consistent with this, PHLPP1 depletion in 3T3 fibroblasts, and the subsequent increase in AKT signalling, has been shown to increase PHLPP2 protein levels by a rapaymcin-sensitive mechanism (79).
An important ramification of the ability of PHLPP to directly dephosphorylate and thus inactivate S6K pertains to the well-established negative feedback loop from S6K to IRS-1 (Figure 2). High PHLPP activity results in decreased S6K activity and, in turn reduced phosphorylation and degradation of IRS-1, thus activating PI3K signalling. Indeed, PHLPP knockdown decreases IRS-1 protein levels and consequent insulin-dependent activation of AKT, in addition to promoting phosphorylation of the direct down-stream target of S6K, ribosomal protein S6 and protein translation (52).
C. Proteolysis
Agonist-regulated proteolysis may be yet another mechanism to regulate the cellular levels of PHLPP. In this regard, Shimizu et al. have reported that treatment of hippocampal neurons with brain derived neurotrophic factor (BDNF) results in a decrease in the cellular levels of PHLPP1β (referred to as SCOP (23)) (80). This agonist-triggered decrease in PHLPP1β levels was prevented by calpain inhibitors, suggesting proteolysis by the Ca2+-regulated calpains. Consistent with calpain-dependent proteolysis, purified calpains promote the proteolysis of PHLPP1β in vitro (80). These findings suggest a role for calpain-mediated proteolysis in regulating the steady state levels of PHLPP1β in neurons (81).
D. Transcriptional regulation
The regulation of PHLPP expression by epigenetic mechanisms presents an attractive target for cancer therapy. However, very little is currently known about how PHLPP transcription is regulated. A recent study reported that PHLPP levels are controlled by histone deacetylases (HDACs), likely via a transcriptional mechanism. Specifically, HDAC3-deficient chondrocytes were shown to have elevated levels of PHLPP1 protein and mRNA; furthermore, treatment of cells with HDAC inhibitors increased the steady state levels of PHLPP1 (82). Histone acetylation is a major mechanism for epigenetic regulation of protein expression levels, and, as such, histone deacetylases (HDACs) are of significant interest as chemotherapeutic targets (83; 84). Whether this will be an attractive way to up regulate PHLPP levels remains to be determined.
E. Polymorphic variant of PHLPP2
PHLPP2 activity is sensitive to a polymorphism that affects 30% of the human population: a single amino acid change (Leu 1016 Ser; see Figure 3A) within the phosphatase domain of PHLPP2 reduces its rate of dephosphorylation of AKT and PKC approximately 5-fold in vitro, thus rendering it ineffective towards AKT and PKC in cells and reducing PHLPP2-dependent apoptosis (85). Whether this polymorphism predisposes to cancer is unclear, however it is noteworthy that in 3 out of 3 heterozygous patients with high-grade breast cancers, the Leu variant (active towards AKT and PKC) was shown to be lost in the tumor. This residue is situated in the predicted substrate-binding cleft of PHLPP2 (see Figure 3B), suggesting that the less abundant Ser variant has a lower affinity for substrates such as AKT and PKC.
4. PHLPP in prostate cancer & therapy
Human analysis and mouse models of prostate cancer have firmly established the pivotal role of the PI 3-Kinase pathway in prostate cancer (86). Classic genetic analysis revealed that PTEN-loss is more often driving the disease than activating mutations in pathway oncogenes. Below, we review how our understanding of PHLPP function and regulation offers potential strategies to fighting the disease.
A. The prostate cancer classification problem
Prostate cancers are unique in that they usually cannot be seen, felt or imaged before invasive diagnosis. The lack of reliable tools to separate the many patients with indolent prostate cancer from the few with potentially aggressive disease presents an enormous health problem: every year one million biopsies lead to over 200,000 newly diagnosed prostate cancers in the US alone. Landmark studies have now shown that this approach results in over-treatment of many men whose tumors would never become life threatening (87); and at the same time men with life-threatening disease can still be missed, due to the random nature of biopsy sampling. Uncertainty in patient stratification mostly stems from inherent problems in today’s standard of care: histopathology based Gleason scoring. Even though it represents the best parameter for patient risk, its problems are three-fold. First, there is well-documented inter-observer as well as intra-observer variability when scoring the same tissue slide. Second, there are large discrepancies in disease outcome that have been observed for men in spite of similar Gleason scores. This represents an acute problem as the majority of men are diagnosed with the intermediate Gleason score of 7. Statistical tools for outcome prediction based on basic tumor and patient information help to ameliorate this problem by making full use of the wealth of available outcome data (88). Thirdly, there are clear limitations in tissue sampling, as biopsies are taken randomly from prostate with an effort to cover most regions but usually without certainty of hitting the most advanced tumor region. Therefore, there is a need to identify sensitive molecular events that help better define the disease. As discussed below, the combination of human genome analysis and basic research into PHLPP function has revealed a major player in this disease.
