Abstract
Autophagy is an important catabolic cellular process that eliminates damaged and unnecessary cytoplasmic proteins and organelles. Basal autophagy occurs during normal physiological conditions, but the activity of this process can be significantly altered in human diseases. Thus, defining the regulatory inputs and signals that control autophagy is essential. Nutrients are key modulators of autophagy. While autophagy is generally accepted to be regulated in a cell autonomous fashion, recent studies suggest nutrients can modulate autophagy in a systemic manner by inducing the secretion of hormones and neurotransmitters that regulate G protein-coupled receptors (GPCRs). Emerging studies show that GPCRs also regulate autophagy by directly detecting extracellular nutrients. We review the role of GPCRs in autophagy regulation, highlighting their potential as therapeutic drug targets.
Keywords: Autophagy, amino acid sensing GPCRs, muscarinic receptor, β-adrenergic receptor, GLP-1 receptor, mTORC1, AMPK
Nutrients regulate autophagy
Autophagy is a catabolic process in which proteins, organelles, and other cytoplasmic contents are engulfed by autophagosomal double membrane vesicles and delivered to lysosomes where they are degraded (Box I). A basal level of autophagy in cells helps to remove damaged proteins and organelles. However, autophagy can be dramatically increased under different physiological conditions, such as hypoxia or nutrient deprivation, an adaptive response critical for cell and organism survival [1]. For example, during starvation, autophagy is induced through the inhibition of the mechanistic target of rapamycin (mTOR), an evolutionary conserved protein kinase and central regulator of cell growth [2]. Dysregulation of autophagy is associated with numerous diseases including cancer, metabolic syndromes and cardiovascular diseases [1] (see Boxes 2 and 3). For example, mouse models of obesity and insulin resistance show decreased autophagy in the liver, and the restoration of autophagy in these mice significantly improves insulin action in the liver, suggesting that the reduction in autophagy is causally related to obesity-induced insulin resistance [3]. The role of autophagy in the development of human disease and how this process may be pharmacologically manipulated will be reviewed throughout this article.
Text Box 1. Regulation of Autophagy.
Autophagy can be initiated when cells undergo metabolic stresses including hypoxia and nutrient limitation. These stresses inhibit mTORCl, resulting in an increase in the activity of UNC51-like kinase 1/2 (ULK1/2). ULK1/2 form a complex with the regulatory proteins Atgl3 and FIP200 [74], which is necessary to initiate autophagy. The type III phosphoinositide-3 kinase (PI3K) VPS34 facilitates the nucleation of the pre-autophagosome (phagophore) by catalyzing the formation of phosphatidylinositol-3-phosphate (PI3P) [75]. An interaction between VPS34 and Beclinl, a Bcl-2-homology (BH)-3 domain only protein, is required to activate VPS34 [76]. Negative regulation of this complex occurs when the anti-apoptotic protein Bcl2 sequesters Beclinl by binding to its BH3 domain. The phosphorylation of Bcl2 by JNK or the phosphorylation of Beclinl by death-associated protein kinase (DAPK) activates autophagy by preventing the Beclinl/Bcl2 interaction, thus, allowing VPS34/Beclinl association [77, 78]. Recent work by Guan and colleagues showed that ULK1 increases VPS34 activity by phosphorylating Beclinl [79]. Another major activator of autophagy is AMPK. A high AMP/ATP ratio activates AMPK, increasing energy levels by inhibiting anabolic and inducing catabolic processes. AMPK enhances autophagy by directly activating ULK1 [80, 81]. AMPK also phosphorylates Beclinl to induce autophagy [82].
Expansion of the phagophore into the mature autophagosome requires two ubiquitin-like conjugation systems. The ubiquitin-like protein Atgl2 is covalently attached to Atg5 by Atg7 and AtglO. The Atgl2/Atg5 conjugate then binds to Atgl6, which is essential for autophagy. The other ubiquitin-like protein LC3 is cleaved by Atg4 and subsequently conjugated to phosphatidylethanolamine (PE) by Atg7 and Atg3 to form LC3-II. LC3-II becomes incorporated in both the inner and outer membranes of autophagosomes. LC3-II, the formation of which is used as a measure of autophagy, is thought to play a role in autophagic cargo recruitment and autophagosome biogenesis. Sequestosome-1 (SQSTM1 or p62) is an adaptor protein that binds polyubiquitinated proteins and LC3-II, recruiting them to the autophagosome for disposal. The final step in autophagy is the fusion of the autophagosome with the lysosome and the degradation of its cargo [1, 83].
Text Box 2. Cancer as a metabolic syndrome and the interplay with GPCRs.
The concept that cancer is a metabolic disorder has been longstanding, with early studies by Warburg suggesting a unique metabolic landscape for tumor cells compared to untransformed cells [84]. Warburg observed that tumor cells preferentially undergo glycolysis instead of oxidative phosphorylation, and his observations on the distinct metabolic phenotype of tumors were further supported by studies revealing their glutamine-dependency [85]. The metabolic changes observed in tumors are likely to be driven by one or more mechanisms operating within the tumor including: (1) micro-environmental conditions which may lead to stabilization and activation of the hypoxia-inducible transcription factor (HIF) pathway due to an anaerobic, improperly-vascularized tumor micro-environment [86], (2) activation of oncogenes such as Ras, myc, Akt, and others which can all adversely regulate the metabolic profile of transformed cells, (3) loss of tumor suppressors, such as p53 whose targets include SCO2 which is required for mitochondrial respiratory chain, and TP53-induced glycolysis and apoptosis regulator (TIGAR) which regulates glycolysis and gluconeogenesis [84, 87], or even mutagenesis of tumor suppressors encoding for mitochondrial proteins such as succinate dehydrogenase (SDH) and fumarate dehydrogenase (FH), which subsequently results in mitochondrial dysfunction and disrupted metabolism [88]. Furthermore, studies have shown that, in a model of stepwise malignant transformation, increasing tumorigenicity correlated with increased sensitivity to glycolytic inhibition [89], and other studies showed inhibition of tumor growth as a result of activation of oxidative phosphorylation and blocking the Warburg effect [90, 91].
