The multi-component mTORC1 complex integrates growth factor and nutrient signals to positively regulate protein synthesis and cell growth through its two major substrates, S6K1 and 4EBP1.1,2 Growth factors, such as insulin, activate mTORC1 through a well established signaling pathway involving the class 1A PI-3 Kinase, Akt, the TSC1/2 complex, and the GTPase Rheb, which binds to and directly activates mTOR.3 Amino acids, on the other hand signal to mTORC1 through mechanisms that are less well understood. A heterodimeric GTPase complex consisting of Rag A/B and Rag C/D was identified as playing an essential role in relaying amino acid signals to mTORC1.4,5 The Rag complex binds to Raptor, the defining component of mTORC1, but unlike Rheb, does not directly stimulate mTORC1 activity.4 Instead, the Rag complex is required for the amino acid-stimulated translocation of mTORC1 to a late endosomal compartment, presumably to facilitate interactions with Rheb.
We have directly examined the role of the endocytic system in mTORC1 signaling, and find that the compartmental integrity of the late endosome is critical for the ability of mTORC1 to respond to amino acids.6 We found that overexpression of constitutively active Rab5 (Rab5 CA) markedly inhibited both insulin and amino acid activation of mTORC1.6 This inhibition was not a general effect of disrupted endocytic trafficking, since specific inhibition of other endocytic steps (clathrin mediated endocytosis, receptor recycling, endosome to trans-Golgi recycling, multivesicular body biogenesis and late endosomal-lysosomal fusion) had no effect on mTORC1 activation by insulin. Instead, the inhibition was due to the fact that constitutively active Rab5CA disrupts early to late endosomal conversion, leading to the formation of hybrid early/late endosomal structures.7 A similar inhibition of mTORC1 signaling was caused by siRNA knockdown of the Rab7 GEF hVps39, which also produces hybrid early/late endosomes. In cells expressing Rab5CA or in which hVps39 expression is suppressed, amino acids stimulate the translocation of mTORC1 to these hybrid endosomes, but mTORC1 cannot signal.
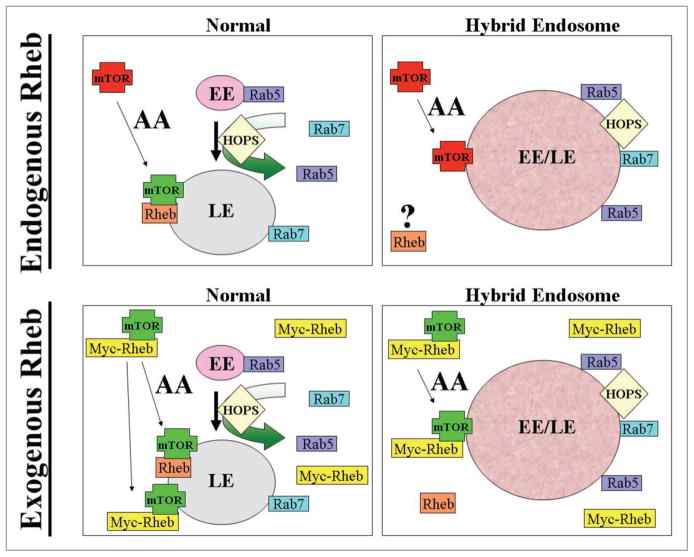
A clue to the mechanism for defective mTORC1 signaling from hybrid endosomes comes from experiments in which Rheb activation was manipulated. Sancak and coworkers had previously suggested that Rheb localizes to late endosomes, and that amino acid-stimulated mTORC1 translocation facilitates mTOR-Rheb interactions.4 We find that in TSC2−/− cells overexpressing Rab5CA, endogenous Rheb is hyperactivated but still cannot activate mTORC1. However, overexpression of exogenous Rheb fully rescues mTORC1 inhibition by Rab5CA. Since overexpressed Rheb can activate mTORC1 even in starved cells,4,8 it presumably finds mTORC1 by mass action and does not need endosomal targeting. Our data suggest that the hybrid early/late endosomes produced by overexpression of Rab5CA or knockdown of hVps39 cannot support mTORC1 signaling due to reduced mTOR-Rheb interactions (Fig. 1).
Figure 1.
Role of the late endosome in regulating mTOR-Rheb interactions. In upper left, a normal cell undergoes early to late endosomal conversion through the action of Rab5 and the HOPS complex. Amino acids can drive the localization of mTOR from cytosol to late endosomes, where mTOR has been suggested to interact with Rheb. In lower left, overexpression of exogenous Rheb leads to Rheb-mTOR interactions even in the absence of amino acids. This activates mTORC1 in an amino acid- and localization-independent manner. In the upper right panel, hybrid early/late endosomes have been formed by Rab5 CA overexpression or hVps39 knockdown. mTOR can still translocate to the hybrid endosomes under amino acid stimulated conditions, but this compartment cannot support mTORC1 signaling, most likely due to reduced Rheb-mTOR interactions. In the bottom right panel, overexpression of exogenous Rheb bypasses the requirement for late endosomes and drives Rheb-mTOR interactions driven by mass action.