B. PHLPP genes are critical for prostate tumor suppression
The recent large scale coordinated effort by the Memorial Sloan Kettering Prostate Oncogenomics Group succeeded in generating the most comprehensive genomic analysis of prostate cancer to date (1). Their approach combined genome wide copy number and gene expression analysis with candidate cancer gene re-sequencing for detection of mutations. This analysis revealed that both PHLPP1 and PHLPP2 genes are deleted at surprisingly high frequency, even rivaling that of PTEN-deletion. In contrast to PTEN however, the PHLPP1 and PHLPP2 deletions on chromosome 18q21 and 16q22, respectively, are typically not focal in nature and encompass hundreds of other genes (79). However, the generation and analysis of Phlpp1 knockout mice revealed highly penetrant neoplasia and cancer in prostate, thus firmly establishing the PHLPP1 protein as a tumor suppressor phosphatase in the pathway (79). Importantly, disease onset was accelerated and outcome exacerbated when Phlpp1-null mice also lost a single copy of Pten, demonstrating strong cooperation between the two tumor suppressors. Of note, both PTEN and PHLPP1 loss in human cells and mouse prostate trigger a feedback surge by PHLPP2 protein, which likely delays but does not prevent the disease phenotype. As a result PHLPP1 and PHLPP2 genes are often co-deleted in metastatic disease (79). Intriguingly, patient prostate genomic analysis revealed that PHLPP2 is even more frequently deleted in primary prostate cancer (15%) than PTEN or PHLPP1 (11% and 7%, respectively, (1)), and furthermore, it is also more frequently deleted in metastatic samples (62%, compared to 51% and 43%, for PTEN and PHLPP1, respectively). Thus, the PHLPP loci on Chromosomes 16q and 18q are emerging as hotspots for molecular prostate cancer classification.
C. Compensatory feedback loops in prostate cancer
Therapeutic progress and understanding of the PI 3-Kinase pathway have been mired by unexpected feedback loops, and it has been realized that mechanistic pathway signaling studies are paramount to make sense of basic results obtained with pathway inhibitors such as the rapalogs (1). As discussed above (see Section 3B), drugs like rapamycin, which block mTORC1 downstream of AKT, can lead to upstream PI 3-kinase and AKT activation through decreased S6 kinase function (89). The PHLPP2 protein was also identified as part of a negative feedback system, which can sense excessive cancer signaling: in prostate, primary mouse cells, and human cell lines, PHLPP2 levels rise upon PI 3-Kinase pathway activation such as PTEN- and/ or PHLPP1- inactivation (see Figure 4). By analogy to S6-kinase activation, the surge in PHLPP2 after PTEN/ PHLPP1 loss is mediated by an mTORC1-dependent mechanism, thus introducing a second major negative pathway feedback (78; 79; 85). This feedback however is not aimed at the RTK but it reverses AKT activation. On top of these two feedback mechanisms, two more have been shown to be critical for tumor suppression: PTEN or PHLPP1 loss activates p53, leading to cellular senescence ((90), see SIDE BAR 2), and similarly, PTEN-loss activates SMAD4 to prevent tumor progression (91). The importance of these feedback controls is revealed by genetic ablation of the fail-safes: while mice with Pten-deficient prostate live a normal life span in spite of prostate lesions, double mutant Pten/Trp53 or Pten/Smad4- null mice die of growths in their prostates, which give rise to masses of several centimeters in diameter. As summarized in Figure 4, these feedbacks are mediated by mTORC1 driven translation, strongly suggesting that rapalogs should only be used after these tumor suppressive fail-safes have been abolished by the cancer. Thus to summarize, multiple lines of evidence have confirmed that a hierarchical organization for compensation among PTEN, PHLPP1, and PHLPP2 exists with PHLPP2 strongly responding to pathway activation including TSC-loss (79). It remains to be seen how efficiently this response delays tumor progression, which can be tested with Phlpp2 knockout mice. The emerging picture of mTORC1 controlling several major tumor suppressors that limit the PI 3-kinase pathway signaling clearly calls for highly refined therapeutic approaches. These must achieve a magic bullet trick and circumvent blocking tumor suppressor activation that depends on mTORC1 while still limiting growth supporting translational functions of the TOR complex. Beyond that, molecular patient tumor analysis will become essential for drawing conclusions on the status of feedback responses to ensure that these will not suffer collateral damage.
Figure 4. Feedback loops and therapy.
mTORC1 activation in cancer results in increased protein translation needed for cell growth and proliferation (black arrows). When mTORC1 signaling is strong, mTORC1 activates three tumor suppressive responses (red arrows) that attenuate the pathway via 1] increasing PHLPP or 2] irreversibly arrest the cell by increasing p53. In prostate, androgen receptor (AR) is activated by RTK signaling and limits AKT signaling through FKBP5-mediated increase in PHLPP activity towards AKT. The pathway’s tumor suppressive feedbacks can be inadvertently blocked by therapeutics (green arrows). First, AR blockade by hormone therapy inhibits PHLPP suppression of AKT, thus activating AKT. Second, mTORC1-targeting kinase inhibitors such as rapalogs block three known negative feedbacks: S6 kinase-dependent negative feedback loop to PI3K signaling via RTKs, mTORC1-dependent increase in PHLPP levels, and mTORC1-dependent increases in p53 levels.
