Abstract
Increased height is a known independent risk factor for atrial fibrillation (AF). However, whether genetic determinants of height influence risk is uncertain. In this candidate gene study, we examined the association of 209 height-associated single-nucleotide polymorphisms (SNPs) with incident AF in 3,309 persons of European descent from the Cardiovascular Health Study, a prospective cohort study of older adults (aged ≥65 years) enrolled in 1989–1990. After a median follow-up period of 13.2 years, 879 participants developed incident AF. The height-associated SNPs together explained approximately 10% of the variation in height (P = 6.0 × 10−8). Using an unweighted genetic height score, we found a nonsignificant association with risk of AF (per allele, hazard ratio = 1.01, 95% confidence interval: 1.00, 1.02; P = 0.06). In weighted analyses, we found that genetically predicted height was strongly associated with AF risk (per 10 cm, hazard ratio = 1.30, 95% confidence interval: 1.03, 1.64; P = 0.03). Importantly, for all models, the inclusion of actual height completely attenuated the genetic height effect. Finally, we identified 1 nonsynonymous SNP (rs1046934) that was independently associated with AF and may warrant future study. In conclusion, we found that genetic determinants of height appear to increase the risk of AF, primarily via height itself. This approach of examining SNPs associated with an intermediate phenotype should be considered as a method for identifying novel genetic targets.
Keywords: atrial fibrillation, cardiovascular disease, genetics, risk factors, risk prediction
Atrial fibrillation (AF) is the most common type of sustained arrhythmia (1), and it has a particular impact in older adults (2). Given its strong relationship to age, AF has often been regarded as a nongenetic disease; however, recent investigations suggest that a genetic component may exist as well. A positive family history has been observed in 15% of patients with lone AF and 5% of all patients with AF (3), and the risk of AF is increased up to 3-fold in persons with at least 1 family member with AF (4–6). A number of gene mutations have been linked to adult-onset AF, with most involving ion channels (7–9), although some have been associated with atrial structure, inflammation, and neurohumoral pathways (10–14).
Although both candidate gene studies and unbiased approaches, such as genome-wide association studies, have identified single-nucleotide polymorphisms (SNPs) associated with risk of AF (14), other approaches may provide additional insight. For example, hybrid approaches based on combining unbiased evaluations (in the form of genetic variants) with a continuously distributed intermediate phenotype linked to a given clinical outcome have the potential to identify novel pathways of association, and thus novel disease mechanisms. Such a “candidate SNP” approach has been used previously to demonstrate the association between polymorphisms associated with low-density lipoprotein cholesterol and risk of myocardial infarction (15).
For AF, height may serve as an attractive intermediate variable. The association between increased height and AF has been observed in a wide range of cohorts. Height was recently examined in the Cardiovascular Health Study (CHS), a cohort study of persons aged 65 years or older, and it was found that even after adjustment for clinical risk factors, as well as atrial and ventricular size, height was independently associated with both incident and prevalent AF (16). A similar association has been observed in other studies (17–24). The possibility that genetically determined height might be associated with AF is attractive, given recent findings from genome-wide association studies that loci associated with increased AF risk, such as those in the paired-like homeodomain 2 gene (PITX2) (25) and the zinc-finger homeobox 3 gene (ZFHX3) (26), are also associated with growth pathways (25–27).
A number of genome-wide association studies have identified genes associated with height (28–34). The largest of these to date was performed in 183,727 people of European ancestry, and it identified 180 loci associated with height (34). To date, no AF-associated loci from genome-wide association studies of AF have been found to also have an association with height or vice versa, although no study has attempted to find a common variant for the two. Such a finding would be interesting, both because height-associated SNPs represent a set of variants that are enriched in plausible candidates and because they may point to specific causal pathways shared by height and AF.
In this study, we examined the role of height-associated genetic variants as risk factors for AF in a well-characterized cohort of older adults. We first examined whether previously identified genetic predictors of height were associated with the risk of AF through creation of a genetic height score. We then used a candidate gene approach with height-associated SNPs to identify individual SNPs that might have pleotropic effects on AF risk.
METHODS
Population
The design and objectives of the CHS have been previously described (35). In brief, the CHS is a longitudinal study of men and women aged 65 years or older who were randomly selected from Medicare lists in Pittsburgh, Pennsylvania; Forsyth County, North Carolina; Sacramento, California; and Hagerstown, Maryland. The original cohort of 5,201 predominantly European-American participants was enrolled in 1989–1990; a second cohort of 687 predominantly African Americans was recruited in 1992–1993. The institutional review board at each study center approved the protocol, and each participant gave informed consent.
