Abstract
We carried out a study of the expression patterns of seven developmental regulatory genes (Lef1, Lhx2, Lhx9, Lhx5, Lmo3, Lmo4, and Prox1), in combination with topological position, to identify the medial pallial derivatives, define its major subdivisions, and compare them between mouse and chicken. In both species, the medial pallium is defined as a pallial sector adjacent to the cortical hem and roof plate/choroid tela, showing moderate to strong ventricular zone expression of Lef1, Lhx2, and Lhx9, but not Lhx5. Based on this, the hippocampal formation (indusium griseum, dentate gyrus, Ammon's horn fields, and subiculum), the medial entorhinal cortex, and part of the amygdalo-hippocampal transition area of mouse appeared to derive from the medial pallium. In the chicken, based on the same position and gene expression profile, we propose that the hippocampus (including the V-shaped area), the parahippocampal area (including its caudolateral part), the entorhinal cortex, and the amygdalo-hippocampal transition area are medial pallial derivatives. Moreover, the combinatorial expression of Lef1, Prox1, Lmo4, and Lmo3 allowed the identification of dentate gyrus/CA3-like, CA1/subicular-like, and medial entorhinal-like comparable sectors in mouse and chicken, and point to the existence of mostly conserved molecular networks involved in hippocampal complex development. Notably, while the mouse medial entorhinal cortex derives from the medial pallium (similarly to the hippocampal formation, both being involved in spatial navigation and spatial memory), the lateral entorhinal cortex (involved in processing non-spatial, contextual information) appears to derive from a distinct dorsolateral caudal pallial sector.
Keywords: medial pallium, hippocampus, dentate gyrus, Ammon's horn fields, entorhinal cortex, dorsolateral caudal pallium, evolution
Introduction
The hippocampal formation is a cortical structure of the telencephalic hemispheres that is essential for spatial navigation and memory formation (Bird and Burgess, 2008). Interest in this region mainly comes from data in mammals showing that: (1) Damage to the hippocampal formation (as it occurs in Alzheimer's disease) produces a decline or have devastating effects in spatial navigation and memory (Bird and Burgess, 2008; Lithfous et al., 2013). (2) The hippocampal formation (in particular, the dentate gyrus) is one of the few brain regions showing adult neurogenesis (Alvarez-Buylla and Lim, 2004; Gould, 2007), which has been related to network plasticity, learning and memory formation, and the ability to adapt to novelty and complexity (Gould et al., 1999; Leuner et al., 2006; Kempermann, 2008; Varela-Nallar and Inestrosa, 2013; Vivar and van Praag, 2013). (3) Dysfunction of the hippocampal formation and dysregulation of adult hippocampal neurogenesis are associated to several mental disorders and neurological diseases (Eisch et al., 2008; DeCarolis and Eisch, 2010; Jun et al., 2012; Mendez-David et al., 2013).
In mammals, the hippocampal formation (HF) comprises three cytoarchitectonically distinct subdivisions, which from lateral to medial are: the subiculum, the hippocampus proper (Ammon's horn fields or cornu ammonis, subdivided in CA1, CA2, and CA3 fields) and the dentate gyrus (reviewed in Witter and Amaral, 2004; Witter, 2012). It also includes a rostral continuation called indusium griseum (Künzle, 2004). Within the HF, each subdivision is unique regarding its histological, neurochemical and connectivity patterns (Witter and Amaral, 2004; Witter, 2012). The projections of the dentate gyrus and CA fields are mostly intrinsic and associational (within the ipsilateral and contralateral HF), while the subiculum (with a small contribution of CA1) is the major output hippocampal center, with projections to several cortical and subcortical regions, hypothalamus, and midline thalamus (Witter and Amaral, 2004). The HF receives input from the medial septum, nucleus of the diagonal band, pallial amygdala, hypothalamus, midline thalamus, and several brainstem monoaminergic cell groups. Most importantly, the HF is reciprocally connected with the entorhinal cortex, which is part of the parahippocampal region (Witter and Amaral, 2004). Due to the strong functional relationship between the HF and the parahippocampal region, both regions are often included as parts of the hippocampal functional complex, although they differ in other respects, such as position, cytoarchitecture, neurochemistry, and connections (Witter and Amaral, 2004; Witter, 2012). The entorhinal cortex is an essential actor for hippocampal functions, and is also extensively and reciprocally connected with the neocortex, pallial and subpallial-extended amygdala, and septum/diagonal band nuclei (Witter and Amaral, 2004).
Classical studies described in detail the development of rat HF subdivisions from distinct progenitor sectors of the medial pallium (Altman and Bayer, 1990a,b,c). More recently, the molecular control of HF development has started to be elucidated. Wnt and BMP signals from the cortical hem, roof plate and/or meninges are essential for HF development (Galceran et al., 2000; Lee et al., 2000; Machon et al., 2007; Choe et al., 2013), and these induce the expression in the medial pallium of several transcription factors important for different aspects of HF development, such as Lhx2 (Porter et al., 1997; Bulchand et al., 2001; Monuki et al., 2001; Vyas et al., 2003), Lef1 (lymphoid enhancer factor 1) and other TCF transcription factors (Galceran et al., 2000; Choe et al., 2013). In particular, Lef1 is crucial for the production of dentate gyrus granule cells, and Lef1 together with other TCFs are necessary for the development of the whole HF, which is not formed following subrogation of their function (Galceran et al., 2000). Wnt and Lef1 induce the expression of another transcription factor specifically in the dentate gyrus, Prox1 (prospero-related homeobox 1 gene), which is involved in the differentiation of granule cells (Zhou et al., 2004; Lavado et al., 2010; Iwano et al., 2012). Interestingly, some of the regulatory genes involved in the development of the HF, such as those encoding some molecules of the Wnt/β catenin pathway, some TCFs (mostly TCF4), and Prox1, continue to be expressed and are functional in the adult dentate gyrus (Shimogori et al., 2004; Karalay et al., 2011), and at least Wnt signaling and Prox1 play important roles in distinct aspects of adult neurogenesis, such as cell proliferation or the differentiation of new granule cells (Karalay et al., 2011; Iwano et al., 2012; Varela-Nallar and Inestrosa, 2013). In addition to its role in granule cell specification, differentiation, and survival (reviewed by Karalay and Jessberger, 2011), recent data in mouse showed that, from late embryonic stages, Prox1 is also expressed in subsets of neocortical and hippocampal interneurons, which derive from the caudolateral ganglionic eminence and the preoptic area of the subpallium (Rubin and Kessaris, 2013). In spite of the abundance of data on HF development, very little is known on the genes that regulate the development of the entorhinal cortex in mammals.
A hippocampal formation, involved in spatial navigation and memory formation, has been identified in a topologically comparable pallial position in non-mammalian amniotes (sauropsids, i.e., birds and reptiles) and in several anamniotes (reviewed by Rodríguez et al., 2002; Reiner et al., 2004). Comparative studies using an evolutionary developmental biology (evodevo) approach are turning extremely useful for understanding not only the origin but also many aspects of both the anatomical and functional organization of brain regions (Puelles and Medina, 2002; Medina and Abellán, 2009; Medina et al., 2011; Abellán et al., 2013). However, very little is known on the regulatory genes involved in the development of the HF and entorhinal cortex in non-mammals. Herein, we carried out a comparative study of the combinatorial mRNA expression patterns of Lef1, several LIM-homeobox (Lhx2, Lhx5, and Lhx9) and LIM-only (Lmo3 and Lmo4) genes, and Prox1 in the developing medial pallium of mouse and chicken. Although the expression of all of these genes was previously studied in the developing dorsomedial pallium of mouse (Galceran et al., 2000; Bulchand et al., 2001, 2003; Zhou et al., 2004; Abellán et al., 2009, 2010; Lavado et al., 2010), herein we analyzed in detail their combinatorial expression patterns in order to: (1) distinguish molecularly the whole ventricular sector of the medial pallium and the different structures it produces in mouse; (2) discern whether the entorhinal cortex develops from the medial pallium or from another embryonic pallial sector; and (3) compare these patterns in mouse with those of the orthologous genes in chicken, as a contribution to understand hippocampal evolution. Our data allowed the identification of dentate gyrus/CA3-like, CA1/subicular-like, and entorhinal-like comparable sectors in mouse and chicken, and point to the existence of mostly conserved molecular networks involved in hippocampal complex development.
Materials and methods
Mouse embryos (Swiss) from embryonic day 11.5 (E11.5) until birth and chicken embryos from embryonic day 6 (E6, HH29) until 2 days after hatching (P2) were used in the present study. All animals were treated according to the regulations and laws of the European Union (86/609/EEC) and the Spanish Government (Royal Decree 1021/2005) for care and handling of animals in research. The protocols used were approved by the Committee for handling and care of research animals of the University of Lleida. The mouse embryos were obtained from pregnant females, and were processed and fixed as previously described (García-López et al., 2008). The chicken embryos were obtained from fertilized eggs bought in a farm, which were incubated in a forced-draft incubator until the desired embryonic stage. The chicken embryos were staged according to Hamburger and Hamilton (1951). Upon extraction, the brains of earlier embryos (E11.5–E15.5 in mouse; 6–11 days incubation in chicken: E6–E11 or HH29-HH37) were dissected and fixed by immersion in 4% paraformaldehyde diluted in 0.1 M phosphate-buffered saline (pH 7.5; PBS) at 4°C during 24 h. Older embryos (E16.5–E18.5 in mouse; from E12 or HH38 to pre-hatching in chicken) and P0–P2 animals were first deeply anesthetized with sodium pentobarbital (Dolethal, 15 mg/kg), and perfused transcardially with NaCl saline solution (0.9% for mouse; 0.75% for chicken), followed by phosphate-buffered 4% paraformaldehyde (pH 7.5). The brains were then dissected and postfixed overnight at 4°C. After fixation, the brains were embedded in 4% agarose in PBS, sectioned at 80–120 μm for in situ hybridization in the transversal or horizontal planes using a vibratome (Leica VT1000S), and were subsequently processed as floating sections.
In situ hybridization
Brain sections were processed for in situ hybridization following a variation of the standard procedure using digoxigenin-labeled riboprobes (Medina et al., 2004; García-López et al., 2008; Abellán and Medina, 2009). The riboprobes were synthesized from cDNAs of different mouse or chicken genes.
The cDNAs from mouse genes were obtained from other laboratories:
Lef1 (Galceran et al., 2000; bp 1–729; Genbank accession no: NM_010703);
Lhx2 (plasmid obtained from S. Rétaux's lab; Rétaux et al., 1999; bp 1–1300; Genbank accession no: NM_010710.3);
Lhx5 (plasmid obtained from H. Westphal's lab; Zhao et al., 1999; bp 1–2226; Genbank accession no: U61155.1);
Lhx9 (plasmid obtained from S. Rétaux's lab; Rétaux et al., 1999; bp 1–1016 [full lenght]; Genbank accession no: AF134761);
Lmo3 (plasmid obtained from J.L.R. Rubenstein's lab; Bulchand et al., 2001; bp 1–2101 [full lenght]; Genbank accession no: NM_207222);
Lmo4 (plasmid obtained from J.L.R Rubenstein's lab; Bulchand et al., 2001; bp 1–498 [full lenght]; Genbank accession no: AF074600).
The cDNAs from chicken genes were purchased [cDNA ESTs purchased from ARK-genomics (Roslin Institute; Midlothian, UK) or Geneservice Limited (Cambridge, UK)], or obtained from other laboratories, as indicated below. The purchased clones were obtained from the BBSRC ChickEST Database (Boardman et al., 2002), and have a corresponding Genbank accesssion number:
cLef1 (bp 1–901; GenBank accession no: CR391621.1; purchased; BBSRC ChickEST Database: clone ChEST891i13);
cLhx2 (Abellán et al., 2009; bp 208–939; Genbank accession no: NM_204889);
cLhx5 (Abellán et al., 2010; bp 49–1042; Genbank accession no: XM_001234552);
cLhx9 (Abellán et al., 2009; bp 596–1502; Genbank accession no: NM_205426);
cLmo3 (Abellán and Medina, 2009; bp 1–666; Genbank accession no: CR406209; purchased; BBSRC ChickEST Database: clone ChEST853b21);
cLmo4 (Abellán and Medina, 2009; purchased; bp 307–1078; Genbank accession no: AF532926; purchased; BBSRC ChickEST Database: clone ChEST54p6);
cProx1 (bp 1–841; GenBank accession no: BU214594; purchased; BBSRC ChickEST Database: clone ChEST49e24).
cWnt8b (641 bp; Hollyday et al., 1995; Garda et al., 2002; Genbank accession no: NC_006093.3).
We used PCR to obtain the DNA template employed for synthesizing the riboprobe. We synthesized the antisense digoxigenin-labeled riboprobes using Roche Diagnostics's (Mannheim, Germany) protocols for the genes mentioned above. Before hybridization, the sections were abundantly washed in PBS containing 0.1% Tween-20 (PBT 1X), prehybridized in hybridization buffer (HB) for 2 h at 58°C, and then hybridized in HB containing the riboprobe overnight at 58°C (0.5–1 μg/ml, depending on the probe and embryo size). The hybridization buffer contained 50% of deionized formamide, 1.3X standard saline citrate (SSC; pH 5), 5 mM ethylene-diamine-tetraacetic acid (EDTA; pH 8.0; Sigma-Aldrich, Steinheim, Germany), 1 mg/ml of yeast tRNA (Sigma-Aldrich), 0.2% Tween-20, 100 μg/ml of heparin (Sigma-Aldrich), completed with water (free of RNAase and DNAase; Sigma-Aldrich). Following hybridization, the sections were washed with a mix 1:1 of MABT 1X (1.2% maleic acid, 0.8% NaOH, 0.84% NaCl, and 0.1% Tween-20) and HB at 58°C during 20 min and washed abundantly at room temperature with MABT 1X (about 2 h). Following this, the sections were blocked with a solution containing blocking reagent (Roche), MABT 1X and sheep serum (Sigma) for 4 h at room temperature, then incubated in an antibody against digoxigenin (alkaline-phosphatase coupled anti-digoxigenin; diluted 1:3500; Roche Diagnostics) overnight at 4°C, later washed with MABT 1X and finally revealed with BM purple (Roche Diagnostics). Sections were then mounted on glycerol gelatine (Sigma).
Immunohistochemistry
Some series of chicken embryonic brain sections (E8–E12) were processed for immunohistochemistry, following a procedure previously described (Abellán and Medina, 2009).
In order to detect radial glial fibers in chicken, we used a monoclonal antibody against chicken vimentin (H5 from Developmental Hybridoma Bank, Iowa, USA; Herman et al., 1993). The specificity of this antibody has beed shown by the manufacturer using Western blot (labeling a band of roughly 52 kDa, corresponding to the protein vimentin).
The immunohistochemical procedure was as follows. After washing in PBS, the sections were incubated in the primary antibody, diluted 1:50 in PBS containing 0.3% Triton X-100, for 2 days at 4°C, under constant and gentle agitation. Then, the sections were washed and incubated in a secondary antiserum for 1 h at room temperature (biotinylated goat anti-mouse IgG; diluted 1:200; Vector, Burlingame, CA, USA). Following this, the sections were washed and incubated in the avidin-biotin complex (ABC kit; Vector; 0.003% dilution) for 1 h at room temperature. Finally, the immunolabeling was revealed by 0.05% diaminobenzidine (DAB; Sigma-Aldrich, Steinheim, Germany) in 0.05 M Tris buffer (pH 7.6), containing 0.03% H2O2.
Digital images and figures
Digital photographs were taken on a Leica microscope (DMR HC) equipped with a Zeiss Axiovision digital camera. Digital images were adjusted for brightness/contrast using Adobe PhotoShop and figures were mounted and labeled using Macromedia FreeHand 10.
Nomenclature
Finally, the nomenclature used in the present study for the chicken telencephalon generally followed that proposed by Reiner et al. (2004), except for developmental units, hippocampal subdivisons, and the entorhinal cortex, for which it followed Redies et al. (2001), Puelles et al. (2007), Abellán et al. (2009). For the mouse embryonic brain we primarily followed Jacobowitz and Abbott (1997), and for the mature mouse hippocampal complex, we followed Paxinos and Franklin (2004) and Witter (2012).
Results
Herein we present data on the expression of Lef1, Lhx2, Lhx9, Lmo3, and Lmo4 in the mouse embryonic medial pallium (summarized in Table 1, and shown in Figures 1–5), and data on the expression of cLef1, cLhx2, cLhx9, cProx1, cLmo3, and cLmo4 in the chicken embryonic medial pallium (summarized in Table 2, and shown in Figures 6–11). The figures are organized according to both the species and the age, showing first those for the mouse and then those for the chicken, and within each species showing first those of early embryonic stages, followed by intermediate stages and finally those for late stages. For comparative purposes we also included published data on Prox1 in mouse in Table 1 (Zhou et al., 2004; Lavado et al., 2010, and the Allen Developing Mouse Brain Atlas). To assist in the distinction of the medial pallial ventricular sector from other pallial sectors, we also analyzed Lhx5 at early developmental stages in mouse and chicken (Tables 1, 2). In addition to its expression in the pallium, and as noted previously (Oosterwegel et al., 1993; Galceran et al., 2000; Gupta et al., 2012; Choe et al., 2013), Lef1 was also expressed in other forebrain regions such as the thalamus (Figures 1D,E), as well as in the mesoderm and pia mater (neural crest-derived part of the meninges) covering the forebrain during development (arrows in Figure 6B), in the developing choroid plexus, and in forebrain blood vessels (Figures 3, 6A–C).
