Abstract
RNAi pathways have evolved as important modulators of gene expression that act in the cytoplasm by degrading RNA target molecules via the activity of short (21-30nt) RNAs1-6 RNAi components have been reported to play a role in the nucleus as they are involved in epigenetic regulation and heterochromatin formation7-10. However, although RNAi-mediated post-transcriptional silencing (PTGS) is well documented, mechanisms of RNAi-mediated transcriptional gene silencing (TGS) and in particular the role of RNAi components in chromatin, especially in higher eukaryotes, are still elusive. Here we show that key RNAi components Dicer-2 (Dcr2) and and Argonaute-2 (AGO2) AGO2 associate with chromatin, with strong preference for euchromatic, transcriptionally active loci and interact with core transcription machinery. Notably Dcr2 and AGO2 loss of function show that transcriptional defects are accompanied by perturbation of Pol II positioning on promoters. Further, both Dcr2 and Ago2 null mutations as well as missense mutations compromising the RNAi activity impair global Pol II dynamics upon heat shock. Finally, AGO2 RIP-seq experiments reveal that, AGO2 is strongly enriched in small-RNAs encompassing promoter as well as other parts of heat shock and other gene loci on both sense and antisense, with a strong bias for antisense, particularly after heat shock. Taken together our results reveal a new scenario in which Dcr2 and AGO2 are globally associated with transcriptionally active loci and may play a pivotal role in shaping the transcriptome by controlling RNA Pol II processivity.
Results
Accumulating evidence indicates that RNAi components and small RNAs act in the nucleus to control heterochromatin formation, repeat-induced gene silencing (RIGS) and transposable element mobilization7-10. However, the global association of RNAi components with chromatin and their role in transcriptional regulation remains to be elucidated. To investigate a role for RNAi in higher eukaryotes chromatin and, possibly, in TGS, we first determined if any of the key RNAi components were present in the nucleus of Drosophila cells. Cellular fractionation of embryonic tissue culture cells (S2 cells) revealed that the miRNA processing factors Dcr1 and AGO1 as well the RNAi component AGO2 are equally distributed between cytoplasm and nucleus (fig. S1a). In contrast, the RNAi protein Dcr2 is greatly enriched in the nuclear fraction (fig. S1a). To evaluate the association of RNAi components with different nuclear compartments, we performed chromatin fractionation experiments 11 (fig S1b,c). Indeed, a substantial portion of Dcr2 and AGO2 are detected in chromatin fractions, along with Pol II, the Negative ELongation Factor-E (NELF-E), Polycomb (Pc), and histone H3 (fig S1c). In contrast, most of Dcr1 and AGO1 are found in the TritonX-100 soluble fraction (S1), along with tubulin, a marker for proteins not associated with chromatin (fig. S1c). Altogether our data indicate that the Dcr2/AGO2 complex is mainly associated with chromatin, while the Dcr1/AGO1 complex is mostly cytoplasmic, in accordance with its cytoplasmic functions (i.e. PTGS) .
To determine if RNAi components associate with chromatin in vivo, Drosophila polytene chromosomes were stained with AGO2 and Dcr2 specific monoclonal antibodies 3,12,13. Both AGO2 and Dcr2 were detected at several hundred sites on polytene chromosomes from wild type larvae (fig. S1d,g; S2a,b). In contrast, little or no staining was detected in an Ago2 or Dcr2 null chromosomes1,2 (fig. S1f,i ;Table S1a). Strikingly, the majority of AGO2 and Dcr2 associated loci correspond to interbands, suggesting that AGO2 and Dcr2 preferentially associates with euchromatic, transcriptionally active loci 14 (fig. S2a,b). In particular, the chromatin binding of Dcr2 requires AGO2 but not vice versa (fig. S3) suggesting that AGO2 acts as the RNAi effector complex also on chromatin.
Interestingly, AGO2 and Dcr2 are present at 87A and 87C cytogenetic loci (fig. S1e,h). These cytogenetic loci contain copies of the hsp70 heat shock gene, thus providing well-characterised inducible candidate genes to investigate the role of Dcr2 and AGO2 in transcription and in particular Pol II pausing regulation 15,16. To determine if Dcr2 and AGO2 affect hsp70 transcription, hsp70 transcript levels were measured in control cells and in cells depleted of Dcr2 or AGO2 (fig. S4). The knock-down of either RNAi components resulted in a significant increase of hsp70 transcripts in non heat shocked cells (fig 1a,b). Similar results were obtained for a second heat shock gene, hsp68 (fig. 1a,b). The increased expression of hsp70 and hsp68 in cells depleted of Dcr2 could be reversed by expression of a flag-tagged wild type copy of Dcr2 (Dcr2-flag) (fig. 1c), indicating that the change in expression was not due to an off-target effect of the Dcr2 RNAi. Interestingly, the levels of the endogenous AGO2 protein increased along with the expression of wild type Dcr2-flag (fig. S4a), suggesting that the Dcr2-flag protein is integrated into the RNAi pathway and that re-establishment of functional levels of Dcr2 and AGO2 rescues the observed transcriptional defects on heat shock genes. However, Dcr2 and AGO2-depletion did not affect the expression of hsp70 and hsp68 during heat shock, suggesting that RNAi activity is not involved in heat shock factor-mediated activation (fig. S5 a,b).