Rapidly proliferating tumors require nutrients and substrates for the biosynthesis of nucleotides, proteins, lipids and other macromolecules. These are provided for, at least in part, by the altered metabolic state of tumor cells. For example, there is an increased dependence of certain tumors on glutaminolysis, which can be used as a source of amino acids but also to produce NADPH for lipid biosynthesis and oxaloacetate for Krebs cycle intermediates [92]. Another mechanism for tumors to obtain nutrients for biosynthetic programs is via autophagy. Autophagy has a dual role in tumors whereby it can play tumor-suppressive or tumor promoting roles in a context-dependent manner [93]. The role of autophagy in providing biosynthetic intermediates is essential for tumor growth, with lipophagy, the degradation of lipids by autophagy [17], degradation of proteins by autophagy to generate amino acids, as well as autophagy-recycled sugars, all contributing essential products to sustain tumor cell growth, in particular under conditions of stress and starvation [93]. Autophagy may therefore play a major role in sustaining the unique metabolic status of tumors, providing valuable by-products as well as relieving cellular stress.
The exact role of autophagy in different tumors makes it difficult to assess the benefit of targeting autophagy, but also provides impetus for studies to clearly define its regulators, as potential targets for anti-autophagic cancer therapeutics. With novel studies showing uncharacterized links between autophagy, GPCRs and nutrient sensing, as well as a large body of literature detailing the significance of GPCRs and downstream signaling from GPCRs in driving tumorigenesis [94], it will be interesting to study the role of autophagy in GPCR-driven tumors, as well as the roles of GPCRs in tumor-promoting or tumor-suppressing, functions of autophagy in cancer.
Text Box 3. Autophagy induction as a therapy for cardiovascular disease.
When blood flow to tissue is reduced during a stroke or myocardial infarction, a rapid increase in autophagy acts to reduce cell death [95, 96]. Importantly, a study by Przyklenk and colleagues suggested that the administration of autophagy inducing drugs can protect cardiac tissue even when administered 30 min into a 45 min cardiac ischemic episode in pigs [97]. Increased autophagy also appears to be beneficial in attenuating left ventricular dysfunction and hypertrophy in mice 1-4 weeks after a myocardial infarction [98]. Thus, autophagy induction might be used as a therapeutic target during acute myocardial infarctions. There are approximately 200 GPCRs expressed in the heart, some of which are known to modulate autophagy [99]. GPCRs are excellent drug targets, as they are the targets for more than 25% of all human pharmaceuticals [100]. Therefore, designing new drugs and using current drugs to target specific GPCRs that regulate autophagy might be therapeutically advantageous in treating cardiovascular diseases.
Numerous mechanisms are used by cells in the surveillance of their environment to properly balance their metabolic decisions. Extracellular nutrients present in the serum are transported into cells by multiple specialized transporter systems. Intracellular nutrient sensors such as AMP-activated kinase (AMPK) or mTOR initiate anabolic processes such as protein and lipid synthesis and reduce catabolic processes like autophagy, when they detect sufficient nutrient levels [4]. Conversely, these same sensors initiate catabolic processes when there is a depletion of nutrients.
GPCRs are direct nutrient sensors that regulate autophagy
Cell surface nutrient receptors, including GPCRs in single cell organisms such as fungi, are well characterized [5]. Traditionally, mammalian non-sensory GPCRs were thought to be receptors for endocrine, paracrine, and autocrine signaling hormones and neurotransmitters produced and secreted in response to physiological cues. However, recent studies have expanded this view by indicating that GPCRs function in the surveillance of the fed state by directly detecting nutrients. Examples of these receptors include the amino acid responsive receptors GPRC6A, taste receptor type 1 member 1 and 3 (T1R1/T1R3), and the calcium receptor (CaR); long chain fatty acid receptors GPR120 and GPR40; and short chain fatty acid receptors GPR41 and GPR43 [6, 7]. There are also many orphan GPCRs that may serve as nutrient sensors. The nutrient receptors are known to be linked to different G protein α subunits including Gs, Gq/G11, Gi, Go, and gustducin. Agonist stimulation causes activation and dissociation of the Gα subunits from the Gβγ subunits, both of which initiate downstream signaling. Gq/G11 activation leads to an increase in phospholipase C (PLC) activity, causing an elevation of intracellular inositol triphosphate (IP3) and diacylglycerol (DAG) concentrations. IP3 binds to the IP3 receptor (IP3R) on the endoplasmic reticulum (ER), which causes the ER to release stored Ca2+ (Figure 1). Gi and gustducin activation lead to decreases in intracellular cyclic AMP (cAMP) concentrations, while Gs activation increases intracellular cAMP levels. Changes in cAMP, IP3 and Ca2+ concentrations regulate a multitude of signaling molecules such as mTOR, AMPK, and the mitogen activated kinases (MAPKs) ERK1/2 that modulate autophagy by a variety of mechanisms (Figure 1).
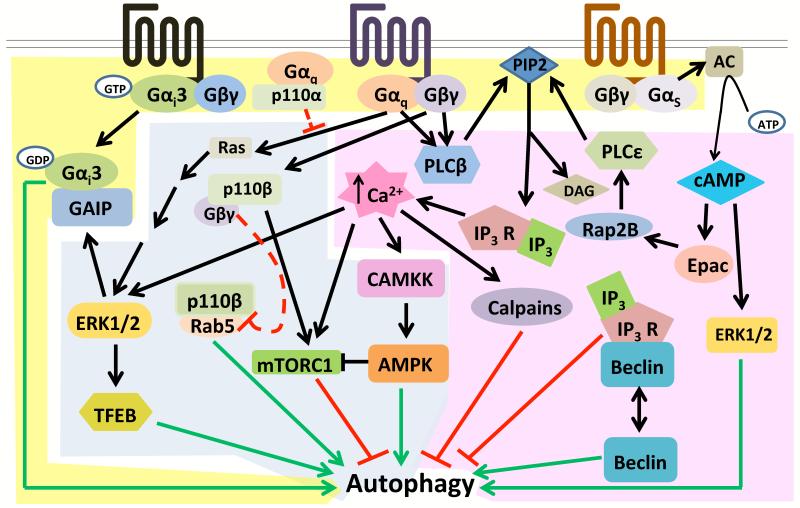
Figure 1. Multiple signals downstream of GPCRs regulate autophagy.