Table 1.
Combinatorial expression of Lef1 and other regulatory genes in the pallial progenitor zones and hippocampal complex primordia of developing mice.
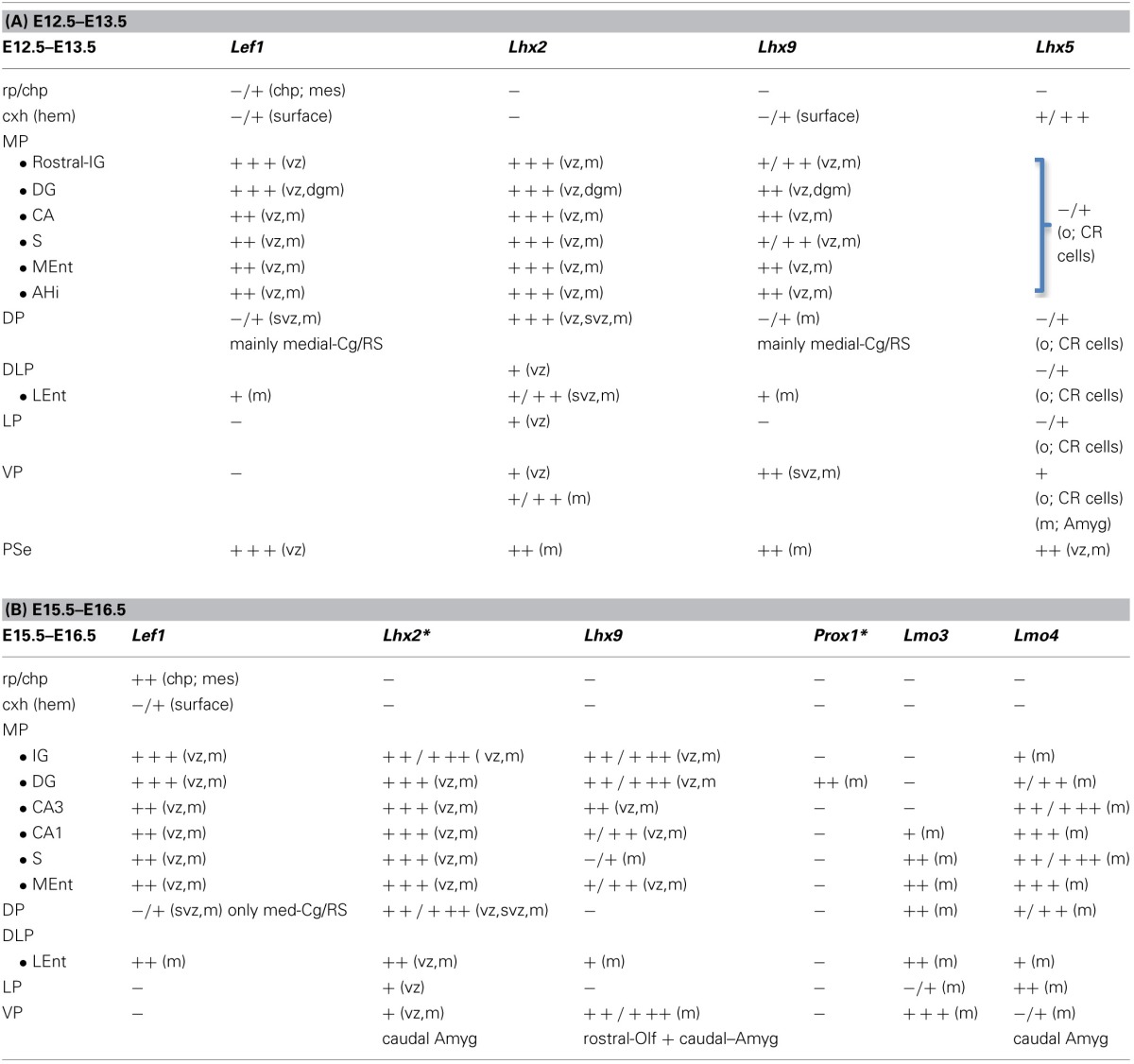
−, no expression; +, weak expression; ++, moderate expression; +++, strong expression.
chp, choroid plexus; dgm, dentate gyrus migratory stream; m, mantle; o, outer or marginal zone; rp, roof plate; svz, subventricular zone; vz, ventricular zone. For other abbreviations see list.
*Based on published data (Prox1: Zhou et al., 2004; Lavado et al., 2010; Lhx2: Allen Brain Atlas web site).
Figure 1.
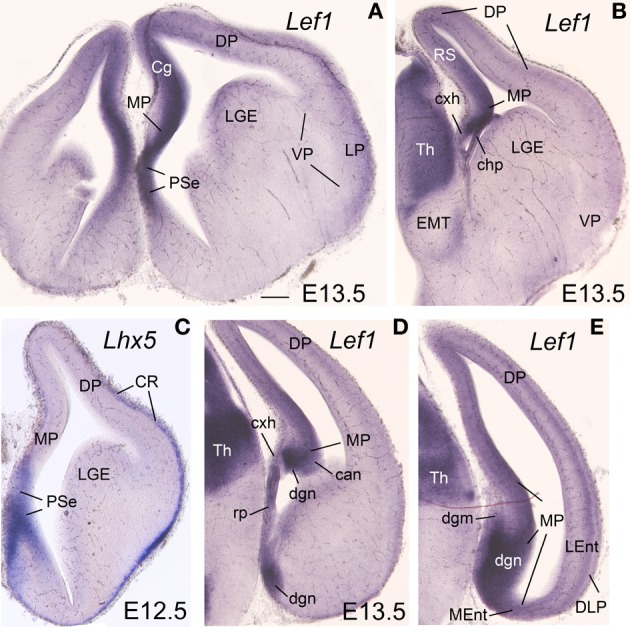
Expression of Lef1 in mouse embryonic telencephalon at E13.5. Digital images of coronal sections of mouse embryonic telencephalon (E13.5), from rostral (A) to caudal (E) levels, hybridized for Lef1. Note the strong expression in the ventricular zone of the medial pallium and pallial septum. For abbreviations see list. Scale bar: (A) = 200 μm (applies to all).
Table 2.
Combinatorial expression of Lef1 and other regulatory genes in the pallial progenitor zones and hippocampal complex primordia of developing chicken.
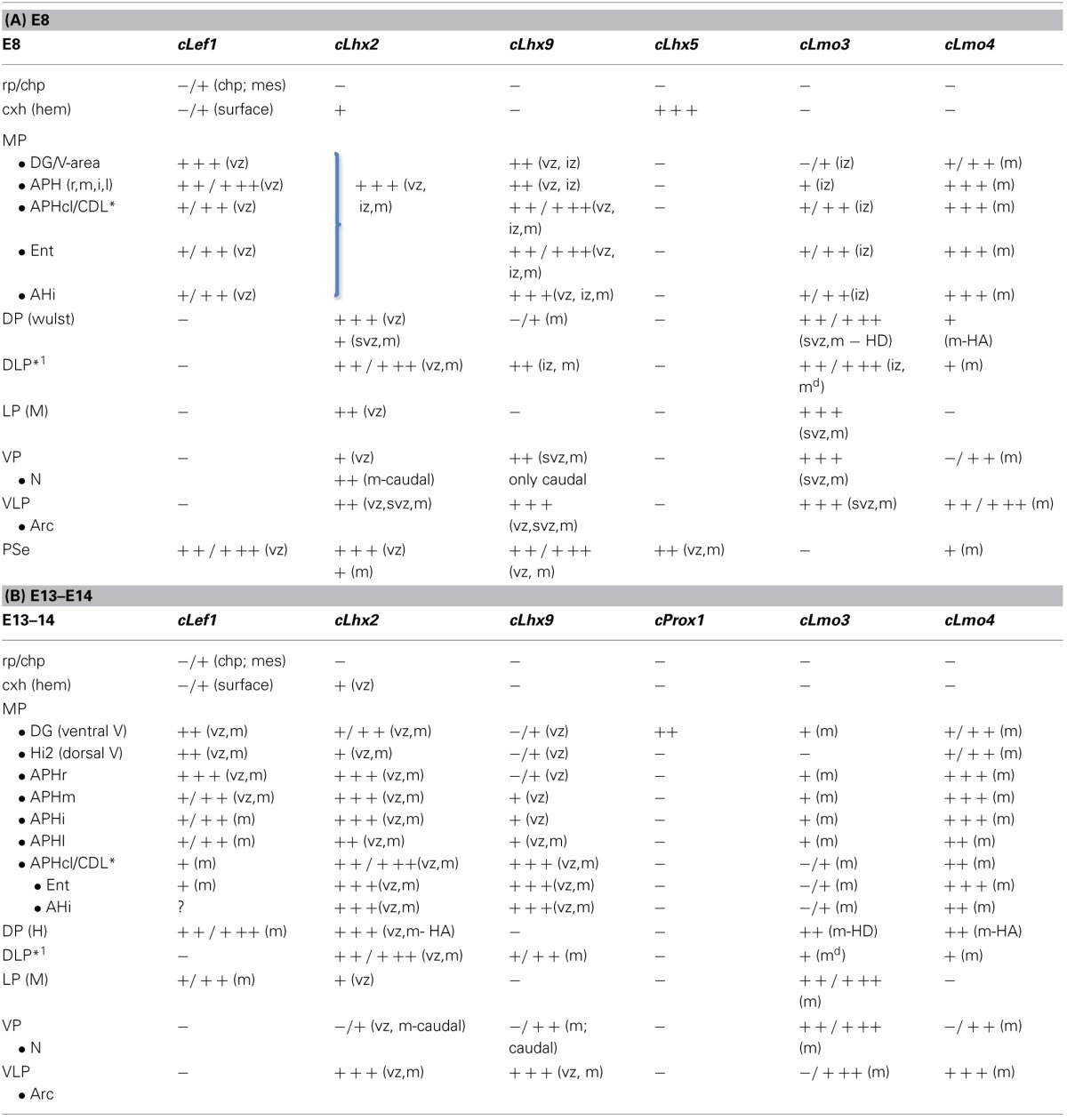
−, no expression; +, weak expression; ++, moderate expression; +++, strong expression.
chp, choroid plexus; iz, intermediate zone; m, mantle; md, deep part of mantle; mes, mesoderm; rp, roof plate; svz, subventricular zone; vz, ventricular zone. For other abbreviations see list.
*APHcl is referred as dorsolateral corticoid area or CDL by the followers of the conclusions of the avian brain nomenclature forum (Reiner et al., 2004; see also Atoji and Wild, 2005). Herein we use APHcl as a preferred term to emphasize its relation to other APH subdivisions (see also Redies et al., 2001; Puelles et al., 2007).
*1DLP is referred as caudal dorsolateral pallium (CDL) in the atlas of the chick brain, by Puelles et al. (2007), and was previously included as part of the avian temporo-parieto-occipital area or TPO (see Discussion in Abellán et al., 2009).
Gene expression patterns with respect to major pallial subdivisions in mouse and chicken
The mRNA expression patterns of all genes analyzed were largely conserved between mouse and chicken, although some differences were also appreciated. During very early development (E11.5 in mouse; E6-E7 in chicken), Lef1 was strongly and distinctly expressed in the ventricular zone (vz) of the medial pallium (MP) and the pallial septum (PSe) (chicken: Figures 6A,B), although it also showed generally weak expression in the vz of other pallial sectors. By E13.5 in mouse (Figures 1A,B,D,E) and E8 in chicken (Figures 6C, 7A), Lef1 became primarily restricted to the vz of the medial pallium and pallial septum (Tables 1A, 2A).
At E13.5–E15.5 in mouse (Figures 1–3) and E8-E9 in chicken (Figures 6, 7), the medial pallial sector was characterized by strong or moderate vz expression of Lef1 (Figures 1, 3, 6C, 7A), Lhx2 (Figures 2A–D, 6D,E), and Lhx9 (Figures 2E–K, 6F, 7B,C), but not Lhx5 (Figures 1C, 6G). This feature allowed the distinction of the medial pallium from other progenitor pallial sectors, such as: the pallial septum (PSe), expressing Lef1 (Figures 1A, 6B), Lhx2 (Figure 6E), Lhx9 (Figure 6F), and also Lhx5 (Figures 1C, 6G) in the vz; the dorsal pallium (DP), expressing strongly Lhx2 in the vz (Figures 2A–D, 6D), but not Lef1 (except its medialmost, cingulate-related area; Figures 1A,B,D), Lhx9 (Figures 2E–G) or Lhx5 (Figure 1C) (data on Lhx2, Lhx9, and Lhx5 in chicken DP is published in Abellán et al., 2009, 2010); and the lateral (LP) and ventral (VP) pallia, showing generally weak expression of Lhx2 in the vz (Figures 2A, 6D,E), but no vz expression of Lef1 (Figures 1A,B, 6C) nor Lhx5 (Figures 1C, 6G) (summarized in Tables 1, 2; for the lateral pallial sector, we followed a recent redefinition done by Puelles, 2014).
Figure 2.
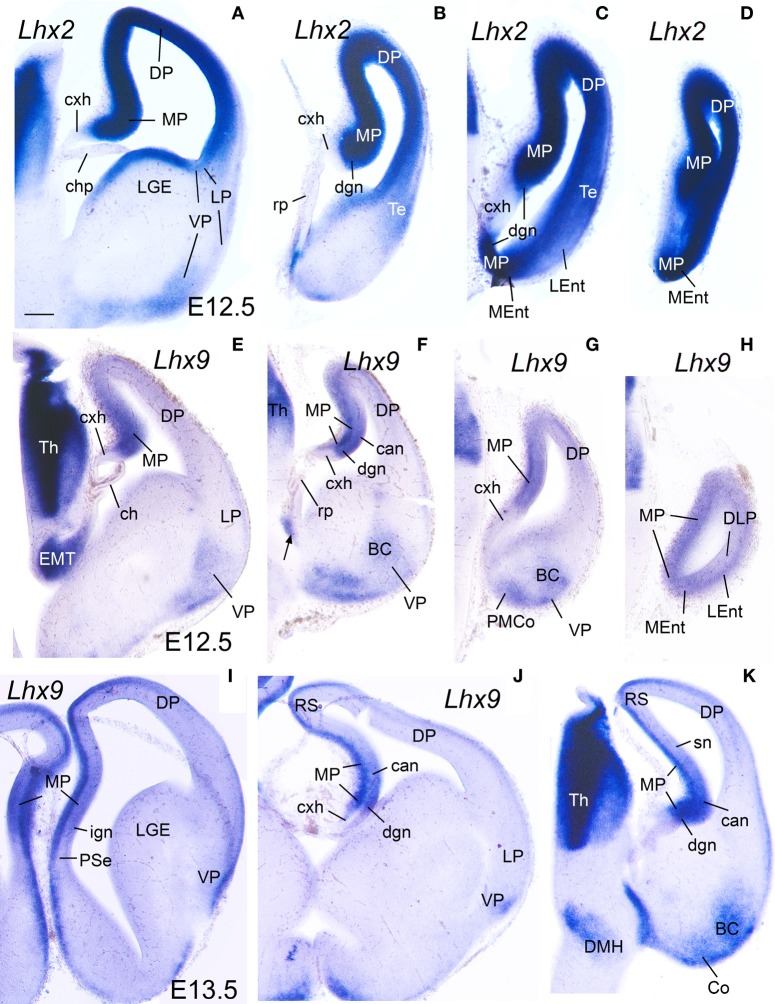
Expression of Lhx2 and Lhx9 in mouse embryonic telencephalon at early stages. Digital images of coronal sections of mouse embryonic telencephalon (A–H: E12.5; I–K: E13.5), from intermediate (left panels) to caudal (right panels) levels, hybridized for Lhx2 (A–D) or Lhx9 (I–K). Note the strong expression in the ventricular zone of the medial pallium. As noted previously, Lhx9 is also distinctly expressed in ventral pallial (VP) derivatives, such as part of the basal amygdalar complex (BC) and cortical amygdalar areas (Co, PMCo). Although weak transient expression is also present in part of the dorsal pallium (DP; Rétaux et al., 1999), this pallial sector is clearly distinguished from MP and VP based on its distinct position and combinatorial genetic profile (Puelles et al., 2000; Abellán et al., 2009). For abbreviations see list. Scale bar: (A) = 200 μm (applies to all).
