Abstract
A promoter polymorphism of the osteopontin (OPN) gene (rs28357094) has been associated with multiple inflammatory states, severity of Duchenne muscular dystrophy (DMD) and muscle size in healthy young adults. We sought to define the mechanism of action of the polymorphism, using allele-specific in vitro reporter assays in muscle cells, and a genotype-stratified intervention in healthy controls. In vitro reporter constructs showed the G allele to respond to estrogen treatment, whereas the T allele showed no transcriptional response. Young adult volunteers (n = 187) were enrolled into a baseline study, and subjects with specific rs28357094 genotypes enrolled into an eccentric muscle challenge intervention [n = 3 TT; n = 3 GG/GT (dominant inheritance model)]. Female volunteers carrying the G allele showed significantly greater inflammation and increased muscle volume change as determined by magnetic resonance imaging T1- and T2-weighted images after eccentric challenge, as well as greater decrement in biceps muscle force. Our data suggest a model where the G allele enables enhanced activities of upstream enhancer elements due to loss of Sp1 binding at the polymorphic site. This results in significantly greater expression of the pro-inflammatory OPN cytokine during tissue remodeling in response to challenge in G allele carriers, promoting muscle hypertrophy in normal females, but increased damage in DMD patients.
INTRODUCTION
Genetic association studies are a powerful tool to identify regions of the human genome associated with specific traits, both in health and disease. A limitation of the approach is the challenges frequently associated with defining the biochemical and physiological consequences of the genetic variants. We hypothesized that targeted interventions in genotype-stratified healthy volunteer groups could provide a means of testing molecular models for specific polymorphisms.
A common polymorphism in the transcriptional promoter of the osteopontin (OPN) gene (rs28357094) has been found to be a strong genetic modifier of muscle size in young adult volunteers (1), and muscle weakness and disease progression in Duchenne muscular dystrophy (DMD) (2). Both studies showed a dominant inheritance model of the polymorphism effect on phenotypes (TT versus GG/GT genotype groups). The T-to-G single nucleotide polymorphism (SNP) is located 66 bp upstream of the OPN transcription initiation site overlapping a specificity protein-1 (SP1) transcription factor-binding site. The minor G allele has shown to significantly inhibit the binding of SP1, and this loss of binding results in an 80% reduction in transcriptional activity in multiple human immortalized cell lines (3). The same rs28357094 polymorphism has been associated with predisposition to autoimmune disorders including systemic lupus erythematosus and juvenile dermatomyositis (4,5).
Osteopontin is an acidic glycoprotein that is a member of the small integrin binding N-linked glycoprotein family that is involved in several biological pathways (6–8). OPN is viewed as an inflammatory cytokine that promotes cellular activation, migration and chemotaxis. The protein has an arginine–glycine–aspartate-binding domain, an adjacent SVVYGLR motif showing potent angiogenic properties (9) and CD44 receptor-binding affinity that allows it to interact with a diverse range of cells (10). It has been extensively studied in a number of physiological processes including bone remodeling, tissue repair, wound healing, autoimmune disorders and numerous inflammatory conditions (11–13).
The OPN protein is at very low or undetectable levels in normal skeletal muscle. However, induction of muscle damage results in a 100-fold increase in OPN gene transcription (1,14). Muscles from Duchenne muscular dystrophy patients, as well as the dystrophin-deficient dog and mouse models also show very high levels of OPN mRNA and protein, consistent with chronic inflammation and myofiber degeneration/regeneration observed in these muscles (2,15). High OPN expression in pre-natal diaphragm of the dystrophin-deficient dogs has been seen in CD11b infiltrating cells prior to post-natal necrosis, and was considered an early biomarker of dystrophin deficiency (16). OPN is considered a pro-inflammatory cytokine in muscle, where it serves as a chemoattractant for macrophages (14), and possibly neutrophils (13), and studies of double knockouts of dystrophin and OPN suggest that OPN can be deleterious to muscle remodeling (17). However, during normal muscle damage and repair, OPN is important for effective neutrophil and macrophage recruitment, clearing of necrotic cells and effective regeneration (18). Thus, increased OPN expression in muscle is seen in both normal and pathological muscle remodeling, and may serve both a normal physiological role and a pathological role (as is the case for many TGFbeta-associated proteins).
The genetic association of the OPN gene promoter polymorphism, rs28357094, with 17% increase in muscle size in healthy young adult females (1), and muscle wasting and weakness in Duchenne muscular dystrophy patients (2) was consistent with an important role of the OPN protein in modulating muscle response to damage and remodeling. However, the effect of the polymorphism on gene expression was difficult to rationalize with the observed phenotype associations. Specifically, the G allele has been shown to cause a 80% reduction in OPN gene promoter activity, and the same G allele was associated with a more severe DMD phenotype in young boys, and larger muscle in female (and not male) adult volunteers. To develop a molecular model for the effect of the polymorphism on muscle, we carried out both in vitro studies of the OPN gene promoter using reporter assays and in vivo studies of an eccentric muscle challenge in adult female volunteers. The in vitro assays used site-directed mutagenesis to remove the possible effects of additional polymorphisms that may be in linkage disequilibrium with the rs28357094 locus and studied estrogen responsiveness of the different alleles. OPN has been previously reported to be estrogen responsive, and the gene promoter contains estrogen-response elements (EREs) (19,20), but the possible interactions of the ERE and the rs28357094 have not been previously studied. Our data show a strong effect of the polymorphism on the response of the OPN gene to estrogen, and the in vitro and in vivo data presented are consistent with a novel molecular model that explains the effects of OPN on muscle remodeling.