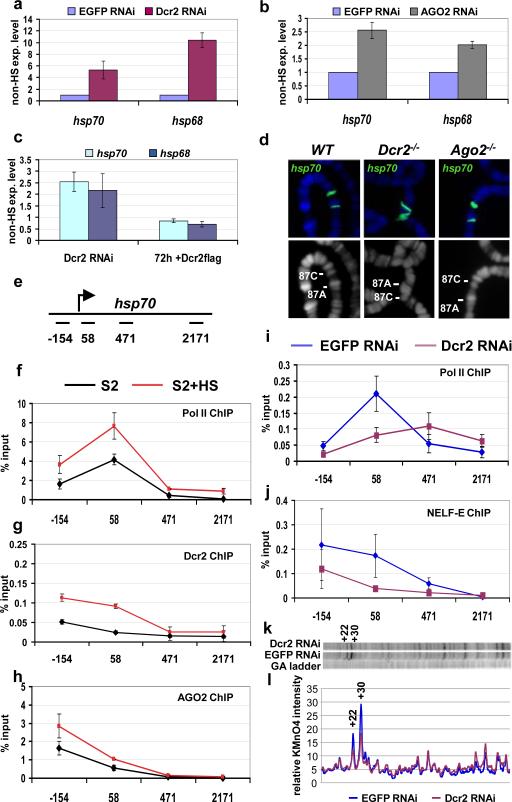
Figure 1. Pol II promoter-proximal pausing on hsp70 is reduced in Dcr2 RNAi cells.
a-c) Quantitative RT-PCR analysis of heat shock gene transcripts. RNA from S2 cells treated with EGFP dsRNA (control) or Dcr2 dsRNA (a), or AGO2 dsRNA (b) were analyzed with primers specific for the indicated heat shock genes. c) Induction of Dcr2-flag transgene is able to revert the phenotype induced by Dcr2-depletion. S2 cells stably transformed with a Dcr2-flag transgene were treated with EGFP dsRNA (control) or Dcr2 dsRNA. The Dcr2-flag expression is induced only in the Dcr2 RNAi sample by the addition of copper. The samples were analyzed before and after 72h induction of the transgene. The transcript levels are shown with respect to the EGFP control (experiment/control ratio). n=3, bars represent the mean±standard deviation. d) hsp70 DNA-FISH on polytene chromosomes from wild type (WT), homozygous Dcr2L811fsX or homozygous Ago2414 mutant larvae; lower pictures show DNA staining; upper pictures show the merge of DNA (blue) and FISH (green) signals. e) Schematic representation of the hsp70 transcription unit with the position of the PCR amplicons used in this study. The numbers indicate the middle of each amplicon with respect to the transcription start site. f-h) ChIP analysis of the hsp70 heat shock gene. Chromatin from S2 cells or S2 cells after exposure to heat shock (HS) was immunoprecipitated with anti-Pol II 4H8 (recognises the Ser5-phosphorylated CTD domain), anti-Dcr2 or anti-AGO2 antibodies. n=3, bars represent the mean± standard deviation. i-j) ChIP analysis of the hsp70 heat shock gene. Chromatin from S2 cells treated with EGFP dsRNA (control) or Dcr2 dsRNA was immunoprecipitated with anti-Pol II 4H8 or anti-NELF-E antibodies. The resulting DNA has been analyzed by quantitative PCR. Protein binding is expressed as a percentage of input subtracted by the background signal. n=3, bars represent the mean± standard deviation. Differences in Pol II ChIP values in 2f and 2i are due to different batches of antibody used in these assays. k) Permanganate mapping of open transcription bubbles on hsp70. Permanganate reacts with single-stranded thymine residues, like those in a open transcription bubble, revealing a transcriptionally engaged RNA polymerase. The autoradiograph includes a G/A ladder, used to determine the position of the bands, and the permanganate reactivity of thymine residues observed in S2 cells treated with EGFP dsRNA (control) or Dcr2 dsRNA. The hyper-reactive thymines +22 and +30 are labeled. Two independent biological samples have been analyzed. Shown is a representative picture l) Quantification of the autoradiograph.
Typically, activation of heat shock genes results in chromosome decondensation of heat shock loci to form large puffs on polytene chromosomes 17. Therefore, we used DNA-FISH to look at heat shock loci chromatin structure in Dcr2L811fsX and Ago2414 null mutant polytene chromosomes. Interestingly, we observed that the 87C locus is partially decondensed in Dcr2L811fsX and Ago2414 mutant chromosomes relative to the wild type controls (fig. 1d). 87C contains 4 copies of hsp70 distributed over 30 kb. Decondensation was not evident at 87A probably because this locus has only two copies of hsp70 encompassed in a region of less than 10 kb.
Since the RT-PCR and cytogenetic data suggested defective maintenance of Pol II pausing, we used chromatin immunoprecipitation (ChIP) to measure the levels of phosphorylated Ser5 Pol II on the hsp70 gene before and after heat shock (fig 1e,f). The primers used for quantitative PCR analysis of the immunoprecipitated DNA span several regions of the hsp70 gene. An upstream primer set centered at –154 detects the heat shock element, the +58 primer set contains the region of the paused polymerase18 and two others lie downstream within the transcription unit extending towards the end of the gene (Fig. 1e). Consistent with previous observations 18, the Pol II profile observed in S2 cells shows a peak in the region of paused Pol II (+58 primer set, fig 1f) and increases after heat shock. Further, ChIP analysis revealed that Dcr2 and AGO2 are present at the hsp70 promoter region (−154/+58, fig 1g,h, fig S6). Interestingly after heat shock both Dcr2 and AGO2 proteins increase on the hsp70 promoter region (fig 1g,h, fig S1i , S6). Consistent with RT-PCR and cytogenetic data, Dcr2 depletion consistently decreased the level of Pol II on hsp70 in the region (primer set +58) of the paused polymerase (fig. 1i). We extended our ChIP analysis to the NELF-E protein, which is part of the transcriptional regulatory complex that causes Pol II pausing 16,19. Depletion of Dcr2 caused the level of NELF on hsp70 to decrease in the +58 region where Pol II pauses (fig. 1 i,j). Pol II and NELF-E protein levels were not altered by Dcr2 depletion (fig. S4b), indicating that the observed diminished level of Pol II and NELF detected on hsp70 is not a consequence of a reduction in the available proteins. Conversely, in Dcr2 depleted cells, we observed a reduction in the level of the AGO2 protein (fig. S4b). This is not due simply to cross-targeting of the dsRNA because the level of the Ago2 transcript was unaffected (fig. S4c). Likewise, the AGO2 dsRNA reduced Dcr2 protein levels (fig. S4d) without affecting Dcr2 mRNA levels (fig. S4e). These results suggest that Dcr2 and AGO2 stabilize each other in a protein complex that is important for repressing heat shock genes in un-induced cells.