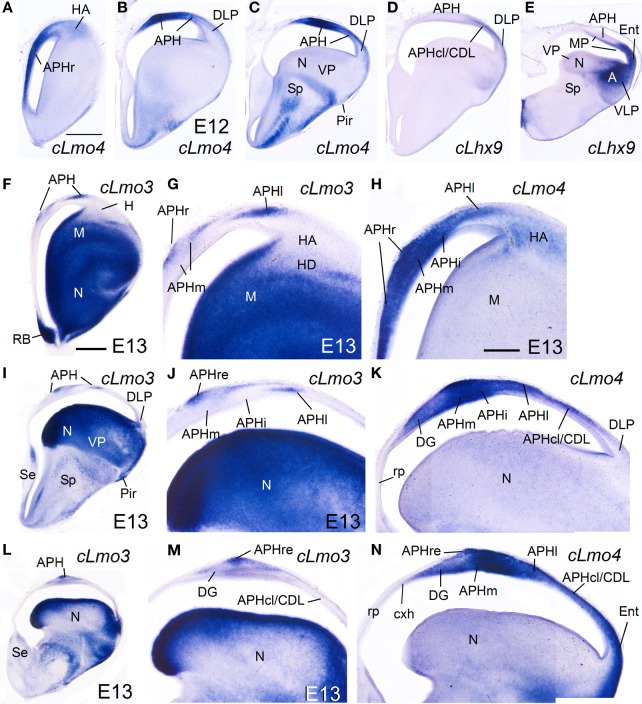
Based on the combinatorial gene expression patterns studied here and on published data (Puelles et al., 2000, 2007; Medina et al., 2004; Abellán et al., 2009; Puelles, 2014), we tentatively distinguished two new pallial sectors, which we named the dorsolateral caudal pallium (DLP) and the ventrolateral caudal pallium (VLP) (Tables 1, 2). The DLP was previously described in chicken as a distinct subdivision belonging to either the lateral pallium (Puelles et al., 2007) or ventral pallium (Abellán et al., 2009), and has been called temporo-parieto-occipital area or pallium externum in some studies (for example, Veenman et al., 1995; Atoji and Wild, 2005). In contrast to the dorsal and lateral pallia, the DLP expressed Lhx9 in the mantle throughout development (Figures 6F, 10A,H,I) and, for this reason, was previously suggested to be part of the ventral pallium (Abellán et al., 2009). However, in contrast to the ventral pallium, the DLP showed abundant vz/mantle expression of Emx1 (chicken: named CDLx in Figure 10 n of Puelles et al., 2000) and Lhx2 (Figure 10B). In addition, in contrast to the medial pallium, the DLP did not express Lef1 (Figure 6C, Table 2). Herein, we tentatively identified a comparable pallial subdivision in the mouse, giving rise to the lateral entorhinal cortex (LEnt), with similar topological position and genetic profile [no expression of Lef1 in vz. (Figures 1E, 3F,G), but expressing Lhx9 in the mantle (Figure 2H) and Emx1 in vz/mantle (see Allen Developing Mouse Brain Atlas); Table 1]. On the other hand, the VLP [for the moment only identified in chicken, and giving rise to the arcopallium (A)] differed from the ventral pallium [giving rise to the nidopallium (N) and piriform cortex (Pir)] for its strong expression of Lhx9 (Figures 7B,C, 9E, 10D,F,J; Abellán et al., 2009), Lhx2 (Figures 7D, 10E), and Emx1 (see Figure 10p in Puelles et al., 2000) in both the ventricular zone and mantle (Table 2).
Figure 3.
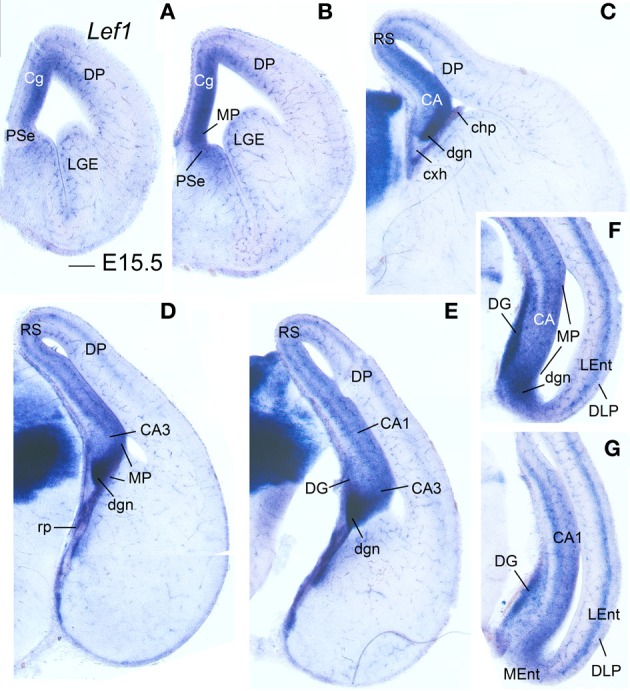
Expression of Lef1 in mouse embryonic telencephalon at E15.5. Digital images of coronal sections of mouse embryonic telencephalon (E15.5), from rostral (A) to caudal (G) levels, hybridized for Lef1. The medial pallial (MP) vz and derivatives show expression of Lef1. Note the lack of Lef1 expression in the vz of the dorsolateral caudal pallium (DLP), giving rise to LEnt. For abbreviations see list. Scale bar: (A) = 200 μm (applies to all).
In addition to the expression in the vz, most derivatives of the medial pallium of mouse and chicken also showed moderate to strong expression of Lef1 and Lhx2 at least during early and intermediate developmental stages, while some or many of them also expressed Lhx9 (see details in next sections; Tables 1, 2). These features, linked to the molecular identity of the medial pallial vz, helped to identify and compare the medial pallial derivatives between mouse and chicken. The results on the combinatorial expression of Lef1 and other developmental regulatory genes in the developing hippocampal complex (including hippocampal formation and entorhinal cortex) are explained in detail below, first for the mouse (Figures 1–5) and then for the chicken (Figures 6–11).
Combinatorial expression of Lef1, Lhx2, Lhx9, Lhx5, Lmo3, and Lmo4 in the developing hippocampal complex of mouse
E12.5–E13.5
During early development, the medial pallial sector (MP) of mouse was distinguished by its moderate to strong expression of Lef1, Lhx2, and Lhx9 in the vz and in postmitotic cells migrating into the mantle (Table 1, Figures 1, 2). Outside the medial pallium, the only additional pallial sector expressing Lef1 in the vz was the pallial septum (PSe) and the adjacent part of the dorsal pallium [DP; cingulate part of neocortical primordium (Cg)] (Table 1A, Figure 1A). Based on the expression of Lef1 (Figures 1B,D,E), Lhx2 (Figures 2A–D), and Lhx9 (Figures 2E–K), the medial pallium appeared to include the progenitor zones of the indusium griseum (rostrally; IG neuroepithelium, ign), the dentate gyrus (DG neuroepithelium, dgn), the CA fields (CA neuropeithelium, can), the subiculum (S neuroepithelium, sn), at least part of the amygdalo-hippocampal transition area (AHi in Figure S1), and a medial and caudal part of the entorhinal cortex (corresponding to the primordium of the so-called medial entorhinal cortex; MEnt). In contrast, the lateral entorhinal cortex (LEnt) appeared to derive from a distinct pallial sector, the DLP, which vz did not express Lef1 (Figure 1E) or Lhx9 (Figure 2H). Nevertheless, the lateral entorhinal cortical plate showed moderate Lef1 expression (Figure 1E), resembling the adjacent part of the neocortex cortical plate.
E15.5–E16.5
Lef1, Lhx2, and Lhx9 continued to be expressed in the vz and mantle of the mouse medial pallium during intermediate development [Table 1B, Figures 3, 4; data on Lhx2 at E15.5 is available in the Allen Developing Mouse Brain Atlas and published elsewhere (Bulchand et al., 2003); for comparative reasons, such data is included in Table 1B]. The expression of Lef1 was moderate to strong in all medial pallial-derived areas [indusium griseum (IG), dentate gyrus (DG), CA fields, subiculum (S), medial entorhinal cortex (MEnt); Figure 3], while Lhx9 remained moderate to strong in most of them but started to be downregulated in the developing CA1 field and, especially, in the developing subiculum (Figures 4A–C).
Figure 4.
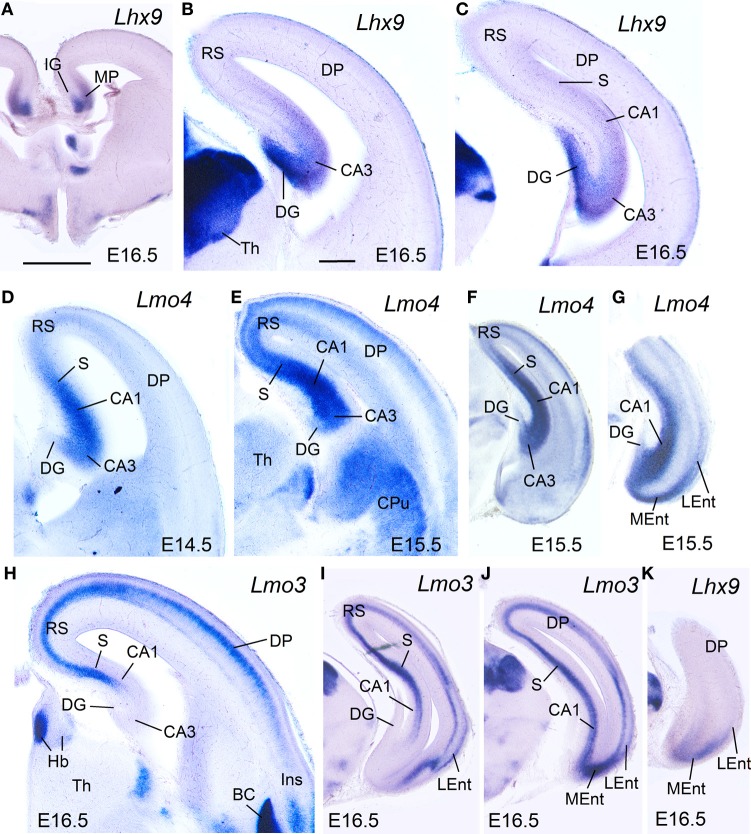
Expression of Lhx9, Lmo3, and Lmo4 in mouse embryonic telencephalon at intermediate stages. Digital images of coronal sections of mouse embryonic telencephalon (E15.5 or E16.5), at intermediate (A–E,H) or caudal (F,G,I–K) levels, hybridized for Lhx9, Lmo3, or Lmo4. For abbreviations see list. Scale bar: (A) = 1 mm (applies to A,F,G,I–K); (B) = 300 μm (applies to B–E,H).
During intermediate development, medial pallial derivatives also showed expression of Lmo4 and Lmo3, with different patterns (Table 1B, Figure 4). Lmo4 showed moderate to strong expression in the developing CA fields, subiculum, and medial entorhinal cortex, while the developing indusium griseum and dentate gyrus only showed weak or weak to moderate Lmo4 expression, respectively (Figures 4D–G). On the other hand, Lmo3 showed weak or moderate expression in the developing CA1, subiculum, and medial entorhinal cortex, but was not expressed in the developing CA3, dentate gyrus and indusium griseum (Figures 4H–J).
At E15.5–E16.5, while the medial entorhinal cortex (MEnt) showed gene expression patterns highly similar to those in other medial pallial derivatives, the lateral entorhinal cortex (LEnt, a DLP derivative) differed in the expression of Lef1 (Figures 3F,G), Lhx9 (Figure 4K), Lmo4 (Figure 4G), and Lmo3 (Figures 4I,J; see also Table 1B). For example, in the DLP/LEnt, expression of Lef1 (Figures 3F,G) and Lhx9 (Figure 4K) was only weak or moderate, and restricted to the mantle. Moreover, in the LEnt, expression of Lmo4 was only weak to moderate (Figure 4G), while Lmo3 showed a bi-layered expression pattern (apparently superficial and deep to the lamina dissecans or layer IV), making it different from the MEnt (Figures 4I,J).
E17.5-P0
During prenatal stages, the expression of Lef1 became weak in most of the medial pallium, and almost disappeared in the CA3 field, with the only exception of the dentate gyrus (DG), where it remained moderate to strong (Figures 5A–D). In contrast, the lateral entorhinal cortex retained moderate expression of Lef1.
Figure 5.
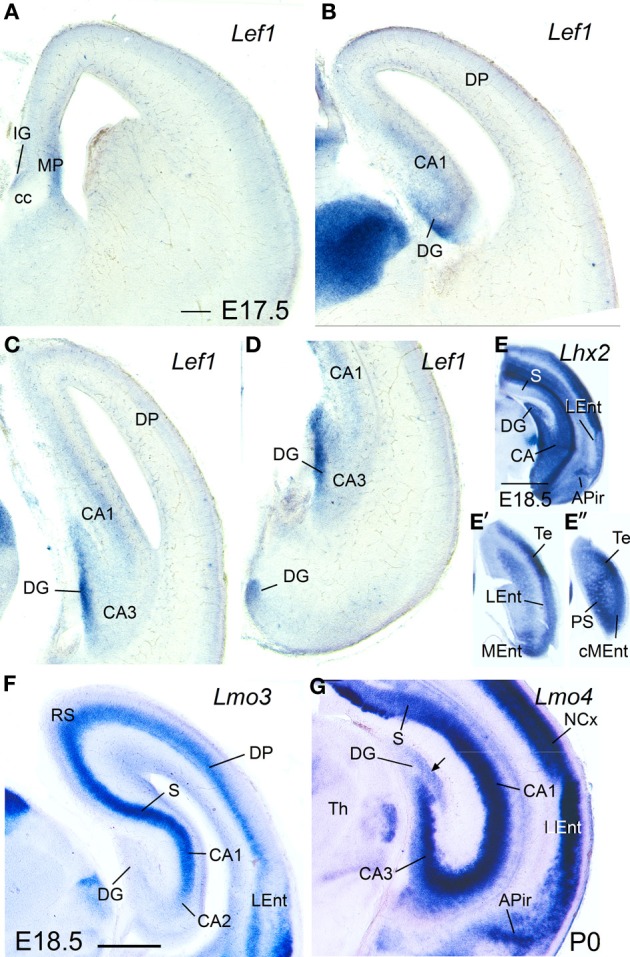
Expression of Lef1, Lhx2, Lmo3, and Lmo4 in mouse embryonic telencephalon at prenatal or neonatal ages. Digital images of coronal sections of prenatal or neonatal mouse telencephalon (E17.5, E18.5, or P0), at intermediate (A,B,F) or caudal (C–E″,G) levels, hybridized for Lef1, Lhx2, Lmo3, or Lmo4. For abbreviations see list. Scale bar: (A) = 200 μm (applies to A–D,F,G). (E) = 1 mm (applies to E–E″).
At perinatal stages, Lhx2 (Figures 5E–E″), Lmo3 (Figure 5F), and Lmo4 (Figure 5G) intensified their expression in the pallium, but retained the specific patterns observed before for the different pallial divisions and subdivisions (Figures 5E–G). The expression of Lhx2 was strong or very strong in most subdivisions of the medial pallium (including vz and mantle; Figures 5E–E″), except the indusium griseum, where the expression was moderate. Lmo3 showed very strong expression in the principal cell layer of the subiculum (S) and CA1, and moderate expression in the indusium griseum (IG) and medial entorhinal cortex (MEnt), but its expression was very weak in CA3 and absent in the dentate gyrus (DG) (Figure 5F). Lmo4 expression was moderate to strong in most medial pallial subdivisions (with the pyramidal cell layer of CA1 showing the strongest expression), except the indusium griseum and the dentate gyrus, which showed only weak expression (Figure 5G). On the other hand, Lhx9 was moderate to strongly expressed in the indusium griseum, dentate gyrus, CA3 field, and medial entorhinal cortex, but appeared completely downregulated in the CA1 and subiculum at E18.5.
Regarding the lateral entorhinal cortex, at perinatal stages continued showing weak to moderate expression of Lhx2 (Figures 5E,E′), Lhx9, and Lmo3 (Figure 5F), as during previous stages. In contrast, expression of Lmo4 became strong at these stages (Figure 5G).
Combinatorial expression of cLef1, cLhx2, cLhx9, cLhx5, cProx1, cLmo3, and cLmo4 in the developing hippocampal complex of chicken
E8
Similarly to the mouse, at E8 the medial pallial sector (MP) of chicken embryos could be distinguished by its moderate to strong expression of cLef1 (Figures 6C, 7A), cLhx2 (Figures 6D,E, 7D), and cLhx9 (Figures 6F, 7B,C) in the vz (Table 2A). The rest of the pallium did not express cLef1 in the vz at E8, and the different pallial subdivisions [dorsal (DP), dorsolateral-caudal (DLP), lateral (LP), ventral (VP), ventrolateral-caudal (VLP)] could additionally be distinguished by a region-specific combinatorial expression of cLhx2, cLhx9, cLmo3, and cLmo4 in the vz, subventricular zone (svz, identified based on Charvet et al., 2009), and/or mantle zones (Table 2A).
Figure 6.