(A) (Highlighted in yellow) Dissociated subunits of heterotrimeric G proteins regulate autophagy: Liberated GDP-bound Gαi3 has been shown to induce autophagy in a VPS34-dependent manner, which is promoted by the RGS family protein GAIP. Gβγ subunits liberated by Gq-coupled receptors activate PLCβ, which cleaves PIP2 to generate IP3 and DAG and IP3 induces calcium release from the ER. (B) (Highlighted in pink) Second messengers generated by GPCR activation modulate autophagy: Gs-coupled receptor stimulation activates adenylyl cyclase (AC) which generates cAMP from ATP. cAMP increases autophagy in some cell types but reduces it in others. While enhanced autophagy can be ERK1/2 dependent, inhibition of autophagy can involve IP3 generated by PLCε. IP3 can negatively regulate autophagy through calcium-activated CAMKKβ signaling and/or via association of IP3R with Beclin-1. Like cAMP, increased cytosolic calcium can decrease autophagy in some contexts but increase it in others. Calcium can induce autophagy via CAMKK and/or ERK1/2 signaling, and reduce autophagy through calpains. (C) (Highlighted in blue) Kinases downstream of GPCRs regulate autophagy: The MAPKs ERK1/2 are stimulated by GPCRs. ERK1/2 can promote autophagy during starvation by phosphorylating the transcription factor TFEB, causing its translocation to the nucleus and accelerated autophagic and lysosomal gene expression. ERK2 also activates GAIP to increase Gαi3-induced autophagy. CAMKKβ activated by Gq/11 and Gs-coupled receptor stimulation can activate the critical autophagy regulator AMPK. The class 1A PI3 kinase p110β binds the endosomal trafficking regulator Rab5 to promote autophagy. Gα/p110α and Gβγ/p110β complexes likely inhibit autophagy.
Amino acid sensing GPCRs
Most of the known amino acid-responsive GPCRs belong to the GPCR class C, which also includes the sweet taste receptor (T1R2/T1R3), metabotropic glutamate receptors, the GABAB receptor, the Ca2+-sensing receptor (CaSR), GPRC6A, and a few orphan receptors. Most members of class C are hetero- or homodimers and contain a large extracellular segment called the Venus Flytrap module that is involved in agonist binding [7, 8]. T1R1 and T1R3 dimerize to form a receptor that binds L-amino acids, specifically L-glutamate, thus enabling the body to sense the umami (savory) flavor [9]. The amino acid-responsive GPCRs have been reported to be coupled to gustducin, Gi, Gs, or Gq depending on the tissue in which they are expressed [7].
T1R1/T1R3 is expressed in most tissues [10, 11] where it is responsive to most of the 20 amino acids with varying affinities, thus acting as a direct sensor of the fed state and amino acid availability [7]. It was recently shown that amino acids signal through T1R1/T1R3 to activate the mTOR protein complex 1 (mTORCl), which consists of mTOR, mLST8 and regulatory associated protein of mTOR (Raptor), and functions as a master regulator of metabolism controlling protein synthesis and other processes. Amino acid deprivation inhibits mTORCl activity in most cells and silencing T1R3 causes a significantly greater decrease in mTORCl activity, upon starvation, in several cell lines. These data suggest that amino acid signaling through T1R1/T1R3 is necessary for optimal mTORCl activation [11]. In light of the fact that amino acid-induced activation of mTORCl is an important mechanism for reducing autophagy, the effect of T1R1/T1R3 on autophagy was investigated. T1R1/T1R3 reduction resulted in increased autophagy in cell culture, even in amino acid replete medium. Autophagy was also increased in the hearts, livers and skeletal muscles of fasted T1R3−/− mice [11]. These observations highlight the importance of nutrient sensing GPCRs on modulating autophagy.
Other studies have also demonstrated that the direct sensing of amino acids by GPCRs regulates autophagy. Kang and Avery used a starvation-hypersensitive C. elegans mutant to identify individual amino acids that could rescue death caused by amino acid deprivation. They found that Leu, Gln, Ala, Val, and Ile, individually reduced starvation-induced death, while other amino acids either had no effect or actually increased death. They further showed that C. elegans homologs of metabotropic glutamate receptors, MGL-1 and MGL-2L are necessary for the ability of leucine to prevent autophagy and death in starved mutant worms [12].
Fatty acid sensing GPCRs
The participation of other nutrient receptors, such as the long chain fatty acid receptor GPR120, in the control of autophagy has yet to be reported, but might be anticipated due to the important role that these receptors play in detecting nutrient availability. GPR120 is an important regulator of metabolism, as GPR120-deficient mice are more prone to obesity, fatty liver development, and glucose intolerance when fed high fat diets [13]. Mutations in GPR120 that inhibit its signaling activity were found to increase the risk of obesity in humans [14]. Ruvkun and colleagues discovered that ω-6 polyunsaturated fatty acids (PUFAs) induce autophagy in both C. elegans and HeLa cells [15]. Thus, because GPR120 is an important mediator of PUFAs, it will be important to determine whether GPR120 activation regulates autophagy.
Nutrient fluctuations induce the secretion of hormones and neurotransmitters that modulate autophagy through GPCRs
Recent studies have begun to illuminate the mechanisms by which GPCRs control the systemic regulation of autophagy. Due to their importance in regulating autophagy, β-adrenergic, muscarinic, glucagon like peptide-1 (GLP-1), and purinergic GPCRs will be discussed in detail (see table 1).
Table 1. GPCRs in autophagy regulation.
| GPCR | Effect on Autophagy | REF |
|---|---|---|
|
| ||
| Amino acid sensing receptors: | Suppress | [11] |
| T1R1-T1R3 (Heterodimeric taste receptors) |
[12] | |
| MGL-1, MGL-2 (C. elegans metabotropic glutamate receptor homologs) |
||
|
| ||
| Muscarinic receptors: | Promote | [12-21] |
| GAR-3 acetylcholine receptors in C. elegans Acetylcholine receptors in rat H9C2 cardiomyoblasts |
[25] | |
|
| ||
| GLP-1 receptor | Promote | [29-31] |
|
| ||
|
β-adrenergic receptors: Both β1 and β2 adrenergic receptors |
Promote | [16, 18-19] |
|
| ||
|
Purinergic receptors: P2Y13 ADP receptor |
Promote | [36] |
β-adrenergic receptors
Epinephrine is secreted by the adrenal glands when hypothalamic neurons detect a drop in systemic blood glucose. Activation of the β-adrenergic GPCR receptors in peripheral tissues by epinephrine induces the lipolysis of triglycerides stored in lipid droplets through a mechanism involving autophagy [16]. Lizaso et al. recently discovered that β-adrenergic activation by isoproterenol in 3T3-L1 adipocytes leads to an increase in cAMP-mediated autophagy-induced lipolysis. They further showed that β-adrenergic activation does not increase the initiation of autophagy, but enhances autophagic flux by promoting the fusion of autophagosomes with lysosomes [16]. An earlier study by Czaja and colleagues suggests that autophagy plays an important role in hydrolyzing triglycerides by facilitating the delivery of lipid droplets to lysosomes. Treatment of hepatocytes with inhibitors of autophagy or knockdown of the autophagy gene Atg5 increased the size and number of lipid droplets as well as triglyceride levels. Lipid accumulation was significantly elevated in the livers of mice with a liver specific deletion of the autophagy gene Atg7 compared to control mice, suggesting that autophagy reduced lipid accumulation in the liver [17].