To confirm that Dcr2 depletion affected Pol II pausing on hsp70, we performed permanganate footprinting analysis. Permanganate reacts with thymine residues in regions of single-stranded DNA, revealing the presence and location of transcriptionally engaged Pol II. We observed a significant reduction in permanganate reactivity of thymine residues at +22 and +30 in Dcr2 depleted cells (fig. 1 k,l) in agreement with the reduced occupancy of Pol II in this region (primer set +58) shown by ChIP (fig. 1i). Hence Dcr2 is involved in maintaining paused Pol II at the observed hsp70 loci.
Heat shock gene expression and loss of paused Pol II upon depletion of Dcr2 could be a consequence of stress induced by depletion of the RNAi machinery in the cell. Therefore, we evaluated the impact of perturbing the RNAi pathway on transcription of non-heat shock genes. Four non-heat shock genes (CG9008, fz, rho and mfas) that have paused Pol II were analyzed by permanganate footprinting 19,20. The results showed that depletion of Dcr2 also altered the distribution of Pol II in the promoter proximal region of these genes (fig. S7). These changes were also accompanied by differences in transcript levels (fig. S8). Notably, the observed changes in Pol II result both in up-(CG9008, fz, rho) and down-regulation (mfas) of the corresponding transcripts (fig. S8). Thus, the RNAi machinery can influence Pol II pausing at non-heat shock genes, arguing against an unspecific Dcr2-induced heat shock response.
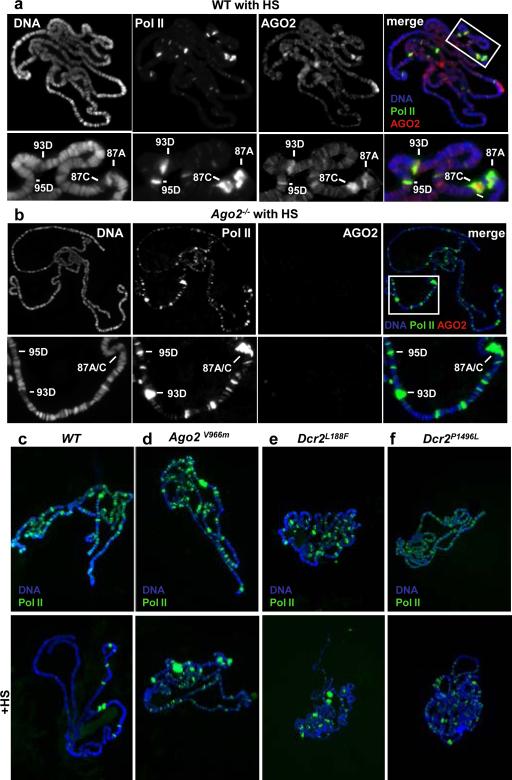
During heat shock, most of the Drosophila genome is transcriptionally quiescent. The elongating Pol II dissociates from euchromatic interbands and accumulates at heat shock loci21. We used this dynamic Pol II relocalization behavior to follow the chromatin binding and distribution of elongating Pol II (Ser2-phosphorylated), AGO2 and Dcr2 in vivo. In agreement with ChIP results (fig. 1h), we found that after heat shock, AGO2 accumulated at heat shock loci (fig. 2a). However, in contrast to Pol II, the association of AGO2 with other loci appears unchanged (fig. 2a). On the other hand, when we analyzed the dynamic chromatin repositioning of Pol II upon heat shock in Ago2 null mutant chromosomes, we found that Pol II is still bound to the heat shock loci (fig 2b). Strikingly, a substantial fraction of elongating Pol II in Ago2414 mutants was retained at many euchromatic sites after heat shock (fig. 2b). The same behavior is observed for Dcr2L811fsX mutant (Table S2). Taken together these results show that AGO2 and Dcr2 associate with many euchromatic sites and their activity is probably required for the correct execution of the global transcriptional repression following heat shock.
Figure 2. Chromatin localization of Pol II and AGO2 after heat shock.
Co-immunolocalization of AGO2 (red) and elongating Pol II (green) in WT (a) and homozygous Ago2414 mutant chromosomes (b) after heat shock (HS). DNA is stained in blue. Single signals are shown in black and white. The bottom row shows a higher magnification of the boxed area. (c) Immunolocalization of elongating Pol II (green) in WT (c), homozygous Ago2V966M (d), homozygous Dcr2L188F (e), homozygous Dcr2P1496L (f), chromosomes shows that missense mutations in Dcr2 or AGO2 influence the behaviour of elongating Pol II. Chromosomes on the lower panel have been exposed to heat shock (HS). DNA is stained in blue.
In order to dissect the role RNAi in Pol II regulation we used three mutants carrying single amino acid substitutions affecting respectively, the helicase (Dcr2L188F), the dicing (Dcr2P1496L) and the AGO2 slicing (Ago2V966M) activities22,23 (see table S1a). To check for Dcr2 and AGO2 specific requirements first we used DNA-FISH to analyse chromosome decondensation at hsp70 loci. As shown in Table S1b, puffing frequencies were increased in all of the above mutants and in particular in the Ago2 slicing mutant (Ago2V966M). In addition, in all these mutants the hsp70A, hsp70B and hsp68 transcript levels were increased before heat shock (fig S9). Interestingly, the transcript increase is also evident at 87A (hsp70A) where the chromatin decondensation is not appreciable at a cytogenetic level (fig. S9, Table S1b). Thus all tested mutants induce transcript up-regulation of the hsp70 genomic region although this is not always accompanied by chromatin decondensation. Uncoupling between chromatin decondensation and transcriptional activation has been previously reported24, although in this case we cannot exclude a RNAi dependent post-transcriptional mechanism influencing the hsp70A transcript levels.