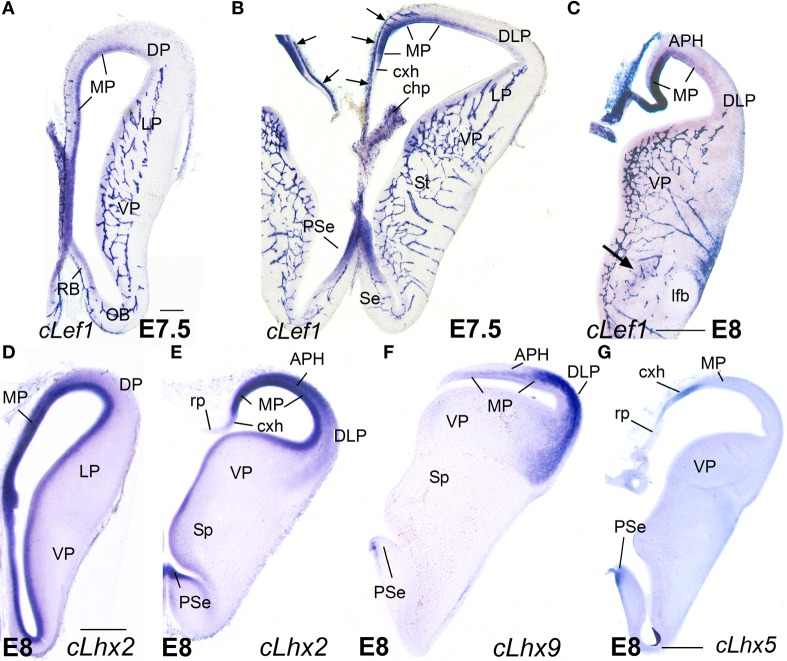
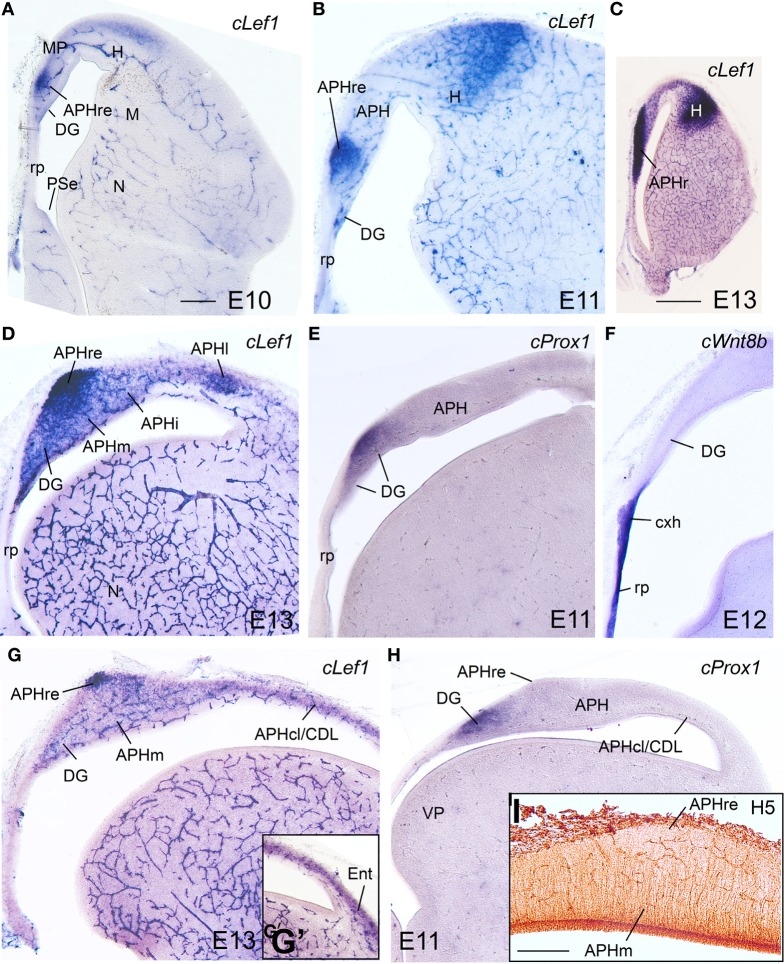
Expression of cLef1, cLhx2, cLhx9, and cLhx5 in chicken embryonic telencephalon at early stages. Digital images of coronal sections of chicken embryonic telencephalon (E7.5 or E8), at rostral (A,D) or intermediate (B,C,E–G) levels, hybridized for cLef1, cLhx2, cLhx9, or cLhx5. Note the moderate to strong expression of cLef1, cLhx2, and cLhx9 in the ventricular zone of the medial pallium (MP) and pallial septum (PSe). The pallial septum also expresses cLhx5. From E8, the dorsolateral pallium (DLP) can be distinguished from MP because it does not express cLef1, but shows moderate to strong expression of cLhx9 in the mantle. Note the expression of cLef1 in the meninges (pia mater; arrows in B), in forebrain blood vessels, and in some cell aggregates around the lateral forebrain bundle (arrow in C). For abbreviations see list. Scale bar: (A) = 200 μm (applies to A,B); (C) = 400 μm; (D) = 500 μm (applies to D–F); (G) = 500 μm.
Figure 7.
Expression of cLef1, cLhx2, cLhx9, cLmo3, and cLmo4 in the chicken embryonic telencephalon at early stages. Digital images of coronal sections of chicken embryonic telencephalon (E8), at intermediate (E,F) or caudal (A–D,G,H) levels, hybridized for cLef1, cLhx2, cLhx9, cLmo3, or cLmo4. The medial pallium is characterized by strong expression of cLef1, cLhx2, and cLhx9 in the ventricular zone, and strong expression of cLhx2 and cLmo4 in the mantle. For abbreviations see list. Scale bar: (A) = 400 μm; (B) = 500 μm (applies to B–H).
Based on the expression of cLef1 (Figures 6C, 7A), cLhx2 (Figures 6D,E, 7E), and cLhx9 (Figures 6F, 7B,C) at the vz, it appeared that the medial pallium in chicken included the progenitor zones of the hippocampus (including the V-shaped area) and the parahippocampal areas (APH), including its caudolateral part (APHcl), which is referred as dorsolateral corticoid area or CDL by some authors (see Discussion; Table 2A). At caudal levels, the avian medial pallium also appeared to include the progenitor zone of the so-called entorhinal cortex (Ent) and the amygdalohippocampal area (AHi, at least its transition part, as defined by Puelles et al., 2007) (Figures 7A,B). Regarding other genes, the chicken medial pallium also showed moderate to strong expression of cLmo4 in the mantle (Figures 7E,H; Table 2A), and expression of cLmo3 in the intermediate zone (a mantle part near the vz, possibly containing migratory neuroblasts) in the APH, with an increasing gradient toward caudolateral levels (Figures 7F,G).
E10–E14
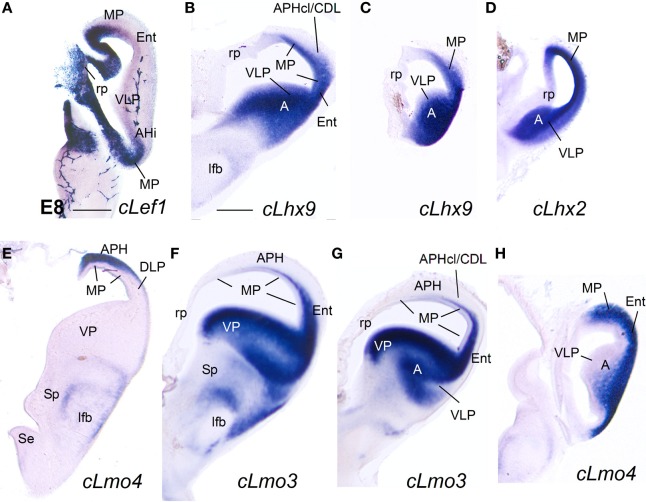
During these stages, the medial pallium continued to show distinct expression of cLef1, cLhx2, and cLhx9, with patterns similar to those found previously (Figures 8–10; Table 2B). As in previous stages, at E10–E14 cLef1 was expressed in the vz of the medial pallium (MP), although only in part of it because it was downregulated in the vz of APHl, APHcl, and entorhinal cortex (Ent) (Figures 8C,D,G,G′). In addition to the vz, the expression of cLef1 now extended into the medial pallial mantle (Figures 8A–D,G,G′; Table 2B). The expression of cLef1 was moderate to strong in the hippocampus (V-shaped area, here named dentate gyrus or DG, as explained below) and the different APH subdivisions, except the APHcl and the Ent where cLef1 expression was only weak (Figures 8A–D,G,G′; Table 2B). The expression of cLef1 allowed distinction of a novel subdivision, called by us the rostral APH (APHr), which showed very strong expression (Figures 8A–D). The APHr may correspond or include the apical part of APH described in the chick brain atlas by Puelles et al. (2007). Comparison of cLef1 with radial glial fiber disposition suggested that APHr vz occupied the rostralmost pole of APH as seen in frontal section (Figure 8C); a group of cLef1-expressing cells appeared to separate from this rostral location, suggesting that they migrated tangentially toward gradually more distant superficial, dorsomedial and caudal positions; we called this migrated part ectopic APHr or APHre (Figures 8B,D,G,I). At intermediate and caudal levels, this extension of APHr (APHre) occupied a small and distinct superficial area at the surface of APHm (Figures 8D,G; compare Figures 8G,I), which appeared to correspond to the parvocellular hippocampal area identified by Atoji and Wild in adult pigeons (2004). During intermediate developmental stages, cLef1 started to be expressed in restricted parts of the mantle of both the dorsal pallium (hyperpallium, H) and the lateral pallium (mesopallium, M) (Figures 8A–C; see also Figure S2).
Figure 8.
Expression of several genes and radial glial fibers in the chicken embryonic medial pallium at intermediate stages. (A–H) Digital images of coronal sections of chicken embryonic telencephalon (E10–E13), at rostral (A–C), intermediate (D–F), or caudal (G–I) levels, hybridized for cLef1, cProx1, or cWnt8b (the latter is used to distinguish the roof plate and cortical hem) Note the strong expression of cLef1 in the rostral APH, which extends caudally to a small area that occupies a superficial position above APHm. cProx1 allows distinction of the dentate gyrus (DG). I: Detail of radial glial fibers in the APH (immunohistochemical staining using H5 antibody). Note that the caudal extension of APHr (ectopic APHr or APHre in G–I) is avoided by fibers. For abbreviations see list. Scale bar: (A) = 200 μm (applies to A,B,D–H); (C) = 1 mm, (I) = 200 μm.
cLhx2, cLhx9, cProx1, cLmo3, and cLmo4 were also expressed in the mantle of the chicken medial pallium at E10–E14, but showed differences between distinct subdivisions (Figures 8–10; Table 2B). In particular, cProx1 was exclusively expressed in a large part of the so-called avian hippocampus, including a large part of the dorsal hippocampus or V-shaped area (dentate gyrus primordium and hippocampal sector 1 or Hi1 of Puelles et al., 2007) and the so-called ventral hippocampus (Figures 8E,H; Table 2B); this makes this chicken medial pallial subdivision comparable by position, embryonic origin, and molecular profile to the mouse dentate gyrus or DG, and we called it accordingly. On the other hand, cLmo4 (Figures 9A–C,H,K,N, 10G) and cLhx2 (Figures 10B,C,E) were moderate to strongly expressed in the whole mantle of all subdivisions of the medial pallium, with the strongest signal levels observed in APHr, APHm, and APHi. In contrast, cLhx9 and cLmo3 expressions in the mantle were restricted to different subdivisions. Thus, cLhx9 was expressed weakly in APHl and strongly in APHcl and entorhinal cortex (Figures 9D,E, 10D,F,H,I). cLmo3 showed generally weak expression in DG, and the superficial layer of APHr, APHm, APHi, and APHl subdivisions (Figures 9F,G,I,J,L,M).
Figure 9.
Expression of several genes in the chicken embryonic telencephalon at intermediate stages. Digital images of coronal sections of chicken embryonic telencephalon (E12, E13), at rostral (A,F–H), intermediate (B–D, I–K) or caudal (E, L–N) levels, hybridized for cLhx9, cLmo3, or cLmo4. Note the strong expression of cLmo4 in medial pallial derivatives, which is remarkable in APHm and APHi. cLhx9 is also expressed in the vz of the medial pallium, and at caudal levels the expression becomes stronger and is additionally present in the mantle. Moreover, cLhx9 is expressed in derivatives of the ventral pallium (VP; in particular, the caudal nidopallium, N), and in both vz and derivatives of the ventrolateral caudal pallium (VLP, which gives rise to the arcopallium). For abbreviations see list. Scale bar: (A) = 1 mm (applies to A–E); (F) = 1 mm (applies to F,I,L); (H) = 500 μm (applies to G,H,J,K,M,N).
Figure 10.
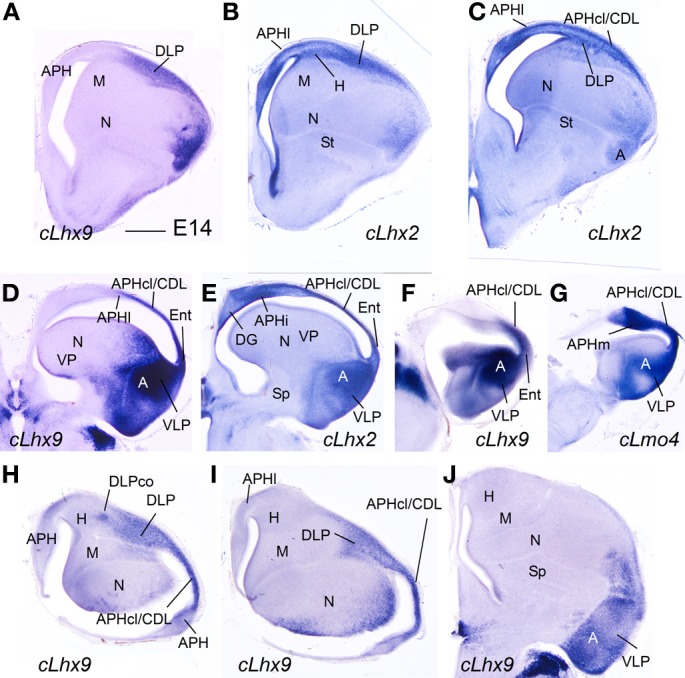
Expression of several genes in the chicken embryonic telencephalon at intermediate stages. (A–G) Digital images of coronal sections of chicken embryonic telencephalon (E14), at intermediate (A–C) or caudal (D–G) levels, hybridized for cLhx2, cLhx9, or cLmo4. (H–J) Digital images of horizontal sections of chicken embryonic telencephalon (E14), from top (H) to bottom (J), hybridized for cLhx9. Note the distinct genetic profile of DLP and VLP. For abbreviations see list. Scale bar: (A) = 1 mm (applies to all).
E16–E18 and hatchlings
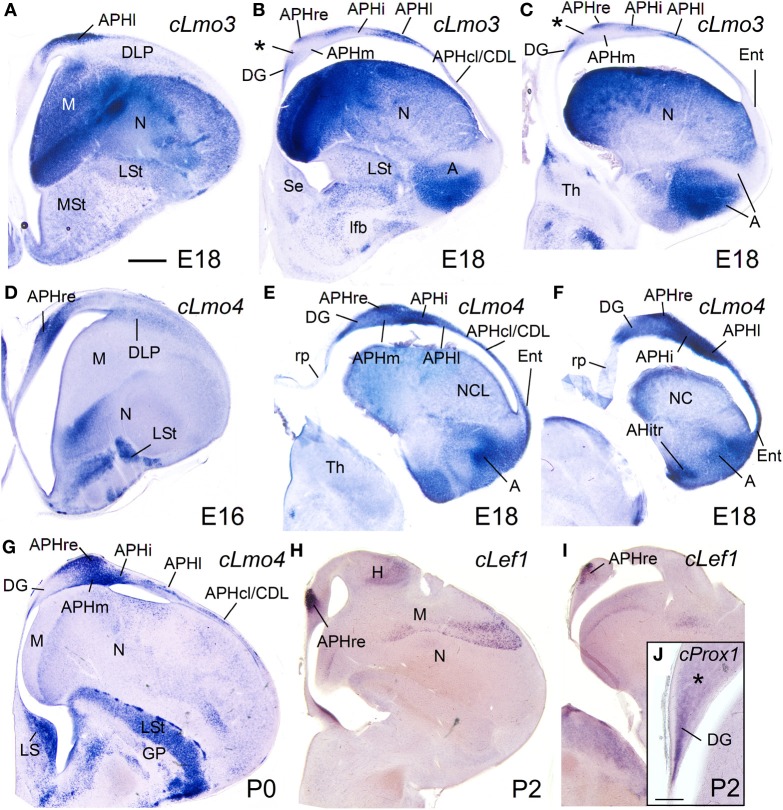
While the expression of cLhx2 remained moderate to strong in the vz and mantle of the whole medial pallium, cLef1 and cLhx9 were completely or almost completely downregulated in the medial pallial vz, and their expression became restricted to only parts of the mantle (Figure 11). cLhx9 retained its expression in the mantle of APHl, APHcl, and entorhinal cortex (not shown here, but seen in Figures 5, 6 in Abellán et al., 2009). cLef1 became downregulated in most medial pallial areas but retained a very strong expression in APHr and its ectopic extension (APHre), which was still visible at P2 (Figures 11H,I). On the other hand, cProx1 retained its distinctive expression in DG at least until P2 (Figure 11J; Figure S2). Finally, the expression patterns of cLmo3 and cLmo4 in the chicken medial pallium during pre-hatching stages were similar to those seen before (E12–E14). By E18, cLmo3 expression was weak in DG and Ent, but moderate in parts of most APH subdivisions, except APHl, where it was strong (Figures 11A–C). The expression pattern of cLmo3 in DG and APH was still similar by P0. In APHi, cLmo3 expression was located deep and superficial to the principal cell layer. However, in APHl and medial APHcl the expression was ample but left empty, free of expression, patches or islands of the cortical plate. By P0, cLmo3 expression became moderate in the entorhinal cortex. On the other hand, at E16–E18, the expression of cLmo4 was moderate to very strong in all medial pallial subdivisions, being remarkable in parts of APH (Figures 11D–F). By P0, cLmo4 expression still was remarkably strong in APHm, APHi, and the ectopic part of APHr (Figure 11G). However, the expression became weak in DG.