SIDE BAR 2.
p53 and senescence
Fully activated oncogenes can trigger cellular senescence, an irreversible form of growth arrest that can protect cells from cancer (121). Similarly, it has been realized that rampant activation of the PI 3-Kinase pathway via loss of tumor suppressors results in senescence arrest (79; 90) that may arguably be exploited therapeutically (122). This response is, however, critically dependent on activation of (non-mutant) p53, achieved by increased translation through mTOR after loss of PHLPP1 (79) or PTEN (123). Thus, mTOR-inhibiting drugs, such as the rapalogs, efficiently block p53 activation after PHLPP-loss. Note, that in contrast mutant Ras activates p53 and senescence through DNA damage response, which is not seen in PHLPP/PTEN-loss induced senescence.
D. PHLPP and PTEN Phosphatases in metastasis
The pathway limiting feedbacks responses pose a progression problem that metastatic cancers must successfully surmount. Indeed, this is exactly what can been observed when metastatic cancer genomes are analyzed. Most of these samples that carry a PTEN alteration also show deletion of all the above-mentioned feedback genes, TP53, PHLPP2, and SMAD4. In addition, most PTEN-deleted metastases lose a copy of the PHLPP1 gene, suggesting that the PI 3-kinase pathway is activated and unchallenged at this stage (1; 79). Note that SMAD4 and PHLPP1 are located close to each other on chromosome 18q and are typically found co-deleted. Intriguingly, genetic dissection of the role of feedback genes in mouse prostate has revealed that loss of Trp53 and Smad4 will not give rise to a phenotype in this tissue (90; 91). This finding is in contrast to the role of the phosphatases, which clearly initiate disease when lost on their own. These findings further emphasize the concept of the feedback genes as being called upon to prevent metastatic disease progression only after the breaks of the PI 3-kinase pathway, PHLPP1/2 and PTEN, have failed. This concept explains why the status of feedback tumor suppressors is critically affecting future disease course. This notion is supported by analysis of the predictive power of loss of the tumor suppressors that are known to trigger a feedback response. Loss or low PTEN and PHLPP1 mRNA expression combined can predict a fast Biochemical Recurrence (BCR, defined by PSA above 0.2ng/ml on two consecutive measurements (79)). At the same time low PHLPP2 mRNA expression is also significantly associated with biochemical relapse of patients after surgery. This finding is consistent with a critical role for PHLPP2 in antagonizing disease progression of PTEN/ PHLPP1 mutant cancers, as discussed in detail above. Furthermore, a PTEN/ SMAD4 based signature for disease progression has also been developed (91). Collectively, these results show that understanding the molecular events and appreciating the natural feedbacks that they cause will greatly enhance our ability to correctly predict a tumor’s future course. This remains an ultimate aim for prostate cancer research, where molecular signatures for the many men without life threatening disease are still not known.
E. PHLPP and hormone therapy
Patients with low or absent PTEN/ PHLPP phosphatase RNA tend to relapse years earlier than those with normal or high RNA levels (79). Thus, it follows that maintaining strong PHLPP/ PTEN protein expression is critical to achieve extended life expectancy in patients who based on their cumulative risk factors have decided to get their prostates removed – at this time roughly 1 million men in the U.S. alone. The standard of care after recurrence is hormone withdrawal therapy (92). Yet strikingly, a recent study has revealed a disturbing connection (93): androgen receptor (AR) inhibitors can lower PHLPP1 protein levels, thus hampering the benefit of this routine approach, at least in patients that have retained PHLPP1 protein in their prostates. Mechanistically, it was shown that the AR target gene, FKBP5, acts as a PHLPP1 chaperone (62). Hence, in addition to cell survival and identity, AR seems to be actively involved in limiting AKT signaling in the prostatic epithelium. Adding to the crosstalk between the PI 3-Kinase pathway and AR signaling, it was found that blocking RTKs could activate AR signaling (see Feedback Figure 4). These data thus suggested that therapeutic intervention in prostate would benefit from a two-pronged approach, which was validated in genetically engineered mice: while the PI 3-kinase pathway inhibitor NVP-BEZ235 effectively reduced disease progression in mice with Pten-deficient prostates, it showed no effect in cancers driven by the c-Myc oncogene. In contrast, combined androgen and PI3K inhibition was highly effective in both models, as well as in mice carrying human prostate cancer xenografts. Collectively, these data demonstrate that standard of care therapy can be mired by unexpected oncogenic survival feedbacks, which must be taken into account to optimize individualized cancer care.
5. PHLPP modulation to fight disease
The central role of PHLPP in AKT-inactivation immediately suggests an entry point for both positive and negative regulation of PI 3-Kinase signaling in disease. Two opposite scenarios can be envisioned: (1) enhancing PHLPP function to block proliferation, and (2) inhibiting PHLPP to support cell survival.