Next we analysed Pol II distribution and dynamics on polytene chromosomes in the same Dcr2 and Ago2 mutants. Remarkably, though with different degree of penetrance, after heat shock all three mutants fail to relocalize elongating Pol II (fig 2c-f; Table S2), suggesting that RNAi enzymatic activity is involved in the global Pol II dynamics upon stress response.
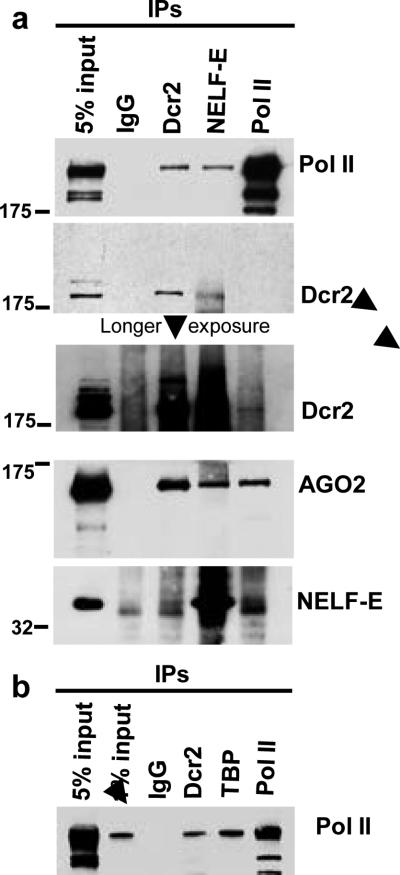
Association of RNAi components with Pol II complexes has been reported in other systems 25,26,27. To investigate the possibility that these proteins are part of a complex that regulates Pol II elongation, thus establishing a link with the observed “pausing” defects, we tested for interactions between Dcr2, NELF-E and Pol II in nuclear extracts derived from Drosophila S2 cells. Immunoprecipitated fractions were evaluated by western blot for the presence of Pol II, Dcr2, AGO2, and NELF-E (Fig. 3a). As expected, Dcr2 interacts with AGO2 28 (fig 3a). In addition, we found Dcr2 and AGO2 interacting with Pol II and NELF-E (fig. 3a). All of these interactions are resistant to RNase treatment, indicating that they are not an indirect consequence of protein trapping associated with emerging RNA molecules (fig. S10a). If compared to other general transcription factors, the amount of Pol II interacting with Dcr2 appears to be in the same range as TBP (TATA-binding protein), confirming the association of Dcr2 with many active loci on polytene chromosomes (fig 3b). When we tested the same interaction in Dcr2 depleted cells we observed a decrease in levels of NELF-E and AGO2 associated with Pol II (fig. S10b). These observations indicate that the Dcr2-AGO2 complex influences the association between NELF-E and Pol II, thereby providing a possible explanation for the observation that the depletion of Dcr2 alters the behavior of Pol II in the promoter proximal region of several genes.
Figure 3. Dcr2 and the RNAi effector protein AGO2 associate with Pol II and NELF.
a) Nuclear extracts from Drosophila S2 cells were immunoprecipitated with the indicated antibodies, and the immunoprecipitated protein complexes were analyzed by western blot for the presence of Pol II, Dcr2, AGO2 and NELF-E proteins. b) Nuclear extracts from S2 cells were immunoprecipitated with the indicated antibodies (TBP: TATA-binding protein) and analyzed by western blot for the presence of Pol II.
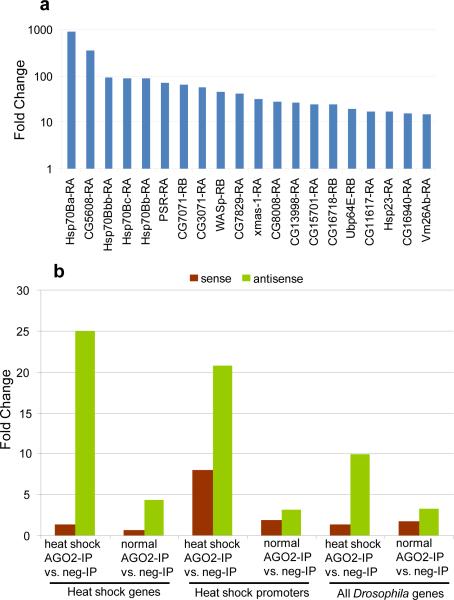
Our data indicate an involvement of the RNAi machinery in the heat shock stress response. In order to evaluate possible heat shock-induced change in the expression signature of AGO2-dependent small RNAs we sequenced small RNAs bound to AGO2 before and after heat shock. Short RNA libraries from S2 cells were generated from RNA fractions obtained by immunoprecipitation with antibodies against AGO2 or control IgG, with or without heat shock (fig. S11). Basic statistics for the four libraries are given in Table S3.. Many sequenced tags map in more than one location to repeat regions (fig. S12), consistent with previous Drosophila deep-sequenced AGO2-IP libraries3,5. Firstly we focused our attention on the promoter region (500bp upstream and 50bp downstream of TSS). We found heat shock loci promoter regions are enriched in short RNAs, showing a dramatic increase upon heat shock. This is most noticeable in hsp23 and all hsp70 loci (Table S4, S5). This observation prompted us to consider short RNA increase across all promoters29. Strikingly, the most enriched promoters are the hsp70B loci and hsp23 is also observed in the top 20, suggesting a direct role for AGO2 at these promoters (fig. 4a). The possible function or role in the heat shock response of other loci present in the top 20 list is not known. Next we tried to gauge shifts in short RNA tags derived from discrete transcribed regions at all gene loci in response to heat shock (fig.S14a). We observed little or no relative enrichment along different length intervals, suggesting tags are not derived exclusively from promoter regions but appear to be equally distributed along the transcription unit (fig. S13). Interestingly, while there is a sharp increase in AGO2 short RNA in the heat shock condition relative to the heat shock condition negative control-IP (confirming the specific association of these tags with AGO2), only a small increase is observed in the heat shock condition AGO2-IP relative to the normal condition AGO2-IP suggesting these transcript-associating tags are present even under normal conditions (fig. S14a). This data suggest a role for AGO2 in regulating Pol II processivity in addition to promoter regulation. Next we analyzed the strand bias of AGO2-associated small RNAs. Sense and antisense tags have a specular distribution within individual libraries, with the ratio of the sums of the sense and antisense short RNA tag counts mapping to heat shock loci and the set of all Drosophila genes equal to approximately 1 (fig. S14b). However, comparison of normalized sense and antisense tag counts across AGO2-IP and negative control-IP libraries reveals a pronounced enrichment in antisense tags and little or no enrichment in sense tags, suggesting antisense tags are highly specific in their association with AGO2 while sense tags are not. Additionally, the observed enrichment implies antisense tag association is strongest in the heat shock condition (fig. 4b). Next we examined cap analysis of gene expression (CAGE) data generated in Drosophila embryo for the modENCODE project29 to survey the extent of antisense transcriptional activity at heat shock loci (Table S6). Antisense transcriptional activity was observed at almost all heat shock loci and surprisingly many hsp70 loci displayed roughly equivalent levels of sense and antisense transcription (Table S6), suggesting a possible source for the observed short RNA tags. Taken together, this data indicates the observed short RNAs are likely derived from dsRNA precursors and sequences antisense to the loci preferentially associate with AGO2, implying sense-targeting.