Figure 11.
Expression of several genes in the chicken embryonic telencephalon at prehatching and early posthatching stages. Digital images of coronal sections of chicken telencephalon (E16, E18, P0, or P2), at intermediate (A,D,G,H) or caudal (B,C,E,F,I,J) levels, hybridized for cLmo3, cLmo4, cLef1, or cProx1. Note the expression of cProx1 in the dentate gyrus, which includes a large part of V-shaped area, but not its dorsal part (asterisk in J). This dorsal part of V-shaped area (hippocampal sector 2) is also free of cLmo3 (asterisk in B,C) and may be comparable to CA3 of mammals. For abbreviations see list. Scale bar: (A) = 1 mm (applies to A–I); (J) = 200 μm.
Discussion
Comparison of the hippocampal formation of mouse and chicken based on combinatorial gene expression patterns
In this study, we used the combinatorial expression patterns of seven developmental regulatory genes (Lef1, Lhx2, Lhx9, Lhx5, Lmo3, Lmo4, and Prox1), together with analysis of topological position and published data on these or other genes, to identify the medial pallial derivatives and define its major subdivisions in mouse and chicken, and to compare such subdivisions between both species. In both mouse and chicken, the medial pallium is defined as a pallial sector adjacent to the cortical hem (expressing cLhx5 and cWn8b; Figures 6G, 8F) and roof plate/choroid tela (expressing cWnt8b, Figure 8F), showing moderate to strong vz expression of Lef1, Lhx2, and Lhx9, but not Lhx5, at least during early developmental stages. The topological position and the combination of genes at early developmental stages make the medial pallium different from other neighboring pallial sectors, such as the pallial septum (which additionally expresses Lhx5), the dorsal pallium (which does not express Lhx9 in the vz, and is mostly devoid of Lef1 except its medialmost, cingulate/retrosplenial areas), the dorsolateral caudal pallium (which does not express Lef1 nor Lhx9 in the vz), and the ventral/ventrolateral-caudal pallia (which do not express Lef1 in the vz). Based on this gene combination, often also present in the mantle, we propose that the indusium griseum, the hippocampal formation (DG, CA fields, and subiculum), the medial entorhinal cortex, and part of the amygdalo-hippocampal transition area of mouse are medial pallial derivatives. It is likely that the presubiculum and parasubiculum also derive from the medial pallium (see these areas expressing Lhx2 in Figure 5E″), although our data were insufficient to clearly determine expression of Lef1 in these areas. In the chicken, based on the same position and gene combination, we propose that the hippocampus (including the V-shaped area and the ventral hippocampus), the APH (including its caudolateral part, often called CDL; Reiner et al., 2004; Atoji and Wild, 2005), the entorhinal cortex, and the amygdalo-hippocampal transition area are medial pallial derivatives.
The genes Lhx2, Lhx9, and Lef1 were previously described to be expressed in the developing hippocampal formation of mouse (Lhx2: Porter et al., 1997; Bulchand et al., 2001; Monuki et al., 2001; Vyas et al., 2003; Lhx9: Rétaux et al., 1999; Vyas et al., 2003; Abellán et al., 2009; Lef1: Galceran et al., 2000; Choe et al., 2013). Herein, we provide more details on their expression in other medial pallial derivatives, such as the indusium griseum, part of the amygdalo-hippocampal transition area and the medial entorhinal cortex. The common origin with other parts of the hippocampal formation may explain some of their similar features and connections (see discussion for the entorhinal cortex below).
The present study is the first one that uses the three genes in combination, in a comparative context and in a comprehensive way, for trying to identify the medial pallial derivatives in the chicken. There are previous, separate reports of expression of these genes in the developing medial pallium of chicken, but none of these showed enough detail (cLhx2 and cLhx9: Abellán et al., 2009; this study was centered in the ventral pallium; see also data of Lhx9 in the zebra finch: Chen et al., 2013) and/or signal quality (cLef1: Gupta et al., 2012). Based on the combinatorial expression patterns presented here, the chicken medial pallium is larger than previously thought since it includes not only the hippocampus (including the V-shaped area and the ventral hippocampus) and medial parts of APH (our APHm, APHi, APHl; simply named APH in the proposal of the Avian Brain Nomenclature Forum; Reiner et al., 2004), but also the caudolateral part of APH, the entorhinal cortex, and the amygdalo-hippocampal transition area. The caudolateral APH (APHcl, using the nomenclature of Redies et al., 2001; Puelles et al., 2007) is called the dorsolateral corticoid area by many authors (CDL; Reiner et al., 2004; Atoji and Wild, 2005). Its medial pallial origin possibly explains its three-layered cytoarchitecture similar to other APH areas (Redies et al., 2001; Puelles et al., 2007), and its extensive connections with other parts of the APH, as shown in pigeons (Atoji and Wild, 2005). Based on its connections, Atoji and Wild (2005) proposed that the APHcl/CDL is comparable to the cingulate cortex of mammals, which primordium also expresses Lef1 during development (present data). However, while the APHcl/CDL derives from the medial pallium (having vz expression of Lef1, Lhx2, and Lhx9), the cingulate cortex and other parts of the neocortex derive from the dorsal pallium (showing lack of expression of Lhx9 at the vz), which disfavors the homology of these two structures. Our data support that APHcl/CDL is really a medial pallial derivative and, as such, part of the avian hippocampal complex; therefore, we recommend to call it simply APHcl and to abandon the term CDL, which is confusing because it is also employed by some authors to refer to the DLP (see, for example, Puelles et al., 2007; Belgard et al., 2013). In addition, we found a novel cell group, the APHr (maybe comparable to the apical APH of Puelles et al., 2007), which could be distinguished by its intense expression of Lef1 from E10 onwards. While Lef1 started to be downregulated in most of the medial pallium, its expression was intensified in APHr during intermediate and late embryonic development, and was still seen defining this cell group after hatching (P2). As discussed later, an ectopic migrated part of APHr appears to reach intermediate and caudal hippocampal formation levels, where it appears to correspond to the so-called parvocellular region of Atoji and Wild (2005).
The present data agree with previous claims of homology of the avian hippocampus and APH with the hippocampal formation of mammals (reviews in Dubbeldam, 1998; Reiner et al., 2004; Striedter, 2005; Atoji and Wild, 2006; Papp et al., 2007), which were based on identical topological position and embryological origin (Ariens-Kapper et al., 1936; Källén, 1962; Redies et al., 2001; Puelles et al., 2007), and some similarities in cyto- and chemo-architecture (Erichsen et al., 1991; Montagnese et al., 1996; Tömböl et al., 2000; see also Herold et al., 2014), connections (Benowitz and Karten, 1976; Casini et al., 1986; Székely and Krebs, 1996; Székely, 1999; Atoji et al., 2002; Atoji and Wild, 2004), and function (Bingman et al., 1984, 2003; Sherry et al., 1992; Colombo and Broadbent, 2000; Clayton et al., 2003; Mayer et al., 2013). Similarly to that of mammals, the avian hippocampal formation is involved in episodic and spatial memory, and contains location-specific and other types of cells involved in spatial navigation (Clayton et al., 2003; Bingman and Sharp, 2006). It also shows oscillatory activity similar to the theta rhythms (Siegel et al., 2000), LTP and LTD synaptic plasticity involved in learning and memory, synaptic modification after training, and evidence of adult neurogenesis (reviewed by Papp et al., 2007). Crucial for the argument of homology is that the hippocampal formation has also been identified in reptiles, and was likely present in the common ancestor of amniotes (reviewed by Rodríguez et al., 2002; Papp et al., 2007; Medina and Abellán, 2009).
Our data also agree with more recent proposals of homology based on massive gene expression data in the adult pallium, which show a striking similarity of mouse and chicken medial pallial derivatives regarding their gene expression profile (for example, Belgard et al., 2013). However, some recent studies have revealed that the hippocampal formation of different avian species also shows a genetic expression profile similar to that of the arcopallium during development (Chen et al., 2013) and in the adult (Jarvis et al., 2013). This particularly refers to the expression of the transcription factors Lhx9 and ER81. However, Lef1, which in mammals has been shown to be essential for the development of the hippocampal formation (see above), is expressed in the developing hippocampal formation of chicken (E8–E9), but not in the arcopallium. After E10, Lef1 also starts to be expressed in parts of the mantle of the chicken dorsal pallium (hyperpallium) and lateral pallium (mesopallium). It is important to remember that most developmental regulatory genes are expressed in more than one region; for example, this is so for Emx1, Emx2, Pax6, Lhx2, Lhx9, and ER81, expressed in several pallial subdivisions, but some also in the subpallium and outside the telencephalon, and even outside the nervous system (Puelles et al., 2000; Abellán and Medina, 2009; Abellán et al., 2009; Tzchori et al., 2009; Chen et al., 2013; present data). Their function is region, time, and context dependent. This also applies to Lef1, which is expressed in the brain and other tissues, in complex patterns that change throughout embryonic development and postnatally (present data; Oosterwegel et al., 1993; Nagalski et al., 2013), having roles that are context-dependent (Mao and Byers, 2011). The context relies on the molecular networks present in the tissue, which change between regions and with time. The molecular network present in the tissue at any time is essential for understanding both the interactions between transcription factors or other regulatory proteins and their region- and time-specific function. For this reason, we pay special attention to both the topological position (Nieuwenhuys, 1998, 2009; Striedter, 2005) and the combinatorial expression patterns of regulatory genes seen during early development (see also discussion in Puelles and Medina, 2002; Puelles and Ferran, 2012; Medina et al., 2013). Studies using knockout mice have shown that Lef1 is one of the key actors involved at early stages in the development of the hippocampal formation, but this transcription factor acts in combination with other regulatory proteins, such as Wnt and BMP proteins, produced at the cortical hem and/or roof plate (Galceran et al., 2000; Choe et al., 2013). How Lef1 interacts with Lhx2 (also essential for hippocampal development; Bulchand et al., 2001; Vyas et al., 2003), Lhx9, and ER81 during medial pallial development is unknown. The role of Lef1 in the development of other brain regions outside the medial pallium (such as the hyperpallium/dorsal pallium or the thalamus) is also unknown. Due to its far lateroventral position, the arcopallium appears to be out of the effect of roof plate/cortical hem BMP/Wnt signals (if existent, such effect is likely very weak; see also Medina and Abellán, 2009; Aboitiz and Zamorano, 2013). A partially different network of transcription factors (without the implication of Lef1) is important for arcopallial development (such as Lhx9, ER81, and other), although the hierarchy, interactions and functions of the different factors within the network are still unknown.
Hippocampal formation subdivisions in mouse and chicken
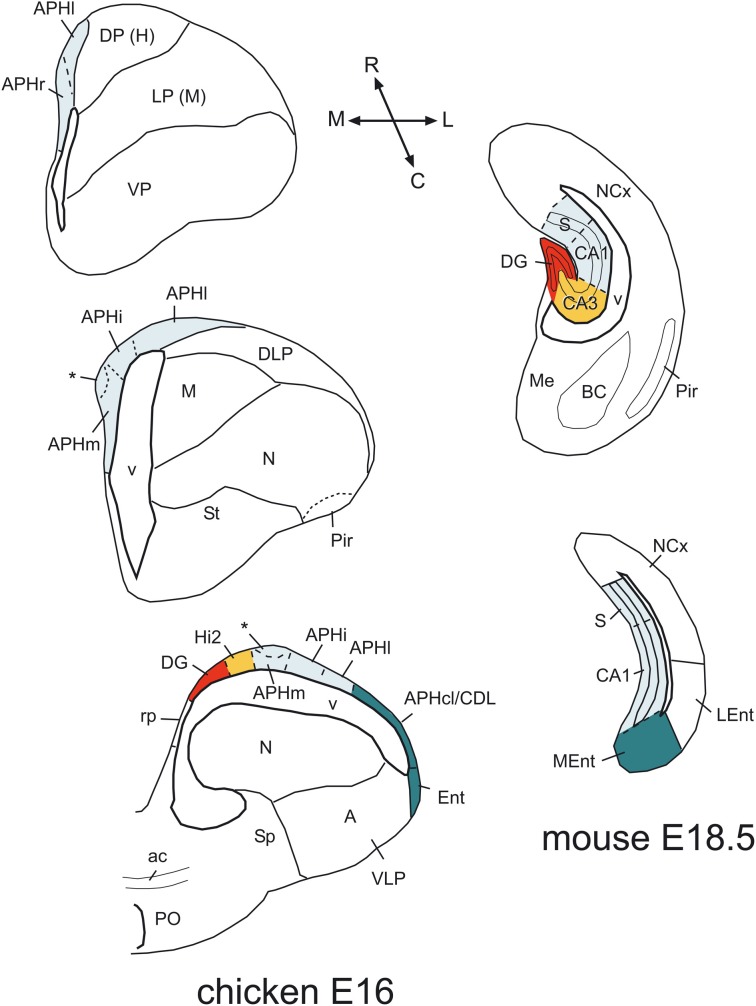
The combinatorial expression of Lef1, Lhx2, Lhx9, Prox1, Lmo4, and Lmo3 was useful for defining some molecular features of the major subdivisions of the hippocampal formation, and for comparative purposes. Below we discuss the evidence suggesting the comparison of specific chicken subdivisions with the mammallian DG/CA3 and the CA1/subiculum (Figure 12).
Figure 12.
Comparison of chicken and mouse medial pallial subdivisions. Schematic drawings of frontal sections through the telencephalon of a chicken (at E16) and a mouse (at E18.5), at rostral intermediate, or caudal levels, showing the major subdivisions of the medial pallium. A color code is used to compare these subdivisions between species. In these schemes, dorsal is to the top and medial is to the left. In the chicken, the rostralmost part is represented by the APHr. The asterisk points to an ectopic part of chicken APHr (possibly a group tangentially migrated cells), observed at the surface of APHm at intermediate and caudal levels of the medial pallium. The rostralmost part of mouse is not represented here, but appears to include the indusium griseum. For abbreviations see list. See text for more details.
Dentate gyrus and CA3
The mouse DG, occupying the medialmost topological position within the medial pallium, typically showed moderate to strong expression of Lef1, Lhx2, Lhx9, and Prox1. Of these, Lef1 and Prox1 have been shown to be of crucial importance. Lack of Lef1 function in mouse leads to agenesis of the DG (Galceran et al., 2000), while conditional inactivation of Prox1 in mouse showed that this transcription factor is essential for specification and maturation of DG granule cells, and maintenance of their cell identity throughout life (Lavado et al., 2010; Karalay et al., 2011; Iwano et al., 2012). In the DG of adult mice, Prox1 is also involved in intermediate progenitor maintenance and maturation of new granule cells (Lavado et al., 2010; Karalay et al., 2011).
In contrasts, there is much controversy on the location or existence of a DG in birds and reptiles. Based on different data, there are diverse opinions on possible areas homologous to DG in birds: V-shaped area or part of it, vs. part of APH (for example, Montagnese et al., 1996; Székely, 1999; Atoji et al., 2002; Atoji and Wild, 2004; Suárez et al., 2006; Puelles et al., 2007; Herold et al., 2014). Moreover, some authors claim that DG may be a novel acquisition of mammals (Papp et al., 2007), which would imply that there is no homolog in birds. However, our data on Lef1 and Prox1 strongly suggest that a large part of the so-called avian hippocampus, including its dorsal (the part of V-shaped area encompassing the dentate gyrus primordium and hippocampal sector 1 of Puelles et al., 2007) and ventral parts (Atoji et al., 2002), could be homologous to mammalian DG, if confirmed its presence in reptiles. Gupta et al. (2012) reached a similar conclusion based on Prox1 in V-shaped area during early/intermediate development (E8–E14), although these authors included the whole V-shaped area and did not mention the ventral hippocampus. Our data show that the dorsalmost part of V-shaped area (hippocampal sector 2 or Hi2 of Puelles et al., 2007; Table 2B) does not express Prox1 at E10–E12 or later (Figures 8H, 11J), raising doubts on the homology of this dorsal part. Gupta et al. (2012) showed that chicken DG cells are born between E6 (the majority) and E10 from the vz deep to the V-shaped area, and start to express Prox1 4 days later (from E8 on). Our data show that Prox1 continues to be expressed in chicken DG after hatching (at least until P2), but Lef1 is downregulated, similarly to the findings in mouse (Nagalski et al., 2013).