The baseline examination included a standardized questionnaire assessing a variety of risk factors, including smoking, alcohol intake, history of stroke, coronary heart disease, and heart failure, self-reported health status, and medication use upon enrollment. Methods of determining prevalent cardiovascular disease have previously been validated (36). The physical examination included measurements of standing height, weight, and seated blood pressure (measured with a random-zero sphygmomanometer) (36), as well as a resting 12-lead electrocardiogram. Fasting laboratory measurements included total cholesterol, high-density lipoprotein cholesterol, glucose, C-reactive protein, and serum creatinine (37).
Determination of incident AF
Participants were contacted every 6 months for follow-up, alternating between a telephone interview and a clinic visit until 1999 and by telephone interview only after that. An annual resting electrocardiogram was obtained yearly until 1999, and discharge diagnoses for all hospitalizations continue to be collected. We identified cases of AF in 2 ways. Annual study electrocardiograms were interpreted by staff of the Epidemiology Coordinating and Research (EPICORE) Centre at the University of Alberta (Edmonton, Alberta, Canada), where the diagnosis of AF or atrial flutter was verified (24). Hospital discharge diagnoses that included codes for AF and atrial flutter were also included, although AF or flutter diagnoses that were made during the same hospitalization as coronary artery bypass surgery or heart valve surgery were not counted. In a prior evaluation in the CHS, the positive predictive value of hospital discharge diagnoses was determined to be 98.6% for diagnosis of AF (24), and a Holter substudy identified that only 1 in 819 subjects (0.1%) had persistent or intermittent AF not identified by the above measures (38).
Of the original cohort of 5,201 individuals, 3,388 persons of European ancestry were successfully genotyped for the analysis, as previously described (39). We excluded 71 participants with prevalent AF and 8 who were missing standing height measurements, leaving a study sample of 3,309 European Americans. A total of 607 persons from both the original cohort and the later African-ancestry cohort underwent successful genotyping. We excluded 10 persons with prevalent AF, leaving a study sample of 597 African Americans. For all analyses, we excluded participants who did not provide written informed consent for DNA testing or for whom DNA was not available.
Height-related polymorphisms
Prior studies have found height-associated loci explaining 0.3% (29) to 20% (34) of the heritable variation in height. We compiled a total of 289 SNPs associated with height from 10 previous studies (28–34, 40, 41), with the majority (n = 180) coming from the Genetic Investigation of Anthropometric Traits (GIANT) Consortium (34) and the second-greatest number (n = 64) coming from the meta-analysis by Lanktree et al. (41). Across these 10 studies, we found 14 duplicate SNPs. Web Table 1 (available at http://aje.oxfordjournals.org/) lists the 289 SNPs from these studies, including the effect allele, minor allele frequency, and reported effect size on predicted height for each SNP.
To minimize linkage disequilibrium and multiple testing among SNPs, we a priori pruned 79 SNPs within 1 megabase (1 × 106 base pairs) of another height-related SNP, leaving 210 SNPs. One SNP was not genotyped in the CHS population and was excluded, leaving a total of 209 height-associated SNPs for analysis (see Web Table 1).
Analysis
We imputed data for SNPs and missing covariates as previously described (42, 43). The primary analysis was performed among persons of European ancestry, with the African-ancestry cohort combined with them as a partial validation cohort (see below). First, we reconfirmed the association of height in 10-cm increments with risk of incident AF in Cox proportional hazards regression models with age- and sex-specific hazard functions, as previously described (16). We then performed linear regression with all 209 SNPs and height to examine the total height variance explained by the 209 SNPs. As an initial approach to avoiding issues associated with overfitting of effect measures derived from the same population, we examined the effect of genetically predicted height on risk of incident AF by creating an unweighted genetic height score. We created the score by summing the number of effect alleles associated with increased height at each locus in an additive model. We confirmed the association between this unweighted height score and measured height using linear regression and then examined the association between the unweighted height score and risk of AF alone and with inclusion of measured height as a covariate. We examined the improvement in the model with inclusion of genetic information, as well as by calculating Akaike's Information Criterion.
We then performed an instrumental variable analysis, in which we used a logistic regression model with incident AF as the outcome and height score as an instrument for measured height, with adjustment for age and sex and robust standard errors (the “qvf” procedure in Stata 12; StataCorp LP, College Station, Texas). Using these models, we calculated the odds ratio for a 10-cm increment in genetically predicted height. We compared associations of incident AF with genetic and predicted height using Wald tests from similar probit models (the “ivprobit” procedure in Stata 12).
We then created a weighted genetic height model based on the effect sizes in the CHS and examined this model in the same manner as the unweighted model above, using age- and sex-specific hazard functions, with and without inclusion of actual height. We also repeated the instrumental variable analysis as above on the weighted genetic height model.