Figure 4. Features of AGO2-associated small RNAs.
a) Top 20 short RNAs most-enriched promoters in heat shock vs. no heat shock conditions. Comparison of the short RNA fold enrichment in the AGO2-IP libraries across the normal and heat shock condition for all promoters. y-axis is shown in log scale. b) Relative enrichment calculated as fold change (y-axis) for the sum of all sense or antisense tags across different conditions. Gene definitions and conditions labeled below the chart. Sense tag enrichment colored in brown (little to no enrichment), antisense tags enrichment colored in green (strongest enrichment in heat shock conditions).
In this work we show that in Drosophila RNAi components act in the nucleus and associate with transcriptionally active rather then inactive gene loci, interact with RNA Pol II and contribute to transcriptional control of higher eukaryotic cells, in particular during heat shock stress response. The latter observation strongly suggests that RNAi function in the nucleus, like in the cytoplasm, may have evolved as a key mechanism that operates in the context of pervasive sense/antisense transcription to regulate RNA level homeostasis in the cell (see also Supplementary discussion).
Methods
Fly Stocks
Flies were maintained under standard procedures. Canton-S or w1118 were used as wild type strains. The Dicer-2L811fsX and Ago2V966M stocks2,23 are from R.W. Carthew. The Ago-2414 stock 1 is from Eric Lai. The Dcr2L188Fand Dcr2P1496L stocks22 are fromY.Sik Lee.
Cytoplasm and nuclear fractionation
S2 cells were washed twice in ice cold PBS, resuspended in solution I (10mM HEPES pH7.9, 10mM KCl, 0.1mM MgCl2, 0.1mM EDTA, 0.1mM DTT, 0.5mM PMSF, Roche protease inhibitor cocktail) and passed 7 times through a 25G syringe. After centrifugation (2000rpm, 10min, 4°C) the supernatant was collected as the cytoplasmic fraction. The pellet was washed in solution I four times and resuspended in solution II (10mM HEPES pH7.9, 400mM NaCl, 1.5mM MgCl2, 0.1mM EDTA, 0.1mM DTT, 0.5mM PMSF, 5%glycerol, Roche protease inhibitor cocktail) and incubated on ice for 30 min with occasional flicking. After centrifugation at 12000rpm 20min 4°C the supernatant was collect as nuclear fraction; 20μg of each sample was analyzed by western blot.
Preparation of Nuclear extract and immunoprecipitation
Nuclear extract from S2 cells and immunoprecipitation were performed according to Lupo et al 30 with minor modifications. Briefly for each immunoprecipitation 300 μl of nuclear extract (600-800μg) was mixed with 300μl TEA150 (10mM Tris-HCl pH 8, 150mM NaCl, 1mM EDTA, 1 mMDTT, 1mM PMSF, 0.1%NP40, proteinase inhibitors leupeptine, aprotinin and pepstatin all 2μg/ml) and 30 μl Protein A/G Plus-agarose beads (Santa Cruz Biotechnology), incubated for 1 hour at 4°C for pre-clearing, The supernatant was transferred to a new tube, the appropriate amount of antibody (abcam anti-Dcr2 5μg and/or 2μg of mouse anti-Dcr2; anti-PolII 4H8 2μg; anti-NELF-E 10μl, anti-TBP 1-2μl; rabbit IgG 2μg) was added to the supernatant and the samples were incubated for 3 hours on a wheel at 4°C. Then 40 μl of A/G Plus-agarose beads were added. After incubation for 2 hours the beads were washed 5× with 500 μl of TEA150 buffer, then resuspended in SDS-PAGE loading buffer and analyzed by western blot. Proteins were detected with the SuperSignal West-Dura substrate (Pierce). For single-strand RNase treatment 2μl of a RNase cocktail (RNase A+T1, Ambion) was added during one of the washing steps and the samples were incubated for 20 minutes at 30°C.