A. Supporting PHLPP function in cancer and its therapy
As discussed in Section 3, basic research has afforded us with a wealth of mechanistic insights into the control of PHLPP levels. Since the vast majority of cancer genome studies report only single copy deletion of the PHLPP genes (source: cBioPortal for Cancer Genomics: http://www.cbioportal.org/public-portal/cross_cancer.do), the approach of boosting the output of the remaining allele has great potential in several tumor types. Moreover, the restoration of PHLPP function may benefit cancer therapy even in tumors that are not driven by PHLPP-loss. It has been shown, that the successful response to several RTK target therapies correlates well with intact PTEN gene status and function ((94; 95). These studies reveal that small molecule RTK inhibitors cooperate with the PI 3-Kinase pathway tumor suppressor PTEN. Thus, there is a strong rationale for supporting PHLPP function to enhance the outcome of target therapy with, for example, Herceptin/ Trastuzumab in breast cancer.
Enhancing PHLPP function may also be helpful in boosting immune function. PHLPP suppresses AKT signaling in T regulatory cells, a suppression that is important for the development and function of these cells (96). The circadian clock also benefits from regulation by PHLPP: deletion of PHLPP1 has been shown to interfere with light-induced resetting of the circadian clock (97). Thus mechanisms to enhance PHLPP activity are likely to have multiple therapeutic benefits.
B. Inhibiting PHLPP function in disease
Impaired AKT signaling underlies a number of diseases in which cell survival pathways are inhibited, such as heart disease and diabetes. Thus, inhibition of PHLPP provides an attractive mechanism to enhance AKT signaling in such diseases. In this regard, first-generation inhibitors of PHLPP have been identified by chemical and virtual screening of the NCI Diversity Set, suggesting that PHLPP is a druggable target with potential as a therapeutic target (48; 98).
PHLPP is a potential target for treating metabolic disorders such as diabetes, where AKT signaling is severely dampened and insulin resistance occurs. By inhibiting PHLPP, AKT signaling would be enhanced, potentially overcoming insulin resistance. Consistent with this, the levels of PHLPP1 protein are highly elevated in skeletal muscle of obese humans, with a striking correlation between increased body mass index, insulin resistance, and increasing levels of PHLPP1 (99). Additionally, PHLPP1 mRNA levels are elevated in muscle cells from diabetic patients (100).
Neurons are very sensitive to changes in the PI 3-Kinase pathway. In contrast to the situation in cancer however, these cells often depend on AKT signaling for survival and recovery from insults: genetic experiments in mouse have demonstrated that AKT signaling is critical for neuronal branching and synaptic contact formation during development (101; 102). In the adult brain, AKT signaling is needed for survival after transient insults or in chronic conditions such as epilepsy and autism (103; 104). Transient pathway inhibition using small molecule antagonists such as rapamycin blocks regrowth and recovery of severed neurons, while deletion of Pten has the opposite effect (105; 106). Furthermore, recovery from stroke also requires transient AKT signaling for neuron survival and reduction of infarct size (107; 108). There is a wealth of literature on the benefits for both naturally occurring or pharmacologically induced PTEN-inhibition in restoring brain function (recently reviewed in (103)). Importantly, loss or knockdown of Phlpp1 has been shown to similarly protect neurons from death in a mouse model of stroke (109), strongly suggesting that PHLPP-inhibitors could reduce infarct size. These findings highlight the potential of PHLPP1/2 inhibition in chronic or acute brain disorders. Intriguingly, Phlpp1-inhibition might even be preferable over the use of PTEN-inhibitors: Phlpp1-deficient mice only show a slight change in brain size, and to date no overt brain cancers have been reported. In contrast, the various mouse models with Pten-deficient neurons suffer from macrocephaly, seizures, autism related disorders and are highly cancer prone (110; 111). Thus, PHLPP-inhibitors may be much safer and better tolerated than inhibitors of PTEN.
Similar to the findings in brain, PHLPP1 has been shown to block cell survival in the heart after ischaemia (112). Thus, transient administration of PHLPP1 inhibitors could also be used to limit the consequences of a heart attack.
6. Summary
By suppressing AKT signaling, PHLPP is poised as a major therapeutic target. However, given the central role of this novel phosphatase family in terminating cell signaling pathways, mechanistic insight into its precise cellular roles is essential before it is targeted in therapies. The ability to precisely control specific pathways suppressed by PHLPP will be essential to developing therapies to specific diseases. Perhaps the most promising avenues will be those involving altering the function of PHLPP on specific protein scaffolds. For example, one could envision AKT2 function being selectively enhanced by perturbing the binding of PHLPP1 to the scaffold coordinating AKT2-PHLPP1 signaling, providing a promising target for insulin resistance. As more mechanistic studies shed light on the substrates and signaling pathways involving PHLPP, strategies to specifically modulate specific functions will be designed.
Acknowlegements
We thank Bill Sinko for Figure 3B and members of the Newton and Trotman labs for helpful suggestions. This work was supported by NIH GM067946 (A.C.N.) and NIH CA137050 (L.C.T).