RESULTS
The −66 bp OPN polymorphism shows differential effects on baseline and estrogen-responsive transcriptional regulation in muscle cells
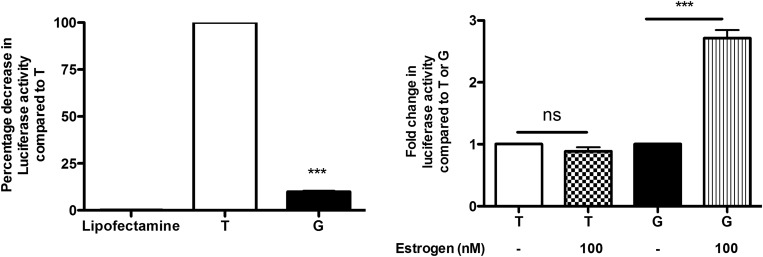
Allele-specific OPN promoter fragments (−899 to +109 bp) were cloned into pGL4.15 luciferase reporter vectors to examine the effects of the −66 bp T/G SNP on OPN promoter activity. The constructs were then transfected into human skeletal muscle myoblasts, and baseline luciferase activity was then determined. The minor G allele caused an 80% reduction in baseline luciferase expression in transfected cells compared with the ancestral T allele (Fig. 1A). These results correspond to established data that suggest that the G allele causes a significant reduction in OPN expression (3).
Figure 1.
The G allele of the rs28357094 polymorphism shows decreased basal promoter activity, but higher transcriptional response to estrogen hormone treatment. (A) Allele-specific promoter reporter constructs were transfected into human myoblasts and assayed for baseline luciferase expression. The G allele showed an 80% reduction in baseline expression (P < 0.001), consistent with previous reports of loss of Sp1 binding and lower baseline expression in other cell types (3). (B) The same constructs were then tested in the presence of estrogen (100 nm). The T allele construct was unresponsive to estrogen, while the G allele construct showed a 3-fold increase in luciferase expression (P < 0.001).
Cells transfected with the allele-specific constructs were then treated with 100 nm of estrogen in serum-free media and a luciferase assay was performed 24 h after the estrogen treatment (Fig. 1B). Myoblasts transfected with constructs containing the G allele and treated with estrogen showed a 3-fold increase in luciferase activity compared with untreated cells. Cells transfected with constructs containing the T allele did not show any significant differences in expression when treated with estrogen (Fig. 1B).
Eccentric exercise challenge in AA adult females
Subjects were enrolled into the Assessing Inherited Metabolic Markers in the Young (AIMM Young) at Howard University (21). As part of the AIMMY IRB consent, subjects were asked if they could be re-contacted for future studies. Genomic DNAs of 187 African-American (AA) participants in the AIMM Young study were genotyped for the −66 T/G OPN variant (rs28357094). The G allele showed an allele frequency of 8.8% in the AA group, with 17% of subjects showing GT or GG genotype (dominant inheritance model). The allele frequency observed in the AA participants was about half that observed in Caucasian populations (1). We limited the current study to the analysis of young adult females due to the female-specific effect of genotype in our previous studies of young adult Caucasian college students (1). Of the 23 AA females with the minor G allele, 14 were re-contacted and 6 females volunteered for the eccentric challenge protocol (3 GG/GT and 3 TT) (demographics in Table 1).
Table 1.
Participants in the eccentric muscle challenge intervention
| ID | OPN genotype | Age (years) | Height (cm) | Weight (lbs) | Neck circumference (cm) | Mid-waist circumference (cm) | Hip circumference (cm) | BMI (lbs/in2) | Eccentric exercise reps |
|---|---|---|---|---|---|---|---|---|---|
| 101 | GG | 23 | 171.9 | 251.8 | 14.3 | 41.1 | 50.1 | 38.6 | 11 |
| 102 | TT | 20 | 176.0 | 223.9 | 14.5 | 41.7 | 44.0 | 32.8 | 16 |
| 103 | GG | 19 | 168.9 | 186.9 | 13.8 | 32.9 | 45.8 | 29.7 | 10 |
| 104 | TT | 19 | 175.0 | 202.3 | nd | nd | nd | 30.0 | 10 |
| 105 | GT | 21 | 171.9 | 172.8 | 13.2 | 32.9 | 45.9 | 26.5 | 12 |
| 106 | TT | 21 | 178.1 | 160.8 | 13.3 | 30.2 | 39.2 | 23.0 | 16 |
Phenotypic features of the six AA females participating in the eccentric challenge intervention are shown. One participant (104) declined some measurements (nd, not done).
The eccentric challenge protocol consisted of nine visits (Fig. 2). The initial visit included a description of the proposed study, informed consent, anthropometric assessments and DXA. Although individuals were from a population-based recruitment strategy, five out of six of the participants had a BMI >26 categorizing them as overweight or obese (Table 1); however, this reflected the overall AA female student population as we recently described (21). Visit 2 included a baseline magnetic resonance imaging (MRI) of upper arm of both the dominant and non-dominant arms. All MRI assessments were done using a pre-formed cast set at 120° angle, and images obtained in a knee coil. All images were done on the same MRI unit by the same staff. Visits 3 and 4 consisted of assessments of baseline isometric strength test and baseline pain assessments.
Figure 2.
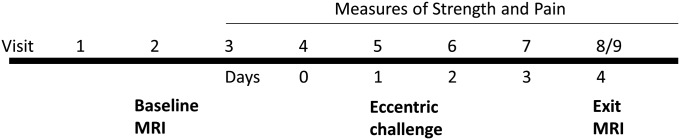
Protocol design for eccentric contraction challenge of the biceps muscle in young adult female volunteers. Initial visits were for informed consent, anthropomorphic measures and assessments of baseline phenotypes including MRI of both the dominant and non-dominant upper arm. Eccentric challenge of the non-dominant arm was at visit 4 (Day 0) with exit MRI at Day 4. Eccentric challenge was done only once on visit 4 (Day 0), with all subsequent visits done to assess the delayed effects of this challenge on strength and MRI changes.