Wang et al. observed that inhibition of β1-adrenergic signaling in rats using anti-β1-adrenergic receptor autoantibodies induces cardiac dysfunction and inhibits autophagy, which was be reversed by treatment with the mTOR inhibitor rapamycin, suggesting that blockade of β1-adrenergic signaling induces heart damage by inhibiting autophagy [18]. Lastly, another study showed that the β2-adrenergic specific agonist salbutamol increased autophagic flux in cardiac fibroblasts [19]. These data suggest that autophagy regulation should be added to the extensive list of β-adrenergic receptor functions. Since numerous therapeutics are used to target the β-adrenergic receptors, it will be important to determine which of the beneficial effects or negative side effects of these drugs are due the modulation of autophagy.
Muscarinic receptors
Muscarinic signaling has been shown to regulate starvation-induced autophagy. Avery and colleagues observed that amino acid deprivation increases the activation of GAR-3, a muscarinic acetylcholine Gq-coupled GPCR in C. elegans. GAR-3 activation promotes MAPK signaling in the pharyngeal muscle, causing an increase in pumping rate and a consequent increase in food intake [20]. Mutant worms lacking G-Protein b subunit (GPB-2), a protein critical for inhibition of Go and Gq signaling by regulators of G protein signaling (RGS), are hypersensitive to starvation [21]. These worms cannot survive muscarinic activation induced by amino acid deprivation. Death is likely caused by excessive autophagy in the pharyngeal muscle [21]. Muscarinic receptor activation has also been found to induce autophagy in H9C2 rat cardiomyoblast cells. Treatment with acetylcholine increases survival of H9C2 cells after hypoxia and reoxygenation, possibly due to increased autophagy [22]. Numerous reports suggest that muscarinic receptor stimulation activates AMPK to regulate other biological processes such as glucose uptake in muscle cells and mitochondria biogenesis in cardiac myoblasts [22-25]. It will be important to determine whether muscarinic receptor-induced AMPK activation is important in the context of nutrient deprivation-induced autophagy.
GLP-1 receptors
Nutrients can also induce the secretion of the incretin glucagon-like peptide-1 (GLP-1). GLP-1 is secreted from L cells in the intestine in response to food intake to promote insulin secretion from the β cells in the pancreas. GLP-1 binds to and activates its Gs-coupled receptor (GLP-1R) not only in pancreatic β cells, but also liver, kidney, lung, CNS, peripheral nervous system, lymphocytes, blood vessels, and heart [26, 27]. While the best studied role of GLP-1 is its function as a stimulator of insulin secretion, agonists of GLP-1R have been reported to reduce hypertension, obesity, fatty liver, dyslipidemia, cardiovascular disease, and other diseases [28]. Anania and colleagues observed that GLP-1 analogs inhibit hepatosteatosis by increasing autophagic flux [29]. Liraglutide, a GLP-1R agonist, has been found to improve cardiac function in mice fed high fat diets. Increased levels of the lipid-modified microtubule-associated protein 1 light chain 3 β (LC3-II) (see Box 1) in the left ventricles of mice treated with liraglutide suggest that autophagy regulation might play a role in this improvement of cardiac function [30]. Data from another study suggest that liraglutide can protect INS-1 pancreatic β cells from fatty acid-induced apoptosis by inducing autophagy [31]. Future studies are needed to determine if some of the beneficial effects of GLP-1R agonists in diseases are due to increased autophagic flux.
Nutrient receptors play an important role in the induction of GLP-1 and insulin secretion. T1R2/T1R3, fatty acid receptors, GPRC6A, and the calcium sensing receptor (CaR) have been shown to cause release of GLP-1 from intestinal L cells in response to glucose, long chain free fatty acids, and amino acids respectively [32]. Thus, in addition to the potential role in cell autonomous autophagy regulation, they could also modulate systemic autophagy by regulating the secretion of GLP-1.
Purinergic receptors
During hyperglycemia, organisms try to regain normal glucose homeostasis by numerous methods. Hyperglycemia induces autophagy in pancreatic β cell causing the removal of damaged mitochondria in the beta cells, thus preventing oxidative stress and allowing them to survive [33, 34]. Hyperglycemia can cause increases in circulating nucleotides such as ADP which can signal through the P2Y purinergic receptors [35]. Recent data suggest that the activation of the P2Y13 GPCR by ADP in HepG2 cells induces autophagy [36]. The widely prescribed antithrombotic clopidogrel (Plavix) is a P2Y12 antagonist, and there is interest in designing drugs to target P2Y13 and the other P2Y receptors for the treatment of diabetes and cardiovascular diseases [37]. The possibility that autophagy modulation could at least be a partial mechanism of action of some of the P2Y-targeted drugs should be investigated.
Signaling molecules downstream of GPCRs that regulate autophagy
mTOR
mTORC1 activation, which is regulated by nutrients, cellular energy status, and growth factors, is an important mechanism by which cells modulate autophagy (see Box 1). In addition to the T1R1/T1R3 amino acid receptor, research suggests that the Gq-coupled GPCRs including the muscarinic M3 (M3R) and the endothelin ET-B receptor (ETR) also can activate mTORC1 [11, 38, 39]. Another study suggests that the bacterial Pasteurella multocida toxin activates mTORC1 by causing Gq subunits to activate PLCβ and protein kinase C [40]. The activation of mTORC1 by the Gs-coupled adenine receptor and the β2-adrenergic receptor, as well as the Gi-coupled CXCR4 receptor was not observed [39]. While this suggests that only Gq-coupled receptors can activate mTORC1, Bennett and colleagues observed that the Gi inhibitor pertussis toxin prevents mTORC1 activation, suggesting the Gi-coupled receptors might activate this kinase complex [41]. It was recently reported that overexpressed Gβγ subunits interact with the carboxyl terminus of mTOR in a yeast two hybrid system and in HEK-293 cells [42]. While this interaction increased mTOR activity, further studies are needed to determine if this occurs in a physiologically relevant manner. It is possible different cellular contexts determine which G protein subunits can activate mTORC1.
cAMP
Many signaling pathways used by GPCRs intersect with those regulating autophagy. Gs-coupled receptor activation increases intracellular cAMP concentrations. Elevated cAMP has been found to increase autophagic flux in adipocyte-derived mesenchymal stem cells treated with prostaglandin E2 (PGE2), an agonist of the Gs-coupled EP2 and EP4 receptors. cAMP-induced autophagy appears to be dependent on the activation of ERK1/2, but independent of the reduction in mTORC1 activity [43]. In contrast, other studies suggested that cAMP inhibits autophagic flux in neuronal cells. In this case, it appears that cAMP inhibits autophagic flux by activating the guanine nucleotide exchange factor Epac, which activates Rap2B. Rap2B then activates PLCε, thereby increasing inositol 1,4,5-triphosphate (IP3) levels and inhibiting autophagy [44, 45]. IP3 is thought to modulate autophagy by multiple mechanisms, which will be discussed in detail below. Further studies are needed to better elucidate the mechanisms underlying this differential, cell type-specific effect of cAMP on autophagy. Additionally, it would be interesting to determine whether elevation of cAMP is responsible for the induction of autophagy through the GLP-1R and other Gs-coupled receptors.