REFERENCES
- 1.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, et al. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 4.Toker A. Protein kinases as mediators of phosphoinositide 3-kinase signaling. Mol Pharmacol. 2000;57:652–658. [PubMed] [Google Scholar]
- 5.Chan TO, Rittenhouse SE, Tsichlis PN. AKT/PKB and other D3 phosphoinositide-regulated kinases: kinase activation by phosphoinositide-dependent phosphorylation. Annu Rev Biochem. 1999;68:965–1014. doi: 10.1146/annurev.biochem.68.1.965. [DOI] [PubMed] [Google Scholar]
- 6.Martelli AM, Tabellini G, Bressanin D, Ognibene A, Goto K, et al. The emerging multiple roles of nuclear Akt. Biochim Biophys Acta. 2012;1823:2168–2178. doi: 10.1016/j.bbamcr.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J Biol Chem. 2005;280:5581–5587. doi: 10.1074/jbc.M411534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altomare DA, Khaled AR. Homeostasis and the importance for a balance between AKT/mTOR activity and intracellular signaling. Curr Med Chem. 2012;19:3748–3762. doi: 10.2174/092986712801661130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memmott RM, Dennis PA. Akt-dependent and -independent mechanisms of mTOR regulation in cancer. Cell Signal. 2009;21:656–664. doi: 10.1016/j.cellsig.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannan KM, Sanij E, Hein N, Hannan RD, Pearson RB. Signaling to the ribosome in cancer--It is more than just mTORC1. IUBMB Life. 2011;63:79–85. doi: 10.1002/iub.428. [DOI] [PubMed] [Google Scholar]
- 12.Fayard E, Tintignac LA, Baudry A, Hemmings BA. Protein kinase B/Akt at a glance. J Cell Sci. 2005;118:5675–5678. doi: 10.1242/jcs.02724. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Paluch BE, Wang X, Jiang X. PTEN at a glance. J Cell Sci. 2012;125:4687–4692. doi: 10.1242/jcs.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 15.Maehama T, Dixon JE. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 16.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. Embo J.Embo J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 18.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 20.Zick Y. Uncoupling insulin signalling by serine/threonine phosphorylation: a molecular basis for insulin resistance. Biochem Soc Trans. 2004;32:812–816. doi: 10.1042/BST0320812. [DOI] [PubMed] [Google Scholar]
- 21.Harrington LS, Findlay GM, Lamb RF. Restraining PI3K: mTOR signalling goes back to the membrane. Trends Biochem Sci. 2005;30:35–42. doi: 10.1016/j.tibs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 23.Shimizu K, Okada M, Takano A, Nagai K. SCOP, a novel gene product expressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Lett. 1999;458:363–369. doi: 10.1016/s0014-5793(99)01190-4. [DOI] [PubMed] [Google Scholar]
- 24.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Goel A, Arnold CN, Niedzwiecki D, Chang DK, Ricciardiello L, et al. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 2003;63:1608–1614. [PubMed] [Google Scholar]
- 26.Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakha EA, Green AR, Powe DG, Roylance R, Ellis IO. Chromosome 16 tumor-suppressor genes in breast cancer. Genes, chromosomes & cancer. 2006;45:527–535. doi: 10.1002/gcc.20318. [DOI] [PubMed] [Google Scholar]
- 28.Safford SD, Goyeau D, Freemerman AJ, Bentley R, Everett ML, et al. Fine mapping of Wilms' tumors with 16q loss of heterozygosity localizes the putative tumor suppressor gene to a region of 6.7 megabases. Ann Surg Oncol. 2003;10:136–143. doi: 10.1245/aso.2003.03.038. [DOI] [PubMed] [Google Scholar]
- 29.Torring N, Borre M, Sorensen KD, Andersen CL, Wiuf C, Orntoft TF. Genome-wide analysis of allelic imbalance in prostate cancer using the Affymetrix 50K SNP mapping array. Br J Cancer. 2007;96:499–506. doi: 10.1038/sj.bjc.6603476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patael-Karasik Y, Daniely M, Gotlieb WH, Ben-Baruch G, Schiby J, et al. Comparative genomic hybridization in inherited and sporadic ovarian tumors in Israel. Cancer Genet Cytogenet. 2000;121:26–32. doi: 10.1016/s0165-4608(00)00224-7. [DOI] [PubMed] [Google Scholar]
- 31.Tsuda H, Zhang WD, Shimosato Y, Yokota J, Terada M, et al. Allele loss on chromosome 16 associated with progression of human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990;87:6791–6794. doi: 10.1073/pnas.87.17.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker DA, Kelly JM. Structure, function and evolution of microbial adenylyl and guanylyl cyclases. Mol Microbiol. 2004;52:1229–1242. doi: 10.1111/j.1365-2958.2004.04067.x. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, et al. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21:7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiganis T, Bennett AM. Protein tyrosine phosphatase function: the substrate perspective. Biochem J. 2007;402:1–15. doi: 10.1042/BJ20061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang L, Weng X, Hofer F, Martin GS, Kim SH. Three-dimensional structure of the Ras-interacting domain of RalGDS. Nat Struct Biol. 1997;4:609–615. doi: 10.1038/nsb0897-609. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu K, Okada M, Nagai K, Fukada Y. Suprachiasmatic nucleus circadian oscillatory protein, a novel binding partner of K-Ras in the membrane rafts, negatively regulates MAPK pathway. J Biol Chem. 2003;278:14920–14925. doi: 10.1074/jbc.M213214200. [DOI] [PubMed] [Google Scholar]