The eccentric muscle challenge was done at visit 4 (Day 0 for post-challenge assessments; Fig. 2) as previously described (22). The subject was positioned with their non-dominant arm at 90° angle and then instructed to resist extension of the arm with maximal voluntary elbow (biceps) contraction, with the extension force generated by the evaluator with a bar and chain connected to the modified preacher's bench. The arm was gradually lengthened from 90° to 180° over a 5 s time. Repeated bouts were done with a 15 s rest period between bouts, until exhaustion (fatigue; average 12 bouts) (Table 1). Measures of isometric strength test were done for both the non-dominant (eccentric bout) and dominant (non-exercised) arms for 4 consecutive days following the intervention. Assessments of pain were also done at each visit. The post-bout MRI was done subsequent to baseline strength and pain assessments, 4 days after the bout of eccentric contractions (visit 9) (Fig. 2). This time point was previously shown to exhibit the greatest degree of muscle swelling (22).
The G allele is associated with greater loss of strength after eccentric challenge
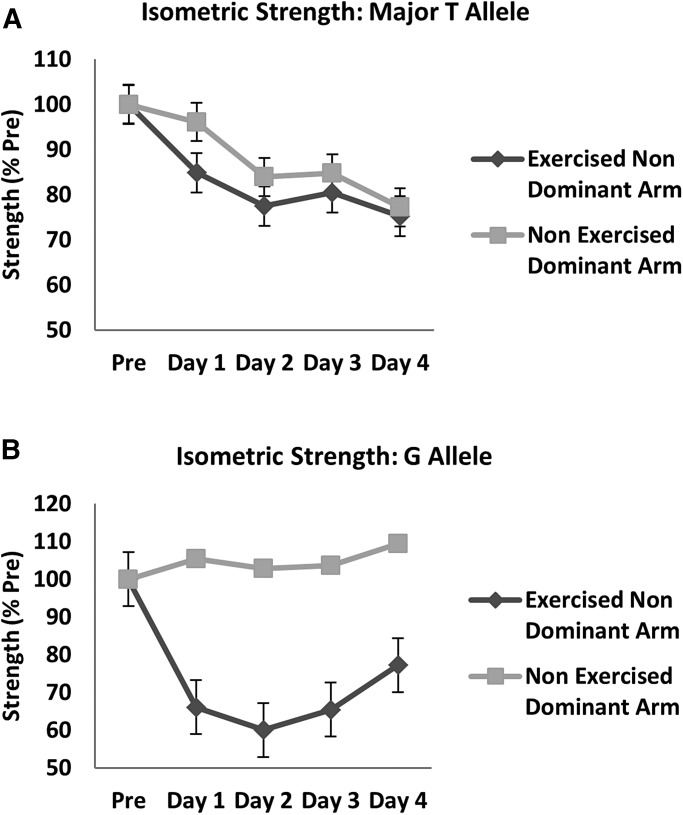
Measures of isometric strength test were done before the bout, and at each of 4 days after the bout on both the exercised non-dominant arm and non-exercised dominant arm. Volunteers with the common TT genotype showed a time-related drop in force of the exercised non-dominant arm, but also showed a drop in the non-exercised arm such that the difference in arms was not significant (Fig. 3A). The volunteers carrying the G allele showed a more substantial drop in force of the exercised non-dominant arm, whereas the non-exercised arm showed no decrease in strength (Fig. 3B). The difference in percentage drop in force between the exercised and non-exercised arms for the G allele carriers was highly statistically significant considering both d0-d2 (P < 0.001) and d0-d4 (P < 0.001) as was the interaction between time and arm (P = 0.02).
Figure 3.
Measures of maximal isometric strength in genotype-stratified young adult female volunteers. Isometric strength of both the non-dominant arm (eccentric bout) and dominant arm (non-exercised control) was measured in TT homozygotes (n = 3) (A), and G allele carriers (GG and GT; n = 3) (B). Pre is prior to the eccentric exercise challenge, with measures then subsequent days after the bout. TT homozygotes showed a small drop in force that was similar in both arms (A). G allele carriers showed a 40% drop in muscle strength in the exercised arm (P < 0.001), with a small increase in strength in the non-exercised arm (B).
The G allele is associated with greater muscle swelling and inflammation
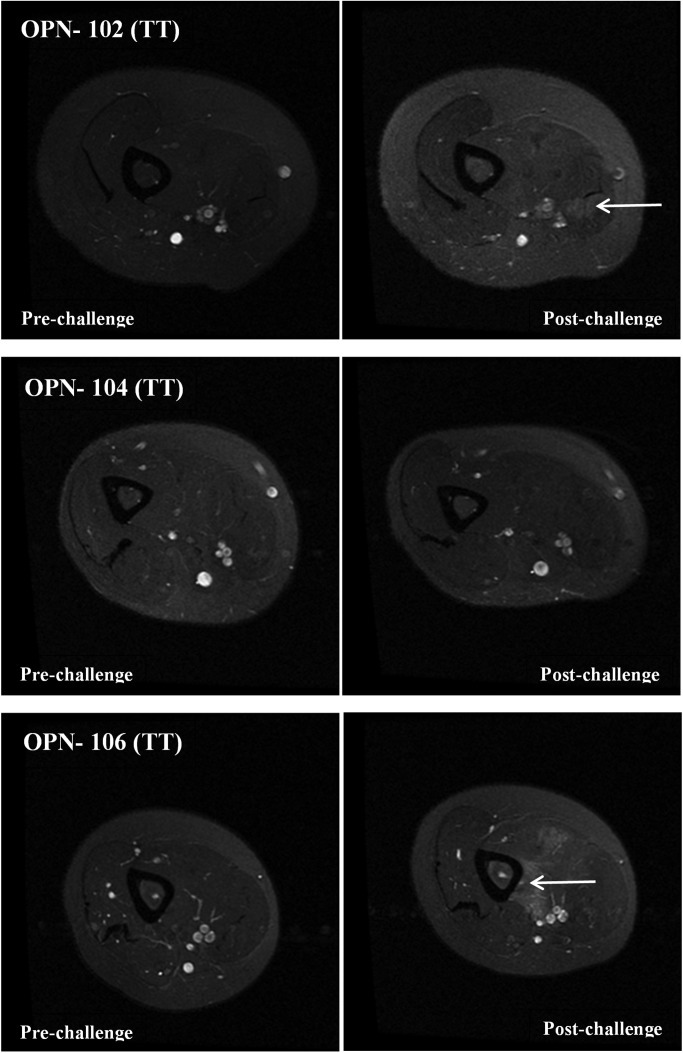
T2 images were taken of both dominant and non-dominant arms before the eccentric challenge and 4 days after the challenge. The non-exercised dominant arm showed similar features in all subjects, with no evidence of inflammation or muscle swelling (Figs 4 and 5). In the TT genotype volunteers, the post-exercise non-dominant arm showed hyper-intense regions of the biceps evident in OPN-102 and OPN-106 (Fig. 4, white arrows). Subject OPN-104 showed no obvious hyper-intense T2 regions.