Previous work has strongly suggested links between mTOR and the endocytic system. TOR has been localized to endocytic membranes in yeast, fly and in mammalian cell culture.4,9,10 Several studies show that TOR regulates nutrient uptake through effects on endocytic trafficking. In yeast, the Rag homologues, Gtr1p and Gtr2p, together with two vacuole associated proteins, Ego1 and Ego3, form a complex, EGOC, that mediates the nutrient driven movement of amino acid permeases from endosomes to plasma membrane.11 Similarly, in Drosophila, dTOR regulates the endocytic trafficking of nutrient transporters.12 Additional data in yeast show that the endocytic system may regulate TOR. Yeast EGOC is required for re-initiation of cell growth following arrest with rapamycin treatment. 13 The homotypic fusion and vacuole protein sorting (HOPS) complex is required for amino acid homeostasis and nutrient regulated TOR signaling.14 Nutrient dependent TOR signaling to Gln3, a cell growth inhibitory transcription factor, occurs on endocytic membranes and requires Golgi-to-endosome trafficking.15 Finally, the HOPS member Vps39/Vam6 has been recently shown to act as a GEF for Gtr1p, potentially linking the HOPS and EGOC complexes in nutrient regulation of TOR signaling.16
It is not yet clear why the late endosome is required for mTORC1 signaling. The hybrid endosomes present in Rab5CA cells may disrupt Rheb-mTOR interactions by altering Rheb targeting or retention time at late endosomes. Alternatively, changes in endosomal composition might lead to conformational changes in endosomal proteins that facilitate mTOR-Rheb interactions. Late endosomes maintain a unique lipid composition, which includes cholesterol levels that are significantly higher than in the ER.17 Late endosomal cholesterol levels are regulated by a unique lipid, lysobisphosphatitic acid (LBPA).18,19 In addition, late endosomes maintain a lower intraluminal pH than early endosomes.20–22 Maintenance of the lipid composition and/or intraluminal pH of the late endosome might be important in the regulation of mTORC1 signaling. Resolving how late endosomes relay amino acid signals to mTORC1 will greatly enhance our understanding of nutrient regulation of mTORC1 signaling and might have implications for therapeutic intervention on the mTORC1 signaling pathway.
Acknowledgments
This work was supported by NIH grant DK07069 and a grant from the Janey Fund (J.M.B.), T32 GM07491 (R.J.F.), and the Diabetes Research Center (DK020541) and Cancer Center (P30 CA013330) at Albert Einstein College of Medicine.
References
- 1.Sarbassov DD, et al. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Hay N, et al. Genes Dev. 2004;18:1926–45. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 3.Avruch J, et al. Oncogene. 2006;25:6361–72. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- 4.Sancak Y, et al. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim E, et al. Nat Cell Biol. 2008;10:935–45. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flinn RJ, et al. Mol Biol Cell. 2010;21:833–41. doi: 10.1091/mbc.E09-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rink J, et al. Cell. 2005;122:735–49. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 8.Tee AR, et al. Curr Biol. 2003;13:1259–68. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 9.Kunz J, et al. J Biol Chem. 2000;275:37011–20. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- 10.Wedaman KP, et al. Mol Biol Cell. 2003;14:1204–20. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao M, et al. Nat Cell Biol. 2006;8:657–67. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 12.Hennig KM, et al. J Cell Biol. 2006;173:963–74. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubouloz F, et al. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Zurita-Martinez SA, et al. Genetics. 2007;176:2139–50. doi: 10.1534/genetics.107.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puria R, et al. Proc Natl Acad Sci USA. 2008;105:7194–9. doi: 10.1073/pnas.0801087105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binda M, et al. Mol Cell. 2009;35:563–73. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 17.Mesmin B, et al. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbalip.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo H, et al. Science. 2004;303:531–4. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 19.Chevallier J, et al. J Biol Chem. 2008;283:27871–80. doi: 10.1074/jbc.M801463200. [DOI] [PubMed] [Google Scholar]
- 20.Murphy RF, et al. J Cell Biol. 1984;98:1757–62. doi: 10.1083/jcb.98.5.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashiro DJ, et al. J Cell Biol. 1987;105:2723–33. doi: 10.1083/jcb.105.6.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashiro DJ, et al. J Cell Biol. 1987;105:2713–21. doi: 10.1083/jcb.105.6.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]