Regarding CA3, in mouse this area shares some features with DG, such as lack of Lmo3 expression (see Table 1B), but it does not express Prox1. In chicken, the dorsal part of V-shaped area, with no expression of Prox1, does not express Lmo3 either, and may be comparable to CA3 (asterisks in Figures 11B,C). Curiously, following postmitotic inactivation of Prox1 in mouse, immature neurons of DG lose their granule cell identity and differentiate into CA3 pyramidal neurons (Iwano et al., 2012). This means that DG immature neurons have the potential of becoming either granule cells or CA3 pyramidal cells. Moreover, recent data have shown that, in rats, CA3 field includes a subpopulation of granule cells, which contain calbindin and Prox1 as those of DG (Szabadics et al., 2010). These interesting observations have implications for understanding hippocampal evolution, since perhaps both DG and CA3 evolved from a common field, which splits into two separate fields either by downregulation of Prox1 in one part (the CA3) or by novel expression of Prox1 in one of the parts (the DG). To know what was the ancestral situation in amniotes, it is mandatory to study Lef1 and Prox1 in different reptiles, including lizards and other Squamates, which are currently considered a sister group of Archosauria (birds, crocodiles and perhaps turtles) and, as such, excellent models for understanding the basal condition in sauropsids (Zardoya and Meyer, 1998; Meyer and Zardoya, 2003; Fong et al., 2012). Comparison of the chicken, crocodile, and lizard hippocampal formation (Nissl images in Papp et al., 2007, for lizard and crocodile; Puelles et al., 2007, for chicken) points to the striking topological and cytoarchitectonic similarity of the chicken ventral hippocampus and the lizard/crocodile medial cortex, and the chicken V-shaped area (especially its dorsal part) to the lizard/crocodile dorsomedial cortex. Although some authors have suggested that the reptilian medial cortex is comparable to mammalian DG and the reptilian dorsomedial cortex is comparable to CA3 (Martínez-Guijarro et al., 1990), other authors suggested that the reptilian medial cortex is comparable to the mammalian indusium griseum (Künzle, 2004), or that both reptilian cortices maybe like mammalian CA3 (Papp et al., 2007). The possible common origin of DG and CA3 may explain why the connections of the avian V-shaped area and the reptilian medial/dorsomedial cortices are a mixture of those of mammalian DG and CA3 [reciprocal connections with the septum, and both ipsi- and contralateral (commissural) projections to other parts of the hippocampal formation; birds: (Casini et al., 1986; Atoji and Wild, 2004; Montagnese et al., 2004); for mammals see (Witter and Amaral, 2004); reptiles: (Lopez-Garcia and Martinez-Guijarro, 1988; Olucha et al., 1988; Martínez-Guijarro et al., 1990; Hoogland and Vermeulen-VanderZee, 1993)]. In any case, it is clear that the avian hippocampal formation has undergone partial divergence during the hundreds of millions of years of separate evolution (Striedter, 2005), which explains why some of the hippocampal subdivisions and features found in extant birds do not really fit well with any of those found in reptiles or mammals (see also Papp et al., 2007; Herold et al., 2014).
In addition to its role in DG granule cell specification, differentiation, and survival (reviewed by Karalay and Jessberger, 2011), recent data in mouse showed that, from late embryonic stages, Prox1 is also expressed in subsets of neocortical and hippocampal interneurons, which derive from the caudolateral ganglionic eminence and the preoptic area of the subpallium (Rubin and Kessaris, 2013). However, our data in chicken did not allow to discriminate the presence of Prox1-expressing interneurons.
CA1 and subiculum
During early (chicken) and/or intermediate (mouse and chicken) development, in addition to Lef1 and Lhx2, most medial pallial derivatives also show moderate to strong expression of Lmo4, while some of them (including CA1 and subiculum) also show Lmo3 expression in an area- and layer-specific way. In the mouse, the strongest expression of Lmo4 occurs in the CA1, while the strongest expression of Lmo3 is seen in the subiculum. In the chicken, the strongest Lmo4 expression is seen in APHm and APHi [roughly corresponding to the dorsomedial APH sector (DM) of Atoji and Wild, 2004], while the strongest Lmo3 expression is seen in APHl [corresponding to the dorsolateral APH sector (DL) of Atoji and Wild, 2004; for comparison see Suárez et al., 2006]. These subdivisions show associational connections with other hippocampal areas, as well as descending projections to the septum, the nucleus accumbens, the pallial amygdala, the extended amygdala, and the hypothalamus, including the mammillary region (Atoji et al., 2002, 2006; Atoji and Wild, 2004, 2005). Importantly, the APHm,i,l (DM and DL fields) are extensively and reciprocally connected with the DG/CA3 area (V-shaped area) (Atoji et al., 2002), thus establishing the basis for the recurrent, associational architecture typical of the hippocampal formation in mammals, and needed for memory acquisition (Papp et al., 2007). Based on their topological position, embryonic origin, genetic profile, and connectivity patterns, these APH subdivisions together appear comparable to the CA1/subiculum of mammals (see also Atoji and Wild, 2004; Suárez et al., 2006).
Radial vs. tangential cell migrations within the medial pallium: the cases of the APHr and DG
Data in chicken and in different mammalian species show that the majority of the neurons of the hippocampal formation migrate radially (following radial glial fibers) from the medial pallium neuroepithelium (mammals: Eckenhoff and Rakic, 1984; Rickmann et al., 1987; Altman and Bayer, 1990a,b; Li and Pleasure, 2005; chicken, Gupta et al., 2012). The exception to this rule is the case of the GABAergic interneurons that populate the hippocampal formation, which migrate from the subpallium (Pleasure et al., 2000; Cobos et al., 2001). In addition, in chicken a part of the cells of APHr (the ectopic APHr or APHre) appears to migrate tangentially within the medial pallium to occupy more caudal, dorsomedial, and superficial positions (present results). Based on Lef1 expression and radial glial fiber disposition, the APHr vz appears to be located at very rostral APH levels (maybe corresponding to the apical APH of Puelles et al., 2007), where Lef1 occupies the whole mantle (Figure 8C; Figure S2A). A band of Lef1-expressing cells (the APHre) appears to extend from this origin, and progressively occupies more superficial, dorsolateral and caudal positions. At intermediate and caudal levels, the Lef1 expression domain related to APHre lies at the surface of APHm (Figures 8D, 9J, 11G) and appears to correspond to the so-called parvocellular region of the hippocampal formation (Atoji and Wild, 2005). Thus, this observation suggests that the neurons of the parvocellular region arrive at their final destination by tangential migration. Supporting this proposal, this region is avoided by radial glial fibers that produce the underlying APHm (Figure 8I). The relation of APHr/APHre to other hippocampal subdivisions of chicken or other amniotes and the function of this cell group remain unknown.
In mammals, the DG granule neurons follow a special type of radial migration due to deformation of the radial glial fibers at the medialmost pallial edge, during the pallial growth that occurs in later developmental stages (Eckenhoff and Rakic, 1984; Rickmann et al., 1987; Li and Pleasure, 2005; note that some authors do not consider this migration to be radial: Altman and Bayer, 1990a). Such deformation of the radial glial fibers is not visible in the medial pallium of chicken (Gupta et al., 2012; present results of radial glial fibers), possibly because it does not grow as much as in mammals.
Entorhinal cortex: two divisions, two embryonic origins
Our data in mouse suggest that the two major divisions described in the entorhinal cortex of different mammals originate in separate pallial domains, the MEnt (caudomedially located) from the medial pallium, and the LEnt (rostrolaterally located) from the dorsolateral caudal pallium (Figure 12). In particular, based on the combinatorial expression of Lef1, Lhx2, Lhx9, and Lmo4, the MEnt appears to derive from the same embryonic domain that produces the hippocampal formation. This may explain some of the distinct features found in MEnt (but not LEnt) (Sewards and Sewards, 2003), such as the presence of cells involved in processing spatial cues (grid cells, head-direction cells, and border cells, which respond to specific position, direction and orientation, and are able to precisely map the spatial environment), and its implication in transmitting information on the spatial context of an experience to the hippocampal formation (Leutgeb et al., 2005; Knierim et al., 2013; Zhang et al., 2013). Moreover, lesion experiments have shown that MEnt (but not LEnt) is involved in spatial learning (Sewards and Sewards, 2003). In contrast, LEnt transmits non-spatial information to the hippocampal formation, related to the content of an experience, and is involved in non-spatial learning and memory retrieval (Knierim et al., 2013; Stouffer and Klein, 2013; Tanninen et al., 2013). While the MEnt receives input from the CA1, subiculum, presubiculum/parasubiculum (all of which also contain place or grid cells; Boccara et al., 2010), and from visual neocortical areas related to the dorsal visual stream (the “where” pathway) involved in processing spatial visual information on object location (Wang et al., 2011), the LEnt receives input from visual areas of the temporo-occipital neocortex and/or perirhinal cortex (including area 35) related to the ventral visual stream (the “what” pathway), involved in object identification and recognition (Sewards and Sewards, 2003; Canto et al., 2008; Wang et al., 2011). In both rodents and cats, both the LEnt and MEnt receive direct olfactory bulb input, although the LEnt is the preferential target (Room et al., 1984; Witter and Amaral, 2004). In rodents, LEnt shows important reciprocal connections with the pallial amygdala and possibly provides the amygdala with complex “contextual” information relevant for behavior (McDonald and Mascagni, 1997), but the involvement of MEnt in such connections is very modest (Sewards and Sewards, 2003). Both MEnt and LEnt receive weak auditory input from the temporal neocortex, and are reciprocally connected with areas of the cingulate, retrosplenial, and frontal neocortex (Sewards and Sewards, 2003; Witter and Amaral, 2004).
Are these two entorhinal cortex divisions present in birds? Current data suggest that the so-called entorhinal cortex of birds (Puelles et al., 2007; Abellán et al., 2009; present work) may be comparable to mammalian MEnt (Figure 12). This cortical area, located laterally to the APHcl/CDL, receives olfactory input (Reiner and Karten, 1985; Atoji and Wild, 2014), and has often been considered a caudal continuation of the piriform cortex (for example, Atoji and Wild, 2014). However, in both mouse and chicken, the piriform cortex shows a genetic profile different from that of this avian cortical field: the piriform cortex is characterized by strong expression of Lmo3, Lmo4, and Cdh10, very weak expression of Lhx9, and no expression of Lhx2 and Lef1; in contrast, the avian entorhinal cortex shows moderate to strong expression of Lmo4, Lhx9, Lhx2, and Lef1, while its cortical plate is nearly free of Lmo3 and Cdh10 (Vyas et al., 2003; Abellán et al., 2009; present data). Also, while the piriform cortex is at the surface of the nidopallium and derives from the ventral pallium (Puelles et al., 2007), the so-called avian entorhinal cortex is adjacent to the APH and lateral horn of the lateral ventricle, and appears to derive from the medial pallium (based on position and expression of Lef1 during early development). Based on its embryonic origin, the so-called avian entorhinal cortex may be comparable to mammalian MEnt. As noted above, MEnt also receives a minor direct input from the olfactory bulb at least in some mammals (Room et al., 1984; Witter and Amaral, 2004).
In addition, the avian APHcl/CDL may also be comparable to mammalian MEnt (Figure 12). In pigeon, the APHcl/CDL is reciprocally and extensively connected with the various areas of the hippocampal formation, i.e., DG/CA3 area, APHm, APHi, and APHl (V-shaped area, DM and DL in Atoji and Wild, 2005). Moreover, large lesions involving the CDL produce visuospatial deficits suggesting a similar role to that of mammalian MEnt, although this needs confirmation by smaller lesions or specific inactivation of APHcl/CDL (discussion in Atoji and Wild, 2005). The observed deficits are consistent with the inputs to APHcl/CDL from the visual hyperpallium (Figure 5 of Atoji and Wild, 2005), which has been involved in the “where” analysis of the information (Watanabe et al., 2011). Moreover, the lateral part of APHcl/CDL also receives direct input from the olfactory bulb (Reiner and Karten, 1985; Atoji and Wild, 2014), and for this reason it has been compared to the entorhinal cortex of mammals (Redies et al., 2001; Suárez et al., 2006). Curiously, both the avian APHcl/CDL and the mammalian MEnt include cell aggregates or patches showing neurochemical features different from the surrounding area (birds: Redies et al., 2001; Kovjanic and Redies, 2003; Suárez et al., 2006; mammals: Witter and Amaral, 2004). These patches were also evident in our chicken material at P0 as areas of the cortical plate free of cLmo3 expression. They appear to be formed by cells having the same embryonic birth date and expressing the same types of cell adhesion-mediating cadherins (Redies et al., 2001; Kovjanic and Redies, 2003; discussed by Suárez et al., 2006). The connections and functional significance of these patches remain unknown.
On the other hand, it is uncertain whether the avian field called dorsolateral caudal pallium (DLP) is or is not comparable to the dorsolateral caudal pallial field that produces LEnt in mammals, even if they occupy similar topological positions and share some similar molecular features (for example, in general weak or moderate expression of Lhx2, Lhx9, Lmo3, and Lmo4; present data; Figure 12). The avian DLP is relatively large, and has a cortical-like area at its surface that extends ventrally (Puelles et al., 2007; called caudodorsolateral pallium or CDL by these authors), but apparently does not receive any direct olfactory input (Atoji and Wild, 2014; called temporo-occipito-mesencephalic area or TPO by these authors). This general field receives different types of information from several pallial areas, including the entopallial belt (involved in the what analysis of the information, Watanabe et al., 2011) and the mesopallium (Figure 9 of Atoji and Wild, 2005), and it projects to the avian pallial amygdala (in particular, the caudolateral nidopallium and the arcopallium) and the basal ganglia (Veenman et al., 1995; Kröner and Güntürkün, 1999). It is also connected reciprocally with the hippocampal formation, but less so than the APHcl/CDL (Atoji and Wild, 2005). It would be interesting to investigate whether there is a structure comparable to avian DLP in reptiles, which would contribute to understand its homology across amniotes. For this purpose, it is necessary to have a molecular marker (or a clear combination of them) specific of this pallial sector.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Luis Puelles (Univ. of Murcia, Spain) and the reviewers for their useful comments on the manuscript. Supported by a grant to Loreta Medina from the Spanish Ministry of Economy and Competitivity (MINECO) and Fondo Europeo de Desarrollo Regional (FEDER): grant no. BFU2012-33029.
Glossary
Abbreviations
- A
arcopallium (part of pallial amygdala; VLP derivative) (chicken)
- ac
anterior commissure
- Amyg
amygdala
- AHi
amygdalo-hippocampal transition area
- AHitr
amygdalo-hippocampal transition area (chicken)
- APH
parahippocampal area (chicken)
- APHcl
caudolateral APH (same as CDL)
- APHi
intermediate APH
- APHl
lateral APH
- APHm
medial APH
- APHr
rostral APH
- APHre
ectopic part of APHr
- APir
amygdalo-piriform transition area
- BC
basal amygdalar complex (part of pallial amygdala)
- CA
Ammon's horn fields (CA1, CA2, CA3)
- can
CA neuroepithelium
- cc
corpus callosum
- CDL
corticoid dorsolateral area (same as APHcl) (chicken)
- chp
choroid plexus
- Cg
cingulate neocortex
- cMEnt
caudal part of MEnt
- CPu
caudoputamen
- CR
Cajal-Retzius cells
- cxh
cortical hem
- DG
dentate gyrus
- dgm
dentate gyrus migratory cells
- dgn
DG neuroepitleium
- DLP
dorsolateral caudal pallium
- DLPco
dorsolateral caudal pallium, core nucleus (chicken)
- DMH
dorsomedial hypothalamus
- DP
dorsal pallium
- EMT
prethalamic eminence
- Ent
entorhinal cortex
- GP
globus pallidus
- H
hyperpallium (DP derivative) (chicken)
- Hb
habenula
- Hi2
hippocampal area 2 (dorsal part of V-shaped area; defined by Puelles et al., 2007)
- ic
internal capsule
- IG
indusium griseum
- Ins
insular cortex
- iz
intermediate zone (deepest part of the mantle, containing migratory cells)
- LEnt
lateral entorhinal cortex
- lfb
lateral forebrain bundle
- LGE
lateral ganglionic eminence
- LP
lateral pallium
- LS
lateral septum
- LSt
lateral striatum (chicken)
- m
mantle
- md
deep part of the mantle
- M
mesopallium (LP derivative) (chicken)
- Me
medial amygdala
- mes
mesoderm
- MEnt
medial entorhinal cortex
- MGE
medial ganglionic eminence
- MP
medial pallium
- N
nidopallium (VP derivative) (chicken)
- NCL
caudolateral nidopallium
- NCx
neocortex
- o
outer or marginal zone
- OB
olfactory bulb
- Olf
olfactory areas
- Pir
piriform cortex
- PMCo
posteromedial cortical amygdalar area (part of pallial amygdala)
- PO
preoptic area
- PSe
pallial septum
- RB
retrobulbar area
- rp
roof plate
- RS
retrosplenial neocortex
- S
subiculum
- Se
septum
- sn
subiculum neuroepithelium
- Sp
subpallium
- St
striatum
- svz
subventricular zone
- tch
choroid tela
- Te
temporal neocortex
- Th
thalamus
- v
ventricle
- V
V-shaped area (chicken)
- VLP
ventrolateral caudal pallium
- VP
ventral pallium
- vz
ventricular zone.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fnana.2014.00059/abstract
We provide two supplementary figures with additional details in the expression of Lef1 and Prox1 in the mouse and chicken forebrain.