Figure 4.
T2-weighted MRI images of young adult female volunteers having TT OPN genotype. Shown are MRI T2-weighted images of the non-dominant upper arm prior to eccentric challenge (pre-challenge) and 4 days after eccentric challenge (post-challenge). Subjects OPN-102 and OPN-106 show areas with an increase of T2 signal in the biceps in the post-challenge images consistent with increased water content (swelling) (white arrows). Areas of hyper-intensity were not evident in OPN-104.
Figure 5.
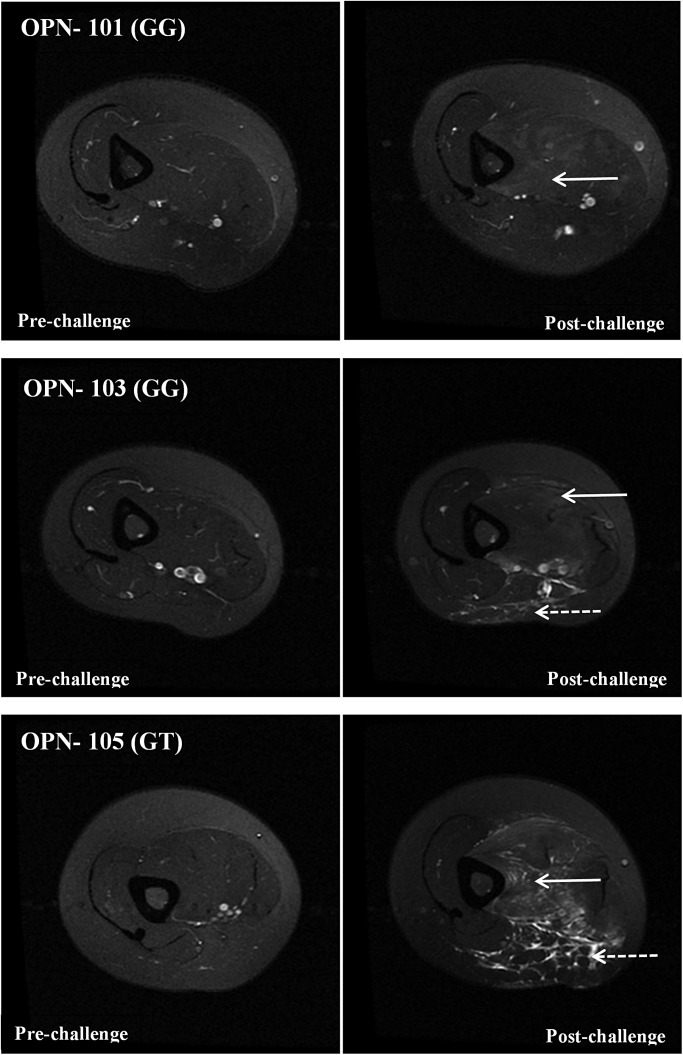
T2-weighted MRI images of young adult females carrying the G allele of the OPN promoter polymorphism. Shown are MRI images using the same methods and time points as in Figure 4. The three subjects carrying the G allele show extensive hyper-intensity throughout much of the biceps muscle (white solid arrows). Subjects OPN-103 and OPN-105 also show evidence of inflammation (stippled arrows) surrounding the brachial artery/nerve area. The inflammation extended into the biceps, and also into the ventral subcutaneous fat tissue.
In the G allele carriers, there was striking hyper-intense regions involving the majority of the biceps in all three subjects (Fig. 5, white solid arrows). In two of the subjects (OPN-103, OPN-105) there was markedly hyper-intense regions at the ventral region of the biceps extending into the underlying subcutaneous fat (Fig. 5, stippled arrows). The area of inflammation appeared centered on the brachial artery and included apparent increased T2 signal in the veins and arteries, but not the brachial plexus.
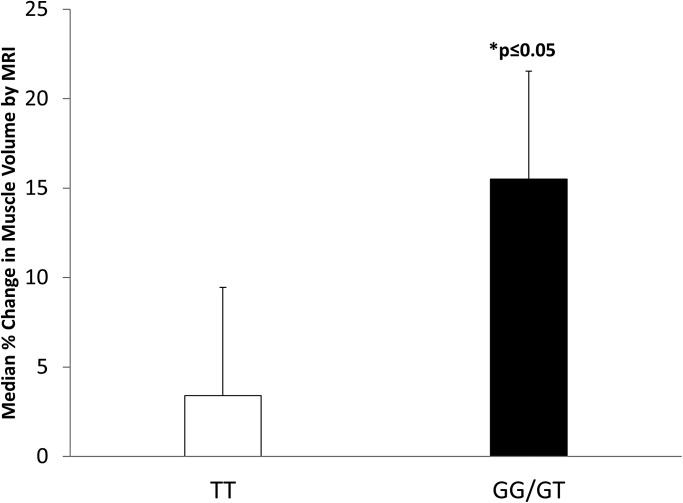
The cross-sectional area of each biceps muscle was determined and the percent change in area of the exercised non-dominant arm determined between the pre-challenge and post-challenge images (Fig. 6). The TT homozygote subjects showed an average of 3% increase in biceps cross-sectional area, whereas the G allele carriers showed an average of 15% increase in size. The difference in swelling between the TT and G genotype carriers was statistically significant (P < 0.05).