RNA interference in S2 cells
S2 cells were grown in serum free insect culture medium (HyQ SFX, Hyclone). Production of dsRNAs (400-500 bp long) of EGFP, Dcr2 and AGO2 was performed in vitro by T7 transcription. Sense and antisense RNAs were denatured by heating, and re-annealed to dsRNA in water. RNAi was performed as described previously 31 and lasted 8-10 days. Primers for production of T7 templates: EGFP-F ACGTAAACGGCCACAAGTTC; EGFP-R TGCTCAGGTAGTGGTTGTCG; DCR2-F GTTCCGCTTTGGTCAACAAT; DCR2-R-TGATCGTCTTTTCCATGCAG; DCR2UTR-FATGATGATTCCAGCCCAGTC; DCR2UTR-R-TTATTTCGACCCAAGGTAAC; AGO2-FGCTGCAATACTTCCAGCACA; AGO2-R-CTCGGCCTTCTGCTTAATTG.
Antibodies
Mouse anti-Pol II 4H8 (Abcam ab5408), recognises the Ser5-phosphorylated CTD domain 32; mouse anti-Pol II H5 (Covance), recognises the elongating Pol II phosphorylated at Ser2 in the CTD domain 18. Rabbit anti-Dicer-1 (ab4735), anti-Dicer-2 (ab4732), anti-histone H3 (ab1791) and mouse anti-actin (ab8224) antibodies were from Abcam; mouse anti-βtubulin (E7) was from the Hybridoma Bank at the University of Iowa; mouse anti-Flag (M2) antibody was purchased from Sigma; mouse anti-AGO1, anti-AGO2 and anti-Dcr2 antibodies have been previously described 3,12,13 ; rabbit anti-Polycomb 33 was provided by R. Paro; rabbit anti-TBP34 was a gift of J.T Kadonaga; anti-NELF-E antibody was previously described 16; in western blot experiments also a non commercial anti-Dicer-2 antibody, kindly provided by Q. Liu 35, was employed.
ChIP
Chromatin was prepared as described 36 from non-treated and heat shocked (as described below) S2 cells with a fixation step of 15 to 25 minutes at room temperature. The following antibodies were employed for imunoprecipitation (approximately 8×106 cell/IP): anti-Pol II 4H8 (2 μl), anti-NELF-E (5μl), anti-AGO2 (9D6, 3 μl), mouse anti-Dcr2 (4 μl) and anti-Flag M2 (as negative control, 4 μl). Specificity of the ChIP-grade anti-AGO2 (9D6) antibody are shown in fig. S6. The anti-Dcr2 antibody used for the ChIP experiment is the same one used for IF experiments shown in fig. S1i. Quantitative-PCR was performed in a DNA Engine OPTICON 2 (MJ Reasearch, Bio-Rad) instrument using the QuantiTect SYBR Green PCR Kit (Qiagen) or a Roche LightCycler 480 using the Absolute QPCR SYBR Green Mix (Thermo Scientific) according to manufacturer's instructions. Relative enrichments were calculated as percentage of the input. Hsp70 primer sequences: -154 and +58 primers were described previously18; +471forward-GATCTGGGCACCACCTACTC; 471reverse-TGGGAGTCGTTGAAGTAGGC; +2171-forward-CACGATCAAGAACGACAAGG; +2171reverse-CTTTGGCCTTAGTCGACCTC. The names of the primer pairs indicate the distance of the middle of each amplicon from the hsp70 transcription start site.
Heat shock induction
Third-instar larvae were transferred to preheated 1.5-ml microcentrifuge tube that was submerged in a 37°C water bath for 40 min. S2 cells were transferred to pre-heated medium and submerged in a 37°C water bath for 40 min.
Quantitative RT-PCR analysis
Total RNA from S2 cells or larvae was isolated with the Trizol reagent (Invitrogen). RNA from each sample was subjected to cDNA synthesis using a Quantitect reverse transcription kit (Qiagen). All primers were annealed at 60 °C. Real-time PCR was performed with the DNA Engine Opticon 2 (MJ). Quantification was normalised to the housekeeping gene GAPDH1, and relative expression levels were calculated using the following equation: A=2[Ct(ref)-Ct(ref-control)]-[Ct(sample)-Ct(sample-control)]. Primer sequences are available on request.
FISH analysis
Cytological preparations and fluorescence in situ hybridization (FISH) were carried out as previously described 37. FISH signals were quantified by densitometric analysis using the Leica QFluor software. Any deviation of 33% from the normal average relative puffing ratio existing in wild type at the 87C/87A loci was considered an increase in puffing at the 87C relative to 87A in the Ago2 and Dcr2 mutants analyzed.
Immunostaining of salivary gland polytene chromosomes
Polytene chromosomes were prepared from third-instar larvae grown at 18°C. Single and double immunostaining were carried out as previously described 38. The Fab-fragment blocking method was used to stain the chromosomes with AGO2 and Pol II antibodies raised both in mouse. Antibodies were used at the following dilutions: anti-Pol II (H5) 1:200; anti-AGO2 1:50; anti-AGO2 (9D6) 1:50; mouse anti-Dcr2 1:100; Fab-fragment (anti-mouse) 1:100. Given the specificity of anti-AGO2 and anti-Dcr2 antibodies on polytene staining, the cytoplasmic and nucleoplasmic background signals were removed (using the Photoshop Lasso tool), in order to highlight AGO2 and Dcr2 chromatin binding.
Permanganate footprinting
S2 cells (2ml, approximately 4×106 cells/ml) were treated with permanganate essentially as previously described 19. The permanganate cleavage pattern was analyzed as previously described 39. Primer sequences are available upon request. Quantification of the autoradiographs was done using the Image Quant software (Biorad).
Dicer-2 flag rescue assay
In order to have selective inhibition of the endogenous Dicer-2 gene, a specific dsRNA targeting the 3’UTR sequence missing in the Dicer-2-flag transgene was used. After eight days with dsRNA treatment the expression of the fusion protein was induced by adding 1mM CuSO4.