We then examined all 209 loci together and performed a global test of the overall association between height alleles and AF risk. From among these SNPs, we then examined those from the 209 candidate SNPs that met experiment-wide significance criteria using Bonferroni correction (P < 0.00024 (0.05/209)) and confirmed Hardy-Weinberg equilibrium using a χ2 test, with violation set at P < 0.05. As a form of internal validation, we evaluated the individual SNPs among African-American participants and in the combined cohort. We also checked for correlation between rs1046934 and the other SNPs in the CHS using Spearman coefficients, with adjustment for multiple testing, and we checked for linkage disequilibrium of this SNP with those in the 1,000 Genomes Project data set using the online SNP Annotation and Proxy Search (SNAP) tool (Broad Institute, Cambridge, Massachusetts (http://www.broadinstitute.org/mpg/snap/)) (44). SAS, version 9.2 (SAS Institute, Inc., Cary, North Carolina), was used for all analyses unless otherwise stated.
RESULTS
Table 1 shows baseline population characteristics. Web Table 2 also displays baseline characteristics according to incident AF. The median follow-up time was 13.2 years (range, 0.05–18.0 years), during which time 879 of the 3,309 persons of self-reported European descent developed incident AF. Using a Cox regression model stratified by age and sex, we confirmed the prior finding in the CHS that height was significantly associated with the risk of incident AF (per 10 cm, hazard ratio (HR) = 1.38, 95% confidence interval (CI): 1.24, 1.54; P < 0.0001).
Table 1.
Baseline Characteristics of Participants by Race/Ethnicity, Cardiovascular Health Study, 1989–1990
| European Americans (n = 3,309) |
African Americans (n = 597) |
|||||
|---|---|---|---|---|---|---|
| Mean (SD) | No. | % | Mean (SD) | No. | % | |
| Age, years | 72.3 (5.4) | 72.7 (5.6) | ||||
| Female sex | 2,007 | 61 | 382 | 64 | ||
| Height, cm | 164.6 (9.3) | 164.5 (9.2) | ||||
| Body mass indexa | 26.4 (4.5) | 28.6 (5.5) | ||||
| Use of antihypertension medication | 1,196 | 36 | 264 | 44 | ||
| History of coronary heart disease | 68 | 2 | 20 | 3 | ||
| History of cerebrovascular accident or stroke | 8 | 0.2 | 5 | 0.8 | ||
| Smoking status | ||||||
| Current smoker | 371 | 11 | 98 | 16 | ||
| Prior smoker | 1,360 | 41 | 217 | 36 | ||
| Never smoker | 1,578 | 48 | 282 | 47 | ||
| Diabetes mellitus | 401 | 12 | 129 | 22 | ||
| Impaired fasting glucose level | 398 | 12 | 75 | 13 | ||
| Systolic blood pressure, mm Hg | 135.1 (21.0) | 141.7 (22.6) | ||||
| Creatinine concentration, mg/dL | 1.01 (0.28) | 1.07 (0.32) | ||||
| C-reactive protein concentration, mg/L | 3.1 (5.6) | 4.4 (6.1) | ||||
| Incident atrial fibrillation | 879 | 27 | 101 | 17 | ||
Abbreviation: SD, standard deviation.
a Weight (kg)/height (m)2.
We first performed a regression with all 209 SNPs on adult height, and we found that collectively they accounted for 10.0% of the variability in height (P = 6.0 × 10−8). We then examined a Cox regression model stratified by age and sex that included all 209 SNPs and evaluated this model with and without the inclusion of adult height. Since only 10% of the variance in height was explained by our genetic model, we found that the effect of adult height itself was minimally attenuated by inclusion of the 209 SNPs in the model (for height alone in the model, HR per 10 cm = 1.38, 95% CI: 1.24, 1.54 (P = 1.13 × 10−8); for height with all 209 SNPs included, HR per 10 cm = 1.38, 95% CI: 1.22, 1.55 (P = 1.35× 10−7)). Overall, inclusion of all 209 SNPs together significantly improved the model (Akaike's Information Criterion increased from 6,993 to 7,135; P = 0.001).
We then created an unweighted genetic additive height score using the 209 SNPs to predict height, with a predicted effect of 0.17 cm (standard error, 0.01 cm) per effect allele (P = 1.04 × 10−48). Using this model, we found that there was a nonsignificant association with the risk of developing AF, with a hazard ratio per associated allele of 1.01 (95% CI: 1.00, 1.02; P = 0.06). Inclusion of body mass index (weight (kg)/height (m)2) in the model did not change the estimates. This association was completely null with inclusion of actual height in the model (HR = 1.00, 95% CI: 0.99, 1.01; P = 0.65).