References
- Abellán A., Desfilis E., Medina L. (2013). The olfactory amygdala in amniotes: an evo-devo approach. Anat. Rec. (Hoboken) 296, 1317–1332 10.1002/ar.22744 [DOI] [PubMed] [Google Scholar]
- Abellán A., Legaz I., Vernier B., Rétaux S., Medina L. (2009). Olfactory and amygdalar structures of the chicken ventral pallium based on the combinatorial expression patterns of LIM and other developmental regulatory genes. J. Comp. Neurol. 516, 166–186 10.1002/cne.22102 [DOI] [PubMed] [Google Scholar]
- Abellán A., Medina L. (2009). Subdivisions and derivatives of the chicken subpallium based on expression of LIM and other regulatory genes and markers of neuron subpopulations during development. J. Comp. Neurol. 515, 465–501 10.1002/cne.22083 [DOI] [PubMed] [Google Scholar]
- Abellán A., Vernier B., Rétaux S., Medina L. (2010). Similarities and differences in the forebrain expression of Lhx1 and Lhx5 between chicken and mouse: insights for understanding telencephalic development and evolution. J. Comp. Neurol. 518, 3512–3528 10.1002/cne.22410 [DOI] [PubMed] [Google Scholar]
- Aboitiz F., Zamorano F. (2013). Neural progenitors, patterning and ecology in neocortical origins. Front. Neuroanat. 7:38 10.3389/fnana.2013.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J., Bayer S. A. (1990a). Mosaic organization of the hippocampal neuroepithelium and the multiple germinal sources of dentate granule cells. J. Comp. Neurol. 301, 325–342 10.1002/cne.903010302 [DOI] [PubMed] [Google Scholar]
- Altman J., Bayer S. A. (1990b). Prolonged sojourn of developing pyramidal cells in the intermediate zone of the hippocampus and their settling in the stratum pyramidale. J. Comp. Neurol. 301, 343–364 10.1002/cne.903010303 [DOI] [PubMed] [Google Scholar]
- Altman J., Bayer S. A. (1990c). Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. J. Comp. Neurol. 301, 365–381 10.1002/cne.903010304 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A., Lim D. A. (2004). For the long run: maintaining germinal niches in the adult brain. Neuron 41, 683–686 10.1016/S0896-6273(04)00111-4 [DOI] [PubMed] [Google Scholar]
- Ariens-Kapper C. U., Huber G. C., Crosby E. C. (1936). The Comparative Anatomy of the Nervous System of Vertebrates, Including Man. New York, NY: Hafner Publishing [Google Scholar]
- Atoji Y., Saito S., Wild J. M. (2006). Fiber connections of the compact division of the posterior pallial amygdala and lateral part of the bed nucleus of the stria terminalis in the pigeon (Columba livia). J. Comp. Neurol. 499, 161–182 10.1002/cne.21042 [DOI] [PubMed] [Google Scholar]
- Atoji Y., Wild J. M. (2004). Fiber connections of the hippocampal formation and septum and subdivisions of the hippocampal formation in the pigeon as revealed by tract tracing and kainic acid lesions. J. Comp. Neurol. 475, 426–461 10.1002/cne.20186 [DOI] [PubMed] [Google Scholar]
- Atoji Y., Wild J. M. (2005). Afferent and efferent connections of the dorsolateral corticoid area and a comparison with connections of the temporo-parieto-occipital area in the pigeon (Columba livia). J. Comp. Neurol. 485, 165–182 10.1002/cne.20490 [DOI] [PubMed] [Google Scholar]
- Atoji Y., Wild J. M. (2006). Anatomy of the avian hippocampal formation. Rev. Neurosci. 17, 3–15 10.1515/REVNEURO.2006.17.1-2.3 [DOI] [PubMed] [Google Scholar]
- Atoji Y., Wild J. M. (2014). Efferent and afferent connections of the olfactory bulb and prepiriform cortex in the pigeon (Columba livia). J. Comp. Neurol. 522, 1728–1752 10.1002/cne.23504 [DOI] [PubMed] [Google Scholar]
- Atoji Y., Wild J. M., Yamamoto Y., Suzuki Y. (2002). Intratelencephalic connections of the hippocampus in pigeons (Columba livia). J. Comp. Neurol. 447, 177–199 10.1002/cne.10239 [DOI] [PubMed] [Google Scholar]
- Belgard T. G., Montiel J. F., Wang W. Z., García-Moreno F., Margulies E. H., Ponting C. P., et al. (2013). Adult pallium transcriptomes surprise in not reflecting predicted homologies across diverse chicken and mouse pallial sectors. Proc. Natl. Acad. Sci. U.S.A. 110, 13150–13155 10.1073/pnas.1307444110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz L. I., Karten H. J. (1976). The tractus infundibuli and other afferents to the parahippocampal region of the pigeon. Brain Res. 102, 174–180 10.1016/0006-8993(76)90584-9 [DOI] [PubMed] [Google Scholar]
- Bingman V. P., Bagnoli P., Ioalè P., Casini G. (1984). Homing behavior of pigeons after telencephalic ablations. Brain Behav. Evol. 24, 94–108 10.1159/000121308 [DOI] [PubMed] [Google Scholar]
- Bingman V. P., Hough G. E., 2nd., Kahn M. C., Siegel J. J. (2003). The homing pigeon hippocampus and space: in search of adaptive specialization. Brain Behav. Evol. 62, 117–127 10.1159/000072442 [DOI] [PubMed] [Google Scholar]
- Bingman V. P., Sharp P. E. (2006). Neuronal implementation of hippocampal-mediated spatial behavior: a comparative evolutionary perspective. Behav. Cogn. Neurosci. Rev. 5, 80–91 10.1177/1534582306289578 [DOI] [PubMed] [Google Scholar]
- Bird C. M., Burgess N. (2008). The hippocampus and memory: insights from spatial processing. Nat. Rev. Neurosci. 9, 182–194 10.1038/nrn2335 [DOI] [PubMed] [Google Scholar]
- Boardman P. E., Sanz-Ezquerro J., Overton I. M., Burt D. W., Bosch E., Fong W. T., et al. (2002). A comprehensive collection of chicken cDNAs. Curr. Biol. 12, 1965–1969 10.1016/S0960-9822(02)01296-4 [DOI] [PubMed] [Google Scholar]
- Boccara C. N., Sargolini F., Thoresen V. H., Solstad T., Witter M. P., Moser E. I., et al. (2010). Grid cells in pre- and parasubiculum. Nat. Neurosci. 13, 987–994 10.1038/nn.2602 [DOI] [PubMed] [Google Scholar]
- Bulchand S., Grove E. A., Porter F. D., Tole S. (2001). LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech. Dev. 100, 165–175 10.1016/S0925-4773(00)00515-3 [DOI] [PubMed] [Google Scholar]
- Bulchand S., Subramanian L., Tole S. (2003). Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev. Dyn. 226, 460–469 10.1002/dvdy.10235 [DOI] [PubMed] [Google Scholar]
- Canto C. B., Wouterlood F. G., Witter M. P. (2008). What does the anatomical organization of the entorhinal cortex tell us? Neural Plast. 2008:381243 10.1155/2008/381243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casini G., Bingman V. P., Bagnoli P. (1986). Connections of the pigeon dorsomedial forebrain studied with WGA-HRP and 3H-proline. J. Comp. Neurol. 245, 454–470 10.1002/cne.902450403 [DOI] [PubMed] [Google Scholar]
- Charvet C. J., Owerkowicz T., Striedter G.F. (2009). Phylogeny of the telencephalic subventricular zone in sauropsids: evidence for the sequential evolution of pallial and subpallial subventricular zones. Brain Behav. Evol. 73, 285–294 10.1159/000230673 [DOI] [PubMed] [Google Scholar]
- Chen C. C., Winkler C. M., Pfenning A. R., Jarvis E. D. (2013). Molecular profiling of the developing avian telencephalon: regional timing and brain subdivision continuities. J. Comp. Neurol. 521, 3666–3701 10.1002/cne.23406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe Y., Kozlova A., Graf D., Pleasure S. J. (2013). Bone morphogenic protein signaling is a major determinant of dentate development. J. Neurosci. 33, 6766–6775 10.1523/JNEUROSCI.0128-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton N. S., Yu K. S., Dickinson A. (2003). Interacting Cache memories: evidence for flexible memory use by Western Scrub-Jays (Aphelocoma californica). J. Exp. Psychol. Anim. Behav. Process. 29, 14–22 10.1037/0097-7403.29.1.14 [DOI] [PubMed] [Google Scholar]
- Cobos I., Puelles L., Martínez S. (2001). The avian telencephalic subpallium originates inhibitory neurons that invade tangentially the pallium (dorsal ventricular ridge and cortical areas). Dev. Biol. 239, 30–45 10.1006/dbio.2001.0422 [DOI] [PubMed] [Google Scholar]
- Colombo M., Broadbent N. (2000). Is the avian hippocampus a functional homologue of the mammalian hippocampus? Neurosci. Biobehav. Rev. 24, 465–484 10.1016/S0149-7634(00)00016-6 [DOI] [PubMed] [Google Scholar]
- DeCarolis N. A., Eisch A. J. (2010). Hippocampal neurogenesis as a target for the treatment of mental illness: a critical evaluation. Neuropharmacology 58, 884–893 10.1016/j.neuropharm.2009.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbeldam J. L. (1998). Birds, in The Central Nervous System of Vertebrates, Vol. 3, eds Nieuwenhuys R., ten Donkelaar H. J., Nicholson C. (Berlin: Springer; ), 1525–1636 [Google Scholar]
- Eckenhoff M. F., Rakic P. (1984). Radial organization of the hippocampal dentate gyrus: a Golgi, ultrastructural, and immunocytochemical analysis in the developing rhesus monkey. J. Comp. Neurol. 223, 1–21 10.1002/cne.902230102 [DOI] [PubMed] [Google Scholar]
- Eisch A. J., Cameron H. A., Encinas J. M., Meltzer L. A., Ming G. L., Overstreet-Wadiche L. S. (2008). Adult neurogenesis, mental health, and mental illness: hope or hype? J. Neurosci. 28, 11785–11791 10.1523/JNEUROSCI.3798-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erichsen J. T., Bingman V. P., Krebs J. R. (1991). The distribution of neuropeptides in the dorsomedial telencephalon of the pigeon (Columba livia): a basis for regional subdivisions. J. Comp. Neurol. 314, 478–492 10.1002/cne.903140306 [DOI] [PubMed] [Google Scholar]
- Fong J. J., Brown J. M., Fujita M. K., Boussau B. (2012). A phylogenomic approach to vertebrate phylogeny supports a turtle-archosaur affinity and a possible paraphyletic lissamphibia. PLoS ONE 7:e48990p 10.1371/journal.pone.0048990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galceran J., Miyashita-Lin E. M., Devaney E., Rubenstein J. L. R., Grosschedl R. (2000). Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development 127, 469–482 [DOI] [PubMed] [Google Scholar]
- García-López M., Abellán A., Legaz I., Rubenstein J. L. R., Puelles L., Medina L. (2008). Histogenetic compartments of the mouse centromedial and extended amygdala based on gene expression patterns during development. J. Comp. Neurol. 506, 46–74 10.1002/cne.21524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garda A. L., Puelles L., Rubenstein J. L. R., Medina L. (2002). Expression patterns of Wnt8b and Wnt7b in the chicken embryonic brain suggest a correlation with forebrain patterning centers and morphogenesis. Neuroscience 113, 689–698 10.1016/S0306-4522(02)00171-9 [DOI] [PubMed] [Google Scholar]
- Gould E. (2007). How widespread is adult neurogenesis in mammals? Nat. Rev. Neurosci. 8, 481–488 10.1038/nrn2147 [DOI] [PubMed] [Google Scholar]
- Gould E., Beylin A., Tanapat P., Reeves A., Shors T. J. (1999). Learning enhances adult neurogenesis in the hippocampal formation. Nat. Neurosci. 2, 260–265 10.1038/6365 [DOI] [PubMed] [Google Scholar]
- Gupta S., Maurya R., Saxena M., Sen J. (2012). Defining structural homology between the mammalian and avian hippocampus through conserved gene expression patterns observed in the chick embryo. Dev. Biol. 366, 125–141 10.1016/j.ydbio.2012.03.027 [DOI] [PubMed] [Google Scholar]
- Hamburger H., Hamilton H. L. (1951). A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92 10.1002/jmor.1050880104 [DOI] [PubMed] [Google Scholar]
- Herman J. P., Victor J. C., Sanes J. R. (1993). Developmentally regulated and spatially restricted antigens of radial glial cells. Dev. Dyn. 197, 307–318 10.1002/aja.1001970408 [DOI] [PubMed] [Google Scholar]
- Herold C., Bingman V. P., Ströckens F., Letzner S., Sauvage M., Palomero-Gallagher N., et al. (2014). Distribution of neurotransmitter receptors and zinc in the pigeon (Columba livia) hippocampal formation: a basis for further comparison with the mammalian hippocampus. J. Comp. Neurol. 522, 2553–2575 10.1002/cne.23549 [DOI] [PubMed] [Google Scholar]
- Hollyday M., McMahon J. A., McMahon A. P. (1995). Wnt expression patterns in chick embryo nervous system. Mech. Dev. 52, 9–25 10.1016/0925-4773(95)00385-E [DOI] [PubMed] [Google Scholar]
- Hoogland P. V., Vermeulen-VanderZee E. (1993). Medial cortex of the lizard Gekko gecko: a hodological study with emphasis on regional specialization. J. Comp. Neurol. 331, 326–338 10.1002/cne.903310304 [DOI] [PubMed] [Google Scholar]
- Iwano T., Masuda A., Kiyonari H., Enomoto H., Matsuzaki F. (2012). Prox1 postmitotically defines dentate gyrus cells by specifying granule cell identity over CA3 pyramidal cell fate in the hippocampus. Development 139, 3051–3062 10.1242/dev.080002 [DOI] [PubMed] [Google Scholar]
- Jacobowitz D. M., Abbott L. C. (1997). Chemoarchitectonic Atlas of the Developing Mouse Brain. Boca Raton, FL: CRC Press [Google Scholar]
- Jarvis E. D., Yu J., Rivas M. V., Horita H., Feenders G., Whitney O., et al. (2013). Global view of the functional molecular organization of the avian cerebrum: mirror images and functional columns. J. Comp. Neurol. 521, 3614–3665 10.1002/cne.23404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun H., Mohammed Qasim Hussaini S., Rigby M. J., Jang M. H. (2012). Functional role of adult hippocampal neurogenesis as a therapeutic strategy for mental disorders. Neural Plast. 2012:854285 10.1155/2012/854285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källén B. (1962). Embryogenesis of brain nuclei in the chick telencephalon. Ergeb. Anat. Entwicklungsgesch. 36, 62–82 [PubMed] [Google Scholar]
- Karalay O., Doberauer K., Vadodaria K. C., Knobloch M., Berti L., Miquelajauregui A., et al. (2011). Prospero-related homeobox 1 gene (Prox1) is regulated by canonical Wnt signaling and has a stage-specific role in adult hippocampal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 5807–5812 10.1073/pnas.1013456108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalay O., Jessberger S. (2011). Translating niche-derived signals into neurogenesis: the function of Prox1 in the adult hippocampus. Cell Cycle 10, 2239–2240 10.4161/cc.10.14.15850 [DOI] [PubMed] [Google Scholar]
- Kempermann G. (2008). The neurogenic reserve hypothesis: what is adult hippocampal neurogenesis good for? Trends Neurosci. 31, 163–169 10.1016/j.tins.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Knierim J. J., Neunuebel J. P., Deshmukh S. S. (2013). Functional correlates of the lateral and medial entorhinal cortex: objects, path integration and local-global reference frames. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130369 10.1098/rstb.2013.0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovjanic D., Redies C. (2003). Small-scale pattern formation in a cortical area of the embryonic chicken telencephalon. J. Comp. Neurol. 456, 95–104 10.1002/cne.2158 [DOI] [PubMed] [Google Scholar]
- Kröner S., Güntürkün O. (1999). Afferent and efferent connections of the caudolateral neostriatum in the pigeon (Columba livia): a retro- and anterograde pathway tracing study. J. Comp. Neurol. 407, 228–260 [DOI] [PubMed] [Google Scholar]
- Künzle H. (2004). The hippocampal continuation (indusium griseum): its connectivity in the hedgehog tenrec and its status within the hippocampal formation of higher vertebrates. Anat. Embryol. (Berl.) 208, 183–213 10.1007/s00429-004-0384-3 [DOI] [PubMed] [Google Scholar]
- Lavado A., Lagutin O. V., Chow L. M., Baker S. J., Oliver G. (2010). Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol. 8:e1000460 10.1371/journal.pbio.1000460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. M., Tole S., Grove E., McMahon A. P. (2000). A local Wnt-3a signal is required for development of the mammalian hippocampus. Development 127, 457–467 [DOI] [PubMed] [Google Scholar]