- 38.Misra S, Miller GJ, Hurley JH. Recognizing phosphatidylinositol 3-phosphate. Cell. 2001;107:559–562. doi: 10.1016/s0092-8674(01)00594-3. [DOI] [PubMed] [Google Scholar]
- 39.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 40.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, et al. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 41.Park WS, Heo WD, Whalen JH, O'Rourke NA, Bryan HM, et al. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell. 2008;30:381–392. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, et al. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- 43.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 44.Cohen PT. Novel protein serine/threonine phosphatases: variety is the spice of life. Trends Biochem Sci. 1997;22:245–251. doi: 10.1016/s0968-0004(97)01060-8. [DOI] [PubMed] [Google Scholar]
- 45.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 46.Brognard J, Newton AC. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol Metab. 2008;19:223–230. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. Embo J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- 48.Sierecki E, Sinko W, McCammon JA, Newton AC. Discovery of small molecule inhibitors of the PH domain leucine-rich repeat protein phosphatase (PHLPP) by chemical and virtual screening. J Med Chem. 2010;53:6899–6911. doi: 10.1021/jm100331d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell communication and signaling : CCS. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao T, Brognard J, Newton AC. The phosphatase PHLPP controls the cellular levels of protein kinase C. J Biol Chem. 2008;283:6300–6311. doi: 10.1074/jbc.M707319200. [DOI] [PubMed] [Google Scholar]
- 52.Liu J, Stevens PD, Li X, Schmidt MD, Gao T. PHLPP-mediated dephosphorylation of S6K1 inhibits protein translation and cell growth. Mol Cell Biol. 2011 doi: 10.1128/MCB.05799-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Molecular cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 54.Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 55.Qiao M, Wang Y, Xu X, Lu J, Dong Y, et al. Mst1 is an interacting protein that mediates PHLPPs' induced apoptosis. Mol Cell. 2010;38:512–523. doi: 10.1016/j.molcel.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 56.Scott JD, Newton AC. Shedding light on local kinase activation. BMC biology. 2012;10:61. doi: 10.1186/1741-7007-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michel JJ, Scott JD. AKAP mediated signal transduction. Annu Rev Pharmacol Toxicol. 2002;42:235–257. doi: 10.1146/annurev.pharmtox.42.083101.135801. [DOI] [PubMed] [Google Scholar]
- 58.Beaulieu JM, Sotnikova TD, Marion S, Lefkowitz RJ, Gainetdinov RR, Caron MG. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell. 2005;122:261–273. doi: 10.1016/j.cell.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 59.Fritz RD, Varga Z, Radziwill G. CNK1 is a novel Akt interaction partner that promotes cell proliferation through the Akt-FoxO signalling axis. Oncogene. 2010;29:3575–3582. doi: 10.1038/onc.2010.104. [DOI] [PubMed] [Google Scholar]
- 60.Fritz RD, Radziwill G. CNK1 and other scaffolds for Akt/FoxO signaling. Biochim Biophys Acta. 2011;1813:1971–1977. doi: 10.1016/j.bbamcr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 61.Gao X, Lowry PR, Zhou X, Depry C, Wei Z, et al. PI3K/Akt signaling requires spatial compartmentalization in plasma membrane microdomains. Proc Natl Acad Sci U S A. 2011;108:14509–14514. doi: 10.1073/pnas.1019386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nitsche C, Edderkaoui M, Moore RM, Eibl G, Kasahara N, et al. The phosphatase PHLPP1 regulates Akt2, promotes pancreatic cancer cell death, and inhibits tumor formation. Gastroenterology. 2012;142:377–387. e1–e5. doi: 10.1053/j.gastro.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takahashi Y, Morales FC, Kreimann EL, Georgescu MM. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. Embo J. 2006;25:910–920. doi: 10.1038/sj.emboj.7600979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Molina JR, Agarwal NK, Morales FC, Hayashi Y, Aldape KD, et al. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene. 2012;31:1264–1274. doi: 10.1038/onc.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kunkel MT, Garcia E, Hall R, Newton AC. PDZ interactions between PHLPP phosphatases and the NHERF scaffold. FASEB J. 2012;26 [Google Scholar]
- 67.Weinman EJ, Hall RA, Friedman PA, Liu-Chen LY, Shenolikar S. The association of NHERF adaptor proteins with g protein-coupled receptors and receptor tyrosine kinases. Annu Rev Physiol. 2006;68:491–505. doi: 10.1146/annurev.physiol.68.040104.131050. [DOI] [PubMed] [Google Scholar]