We examined the association of this unweighted genetic height as an instrument score with risk of AF and confirmed that there was a similar but nonsignificant effect, with a hazard ratio of 1.52 per 10 cm of predicted height (95% CI: 0.98, 2.36; P = 0.06). We then used this same unweighted model in a formal instrumental variable analysis, in which genetically predicted height had a comparable odds ratio for association with incident AF (odds ratio = 1.43, 95% CI: 0.86, 2.38; P = 0.16), albeit with wide confidence intervals and thus a nonsignficant P value. We did not find evidence that the genetic and measured associations of height with AF differed (P = 0.91).
We then examined the association of AF with predicted height based upon the 209 SNPs, using their observed associations with measured height within the CHS. In this weighted model, we found a significant association between genetically predicted height and risk of AF, with a hazard ratio per 10 cm of 1.30 (95% CI: 1.03, 1.64; P = 0.03). As shown in Figure 1, there was a linear association between genetically predicted height and the risk of AF. The weighted genetic height score was no longer significantly associated with height when measured height was included in the model, which itself remained significantly associated with risk of AF (for weighted genetic height score, HR per 10 cm = 1.00, 95% CI: 0.80, 1.32 (P = 0.85); for measured height, HR per 10 cm = 1.38, 95% CI: 1.22, 1.55 (P < 0.0001)). In the instrumental variable analysis, we found that the hazard ratio for a 10-cm increment in height was 1.61 (95% CI: 1.23, 2.12; P < 0.001), while the difference from the actual effect of height was also not significant at P = 0.45.
Figure 1.
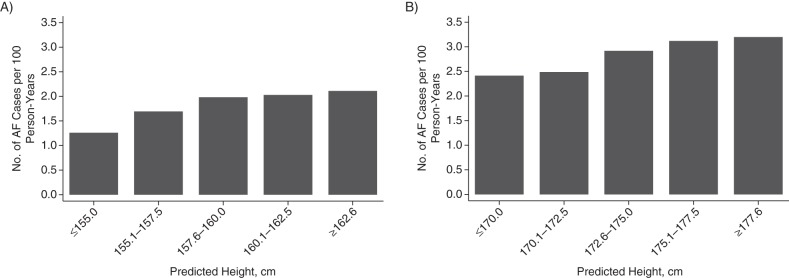
Association between genetically predicted height and risk of atrial fibrillation (AF) among European-American women (A) and men (B) in the Cardiovascular Health Study, 1989–1990. Genetically predicted height for European Americans was based on predicted height using 209 candidate single-nucleotide polymorphisms (see text for details). There were a total of 39,815.5 person-years at risk. Number of cases/total number of participants in each height subgroup: women—≤155.0 cm, 26/157; 155.1–157.5 cm, 94/434; 157.6–160.0 cm, 170/681; 160.1–162.5 cm, 133/517; ≥162.6 cm, 59/218; men—≤170.0 cm, 63/227; 170.1–172.5 cm, 86/312; 172.6–175.0 cm, 121/387; 175.1–177.5 cm, 91/269; ≥177.6 cm, 36/107.
Only 1 SNP was associated with AF after Bonferroni correction for multiple testing, when included in a model with all other SNPs: rs1046934 (Web Table 3). We checked for correlation between rs1046934 and the other 208 SNPs in the CHS, and none were correlated at the Bonferroni-adjusted significance level (P < 0.00024). In the CHS cohort of European Americans, the minor allele frequency for rs1046934 was 0.33, which is similar to what has been observed in persons of European ancestry generally (45) (A/A: 46%; A/C: 43%; C/C: 11% (Table 2)). This SNP did not violate Hardy-Weinberg equilibrium (P = 0.45). When results were modeled separately by zygosity, the hazard ratio for heterozygotes was 1.18 (95% CI: 1.02, 1.37; P = 0.02) and that for homozygotes was 1.46 (95% CI: 1.18, 1.80; P = 0.0004). When we adjusted for clinical risk factors for AF, we observed no attenuation of the risk estimates for rs1046934 in the risk of AF (per C allele, HR = 1.21, 95% CI: 1.10, 1.34 (P = 0.0001); Web Table 4).
Table 2.