Chromatin binding assay
The procedure was used essentially as previously described 11. S2 cells (60ml approximately 3-6x106/ml) were washed with cold PBS. One tenth of the cell suspension (control fraction, C) was resuspended in RIPA buffer (150 mMTris–HCl, (pH 8.0), 150 mM NaCl, 0.5% DOC, 0.1% (w/v) SDS, 1% (v/v) NP-40 protease inhibitors leupeptine, aprotinin and pepstatin all 2μg/ml, 1 mM PMSF and phosphatase inhibitors NaF, Sodium orthovanadate 1mM) and left 30 min on ice. The remaining fraction was lysed for 15 min on ice in cold CSKI buffer (10 mM Pipes, pH 6.8, 100 mM NaCl, 1 mM EDTA, 300 mM sucrose, 1 mM MgCl2, 1 mM DTT, 0.5% (v/v) Triton X-100, protease inhibitors leupeptine, aprotinin and pepstatin all 2μg/ml, 1 mM PMSF and phosphatase inhibitors NaF, Sodium orthovanadate 1mM). The cell lysate was divided into two portions, which were centrifuged at 500g at 4 °C for 3 min. The supernatants (S1 fraction), which contain Triton-soluble proteins, were further analyzed. One of the pellets, was washed twice in CSKI buffer and then resuspended in RIPA buffer (the P1 fraction). The second pellet, after CSKI washes, was resuspended in CSK II buffer (10 mM Pipes pH 6.8, 50 mM NaCl, 300 mM sucrose, 6 mM MgCl2, 1 mM DTT, proteinase inhibitors leupeptine, aprotinin and pepstatin all 2μg/ml and phospatase inhibitors NaF, Sodium orthovanadate 1mM), treated with DNase for 30 min followed by extraction with 250 mM NH2SO4 for 10 min at 25°C. The sample treated with DNAse (Promega) and salt was then centrifuged at 1200g for 6 min at 4 °C and the supernatant (S2 fraction), and pellet (P2 fraction), were collected. P2 was also resuspended in RIPA buffer. 20μg of all fractions were analyzed by immunoblotting.
Purification of AGO2 associated RNAs
For heat shock, S2 cells were transferred to pre-heated medium at 37 °C and submerged in a 37 °C water bath for 40 min. Immunoprecipitation was performed as previously described 3. Briefly, S2 cells were washed twice in ice-cold PBS, resuspended in an Empigen-containing PBS buffer (1% Empigen, 1 mM EDTA, 100 mM DTT, 2 μg/mL Pepstatin, 2 μg/mL Leupeptin, and 0.5% Aprotinin) and incubated on ice for 10 min to lyse cells. After centrifugation (15,000 rpm for 20 min at 4 °C), the supernatant was collected. Immunoprecipitation was performed using anti-AGO2 antibody (9D6), or mouse non-immune IgG (negative control), immobilized on GammaBind beads (GE Healthcare). The reaction mixture was rocked at 4°C for 2 h and washed five times with Empigen-containing PBS buffer. For silver staining, the washed beads were incubated with 2x sample buffer (20% Glycerol, 100 mM Tris-HCl (pH 6.8), 4% SDS, 0.02 % Bromophenol blue) for 10 min at room temperature and DTT (200 mM final) was added to the mixture. The sample was then incubated at 95 °C for 5 min and resolved by SDS-PAGE. After electrophoresis, protein bands were visualized by SilverQuest (Invitrogen). Small RNAs associated with AGO2 were treated with phenol:chloroform and precipitated with isopropyl alcohol. RNAs were then dephosphorylated with CIP (NEB), labeled with 32P-gamma-ATP with T4 polynucleotide kinase (NEB) and resolved on a 12 % acrylamide denaturing gel.
Short RNAs library sequencing and computational analysis
Short RNA libraries were prepared as described previously with one of two barcodes attached to the libraries generated from the IP experiments, regardless of the presence or absence of heat shock40. Libraries were sequenced using the Genome Analyzer GA-IIx (Illumina) in two lanes. Linkers and barcodes sequences were extracted from raw tags; tags were mapped to the Drosophila dm3 genome assembly using the Nexalign program41. Genome classification of individual tags was determined by overlap with existing definitions culled from publicly available genome tracks in the FlyBase and functional RNA databases42,43. Tags mapping to more than 10 locations in the genome were removed from subsequent analyses; tags counts for remaining tags were distributed evenly across the total number of mapping sites. Tags were normalized to tags per million prior to comparisons across libraries. A summary of the basic statistics for the four libraries is provided in Table S3. The extraction rates were between 70-75% per library (not shown); mapping rates were above 90% (see table S3). As an additional normalization strategy, tags per million miRNA counts were also tabulated for each library (Table S5) because miRNA percentages were largely unaffected by heat shock treatment (Figure S12); fold enrichments calculated across conditions with these values were consistent with tpm normalization (data not shown). Sense-antisense distinctions were decided by Flybase gene definitions; overlapping gene definitions on the same strand were merged and tags mapping to overlapping sense/anti-sense transcripts were included in both sense and anti-sense totals. Clusters of CAGE tags, representative of transcriptional start site activity, were linked to heat shock loci using 5’ and 3’ Flybase gene definitions; each locus was visually inspected in a genome browser and gene definitions were adjusted to account for differences in transcriptional start sites between embryonic tissue and FlyBase gene definitions. Large scale genome comparisons were carried out using the bedtools utilities suite44.