- Leuner B., Gould E., Shors T. J. (2006). Is there a link between adult neurogenesis and learning? Hippocampus 16, 216–224 10.1002/hipo.20153 [DOI] [PubMed] [Google Scholar]
- Leutgeb S., Leutgeb J. K., Moser M. B., Moser E. I. (2005). Place cells, spatial maps and the population code for memory. Curr. Opin. Neurobiol. 15, 738–746 10.1016/j.conb.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Li G., Pleasure S. J. (2005). Morphogenesis of the dentate gyrus: what we are learning from mouse mutants. Dev. Neurosci. 27, 93–99 10.1159/000085980 [DOI] [PubMed] [Google Scholar]
- Lithfous S., Dufour A., Després O. (2013). Spatial navigation in normal aging and the prodromal stage of Alzheimer's disease: insights from imaging and behavioral studies. Ageing Res. Rev. 12, 201–213 10.1016/j.arr.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia C., Martinez-Guijarro F. J. (1988). Neurons in the medial cortex give rise to Timm-positive boutons in the cerebral cortex of lizards. Brain Res. 463, 205–217 10.1016/0006-8993(88)90393-9 [DOI] [PubMed] [Google Scholar]
- Machon O., Backman M., Machonova O., Kozmik Z., Vacik T., Andersen L., et al. (2007). A dynamic gradient of Wnt signaling controls initiation of neurogenesis in the mammalian cortex and cellular specification in the hippocampus. Dev. Biol. 311, 223–237 10.1016/j.ydbio.2007.08.038 [DOI] [PubMed] [Google Scholar]
- Mao C. D., Byers S. W. (2011). Cell-context dependent TCF/LEF expression and function: alternative tales of repression, de-repression and activation potentials. Crit. Rev. Eukaryot. Gene Expr. 21, 207–236 10.1615/CritRevEukarGeneExpr.v21.i3.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Guijarro F. J., Desfilis E., López-García C. (1990). Organization of the dorsomedial cortex in the lizard Podarcis hispanica, in The Forebrain in Nonmammals. New Aspects of Structure and Development, eds Schwerdtfeger W. K., Germroth P. (Berlin: Springer-Verlag; ), 77–92 [Google Scholar]
- Mayer U., Watanabe S., Bischof H. J. (2013). Spatial memory and the avian hippocampus: research in zebra finches. J. Physiol. 107, 2–12 10.1016/j.jphysparis.2012.05.002 [DOI] [PubMed] [Google Scholar]
- McDonald A. J., Mascagni F. (1997). Projections of the lateral entorhinal cortex to the amygdala: a Phaseolus vulgaris leucoagglutinin study in the rat. Neuroscience 77, 445–459 10.1016/S0306-4522(96)00478-2 [DOI] [PubMed] [Google Scholar]
- Medina L., Abellán A. (2009). Development and evolution of the pallium. Semin. Cell. Dev. Biol. 20, 698–711 10.1016/j.semcdb.2009.04.008 [DOI] [PubMed] [Google Scholar]
- Medina L., Abellán A., Desfilis E. (2013). A never-ending search for the evolutionary origin of the neocortex: rethinking the homology concept. Brain Behav. Evol. 81, 150–153 10.1159/000348282 [DOI] [PubMed] [Google Scholar]
- Medina L., Bupesh M., Abellán A. (2011). Contribution of genoarchitecture to understanding forebrain evolution and development, with particular emphasis on the amygdala. Brain Behav. Evol. 78, 216–236 10.1159/000330056 [DOI] [PubMed] [Google Scholar]
- Medina L., Legaz I., González G., De Castro F., Rubenstein J. L. R., Puelles L. (2004). Expression of Dbx1, Neurogenin 2, Semaphorin 5A, Cadherin 8, and Emx1 distinguish ventral and lateral pallial histogenetic divisions in the developing mouse claustroamygdaloid complex. J. Comp. Neurol. 474, 504–523 10.1002/cne.20141 [DOI] [PubMed] [Google Scholar]
- Mendez-David I., Hen R., Gardier A. M., David D. J. (2013). Adult hippocampal neurogenesis: an actor in the antidepressant-like action. Ann. Pharm. Fr. 71, 143–149 10.1016/j.pharma.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Meyer A., Zardoya R. (2003). Recent advances in the (molecular) phylogeny of vertebrates. Annu. Rev. Ecol. Evol. Syst. 34, 311–338 10.1146/annurev.ecolsys.34.011802.132351 [DOI] [Google Scholar]
- Montagnese C. M., Krebs J. R., Meyer G. (1996). The dorsomedial and dorsolateral forebrain of the zebra finch, Taeniopygia guttata: a Golgi study. Cell Tissue Res. 283, 263–282 10.1007/s004410050537 [DOI] [PubMed] [Google Scholar]
- Montagnese C. M., Székely A. D., Adám A., Csillag A. (2004). Efferent connections of septal nuclei of the domestic chick (Gallus domesticus): an anterograde pathway tracing study with a bearing on functional circuits. J. Comp. Neurol. 469, 437–456 10.1002/cne.11018 [DOI] [PubMed] [Google Scholar]
- Monuki E. S., Porter F. D., Walsh C. A. (2001). Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron 32, 591–604 10.1016/S0896-6273(01)00504-9 [DOI] [PubMed] [Google Scholar]
- Nagalski A., Irimia M., Szewczyk L., Ferran J. L., Misztal K., Kuznicki J., et al. (2013). Postnatal isoform switch and protein localization of LEF1 and TCF7L2 transcription factors in cortical, thalamic, and mesencephalic regions of the adult mouse brain. Brain Struct. Funct. 218, 1531–1549 10.1007/s00429-012-0474-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuys R. (1998). Comparative neuroanatomy: place, principles and programme, in The Central Nervous System of Vertebrates, Vol. 1, eds Nieuwenhuys R., ten Donkelaar H. J., Nicholson C. (Berlin: Springer; ), 273–326 [Google Scholar]
- Nieuwenhuys R. (2009). The forebrain of actinopterygians revisited. Brain Behav. Evol. 73, 229–252 10.1159/000225622 [DOI] [PubMed] [Google Scholar]
- Olucha F., Martinez-Garcia F., Poch L., Schwerdtfeger W. K., Lopez-Garcia C. (1988). Projections from the medial cortex in the brain of lizards: correlation of anterograde and retrograde transport of horseradish peroxidase with Timm staining. J. Comp. Neurol. 276, 469–480 10.1002/cne.902760402 [DOI] [PubMed] [Google Scholar]
- Oosterwegel M., van de Wetering M., Timmerman J., Kruisbeek A., Destree O., Meijlink F., et al. (1993). Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development 118, 439–448 [DOI] [PubMed] [Google Scholar]
- Papp G., Witter M. P., Treves A. (2007). The CA3 network as a memory store for spatial representations. Learn. Mem. 14, 732–744 10.1101/lm.687407 [DOI] [PubMed] [Google Scholar]
- Paxinos G., Franklin K. B. J. (2004). The Mouse Brain in Stereotaxic Coordinates, 2nd Edn. San Diego, CA: Academic Press [Google Scholar]
- Pleasure S. J., Anderson S., Hevner R., Bagri A., Marin O., Lowenstein D. H., et al. (2000). Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron 28, 727–740 10.1016/S0896-6273(00)00149-5 [DOI] [PubMed] [Google Scholar]
- Porter F. D., Drago J., Xu Y., Cheema S. S., Wassif C., Huang S. P., et al. (1997). Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 124, 2935–2944 [DOI] [PubMed] [Google Scholar]
- Puelles L. (2014). Development and evolution of the claustrum, in The Claustrum. Structural, Functional, and Clinical Neuroscience, eds Smythies J. R., Edelstein L. R., Ramachandran V. S. (Amsterdam: Elsevier-Academic Press; ), 119–176 [Google Scholar]
- Puelles L., Ferran J. L. (2012). Concept of neural genoarchitecture and its genomic fundament. Front. Neuroanat. 6:47 10.3389/fnana.2012.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L., Kuwana E., Puelles E., Bulfone A., Shimamura K., Keleher J., et al. (2000). Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J. Comp. Neurol. 424, 409–438 [DOI] [PubMed] [Google Scholar]
- Puelles L., Martinez de-la-Torre M., Paxinos G., Watson C., Martinez S. (2007). The Chick Brain in Stereotaxic Coordinates. An Atlas featuring Neuromeric Subdivisions and Mammalian Homologies. Amsterdam: Elsevier-Academic Press [Google Scholar]
- Puelles L., Medina L. (2002). Field homology as a way to reconcile genetic and developmental variability with adult homology. Brain Res. Bull. 57, 243–255 10.1016/S0361-9230(01)00693-1 [DOI] [PubMed] [Google Scholar]
- Redies C., Medina L., Puelles L. (2001). Cadherin expression by embryonic divisions and derived gray matter structures in the telencephalon of the chicken. J. Comp. Neurol. 438, 253–285 10.1002/cne.1315 [DOI] [PubMed] [Google Scholar]
- Reiner A., Karten H. J. (1985). Comparison of olfactory bulb projections in pigeons and turtles. Brain Behav. Evol. 27, 11–27 10.1159/000118717 [DOI] [PubMed] [Google Scholar]
- Reiner A., Perkel D. J., Bruce L. L., Butler A. B., Csillag A., Kuenzel W., et al. (2004). Revised nomenclature for avian telencephalon and some related brainstem nuclei. J. Comp. Neurol. 473, 377–414 10.1002/cne.20118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rétaux S., Rogard M., Bach I., Failli V., Besson M. J. (1999). Lhx9: a novel LIM-homeodomain gene expressed in the developing forebrain. J. Neurosci. 19, 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickmann M., Amaral D. G., Cowan W. M. (1987). Organization of radial glial cells during the development of the rat dentate gyrus. J. Comp. Neurol. 264, 449–479 10.1002/cne.902640403 [DOI] [PubMed] [Google Scholar]
- Rodríguez F., López J. C., Vargas J. P., Broglio C., Gómez Y., Salas C. (2002). Spatial memory and hippocampal pallium through vertebrate evolution: insights from reptiles and teleost fish. Brain Res. Bull. 57, 499–503 10.1016/S0361-9230(01)00682-7 [DOI] [PubMed] [Google Scholar]
- Room P., Groenewegen H. J., Lohman A. H. (1984). Inputs from the olfactory bulb and olfactory cortex to the entorhinal cortex in the cat. I. Anatomical observations. Exp. Brain Res. 56, 488–496 10.1007/BF00237989 [DOI] [PubMed] [Google Scholar]
- Rubin A. N., Kessaris N. (2013). PROX1: a lineage tracer for cortical interneurons originating in the lateral/caudal ganglionic eminence and preoptic area. PLoS ONE 18:e77339 10.1371/journal.pone.0077339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewards T. V., Sewards M. A. (2003). Input and output stations of the entorhinal cortex: superficial vs. deep layers or lateral vs. medial divisions? Brain Res. Brain Res. Rev. 42, 243–251 10.1016/S0165-0173(03)00175-9 [DOI] [PubMed] [Google Scholar]
- Sherry D. F., Jacobs L. F., Gaulin S. J. (1992). Spatial memory and adaptive specialization of the hippocampus. Trends Neurosci. 15, 298–303 10.1016/0166-2236(92)90080-R [DOI] [PubMed] [Google Scholar]
- Shimogori T., VanSant J., Paik E., Grove E. A. (2004). Members of the Wnt, Fz, and Frp gene families expressed in postnatal mouse cerebral cortex. J. Comp. Neurol. 473, 496–510 10.1002/cne.20135 [DOI] [PubMed] [Google Scholar]
- Siegel J. J., Nitz D., Bingman V. P. (2000). Hippocampal theta rhythm in awake, freely moving homing pigeons. Hippocampus 10, 627–631 [DOI] [PubMed] [Google Scholar]
- Stouffer E. M., Klein J. E. (2013). Lesions of the lateral entorhinal cortex disrupt non-spatial latent learning but spare spatial latent learning in the rat (Rattus norvegicus). Acta Neurobiol. Exp. (Wars) 73, 430–437 [DOI] [PubMed] [Google Scholar]
- Striedter G. F. (2005). Principles of Brain Evolution. Sunderland, MA: Sinauer Associates Inc [Google Scholar]
- Suárez J., Dávila J. C., Real M. A., Guirado S., Medina L. (2006). Calcium-binding proteins, neuronal nitric oxide synthase, and GABA help to distinguish different pallial areas in the developing and adult chicken. I. Hippocampal formation and hyperpallium. J. Comp. Neurol. 497, 751–771 10.1002/cne.21004 [DOI] [PubMed] [Google Scholar]
- Szabadics J., Varga C., Brunner J., Chen K., Soltesz I. (2010). Granule cells in the CA3 area. J. Neurosci. 30, 8296–8307 10.1523/JNEUROSCI.5602-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely A. D. (1999). The avian hippocampal formation: subdivisions and connectivity. Behav. Brain Res. 98, 219–225 10.1016/S0166-4328(98)00087-4 [DOI] [PubMed] [Google Scholar]
- Székely A. D., Krebs J. R. (1996). Efferent connectivity of the hippocampal formation of the zebra finch (Taenopygia guttata): an anterograde pathway tracing study using Phaseolus vulgaris leucoagglutinin. J. Comp. Neurol. 368, 198–214 [DOI] [PubMed] [Google Scholar]
- Tanninen S. E., Morrissey M. D., Takehara-Nishiuchi K. (2013). Unilateral lateral entorhinal inactivation impairs memory expression in trace eyeblink conditioning. PLoS ONE 8:e84543 10.1371/journal.pone.0084543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tömböl T., Davies D. C., Németh A., Sebestény T., Alpár A. (2000). A comparative Golgi study of chicken (Gallus domesticus) and homing pigeon (Columba livia) hippocampus. Anat. Embryol. (Berl.) 201, 85–101 10.1007/PL00008235 [DOI] [PubMed] [Google Scholar]
- Tzchori I., Day T. F., Carolan P. J., Zhao Y., Wassif C. A., Li L., et al. (2009). LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136, 1375–1385 10.1242/dev.026476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Nallar L., Inestrosa N. C. (2013). Wnt signaling in the regulation of adult hippocampal neurogenesis. Front. Cell. Neurosci. 7:100 10.3389/fncel.2013.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenman C. L., Wild J. M., Reiner A. (1995). Organization of the avian “corticostriatal” projection system: a retrograde and anterograde pathway tracing study in pigeons. J. Comp. Neurol. 354, 87–126 10.1002/cne.903540108 [DOI] [PubMed] [Google Scholar]
- Vivar C., van Praag H. (2013). Functional circuits of new neurons in the dentate gyrus. Front. Neural Circuits 7:15 10.3389/fncir.2013.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas A., Saha B., Lai E., Tole S. (2003). Paleocortex is specified in mice in which dorsal telencephalic patterning is severely disrupted. J. Comp. Neurol. 466, 545–553 10.1002/cne.10900 [DOI] [PubMed] [Google Scholar]
- Wang Q., Gao E., Burkhalter A. (2011). Gateways of ventral and dorsal streams in mouse visual cortex. J. Neurosci. 31, 1905–1918 10.1523/JNEUROSCI.3488-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Mayer U., Bischof H. J. (2011). Visual Wulst analyses “where” and entopallium analyses “what” in the zebra finch visual system. Behav. Brain Res. 222, 51–56 10.1016/j.bbr.2011.03.035 [DOI] [PubMed] [Google Scholar]
- Witter M. P. (2012). Hippocampus, in The Mouse Nervous System, eds Watson C., Paxinos G., Puelles L. (Amsterdam: Elsevier-Academic Press; ), 112–139 [Google Scholar]
- Witter M. P., Amaral D. G. (2004). Hippocampal formation, in The Rat Nervous System, 3rd Edn, ed Paxinos G. (Amsterdam: Elsevier-Academic Press; ), 635–704 [Google Scholar]
- Zardoya R., Meyer A. (1998). Complete mitochondrial genome suggests diapsid affinities of turtles. Proc. Natl. Acad. Sci. U.S.A. 95, 14226–14231 10.1073/pnas.95.24.14226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. J., Ye J., Couey J. J., Witter M., Moser E. I., Moser M. B. (2013). Functional connectivity of the entorhinal-hippocampal space circuit. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20120516 10.1098/rstb.2012.0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Sheng H. Z., Amini R., Grinberg A., Lee E., Huang S., et al. (1999). Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science 284, 1155–1158 10.1126/science.284.5417.1155 [DOI] [PubMed] [Google Scholar]
- Zhou C. J., Zhao C., Pleasure S. J. (2004). Wnt signaling mutants have decreased dentate granule cell production and radial glial scaffolding abnormalities. J. Neurosci. 24, 121–126 10.1523/JNEUROSCI.4071-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.