Baseline Characteristics of European-American Participants by rs1046934 Genotype,a Cardiovascular Health Study, 1989–1990
| rs1046934 Genotype |
P Value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A/A |
A/C |
C/C |
||||||||
| Mean (SD) | No. | % | Mean (SD) | No. | % | Mean (SD) | No. | % | ||
| Total | 1,515 | 46 | 1,434 | 43 | 360 | 11 | ||||
| Demographic characteristic | ||||||||||
| Age, years | 72.4 (5.4) | 72.2 (5.3) | 72.5 (5.6) | 0.57 | ||||||
| Female sex | 901 | 59 | 882 | 62 | 224 | 62 | 0.43 | |||
| Standing height, cm | 164.6 (9.2) | 164.5 (9.5) | 165.1 (9.2) | 0.60 | ||||||
| Body mass indexb | 26.5 (4.5) | 26.2 (4.4) | 26.5 (4.6) | 0.37 | ||||||
| Current smoking | 189 | 13 | 145 | 10 | 37 | 10 | 0.32 | |||
| Risk factor | ||||||||||
| Diabetes mellitus | 193 | 13 | 165 | 12 | 43 | 12 | 0.82 | |||
| History of myocardial infarction | 15 | 1 | 10 | 1 | 4 | 1 | 0.61 | |||
| History of cerebrovascular accident or stroke | 1 | 0.1 | 6 | 0.4 | 1 | 0.3 | 0.15 | |||
| Use of antihypertension medication | 557 | 37 | 523 | 36 | 116 | 32 | 0.26 | |||
| Physiological measurement | ||||||||||
| Systolic blood pressure, mm Hg | 135.7 (21.2) | 134.9 (20.8) | 133.6 (21.0) | 0.22 | ||||||
| Left atrial diameter, cm | 3.82 (0.61) | 3.81 (0.64) | 3.85 (0.63) | 0.56 | ||||||
| Creatinine concentration, mg/dL | 1.01 (0.27) | 1.01 (0.27) | 1.01 (0.31) | 0.97 | ||||||
| C-reactive protein concentration, mg/L | 3.02 (4.92) | 3.26 (6.52) | 2.89 (4.31) | 0.36 | ||||||
| Heart rate, beats/minute | 64.5 (9.9) | 64.7 (10.3) | 65 (10.2) | 0.70 | ||||||
| Electrocardiographic interval, ms | ||||||||||
| P-R interval | 168.5 (30.9) | 165.3 (31.6) | 167.4 (31.0) | 0.02 | ||||||
| QRS interval | 91.5 (15.9) | 91.2 (15.4) | 91.6 (15.9) | 0.80 | ||||||
| Q-T interval | 416.5 (33.0) | 416.2 (33.1) | 416.7 (33.1) | 0.96 | ||||||
Abbreviations: A, adenine; C, cytosine; SD, standard deviation.
a A is the major allele; C is the minor allele. The frequency of the minor allele in European Americans is 0.33.
b Weight (kg)/height (m)2.
To validate our results, we examined the effect of this SNP using the separate but smaller cohort of 597 self-reported African Americans, of whom 101 developed AF after a median follow-up period of 12.6 years (range, 0.3–18.0 years). The minor allele frequency for rs1046934 in this population was 0.16, which is similar to what has been reported in other African-ancestry samples (45). The genotype frequencies were 69.9% for A/A (417 individuals), 27.6% for A/C (165 individuals), and 2.5% for C/C (15 individuals; see Table 2). In the combined cohort of European Americans and African Americans, the effect was highly significant; per copy of the rs1046934 C allele, the hazard ratio was 1.23 (95% CI: 1.12, 1.35; P = 1.9 × 10−5). In the African-American cohort alone, there was a similar trend that did not reach statistical significance, with a hazard ratio per C allele of 1.31 (95% CI: 0.88, 1.95; P = 0.19).
We examined rs1046934 against the 1,000 Genomes Project database using the SNAP tool available through the Broad Institute (44) and found that 2 additional SNPs are in complete linkage disequilibrium (r2 = 1) with rs1046934: rs2274432 and rs1926872. The latter SNP is in a noncoding region approximately 5,000 base pairs upstream from rs1046934, and the former (rs2274432) is also a nonsynonymous substitution in the coding region of the tRNA-splicing endonuclease 15 gene (TSEN15), about 2,500 base pairs upstream from rs1046934. The variant has a minor allele (A) with a frequency similar to that of rs1046934 (approximately 31%).
DISCUSSION
In this large prospective cohort study, we found that genetic predictors of height were associated with the risk of incident AF, confirming that height itself may play a role in increasing the risk of incident AF, rather than being a marker of pleiotropic factors. We demonstrated this finding by first confirming that the candidate genes selected were associated with height and that the genetic height score itself was associated with risk of AF. We then demonstrated that, as it should, the risk of AF associated with genetically predicted height was completely attenuated with inclusion of adult height in the model, indicating that height (at least in this analysis) was not merely a marker of genetic pleiotropy but was likely causal, at least in part. At the same time, we used this enriched sample of SNPs to identify 1 SNP that was significantly associated with AF risk, even after Bonferroni experiment-wide correction and adjustment for height—indicating that this approach may hold promise for identification of novel genes with pleiotropic effects on AF risk based on other intermediate phenotypes.