Figure 6.
Percent volume change in biceps muscles after an acute eccentric exercise bout. Biceps muscle volume change was measured from MRI images in the exercised arm of genotype-stratified subjects at Day 4 after the bout of eccentric challenge. G allele carriers showed greater biceps swelling compared with TT homozygotes (P < 0.05).
DISCUSSION
The goal of our study was to provide in vitro and in vivo data sufficient to build a unifying model for the action of the −66 bp T/G polymorphism in the Sp1 transcription factor-binding site of the OPN gene promoter in muscle. Our previous findings had presented somewhat of an enigma, where the G allele was associated with larger muscle volume in young adult volunteer females (1), yet weaker muscles in young DMD boys (2). Moreover, previous studies had shown the G allele to result in 80% loss of OPN gene expression in different human immortalized cell lines (3), suggesting that less OPN (an inflammatory cytokine) results in larger muscles in females, yet weaker muscles in DMD boys. Testing of allele-specific reporter constructs in human muscle cells confirmed previous findings in other cell types, with a loss of ∼80% of promoter strength (Fig. 1A).
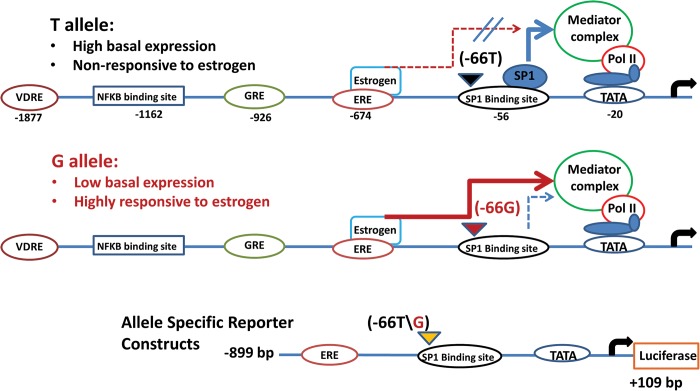
Our search of the OPN gene promoter for enhancer elements found potential enhancer sequences for multiple steroid hormone-binding sites [estrogen receptor (−674 bp), glucocorticoid receptor (−926 bp), vitamin D receptor (−1877 bp)], as well as a potential Nuclear factor kappa beta (NFkB) binding site (−1162 bp) (Fig. 7). Given the female-specific effect of the promoter polymorphism on muscle size, we hypothesized that there may be an allele-specific interaction between the estrogen enhancer and the more proximal Sp1 transcription factor site. Indeed, treatment of muscle cells with allele-specific reporter constructs showed that estrogen induced a 3-fold increase over baseline for the G allele, but no change in expression for the T allele (Fig. 1B).
Figure 7.
Schematic of the OPN gene promoter structure, and proposed model for effect of genotype on OPN gene expression. The top two lines show the OPN gene promoter structure and proposed model for the allele-specific effects of the rs28357094 polymorphism (−66T, −66G) on transcriptional regulation of the OPN gene. The −66 T allele is shown to have high basal expression, but blocks positive transcriptional response to the estrogen enhancer due to residency of the SP1 transcription factor in the intact SP1-binding site (top line of figure). The −66G allele shows reduced basal expression, but is highly responsive to estrogen due to loss of the SP1-binding site (second line of figure) (3). The predicted enhancer elements are in the more distal regions of the OPN gene promoter (VDRE, vitamin D receptor element; GRE, glucocorticoid response element; ERE, estrogen response element). The lower line of the figure shows the allele-specific reporter constructs used for transfection of human muscle cells. The genotype/phenotype associations of this polymorphism with muscle phenotypes have shown data consistent with a dominant model of inheritance, with two genotype groups (TT and GT + GG).
These data suggest a hypothetical model for the sex-specific effects of the polymorphism on muscle size in females. High estrogen levels in young adult females lead to increased damage-induced OPN levels in G allele carriers due to lack of residency of the more proximal Sp1 transcription factor binding. On the other hand, T allele homozygotes have the estrogen (and possibly NFkB) response blocked due to residency of the Sp1 transcription factor near the mediator complex (Fig. 7). This model predicted that young adult females carrying the G allele may show greater sensitivity to muscle damage, and the muscle damage may in turn cause more hypertrophy in these women.
Bouts of eccentric muscle activity (lengthening contractions) are known to induce mild muscle damage and stimulate muscle hypertrophy. We hypothesized that a bout of eccentric activity in young adult females stratified for OPN genotype would show a differential response to an eccentric bout, with the G allele carriers showing greater inflammation and swelling. To test this, female volunteers were recruited from the AIMM Young baseline genotype/phenotype study of young adults (21). Three females of each genotype undertook an eccentric exercise bout of the non-dominant upper arm (biceps), with longitudinal measures of strength and imaging by MRI. This study confirmed our hypothesis. The G allele carriers showed significantly greater muscle swelling at Day 4 after the bout and greater loss of strength (Figs 3–6). Surprising, there was striking inflammatory signal in the MRI in two of the three G allele carriers, centered on the brachial large blood vessels (Fig. 5). The inflammation extended into the biceps, but also into the subcutaneous fat ventral to the brachial complex.