Gαq, Calcium, PLCβ, and IP3
Activation of GPCRs coupled to Gq subunits leads to the activation of phospholipase beta (PLCβ). Optimal amino acid-induced activation of mTORC1 requires PLCβ in pancreatic β cells [11] and C2C12 myoblasts [41]. Thus, it is likely that amino acid sufficiency inhibits autophagy by activating mTORC1 in a PLCβ-dependent manner.
PLCβ generates IP3 and DAG by cleaving phosphatidylinositol 4, 5-bisphosphate (PIP2). Data from Rubinsztein and colleagues suggested that IP3 inhibits autophagy independently of mTORC1 regulation [46]. Other studies have reported that the inhibition or deletion of IP3R by antagonists or genetic manipulation, respectively, initiate autophagy [47, 48]. Beclin 1, a protein involved in autophagy initiation (see Box 1), was observed to co-immunoprecipitate with IP3R, an interaction that was blocked with an IP3 antagonist, leading to the hypothesis that IP3R activation inhibits autophagy by sequestering Beclin 1 away from other autophagic machinery [47, 49, 50]. The antiapoptotic protein Bcl2 also prevents autophagy by sequestering Beclin 1 (see Box 1). It was recently discovered that the nutrient-deprived autophagy factor-1 (NAF-1) interaction with and Bcl2 and Beclin 1 is necessary for Bcl2 to inhibit autophagy. Interestingly, IP3R was found in a complex with NAF-1 and Bcl2, suggesting that the inhibition of autophagy by IP3R and Bcl2 might be mechanistically connected [51].
The IP3 generated by PLCβ induces Ca2+ release from the ER by activating the IP3R located in the ER membrane. One study suggested that IP3 decreased autophagy by increasing intracellular Ca2+ and activating calcium-dependent cysteine proteases (calpains) [44]. A report by Khan and Joseph suggested that Ca2+ released by IP3R activation reduces autophagy by maintaining mTORC1 activity [48]. In contrast, other data suggested that cytosolic Ca2+ increases autophagy through the Ca2+-dependent activation of the calcium/calmodulin-dependent protein kinase kinase β (CAMKKβ). CAMKKβ can activate AMPK which initiates autophagy by multiple mechanisms. Other studies suggested that Ca2+-induced autophagy through protein kinase C theta or ERK1/2 [52]. Future research is needed to identify the mechanisms by which elevated cytosolic Ca2+ can induce or inhibit autophagy in different conditions and cell types.
Gαi
Several studies have implicated Gαi subunits, in particular Gαi3, in the regulation of autophagy. The involvement of Gαi3 was first identified in HT-29 cells, in which a differentiation-dependent increase in autophagy is required for proper trafficking and metabolism of N-linked glycoproteins and sphingoglycolipids [53-55]. The involvement of Gαi3 was abrogated by constitutive binding to GTP or by preventing Gβγ dissociation, but was enhanced by a mutation that increases its intrinsic GTPase-accelerating protein (GAP) activity and affinity for GDP over GTP [56]. This was confirmed by studies where induction of autophagy was also observed when stimulating the activity of two Gαi3 inhibitors: the Gαi3 GTPase activating protein AGS3 (activator of G protein signaling) and the Gαi3 guanine nucleotide dissociation inhibitor (GDI) [55, 57, 58]. This Gαi3-dependent process requires the activation of the class III PI3-kinase VPS34. The mechanism could involve binding of Gαi3 to the VPS15 regulatory subunit of VPS34, as it has a WD40 repeat similar to that in Gβγ subunits and has previously been shown to bind to GDP-bound Gα subunits [59]. Also important is the intracellular localization of Gαi3 at the endoplasmic reticulum (ER) and Golgi, with cytosolic or plasma membrane localization acting as a suppressor of its autophagic role [58]. A recent study shows that stimulation of starved hepatocytes with insulin caused translocation of Gαi3 from autophagosomes to the plasma membrane, resulting in loss of starvation-induced autophagy. This anti-autophagic effect of insulin was abrogated in Gαi3-/- mice. Similarly, phenylalanine stimulation of starved cells led to decreased autophagy coupled to translocation of Gαi3 from autophagosomes to a diffuse cytosolic distribution [60]. These data clearly identify a hormone- and nutrient-sensitive role of Gαi3 in autophagy, although the exact mechanism of action is still undefined. It could be postulated that active GTP-bound Gαi3 normally inhibits autophagy, and that the increase in autophagy caused by Gαi3 binding to GDP is due to a reduction in Gαi3 activity. Gαi3 overexpression experiments could be designed to address this.
Class IA PI3-kinase, p110β and Rab5
The class IA PI3-kinase, p110β, is activated downstream of GPCRs by direct binding to Gβγ subunits [61]. p110β can bind to Ras-related proteins in brain 5 (Rab5) to mediate autophagy in growth factor-depleted conditions [62, 63]. The binding regions of Gβγ and Rab5 subunits on p110β are close, suggesting that the interactions might be mutually exclusive. This may represent a model in which GPCR signaling could inhibit autophagy by recruiting p110β away from Rab5. GPCRs may also negatively regulate the p110α isoform of the class IA PI3-kinases and block their growth-promoting, anti-autophagic signaling. p110α has been shown to interact with Gαq and Gα16 subunits, resulting in inhibition of p110α activity, possibly by displacing Ras binding [64-67]. This could represent a mechanism whereby GPCR activation and heterotrimer dissociation lead to opposing effects on PI3-Kinase signaling and autophagy, with the Gα subunit inhibiting p110α while the Gβγ subunits activate p110β.