- 68.Lazar CS, Cresson CM, Lauffenburger DA, Gill GN. The Na+/H+ exchanger regulatory factor stabilizes epidermal growth factor receptors at the cell surface. Mol Biol Cell. 2004;15:5470–5480. doi: 10.1091/mbc.E04-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maudsley S, Zamah AM, Rahman N, Blitzer JT, Luttrell LM, et al. Platelet-derived growth factor receptor association with Na(+)/H(+) exchanger regulatory factor potentiates receptor activity. Mol Cell Biol. 2000;20:8352–8363. doi: 10.1128/mcb.20.22.8352-8363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X, Yang H, Liu J, Schmidt MD, Gao T. Scribble-mediated membrane targeting of PHLPP1 is required for its negative regulation of Akt. EMBO Rep. 2011;12:818–824. doi: 10.1038/embor.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L, Fridley B, Kalari K, Jenkins G, Batzler A, et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 2008;68:7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dibble CC, Manning BD. A molecular link between AKT regulation and chemotherapeutic response. Cancer Cell. 2009;16:178–180. doi: 10.1016/j.ccr.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 73.Hou J, Wang L. FKBP5 as a selection biomarker for gemcitabine and Akt inhibitors in treatment of pancreatic cancer. PLoS One. 2012;7:e36252. doi: 10.1371/journal.pone.0036252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 76.Li X, Liu J, Gao T. beta-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt. Mol Cell Biol. 2009;29:6192–6205. doi: 10.1128/MCB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Warfel NA, Niederst M, Stevens MW, Brennan PM, Frame MC, Newton AC. Mislocalization of the E3 Ligase, {beta}-Transducin Repeat-containing Protein 1 ({beta}-TrCP1), in Glioblastoma Uncouples Negative Feedback between the Pleckstrin Homology Domain Leucine-rich Repeat Protein Phosphatase 1 (PHLPP1) and Akt. J Biol Chem. 2010;286:19777–19788. doi: 10.1074/jbc.M111.237081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu J, Stevens PD, Gao T. mTOR-dependent regulation of PHLPP expression controls the rapamycin sensitivity in cancer cells. J Biol Chem. 2010;286:6510–6520. doi: 10.1074/jbc.M110.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, et al. Identification of PHLPP1 as a Tumor Suppressor Reveals the Role of Feedback Activation in PTEN-Mutant Prostate Cancer Progression. Cancer Cell. 2011;20:173–186. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shimizu K, Phan T, Mansuy IM, Storm DR. Proteolytic degradation of SCOP in the hippocampus contributes to activation of MAP kinase and memory. Cell. 2007;128:1219–1229. doi: 10.1016/j.cell.2006.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shimizu K, Mackenzie SM, Storm DR. SCOP/PHLPP and its functional role in the brain. Mol Biosyst. 2010;6:38–43. doi: 10.1039/b911410f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bradley EW, Carpio LR, Westendorf JJ. Histone deacetylase 3 suppression increases PH domain and leucine-rich repeat phosphatase (Phlpp)1 expression in chondrocytes to suppress Akt signaling and matrix secretion. J Biol Chem. 2013 doi: 10.1074/jbc.M112.423723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner JM, Hackanson B, Lubbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clinical epigenetics. 2010;1:117–136. doi: 10.1007/s13148-010-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hagelkruys A, Sawicka A, Rennmayr M, Seiser C. The biology of HDAC in cancer: the nuclear and epigenetic components. Handbook of experimental pharmacology. 2011;206:13–37. doi: 10.1007/978-3-642-21631-2_2. [DOI] [PubMed] [Google Scholar]
- 85.Brognard J, Niederst M, Reyes G, Warfel N, Newton AC. Common polymorphism in the phosphatase PHLPP2 results in reduced regulation of Akt and protein kinase C. The Journal of biological chemistry. 2009;284:15215–15223. doi: 10.1074/jbc.M901468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes & development. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thompson IM, Jr, Tangen CM. Prostate cancer--uncertainty and a way forward. The New England journal of medicine. 2012;367:270–271. doi: 10.1056/NEJMe1205012. [DOI] [PubMed] [Google Scholar]
- 88.Touijer K, Scardino PT. Nomograms for staging, prognosis, and predicting treatment outcomes. Cancer. 2009;115:3107–3111. doi: 10.1002/cncr.24352. [DOI] [PubMed] [Google Scholar]
- 89.Manning BD. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. The Journal of cell biology. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8:440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carver BS, Chapinski CC, Wongvipat J, Hieronymus H, Chen Y, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN--deficient prostate cancer. 2011 doi: 10.1016/j.ccr.2011.04.008. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 95.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. British journal of cancer. 2007;97:1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Patterson SJ, Han JM, Garcia R, Assi K, Gao T, et al. Cutting Edge: PHLPP Regulates the Development, Function, and Molecular Signaling Pathways of Regulatory T Cells. J Immunol. 2011 doi: 10.4049/jimmunol.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Masubuchi S, Gao T, O'Neill A, Eckel-Mahan K, Newton AC, Sassone-Corsi P. Protein phosphatase PHLPP1 controls the light-induced resetting of the circadian clock. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0910292107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sinko W, Sierecki E, de Oliveira CA, McCammon JA. Guide to Virtual Screening: Application to the Akt Phosphatase PHLPP. Methods Mol Biol. 2012;819:561–573. doi: 10.1007/978-1-61779-465-0_33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Andreozzi F, Procopio C, Greco A, Mannino GC, Miele C, et al. Increased levels of the Akt-specific phosphatase PH domain leucine-rich repeat protein phosphatase (PHLPP)-1 in obese participants are associated with insulin resistance. Diabetologia. 2011 doi: 10.1007/s00125-011-2116-6. [DOI] [PubMed] [Google Scholar]