Our model suggests that G allele carriers show heightened sensitivity to mild muscle damage, with both exaggerated muscle swelling and inflammatory response, and this leads to greater susceptibility to muscle hypertrophy. In Duchenne muscular dystrophy, patient muscle shows a high degree of activation of NFkB pro-inflammatory pathways soon after birth, many years prior to clinical onset (23). Given the promoter structure of the OPN gene, with multiple predicted steroid hormone enhancers and NFkB promoter element (Fig. 7), our model can be extended to the observed genotype/phenotype relationship in DMD. DMD patients carrying the G allele may show greater NFkB- and/or glucocorticoid-induced transcription of the OPN gene during chronic inflammation, leading to exacerbation of the pro-inflammatory state of muscle and worsening of phenotype, as we have previously reported. Thus, the integration of gene structure, in vitro reporter assays and intervention study in young adult volunteers provides a hypothetical unifying model for the interaction of the OPN promoter SNP and muscle remodeling. However, the complex regulation of the OPN gene in response to multiple inflammatory and steroid hormone promoter elements makes it challenging to predict the precise responses of the gene in vivo.
Limitations to the study presented here include the low number of volunteers enrolled into the eccentric intervention (n = 3 per genotype group), and the lack of muscle biopsies from the volunteers to correlate OPN protein and mRNA levels with respective genotypes and degree of inflammation. Extensive correlations of OPN genotype with OPN protein levels have been previously reported for DMD patient muscle, and there was no association of genotype with OPN levels (24). The argument presented in this previous publication is that the variable degree of inflammation in each biopsy and complex regulation of OPN protein in the extracellular space, both likely obscures the in vivo correlations of OPN genotype and OPN gene expression seen in vitro. Finally, we have not directly addressed the molecular basis of the association of the G allele with muscle weakness in DMD. Our results suggest that this association may be related to glucocorticoid treatment of DMD, but further genetic associations studies of DMD patient populations treated and un-treated with glucocorticoids must be done to test this hypothesis.
MATERIALS AND METHODS
Human subjects
One hundred and eighty-seven AA females previously recruited into the NIH P20 AIMM Young Study from Howard University were genotyped for the rs2857094 polymorphism. AIMM Young is an ongoing project that concentrates on the genetics of metabolic syndrome in young adults (21). It functions as a collaborative between the Georgetown-Howard Universities Center for Clinical and Translational Science and Children's National Medical Center. AIMM Young subjects were re-contacted based on genotype and asked to complete an eccentric muscle challenge with MRI as the final outcome. A total of six females (3 GG/GT and 3 TT) were subsequently enrolled into the eccentric exercise intervention. All females were 19–23 years of age, healthy and either sedentary or only recreationally active. Participants were screened prior to study entry to determine if they had any orthopedic, muscular or other medical conditions that would prohibit them from completing the study.
Genotyping
To determine the specific OPN genotype of rs28357094 (−66 T/G variant), DNA was extracted from the peripheral blood of all 187 individuals within the Howard University AIMM Young Study using standard manufacturer's procedures (Qiagen, Valencia, CA, USA). DNA was then amplified on an ABI 2720 Thermal Cycler using TaqMan rs28357094 primers/probe: forward 5′-AAGTGCTCTTCCTGGATGCTG-3′, reverse 5′-CTCCTGCTGCTGCTGACAAC-3′and sickle cell probe rs334: forward 5′-AGTCAGGGCAGAGCCATCTA-3′, reverse 5′-CTCACCACCAACTTCATCCA-3′.
Biceps isometric strength test
Participants performed isometric strength test of the biceps muscles using a modified arm curl bench (CSN Stores, MA, USA). To assess biceps flexion isometric strength, the arm was positioned at a 90° angle with the curl bar attached to a load cell (Transducer Technologies, Inc., Whittier, CA, USA). During the test the subject was instructed to exert a maximal biceps muscle contraction at the 90° angle and the tension generated was recorded as maximal strength expressed in kilograms. Muscle strength was assessed in both the right and left arms with the subjects performing three maximal contractions on each arm. All statistical analyses utilized the average value for these three strength measurements.
Exercise protocol
Each participant performed one set of maximal lengthening contractions (eccentric) on a modified preachers arm curl machine. The eccentric exercise protocol has been detailed previously (22). To perform this exercise, the active arm of the participant was placed on the padded support in a flexed position, while the passive arm rested on the participant's side. The participant's arm was then guided by the investigator from a completely flexed position to a fully extended position. The modifications in the preacher's bench allowed for maximal resistance to the participant throughout the entire range of motion. Each contraction lasted ∼5 s and was repeated every 15 s. The participant's arm was then brought back to the flexed position to prevent concentric muscle contractions. The procedure was repeated a maximum of 24 times or until the participant fatigued. Subsequent isometric strength tests and a 25 ml blood sample were obtained.
Magnetic resonance imaging
The MRI procedure was performed on a GE Signa HDxt 3.0T High-Field System (General Electric Company, CT, USA) using a GE Signa HDxt 3.0T HD Knee coil (General Electric Company). The participant was placed laterally with arm placed into a plastic mold to secure it comfortably at a 120° angle in the knee coil, and ensure reliability from subject to subject regarding placement of the arm and related images. 64—4 mm slices were taken because of the broad limit of the long axis of the knee coil (∼256 mm). In a coronal view, the slice direction was parallel to the humerus plane and the fourth slice was precisely placed at the vertex of elbow point. In a sagittal view, the slice direction was parallel to the humerus plane. The region of interest (ROI) was adjusted to cover the area from the elbow joint to the limit of the knee coil. The imaging sequence used in the T1-weighted upper arm scan was a Fast Spin Echo sequence: TE = 6 ms, TR = 1000 ms, echo train = 6, FOV = 14 cm, matrix = 320 × 224, slice thickness = 4 mm, slice gap = 0 mm, 64 slices. Two sets of double-echo sequences were used to get the T2 fat suppression images at TE = 7.1, 40, 70, 90 ms. Fast spin echo sequences: (1) acquisition: the TE = 7.1 and 70.1 ms. Parameters: TE = 7.1 and 70.1 ms, TR = 4467 ms, echo train = 18, FOV = 14 cm, matrix = 256 × 192, slice thickness = 4 mm, slice gap = 0 mm, 64 slices. (2) Acquisition: the 40 and 90 ms. Parameters were the same as the 7.1 and 70.1 ms parameters.