In our previous study, despite extensive adjustment for multiple potential clinical mediators, including left atrial size, left ventricular mass, and parameters of diastolic dysfunction, we were unable to detect any substantial attenuation of the height-associated risk of incident AF (16). Pleiotropy was one potential explanation, in which genes associated with height could also affect processes in the heart unrelated to height to increase the risk of AF—an interesting possible explanation given that independent genetic studies of AF have identified genes associated with growth pathways, such as PITX2 (25) and ZFHX3 (26). These possibilities may still exist, although it is apparent from our findings that whatever the mechanism for growth leading to an AF substrate, it operates in parallel with that leading to increased height for the vast majority of height-related SNPs. One potential area of exploration is the association of height with pulmonary vein anatomy, as the pulmonary veins are a well-described trigger of AF (46) and target for therapy.
At the same time, we also found that 1 locus was associated with risk of AF even after adjustment for height. The SNP rs1046934, which was independently associated with risk of AF in our cohort, has not been described in association with AF previously. rs1046934 is found on chromosome 1 and codes a minor allele (C), which is found in approximately 32.2% of persons of European ancestry (1,000 Genomes Project) (45) but is less frequent in African Americans (approximately 10%) (45). The rs1046934 polymorphism is nonsynonymous and codes for a change from glutamine (Q) to histidine (H) at position 256 (Q256H) of the protein encoded by TSEN15, an important component of tRNA-splicing endonuclease. tRNA splicing is a fundamental process required for cell growth and division, and the TSEN15 protein is highly conserved across vertebrates. This process, which involves the removal of introns from pre-tRNA to form “mature” tRNA, is not as well-described as its more common cousin, mRNA splicing, and its role in human disease in general remains poorly understood. tRNA splicing, specifically the tRNA-splicing endonuclease complex, has been linked with pontocerebellar hypoplasia (47), but associations with cardiac disease have not been previously identified. As a requisite to our investigation, the rs1046934 variant has been associated with increased height in large genome-wide association studies (34), although the mechanism behind this association is unknown. Importantly, 2 additional SNPs were in complete linkage disequilibrium (r2 = 1) with rs1046934 in the 1,000 Genomes data set, one of which (rs2274432) is also a nonsynonymous substitution in the coding region of TSEN15. It remains to be determined whether either of these SNPs is causal for AF; however, they suggest that tRNA splicing may play a previously unrecognized role in creating a substrate for AF development, which deserves further investigation.
Among the limitations of our study was the finding that only about 10% of the total variance in height was explained by the selected candidate SNPs. In studies of over 300,000 SNPs, it has been estimated that genes explain about 45% of the variance in height (48), although studies with smaller numbers of loci (n = 190) have estimated that approximately 10%–20% of the variance in height is explainable by genetics (34). That rare variants might contribute to individual height, as well as to AF risk, is a possibility that will require future investigation as these variants are uncovered. Another limitation was our inability to validate the SNP results in a completely separate cohort, which is especially important given that this particular gene has not been associated with cardiac disease previously, although our top SNPs were at least as strongly numerically associated with risk of AF among African-American participants as among European Americans, and we included a large number of cases in both our European- and African-American cohorts.
In conclusion, in this candidate gene analysis of data from a large prospective cohort study, we found that genetic predictors of increased height were significantly associated with the risk of AF. In addition, we found that a candidate SNP associated with the tRNA-splicing protein gene TSEN15 was independently associated with the risk of AF. Future studies are needed to validate these findings and to examine the role of this particular pathway in development of AF.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: VA Boston Healthcare System, Harvard Medical School, West Roxbury, Massachusetts (Michael A. Rosenberg); Center for Human Genetic Research, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts (Michael A. Rosenberg, Christopher Newton-Cheh); Department of Epidemiology and Population Health, Albert Einstein College of Medicine, Yeshiva University, Bronx, New York (Robert C. Kaplan); Cardiovascular Health Research Unit, Departments of Medicine and Health Services, Schools of Medicine and Public Health, University of Washington, Seattle, Washington (David S. Siscovick, Bruce M. Psaty); Group Health Research Institute, Group Health Cooperative, Seattle, Washington (Bruce M. Psaty); Department of Epidemiology, School of Public Health, University of Washington, Seattle, Washington (David S. Siscovick, Bruce M. Psaty, Susan R. Heckbert); and Department of General Medicine and Primary Care, Beth Israel Deaconess Medical Center, Boston, Massachusetts (Kenneth J. Mukamal).
All authors contributed equally to this work.