MAPK
Another major GPCR-activated pathway involves MAPK signaling, with the MAPKs ERK1/2 also having roles in autophagic regulation. For example, ERK2 phosphorylates GAIP to increase its GAP activity on Gαi3, and stimulate Gαi3-dependent autophagy [68]. ERK signaling can regulate the expression of autophagy-related genes by growth factor- and nutrient-dependent phosphorylation of the transcription factor TFEB. ERK1/2-mediated phosphorylation of TFEB prevents its nuclear localization. Under starvation conditions, low ERK1/2 activity and TFEB phosphorylation cause TFEB nuclear localization and enhanced transcription of autophagy and lysosomal biogenesis genes [69]. Furthermore, a recent study suggests that ERK1/2 and components of the MAPK signaling module might co-localize with autophagy-related proteins at the extra-luminal face of autophagosomes and also in the nucleus. The interaction between MAPK pathway components and autophagy proteins is reported to be required for proper ERK1/2 activation under different nutrient and growth factor conditions [70]. While the significance of the interaction of pERK1/2 with autophagosomes or with different autophagy proteins is uncertain, the available data suggest an exciting crosstalk between the MAPK signaling and autophagic pathways that needs to be explored in greater detail.
AMPK
Nutrient deprivation leads to the activation of AMPK. AMPK activates autophagy by reducing the activity of mTORC1 and by directly activating specific autophagy regulatory proteins (see Box 1). While an increase in the AMP/ATP ratio leads to AMPK activation, AMPK can also be activated by direct phosphorylation by the tumor suppressor liver kinase B1 (LKB1), and by CAMKKβ, as mentioned above [71]. Numerous receptors coupled to Gαq/11, Gαs, and Gαi have been shown to activate AMPK [72]. Future work is needed to determine connections between GPCR-induced activation of AMPK and the role of AMPK in regulating autophagy.
Concluding remarks and future directions
GPCRs regulate autophagy by both directly detecting nutrients and by mediating the effects of hormones and neurotransmitters that are secreted in response to organismal nutrient fluctuations. The mechanisms by which GPCRs regulate autophagy include the modulation of second messengers such as cAMP and Ca2+ and other downstream effectors including ERK1/2 and mTORC1. Many studies suggest that increased autophagic flux is beneficial in preventing the detrimental effects of cardiovascular disease, diabetes, metabolic syndromes, and neurodegenerative diseases. Interestingly, a recent study by Levine and colleagues indicates exercise-induced autophagy in skeletal and cardiac muscles is necessary for the beneficial metabolic effects of exercise [73]. Autophagy regulation is emerging as an important therapeutic target in the treatment of human disease. As GPCRs are excellent drug targets, further studies on the mechanisms by which GPCRs regulate autophagy are needed (see Box 4).
Text Box 4. Outstanding questions.
Do amino acid-responsive GPCRs in addition to T1R1/T1R3 regulate autophagy?
How does autophagy caused by reduced signaling through T1R1/T1R3 affect physiology?
Which beneficial effects of GLP-1 agonists are due to autophagy regulation?
Does PLCβ regulate autophagy in an mTORC1-dependent manner?
Do Gα subunits other than Gαi3 play a role in autophagy? Is subcellular localization the only determinant of the involvement of this specific subunit in autophagy regulation?
How do the second messengers cAMP and Ca2+ induce autophagy in certain models while inhibiting autophagy in others?
Do free fatty acids regulate autophagy through FFA-binding GPCRs?
Can AMPK activated by CAMKK initiate autophagy?
Highlights.
G protein coupled receptors (GPCRs) can sense nutrients and regulate autophagy.
GPCRs regulate systemic autophagy by mediating the effects of hormones secreted in response to systemic nutrient fluctuations.
Downstream effectors of GPCRs that regulate autophagy include: cAMP, Ca2+, IP3, PI3-kinase
Acknowlegements
Research supported by grants from the National Institutes of Health R01 DK55310 and from the Cancer Prevention and Research Institute of Texas (CPRIT) to MHC. EMW was supported in part by a mentor-based postdoctoral fellowship from the American Diabetes Association. We thank Andrés Lorente-Rodriguez and Aroon Karra for helpful comments and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11(9):709–30. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15(6):555–64. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang L, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11(6):467–78. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan HX, Xiong Y, Guan KL. Nutrient sensing, metabolism, and cell growth control. Mol Cell. 2013;49(3):379–87. doi: 10.1016/j.molcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue C, Hsueh YP, Heitman J. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev. 2008;32(6):1010–32. doi: 10.1111/j.1574-6976.2008.00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blad CC, Tang C, Offermanns S. G protein-coupled receptors for energy metabolites as new therapeutic targets. Nat Rev Drug Discov. 2012;11(8):603–19. doi: 10.1038/nrd3777. [DOI] [PubMed] [Google Scholar]
- 7.Wauson EM, Lorente-Rodriguez A, Cobb MH. Minireview: Nutrient Sensing by G Protein-Coupled Receptors. Mol Endocrinol. 2013 doi: 10.1210/me.2013-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wellendorph P, Johansen LD, Brauner-Osborne H. Molecular pharmacology of promiscuous seven transmembrane receptors sensing organic nutrients. Mol Pharmacol. 2009;76(3):453–65. doi: 10.1124/mol.109.055244. [DOI] [PubMed] [Google Scholar]
- 9.Nelson G, et al. An amino-acid taste receptor. Nature. 2002;416(6877):199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 10.Foster SR, Roura E, Thomas WG. Extrasensory perception: Odorant and taste receptors beyond the nose and mouth. Pharmacol Ther. 2013 doi: 10.1016/j.pharmthera.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Wauson EM, et al. The G protein-coupled taste receptor T1R1/T1R3 regulates mTORC1 and autophagy. Mol Cell. 2012;47(6):851–62. doi: 10.1016/j.molcel.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang C, Avery L. Systemic regulation of starvation response in Caenorhabditis elegans. Genes Dev. 2009;23(1):12–7. doi: 10.1101/gad.1723409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh DY, et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142(5):687–98. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichimura A, et al. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature. 2012;483(7389):350–4. doi: 10.1038/nature10798. [DOI] [PubMed] [Google Scholar]