After retrieving scans, the total biceps region was identified on T1-weighted images and the area was outlined using ImageJ software. The scans were adjusted so that identical regions were compared each time. ‘Muscle volume’ was defined as the total pixilated region of the outlined area in millimeters. The swelling post-exercise was determined by comparing changes in muscle volume between MRI scans at study entry and post-eccentric exercise.
Plasmid constructs
Osteopontin promoter fragment from −899 to +109 was prepared by PCR amplification from genomic DNA of homozygous individual, cloned into pGL4.15 [luc2P/Hygro] vector (Promega, Medison, WI, USA) and the inserted sequence was verified by Switchgear genomics (Menlo Park, CA, USA). Site-directed mutagenesis was performed according to manufacturer's instruction (Agilent Technologies, Santa Clara, CA, USA) to obtain single site-specific mutation construct at position −66 bp of the promoter region of OPN (T–G). The primers used for the site-directed mutagenesis were as follows: sense primer 5′-CCCAAGGTTGCACAGTTCAGCAGTGACACAG-3′ and antisense primer 5′-GGGTTCCAACGTGTCAAGTCGTCACTGTGTC-3′.
Cell culture and transfection
Human skeletal myoblast cell line (obtained from Vincent Mouly, Institut de Myologie, Paris, France) was maintained in skeletal muscle cell growth medium using skeletal muscle cell growth medium kit (Promocell, Heidelberg, Germany) supplemented with 20% fetal calf serum and penicillin–streptomycin (1% (v/v); all supplied by Life Technology, Carlsbad, CA, USA). Cultures were maintained in this complete growth medium in a humidified atmosphere under 5% CO2 at 37°C.
Cell transfection was performed using Lipofectamine LTX (Life Technology) according to manufacturer's instruction. Cells were plated into 6-well plates at 70–80% confluence. Transfection was performed over a 48 h period in complete growth medium without antibiotic. The amount of DNA used for transfection was 2.5 µg. In the experiments where estrogen (Sigma-Aldrich, St. Louis, MO, USA) was added, the complete growth medium with transfection reagent was removed; cells were washed and replaced with serum-free medium containing 100 nm estrogen for further 24 h. Cell lysates were prepared using lysis buffer consisting of 25 mm Gly (pH 7.8), 1% Triton X-100 (v/v), 15 mm MgSO4 (heptahydrate), 4 mm EGTA and 1 mm DTT in 10 ml water. Luciferase activity was measured using Luciferase Assay system (Promega) with Centro LB 960 microplate luminometer (Berthold Technologies, Oak Ridge, TN, USA). The data obtained were normalized to the protein concentration from each well.
Statistical analysis
Several phenotypes were compared with tests appropriate for the distribution of the dependent variable. A P-value of ≤0.05 was considered statistically significant. As shown in Figure 1, the comparison of % decrease in luciferase activity between TT and G allele cells were performed using a student's t-test and the comparison of fold change between baseline and post-estrogen treatment was performed in TT and G allele cells separately using a paired t-test. Analyses comparing changes in isometric strength between arms (Fig. 3) were performed using a repeated-measure ANOVA including main effects of arm and time and an arm × time interaction. Lastly, the comparison between median % change in muscle volume between TT and G allele individuals was performed using a Wilcoxon rank sum test.
FUNDING
This work was supported by the Georgetown-Howard Universities Center for Clinical and Translational Science Pilot Study Program (5UL1TR000101-04), NIMHD P20MD000198, NIMHD G12MD007597 and the Clark Family Foundation. Funding to pay the Open Access publication charges for this article was provided by the Clark Family Foundation.
ACKNOWLEDGEMENTS
The authors thank Dr Priscilla Clarkson and Dr Kevin O'Fallon for training in the eccentric exercise protocol and Heather Gordish-Dressman, PhD for statistical consultations.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hoffman E.P., Gordish-Dressman H., McLane V.D., Devaney J.M., Thompson P.D., Visich P., Gordon P.M., Pescatello L.S., Zoeller R.F., Moyna N.M., et al. Alterations in osteopontin modify muscle size in females in both humans and mice. Med. Sci. Sports Exerc. 2013;45:1060–1068. doi: 10.1249/MSS.0b013e31828093c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pegoraro E., Hoffman E.P., Piva L., Gavassini B.F., Cagnin S., Ermani M., Bello L., Soraru G., Pacchioni B., Bonifati M.D., et al. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76:219–226. doi: 10.1212/WNL.0b013e318207afeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giacopelli F., Marciano R., Pistorio A., Catarsi P., Canini S., Karsenty G., Ravazzolo R. Polymorphisms in the osteopontin promoter affect its transcriptional activity. Physiol. Genomics. 2004;20:87–96. doi: 10.1152/physiolgenomics.00138.2004. [DOI] [PubMed] [Google Scholar]