Supplementary Material
Acknowledgements
We are deeply grateful Pino Macino for stimulating discussions. We also thank R. Carthew, E. Lai, Q.Liu, R. Paro, Y. Sik Lee, J.T. Kadonaga for reagents. This work was supported by grants from NIH GM47477 to D.S.G; the Deutsche Forschungsgemeinschaft SPP 1356 to A.B.; Grant-in-Aid for Scientific Research (A) No.20241047 and Japan Society for the Promotion of Science (JSPS) through the “Funding Program for Next Generation World-Leading Researchers (NEXT Program),” initiated by the Council for Science and Technology Policy (CSTP) to PC, a Research Grant for RIKEN Omics Science Center from MEXT, Grant-in-Aid from MEXT to K.M., M.C.S and H.S. MCS is also supported by CREST (Core Research for Evolutional Science and Technology) from the JST (Japan Science and Technology Agency); Fondazione Telethon, Giovanni Armenise Harvard Foundation, FIRB-MIUR, Associazione Italiana Ricerca Cancro (AIRC), Fondazione Compagnia San Paolo and EMBO Young investigator program to D.F.V.C.; Fondazione Telethon, Associazione Italiana Ricerca Cancro (AIRC), EU FP6 The Epigenome Network of Excellence and Fondazione Compagnia San Paolo to V.O.; With the contribution of Italian Ministry of Foreign Affairs, “Direzione Generale per la Promozione e la Cooperazione Culturale” to V.O. A.S. is supported by a JSPS fellowship ID P09745. Sequencing was provided by the Genas service (RIKEN Omics Science Center).
Footnotes
Author Contributions
FMC and VO conceived the study. FMC and AB performed the ChIP experiments, FMC and KMP did the chromatin fractionation assays and western blots; FMC did the RT-qPCR on S2 cells; KMP and FLS performed the RT-qPCR on mutant larvae prepared by MCO; FMC performed the coimmunoprecipitations and contributed reagents for the chromosome and permanganate footprinting experiments; MCO performed polytene chromosome experiments; GOK and DSG performed the permanganate footprinting experiments; AMB performed bioinformatic analysis; AS and PC performed the deep sequencing; KM, HS and MCS performed AGO2 RNA pull down ; FMC, MCO, DSG, DFVC and VO designed experiments and interpreted the results; FMC, DSG, DFVC and VO wrote the manuscript with the contribution of MCO, AB, AMB, and inputs from the other coauthors.
Author informations: Data has been deposited in the DNA Data Bank of Japan (DDBJ) under accession code DRA000418. The authors declare no competing financial interest.
References
- 1.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–66. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 3.Kawamura Y, et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–7. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 4.Ghildiyal M, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–81. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamura K, et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–6. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–37. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–20. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teixeira FK, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–4. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 11.Llano M, et al. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006;360:760–73. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 12.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–48. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyoshi K, Okada TN, Siomi H, Siomi MC. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA. 2009;15:1282–91. doi: 10.1261/rna.1541209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 1993;7:2329–44. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- 15.Lis JT. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature. 2007;450:198–202. doi: 10.1038/nature06324. [DOI] [PubMed] [Google Scholar]
- 16.Wu CH, et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–14. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon JA, Sutton CA, Lobell RB, Glaser RL, Lis JT. Determinants of heat shock-induced chromosome puffing. Cell. 1985;40:805–17. doi: 10.1016/0092-8674(85)90340-x. [DOI] [PubMed] [Google Scholar]
- 18.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–37. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C, et al. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilchrist DA, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–33. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai W, et al. RNA polymerase II-mediated transcription at active loci does not require histone H3S10 phosphorylation in Drosophila. Development. 2008;135:2917–25. doi: 10.1242/dev.024927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim do H., Kim J, Kim S, Carthew RW, Lee YS. Functional analysis of dicer-2 missense mutations in the siRNA pathway of Drosophila. Biochem Biophys Res Commun. 2008;371:525–30. doi: 10.1016/j.bbrc.2008.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K, Lee YS, Carthew RW. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2007;13:22–9. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–30. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–7. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 26.Kavi HH, Birchler JA. Interaction of RNA polymerase II and the small RNA machinery affects heterochromatic silencing in Drosophila. Epigenetics Chromatin. 2009;2:15. doi: 10.1186/1756-8935-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–44. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoskins RA, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lupo R, Breiling A, Bianchi ME, Orlando V. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalise with Polycomb and maintain Fab-7 PRE silencing. Mol Cell. 2001;7:127–36. doi: 10.1016/s1097-2765(01)00161-7. [DOI] [PubMed] [Google Scholar]
- 31.Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–5. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- 32.Stock JK, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–35. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 33.Messmer S, Franke A, Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992;6:1241–54. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- 34.Hsu JY, Juven-Gershon T, Marr MT, 2nd, Wright KJ, Tjian R, et al. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 2008;22:2353–8. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;30:1921–5. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 36.Breiling A, O'Neill LP, D'Eliseo D, Turner BM, Orlando V. Epigenome changes in active and inactive polycomb-group-controlled regions. EMBO Rep. 2004;5:976–82. doi: 10.1038/sj.embor.7400260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pimpinelli S, Bonaccorsi S, Fanti L, Gatti M. In: Preparation and analysis of mitotic chromosomes of Drosophila melanogaster. Drosophila: A Laboratory Manual. Sullivan W, Ashburner M, Hawley S, editors. Cold Spring Harbor LaboratoryPress; Cold Spring Harbor, NY: 2000. pp. 1–24. [Google Scholar]
- 38.Corona DF, Armstrong JA, Tamkun JW. Genetic and cytological analysis of Drosophila chromatin-remodeling factors. Methods Enzymol. 2004;377:70–85. doi: 10.1016/S0076-6879(03)77004-9. [DOI] [PubMed] [Google Scholar]
- 39.Cartwright IL, et al. Analysis of Drosophila chromatin structure in vivo. Methods Enzymol. 1999;304:462–496. doi: 10.1016/s0076-6879(99)04028-8. [DOI] [PubMed] [Google Scholar]
- 40.Kawano M, et al. Reduction of non-insert sequence reads by dimer eliminator LNA oligonucleotide for small RNA deep sequencing. Biotechniques. 2010;49:751–5. doi: 10.2144/000113516. [DOI] [PubMed] [Google Scholar]
- 41.de Hoon MJ, et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 2010;20:257–64. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tweedie S, et al. FlyBase Consortium. FlyBase:enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37(Database issue):D555–9. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mituyama TYK, et al. The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 2009;37:D89–92. doi: 10.1093/nar/gkn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.