- 15.O’Rourke EJ, et al. omega-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 2013;27(4):429–40. doi: 10.1101/gad.205294.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lizaso A, Tan KT, Lee YH. beta-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy. 2013;9(8):1228–43. doi: 10.4161/auto.24893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, et al. Decreased Autophagy in Rat Heart Induced by Anti-beta1-Adrenergic Receptor Autoantibodies Contributes to the Decline in Mitochondrial Membrane Potential. PLoS One. 2013;8(11):e81296. doi: 10.1371/journal.pone.0081296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aranguiz-Urroz P, et al. Beta(2)-adrenergic receptor regulates cardiac fibroblast autophagy and collagen degradation. Biochim Biophys Acta. 2011;1812(1):23–31. doi: 10.1016/j.bbadis.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 20.You YJ, et al. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 2006;3(4):237–45. doi: 10.1016/j.cmet.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21(17):2161–71. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M, et al. Acetylcholine mediates AMPK-dependent autophagic cytoprotection in H9c2 cells during hypoxia/reoxygenation injury. Cell Physiol Biochem. 2013;32(3):601–13. doi: 10.1159/000354464. [DOI] [PubMed] [Google Scholar]
- 23.Thornton C, Sardini A, Carling D. Muscarinic receptor activation of AMP-activated protein kinase inhibits orexigenic neuropeptide mRNA expression. J Biol Chem. 2008;283(25):17116–22. doi: 10.1074/jbc.M708987200. [DOI] [PubMed] [Google Scholar]
- 24.Merlin J, et al. The M3-muscarinic acetylcholine receptor stimulates glucose uptake in L6 skeletal muscle cells by a CaMKK-AMPK-dependent mechanism. Cell Signal. 2010;22(7):1104–13. doi: 10.1016/j.cellsig.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Sun L, et al. Cardioprotection by acetylcholine: a novel mechanism via mitochondrial biogenesis and function involving the PGC-1alpha pathway. J Cell Physiol. 2013;228(6):1238–48. doi: 10.1002/jcp.24277. [DOI] [PubMed] [Google Scholar]
- 26.Bullock BP, Heller RS, Habener JF. Tissue distribution of messenger ribonucleic acid encoding the rat glucagon-like peptide-1 receptor. Endocrinology. 1996;137(7):2968–78. doi: 10.1210/endo.137.7.8770921. [DOI] [PubMed] [Google Scholar]
- 27.Ussher JR, Drucker DJ. Cardiovascular biology of the incretin system. Endocr Rev. 2012;33(2):187–215. doi: 10.1210/er.2011-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yabe D, Seino Y. Incretin actions beyond the pancreas: lessons from knockout mice. Curr Opin Pharmacol. 2013;13(6):946–53. doi: 10.1016/j.coph.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Sharma S, et al. GLP-1 analogs reduce hepatocyte steatosis and improve survival by enhancing the unfolded protein response and promoting macroautophagy. PLoS One. 2011;6(9):e25269. doi: 10.1371/journal.pone.0025269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noyan-Ashraf MH, et al. A glucagon-like peptide-1 analog reverses the molecular pathology and cardiac dysfunction of a mouse model of obesity. Circulation. 2013;127(1):74–85. doi: 10.1161/CIRCULATIONAHA.112.091215. [DOI] [PubMed] [Google Scholar]
- 31.Jing Yin J, et al. Liraglutide Improves the Survival of INS-1 Cells by Promoting Macroautophagy. Int J Endocrinol Metab. 2013;11(3):184–90. doi: 10.5812/ijem.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasoamanana R, et al. Nutrient sensing and signalling by the gut. Proc Nutr Soc. 2012;71(4):446–55. doi: 10.1017/S0029665112000110. [DOI] [PubMed] [Google Scholar]
- 33.Han D, et al. Activation of autophagy through modulation of 5′-AMP-activated protein kinase protects pancreatic beta-cells from high glucose. Biochem J. 2010;425(3):541–51. doi: 10.1042/BJ20090429. [DOI] [PubMed] [Google Scholar]
- 34.Jung HS, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8(4):318–24. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Costa G, et al. High glucose changes extracellular adenosine triphosphate levels in rat retinal cultures. J Neurosci Res. 2009;87(6):1375–80. doi: 10.1002/jnr.21956. [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee C, Sparks DL. Extracellular nucleotides inhibit insulin receptor signaling, stimulate autophagy and control lipoprotein secretion. PLoS One. 2012;7(5):e36916. doi: 10.1371/journal.pone.0036916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erlinge D, Burnstock G. P2 receptors in cardiovascular regulation and disease. Purinergic Signal. 2008;4(1):1–20. doi: 10.1007/s11302-007-9078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang X, et al. Muscarinic receptor-mediated activation of p70 S6 kinase 1 (S6K1) in 1321N1 astrocytoma cells: permissive role of phosphoinositide 3-kinase. Biochem J. 2003;374(Pt 1):137–43. doi: 10.1042/BJ20021910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michel G, et al. Plasma membrane translocation of REDD1 governed by GPCRs contributes to mTORC1 activation. J Cell Sci. 2014;127(Pt 4):773–87. doi: 10.1242/jcs.136432. [DOI] [PubMed] [Google Scholar]
- 40.Oubrahim H, et al. Mammalian target of rapamycin complex 1 (mTORC1) plays a role in Pasteurella multocida toxin (PMT)-induced protein synthesis and proliferation in Swiss 3T3 cells. J Biol Chem. 2013;288(4):2805–15. doi: 10.1074/jbc.M112.427351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mercan F, et al. Novel role for SHP-2 in nutrient-responsive control of S6 kinase 1 signaling. Mol Cell Biol. 2013;33(2):293–306. doi: 10.1128/MCB.01285-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robles-Molina E, et al. Gbetagamma interacts with mTOR and promotes its activation. Biochem Biophys Res Commun. 2014;444(2):218–23. doi: 10.1016/j.bbrc.2014.01.044. [DOI] [PubMed] [Google Scholar]
- 43.Ugland H, et al. cAMP induces autophagy via a novel pathway involving ERK, cyclin E and Beclin 1. Autophagy. 2011;7(10):1199–211. doi: 10.4161/auto.7.10.16649. [DOI] [PubMed] [Google Scholar]
- 44.Williams A, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4(5):295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherra SJ, 3rd, et al. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol. 2010;190(4):533–9. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar S, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170(7):1101–11. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vicencio JM, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16(7):1006–17. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 48.Khan MT, Joseph SK. Role of inositol trisphosphate receptors in autophagy in DT40 cells. J Biol Chem. 2010;285(22):16912–20. doi: 10.1074/jbc.M110.114207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Criollo A, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 2007;14(5):1029–39. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 50.Wong A, et al. Regulation of autophagy in cardiomyocytes by Ins(1,4,5)P(3) and IP(3)-receptors. J Mol Cell Cardiol. 2013;54:19–24. doi: 10.1016/j.yjmcc.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 51.Chang NC, et al. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010;29(3):606–18. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.East DA, Campanella M. Ca2+ in quality control: an unresolved riddle critical to autophagy and mitophagy. Autophagy. 2013;9(11):1710–9. doi: 10.4161/auto.25367. [DOI] [PubMed] [Google Scholar]
- 53.Ghidoni R, et al. The metabolism of sphingo(glyco)lipids is correlated with the differentiation-dependent autophagic pathway in HT-29 cells. Eur J Biochem. 1996;237(2):454–9. doi: 10.1111/j.1432-1033.1996.0454k.x. [DOI] [PubMed] [Google Scholar]