- 100.Cozzone D, Frojdo S, Disse E, Debard C, Laville M, et al. Isoform-specific defects of insulin stimulation of Akt/protein kinase B (PKB) in skeletal muscle cells from type 2 diabetic patients. Diabetologia. 2008;51:512–521. doi: 10.1007/s00125-007-0913-8. [DOI] [PubMed] [Google Scholar]
- 101.Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3'-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ning K, Drepper C, Valori CF, Ahsan M, Wyles M, et al. PTEN depletion rescues axonal growth defect and improves survival in SMN-deficient motor neurons. Human molecular genetics. 2010;19:3159–3168. doi: 10.1093/hmg/ddq226. [DOI] [PubMed] [Google Scholar]
- 103.Naguib A, Trotman LC. PTEN plasticity: how the taming of a lethal gene can go too far. Trends in Cell Biology. 2013 doi: 10.1016/j.tcb.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pun RY, Rolle IJ, Lasarge CL, Hosford BE, Rosen JM, et al. Excessive activation of mTOR in postnatally generated granule cells is sufficient to cause epilepsy. Neuron. 2012;75:1022–1034. doi: 10.1016/j.neuron.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun F, Park KK, Belin S, Wang D, Lu T, et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ferguson TA, Son YJ. Extrinsic and intrinsic determinants of nerve regeneration. J Tissue Eng. 2011;2 doi: 10.1177/2041731411418392. 2041731411418392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Howitt J, Lackovic J, Low LH, Naguib A, Macintyre A, et al. Ndfip1 regulates nuclear Pten import in vivo to promote neuronal survival following cerebral ischemia. The Journal of cell biology. 2012;196:29–36. doi: 10.1083/jcb.201105009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lackovic J, Howitt J, Callaway JK, Silke J, Bartlett P, Tan SS. Differential regulation of Nedd4 ubiquitin ligases and their adaptor protein Ndfip1 in a rat model of ischemic stroke. Exp Neurol. 2012;235:326–335. doi: 10.1016/j.expneurol.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 109.Chen B, Van Winkle JA, Lyden PD, Brown JH, Purcell NH. PHLPP1 gene deletion protects the brain from ischemic injury. J Cereb Blood Flow Metab. 2013;33:196–204. doi: 10.1038/jcbfm.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou J, Parada LF. PTEN signaling in autism spectrum disorders. Curr Opin Neurobiol. 2012;22:873–879. doi: 10.1016/j.conb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 111.Endersby R, Baker SJ. PTEN signaling in brain: neuropathology and tumorigenesis. Oncogene. 2008;27:5416–5430. doi: 10.1038/onc.2008.239. [DOI] [PubMed] [Google Scholar]
- 112.Miyamoto S, Purcell NH, Smith JM, Gao T, Whittaker R, et al. PHLPP-1 Negatively Regulates Akt Activity and Survival in the Heart. Circ Res. 2010;107:476–484. doi: 10.1161/CIRCRESAHA.109.215020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boulbes DR, Shaiken T, Sarbassov dos D. Endoplasmic reticulum is a main localization site of mTORC2. Biochem Biophys Res Commun. 2011;413:46–52. doi: 10.1016/j.bbrc.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, et al. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. Embo J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 116.Toker A, Newton A. Cellular Signalling: Pivoting around PDK-1. Cell. 2000;103:185–188. doi: 10.1016/s0092-8674(00)00110-0. [DOI] [PubMed] [Google Scholar]
- 117.Behn-Krappa A, Newton AC. The hydrophobic phosphorylation motif of conventional protein kinase C is regulated by autophosphorylation. Curr Biol. 1999;9:728–737. doi: 10.1016/s0960-9822(99)80332-7. [DOI] [PubMed] [Google Scholar]
- 118.Toker A, Newton AC. Akt/protein kinase B is regulated by autophosphorylation at the hypothetical PDK-2 site. J Biol Chem. 2000;275:8271–8274. doi: 10.1074/jbc.275.12.8271. [DOI] [PubMed] [Google Scholar]
- 119.Feng J, Park J, Cron P, Hess D, Hemmings BA. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J Biol Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- 120.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 121.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 122.Nardella C, Clohessy JG, Alimonti A, Pandolfi PP. Pro-senescence therapy for cancer treatment. Nature reviews. Cancer. 2011;11:503–511. doi: 10.1038/nrc3057. [DOI] [PubMed] [Google Scholar]
- 123.Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120:681–693. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]