- 4.Niewold T.B., Kariuki S.N., Morgan G.A., Shrestha S., Pachman L.M. Gene-gene-sex interaction in cytokine gene polymorphisms revealed by serum interferon alpha phenotype in juvenile dermatomyositis. J. Pediatr. 2010;157:653–657. doi: 10.1016/j.jpeds.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trivedi T., Franek B.S., Green S.L., Kariuki S.N., Kumabe M., Mikolaitis R.A., Jolly M., Utset T.O., Niewold T.B. Osteopontin alleles are associated with clinical characteristics in systemic lupus erythematosus. J. Biomed. Biotechnol., 2011;2011:802581. doi: 10.1155/2011/802581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denhardt D.T., Noda M., O'Regan A.W., Pavlin D., Berman J.S. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J. Clin. Invest. 2001;107:1055–1061. doi: 10.1172/JCI12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimba E.R., Tilli T.M. Human osteopontin splicing isoforms: known roles, potential clinical applications and activated signaling pathways. Cancer Lett. 2013;331:11–17. doi: 10.1016/j.canlet.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Konno S., Kurokawa M., Uede T., Nishimura M., Huang S.K. Role of osteopontin, a multifunctional protein, in allergy and asthma. Clin. Exp. Allergy. 2011;41:1360–1366. doi: 10.1111/j.1365-2222.2011.03775.x. [DOI] [PubMed] [Google Scholar]
- 9.Uchinaka A., Kawaguchi N., Hamada Y., Mori S., Miyagawa S., Saito A., Sawa Y., Matsuura N. Transplantation of myoblast sheets that secrete the novel peptide SVVYGLR improves cardiac function in failing hearts. Cardiovasc. Res. 2013;99:102–110. doi: 10.1093/cvr/cvt088. [DOI] [PubMed] [Google Scholar]
- 10.Weber G.F., Ashkar S., Glimcher M.J., Cantor H. Receptor–ligand interaction between CD44 and osteopontin (Eta-1) Science. 1996;271:509–512. doi: 10.1126/science.271.5248.509. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigues L.R., Teixeira J.A., Schmitt F.L., Paulsson M., Lindmark-Mansson H. The role of osteopontin in tumor progression and metastasis in breast cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:1087–1097. doi: 10.1158/1055-9965.EPI-06-1008. [DOI] [PubMed] [Google Scholar]
- 12.Uede T. Osteopontin, intrinsic tissue regulator of intractable inflammatory diseases. Pathol. Int. 2011;61:265–280. doi: 10.1111/j.1440-1827.2011.02649.x. [DOI] [PubMed] [Google Scholar]
- 13.Yang M., Ramachandran A., Yan H.M., Woolbright B.L., Copple B.L., Fickert P., Trauner M., Jaeschke H. Osteopontin is an initial mediator of inflammation and liver injury during obstructive cholestasis after bile duct ligation in mice. Toxicol. Lett. 2014;224:186–802195. doi: 10.1016/j.toxlet.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirata A., Masuda S., Tamura T., Kai K., Ojima K., Fukase A., Motoyoshi K., Kamakura K., Miyagoe-Suzuki Y., Takeda S. Expression profiling of cytokines and related genes in regenerating skeletal muscle after cardiotoxin injection: a role for osteopontin. Am. J. Pathol. 2003;163:203–215. doi: 10.1016/S0002-9440(10)63644-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turk R., Sterrenburg E., van der Wees C.G., de Meijer E.J., de Menezes R.X., Groh S., Campbell K.P., Noguchi S., van Ommen G.J., den Dunnen J.T., 't Hoen P.A. Common pathological mechanisms in mouse models for muscular dystrophies. FASEB J. 2006;20:127–129. doi: 10.1096/fj.05-4678fje. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura A., Kobayashi M., Kuraoka M., Yuasa K., Yugeta N., Okada T., Takeda S. Initial pulmonary respiration causes massive diaphragm damage and hyper-CKemia in Duchenne muscular dystrophy dog. Sci. Rep. 2013;3:2183. doi: 10.1038/srep02183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vetrone S.A., Montecino-Rodriguez E., Kudryashova E., Kramerova I., Hoffman E.P., Liu S.D., Miceli M.C., Spencer M.J. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J. Clin. Invest. 2009;119:1583–1594. doi: 10.1172/JCI37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uaesoontrachoon K., Wasgewatte Wijesinghe D.K., Mackie E.J., Pagel C.N. Osteopontin deficiency delays inflammatory infiltration and the onset of muscle regeneration in a mouse model of muscle injury. Dis. Model Mech. 2013;6:197–205. doi: 10.1242/dmm.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig A.M., Denhardt D.T. The murine gene encoding secreted phosphoprotein 1 (osteopontin): promoter structure, activity, and induction in vivo by estrogen and progesterone. Gene. 1991;100:163–171. doi: 10.1016/0378-1119(91)90362-f. [DOI] [PubMed] [Google Scholar]
- 20.Xie Q.Z., Qi Q.R., Chen Y.X., Xu W.M., Liu Q., Yang J. Uterine micro-environment and estrogen-dependent regulation of osteopontin expression in mouse blastocyst. Int. J. Mol. Sci. 2013;14:14504–14517. doi: 10.3390/ijms140714504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnold T.J., Schweitzer A., Hoffman H.J., Onyewu C., Hurtado M.E., Hoffman E.P., Klein C.J. Neck and waist circumference biomarkers of cardiovascular risk in a cohort of predominantly African-American College Students: a preliminary study. J. Acad. Nutr. Diet. 2014;114:107–116. doi: 10.1016/j.jand.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nosaka K., Clarkson P.M. Changes in indicators of inflammation after eccentric exercise of the elbow flexors. Med. Sci. Sports Exerc. 1996;28:953–961. doi: 10.1097/00005768-199608000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y.W., Nagaraju K., Bakay M., McIntyre O., Rawat R., Shi R., Hoffman E.P. Early onset of inflammation and later involvement of TGFbeta in Duchenne muscular dystrophy. Neurology. 2005;65:826–834. doi: 10.1212/01.wnl.0000173836.09176.c4. [DOI] [PubMed] [Google Scholar]
- 24.Piva L., Gavassini B.F., Bello L., Fanin M., Soraru G., Barp A., Ermani M., Angelini C., Hoffman E.P., Pegoraro E. TGFBR2 but not SPP1 genotype modulates osteopontin expression in Duchenne muscular dystrophy muscle. J. Pathol. 2012;228:251–259. doi: 10.1002/path.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]