- 54.Houri JJ, et al. Differentiation-dependent autophagy controls the fate of newly synthesized N-linked glycoproteins in the colon adenocarcinoma HT-29 cell line. Biochem J. 1995;309(Pt 2):521–7. doi: 10.1042/bj3090521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogier-Denis E, et al. Evidence for a dual control of macroautophagic sequestration and intracellular trafficking of N-linked glycoproteins by the trimeric G(i3) protein in HT-29 cells. Biochem Biophys Res Commun. 1997;235(1):166–70. doi: 10.1006/bbrc.1997.6727. [DOI] [PubMed] [Google Scholar]
- 56.Ogier-Denis E, et al. Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. J Biol Chem. 1996;271(45):28593–600. doi: 10.1074/jbc.271.45.28593. [DOI] [PubMed] [Google Scholar]
- 57.Pattingre S, et al. The G-protein regulator AGS3 controls an early event during macroautophagy in human intestinal HT-29 cells. J Biol Chem. 2003;278(23):20995–1002. doi: 10.1074/jbc.M300917200. [DOI] [PubMed] [Google Scholar]
- 58.Petiot A, et al. Subcellular localization of the Galphai3 protein and G alpha interacting protein, two proteins involved in the control of macroautophagy in human colon cancer HT-29 cells. Biochem J. 1999;337(Pt 2):289–95. [PMC free article] [PubMed] [Google Scholar]
- 59.Slessareva JE, et al. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126(1):191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 60.Gohla A, et al. An obligatory requirement for the heterotrimeric G protein Gi3 in the antiautophagic action of insulin in the liver. Proc Natl Acad Sci U S A. 2007;104(8):3003–8. doi: 10.1073/pnas.0611434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dbouk HA, et al. G protein-coupled receptor-mediated activation of p110beta by Gbetagamma is required for cellular transformation and invasiveness. Sci Signal. 2012;5(253):ra89. doi: 10.1126/scisignal.2003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dou Z, et al. The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J Cell Biol. 2010;191(4):827–43. doi: 10.1083/jcb.201006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dou Z, et al. Class IA PI3K p110beta subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell. 2013;50(1):29–42. doi: 10.1016/j.molcel.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ballou LM, et al. Galphaq binds to p110alpha/p85alpha phosphoinositide 3-kinase and displaces Ras. Biochem J. 2006;394(Pt 3):557–62. doi: 10.1042/BJ20051493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taboubi S, et al. Gq-coupled purinergic receptors inhibit insulin-like growth factor-I/phosphoinositide 3-kinase pathway-dependent keratinocyte migration. Mol Biol Cell. 2010;21(6):946–55. doi: 10.1091/mbc.E09-06-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taboubi S, et al. G alpha(q/11)-coupled P2Y2 nucleotide receptor inhibits human keratinocyte spreading and migration. FASEB J. 2007;21(14):4047–58. doi: 10.1096/fj.06-7476com. [DOI] [PubMed] [Google Scholar]
- 67.Yeung WW, Wong YH. Galpha16 interacts with Class IA phosphatidylinositol 3-kinases and inhibits Akt signaling. Cell Signal. 2010;22(9):1379–87. doi: 10.1016/j.cellsig.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 68.Ogier-Denis E, et al. Erk1/2-dependent phosphorylation of Galpha-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J Biol Chem. 2000;275(50):39090–5. doi: 10.1074/jbc.M006198200. [DOI] [PubMed] [Google Scholar]
- 69.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martinez-Lopez N, et al. Autophagy proteins regulate ERK phosphorylation. Nat Commun. 2013;4:2799. doi: 10.1038/ncomms3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carling D, Sanders MJ, Woods A. The regulation of AMP-activated protein kinase by upstream kinases. Int J Obes (Lond) 2008;32(Suppl 4):S55–9. doi: 10.1038/ijo.2008.124. [DOI] [PubMed] [Google Scholar]
- 72.Hutchinson DS, Summers RJ, Bengtsson T. Regulation of AMP-activated protein kinase activity by G-protein coupled receptors: potential utility in treatment of diabetes and heart disease. Pharmacol Ther. 2008;119(3):291–310. doi: 10.1016/j.pharmthera.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 73.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511–5. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kihara A, et al. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152(3):519–30. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Furuya N, et al. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1(1):46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 77.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–48. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei Y, et al. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30(6):678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331(6016):456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152(1-2):290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 85.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254(8):2669–76. [PubMed] [Google Scholar]
- 86.Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 87.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17(6):286–91. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 88.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5(11):857–66. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 89.Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci U S A. 2005;102(17):5992–7. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9(6):425–34. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 91.Shim H, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94(13):6658–63. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148(6):1132–44. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Hayre M, et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer. 2013;13(6):412–24. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kubli DA, Gustafsson AB. Cardiomyocyte health: adapting to metabolic changes through autophagy. Trends Endocrinol Metab. 2014;25(3):156–164. doi: 10.1016/j.tem.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Papadakis M, et al. Tsc1 (hamartin) confers neuroprotection against ischemia by inducing autophagy. Nat Med. 2013;19(3):351–7. doi: 10.1038/nm.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sala-Mercado JA, et al. Profound cardioprotection with chloramphenicol succinate in the swine model of myocardial ischemia-reperfusion injury. Circulation. 2010;122(11 Suppl):S179–84. doi: 10.1161/CIRCULATIONAHA.109.928242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buss SJ, et al. Beneficial Effects of Mammalian Target of Rapamycin Inhibition on Left Ventricular Remodeling After Myocardial Infarction. Journal of the American College of Cardiology. 2009;54(25):2435–2446. doi: 10.1016/j.jacc.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 99.Tang CM, Insel PA. GPCR expression in the heart; “new” receptors in myocytes and fibroblasts. Trends Cardiovasc Med. 2004;14(3):94–9. doi: 10.1016/j.tcm.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 100.Heng BC, Aubel D, Fussenegger M. An overview of the diverse roles of G-protein coupled receptors (GPCRs) in the pathophysiology of various human diseases. Biotechnol Adv. 2013;31(8):1676–94. doi: 10.1016/j.biotechadv.2013.08.017. [DOI] [PubMed] [Google Scholar]