This work was funded by contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, and N01HC85086 and grants HL103612, HL080295, HL087652 and HL068986 from the National Heart, Lung, and Blood Institute, with an additional contribution from the National Institute of Neurological Disorders and Stroke. Additional support was provided by grants AG023629 and 1R01AG031890 (to R.C.K.) from the National Institute on Aging. M.A.R. was also supported by the 2013 Postdoctoral Fellowship Award of the Heart Rhythm Society (Washington, DC).
A full list of the principal investigators and institutions in the Cardiovascular Health Study can be found at http://www.chs-nhlbi.org/PI.htm.
Conflict of interest: none declared.
REFERENCES
- 1.Majeed A, Moser K, Carroll K. Trends in the prevalence and management of atrial fibrillation in general practice in England and Wales, 1994–1998: analysis of data from the General Practice Research Database. Heart. 2001;86(3):284–288. doi: 10.1136/heart.86.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Go AS, Hylek EM, Phillips KA, et al. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the AnTicoagulation and Risk Factors in Atrial Fibrillation (ATRIA) study. JAMA. 2001;285(18):2370–2375. doi: 10.1001/jama.285.18.2370. [DOI] [PubMed] [Google Scholar]
- 3.Darbar D, Herron KJ, Ballew JD, et al. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41(12):2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 4.Fox CS, Parise H, D'Agostino RB, Sr, et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291(23):2851–2855. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 5.Ellinor PT, Yoerger DM, Ruskin JN, et al. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118(2):179–184. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 6.Patton KK, Zacks ES, Chang JY, et al. Clinical subtypes of lone atrial fibrillation. Pacing Clin Electrophysiol. 2005;28(7):630–638. doi: 10.1111/j.1540-8159.2005.00161.x. [DOI] [PubMed] [Google Scholar]
- 7.Hong K, Bjerregaard P, Gussak I, et al. Short QT syndrome and atrial fibrillation caused by mutation in KCNH2. J Cardiovasc Electrophysiol. 2005;16(4):394–396. doi: 10.1046/j.1540-8167.2005.40621.x. [DOI] [PubMed] [Google Scholar]
- 8.Xia M, Jin Q, Bendahhou S, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332(4):1012–1019. doi: 10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 9.Olson TM, Alekseev AE, Liu XK, et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15(14):2185–2191. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 10.Fatini C, Sticchi E, Genuardi M, et al. Analysis of minK and eNOS genes as candidate loci for predisposition to non-valvular atrial fibrillation. Eur Heart J. 2006;27(14):1712–1718. doi: 10.1093/eurheartj/ehl087. [DOI] [PubMed] [Google Scholar]
- 11.Fatini C, Sticchi E, Gensini F, et al. Lone and secondary nonvalvular atrial fibrillation: role of a genetic susceptibility. Int J Cardiol. 2007;120(1):59–65. doi: 10.1016/j.ijcard.2006.08.079. [DOI] [PubMed] [Google Scholar]
- 12.Kato K, Oguri M, Hibino T, et al. Genetic factors for lone atrial fibrillation. Int J Mol Med. 2007;19(6):933–939. [PubMed] [Google Scholar]
- 13.Nattel S, Shiroshita-Takeshita A, Cardin S, et al. Mechanisms of atrial remodeling and clinical relevance. Curr Opin Cardiol. 2005;20(1):21–25. [PubMed] [Google Scholar]
- 14.Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44(6):670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg MA, Patton KK, Sotoodehnia N, et al. The impact of height on the risk of atrial fibrillation: the Cardiovascular Health Study. Eur Heart J. 2012;33(21):2709–2717. doi: 10.1093/eurheartj/ehs301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamberlain AM, Agarwal SK, Folsom AR, et al. A clinical risk score for atrial fibrillation in a biracial prospective cohort (from the Atherosclerosis Risk in Communities [ARIC] study) Am J Cardiol. 2011;107(1):85–91. doi: 10.1016/j.amjcard.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnabel RB, Sullivan LM, Levy D, et al. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373(9665):739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki S, Yamashita T, Ohtsuka T, et al. Body size and atrial fibrillation in Japanese outpatients. Circ J. 2010;74(1):66–70. doi: 10.1253/circj.cj-09-0431. [DOI] [PubMed] [Google Scholar]
- 20.Hanna IR, Heeke B, Bush H, et al. The relationship between stature and the prevalence of atrial fibrillation in patients with left ventricular dysfunction. J Am Coll Cardiol. 2006;47(8):1683–1688. doi: 10.1016/j.jacc.2005.11.068. [DOI] [PubMed] [Google Scholar]
- 21.Rosengren A, Hauptman PJ, Lappas G, et al. Big men and atrial fibrillation: effects of body size and weight gain on risk of atrial fibrillation in men. Eur Heart J. 2009;30(9):1113–1120. doi: 10.1093/eurheartj/ehp076. [DOI] [PubMed] [Google Scholar]
- 22.Mont L, Tamborero D, Elosua R, et al. Physical activity, height, and left atrial size are independent risk factors for lone atrial fibrillation in middle-aged healthy individuals. Europace. 2008;10(1):15–20. doi: 10.1093/europace/eum263. [DOI] [PubMed] [Google Scholar]
- 23.Long MJ, Jiang CQ, Lam TH, et al. Atrial fibrillation and obesity among older Chinese: the Guangzhou Biobank Cohort Study. Int J Cardiol. 2011;148(1):48–52. doi: 10.1016/j.ijcard.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Psaty BM, Manolio TA, Kuller LH, et al. Incidence of and risk factors for atrial fibrillation in older adults. Circulation. 1997;96(7):2455–2461. doi: 10.1161/01.cir.96.7.2455. [DOI] [PubMed] [Google Scholar]
- 25.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448(7151):353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 26.Benjamin EJ, Rice KM, Arking DE, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41(8):879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conen D, Tedrow UB, Cook NR, et al. Birth weight is a significant risk factor for incident atrial fibrillation. Circulation. 2010;122(8):764–770. doi: 10.1161/CIRCULATIONAHA.110.947978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weedon MN, Lettre G, Freathy RM, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39(10):1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lettre G, Jackson AU, Gieger C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40(5):584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weedon MN, Lango H, Lindgren CM, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40(5):575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson A, Marroni F, Hayward C, et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum Mol Genet. 2009;18(2):373–380. doi: 10.1093/hmg/ddn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei SF, Tan LJ, Liu XG, et al. Genome-wide association study identifies two novel loci containing FLNB and SBF2 genes underlying stature variation. Hum Mol Genet. 2009;18(9):1661–1669. doi: 10.1093/hmg/ddn405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soranzo N, Rivadeneira F, Chinappen-Horsley U, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5(4):e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1(3):263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 36.Psaty BM, Kuller LH, Bild D, et al. Methods of assessing prevalent cardiovascular disease in the Cardiovascular Health Study. Ann Epidemiol. 1995;5(4):270–277. doi: 10.1016/1047-2797(94)00092-8. [DOI] [PubMed] [Google Scholar]
- 37.Cushman M, Cornell ES, Howard PR, et al. Laboratory methods and quality assurance in the Cardiovascular Health Study. Clin Chem. 1995;41(2):264–270. [PubMed] [Google Scholar]
- 38.Mozaffarian D, Psaty BM, Rimm EB, et al. Fish intake and risk of incident atrial fibrillation. Circulation. 2004;110(4):368–373. doi: 10.1161/01.CIR.0000138154.00779.A5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suchy-Dicey AM, Enquobahrie DA, Heckbert SR, et al. Hemodynamic fluid shear stress response genes and carotid intima-media thickness: a candidate gene association analysis in the Cardiovascular Health Study. Int J Mol Epidemiol Genet. 2012;3(2):174–178. [PMC free article] [PubMed] [Google Scholar]
- 40.Estrada K, Krawczak M, Schreiber S, et al. A genome-wide association study of northwestern Europeans involves the C-type natriuretic peptide signaling pathway in the etiology of human height variation. Hum Mol Genet. 2009;18(18):3516–3524. doi: 10.1093/hmg/ddp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanktree MB, Guo Y, Murtaza M, et al. Meta-analysis of dense genecentric association studies reveals common and uncommon variants associated with height. Am J Hum Genet. 2011;88(1):6–18. doi: 10.1016/j.ajhg.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold AM, Kronmal RA. Multiple imputation of baseline data in the Cardiovascular Health Study. Am J Epidemiol. 2003;157(1):74–84. doi: 10.1093/aje/kwf156. [DOI] [PubMed] [Google Scholar]
- 43.Psaty BM, O'Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson AD, Handsaker RE, Pulit SL, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Center for Biotechnology Information, National Library of Medicine. Bethesda, MD: National Library of Medicine; 2013. Reference SNP (refSNP) Cluster Report: rs1046934. http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=1046934. ). (Accessed March 13, 2013) [Google Scholar]
- 46.Corradi D. Atrial fibrillation from the pathologist's perspective. Cardiovasc Pathol. 2014;23(2):71–84. doi: 10.1016/j.carpath.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Cassandrini D, Biancheri R, Tessa A, et al. Pontocerebellar hypoplasia: clinical, pathologic, and genetic studies. Neurology. 2010;75(16):1459–1464. doi: 10.1212/WNL.0b013e3181f88173. [DOI] [PubMed] [Google Scholar]
- 48.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


